分析DP-GEN任务¶
+加载环境¶
+首先,导入环境:
+from catflow.tesla.dpgen import DPTask
+加载DP-GEN工作目录:
+t = DPTask(
+ path='/path/to/dpgen/',
+ param_file='param.json',
+ machine_file='machine.json',
+ record_file='record.dpgen'
+)
+便可根据所需分析的部分,对训练情况进行分析。
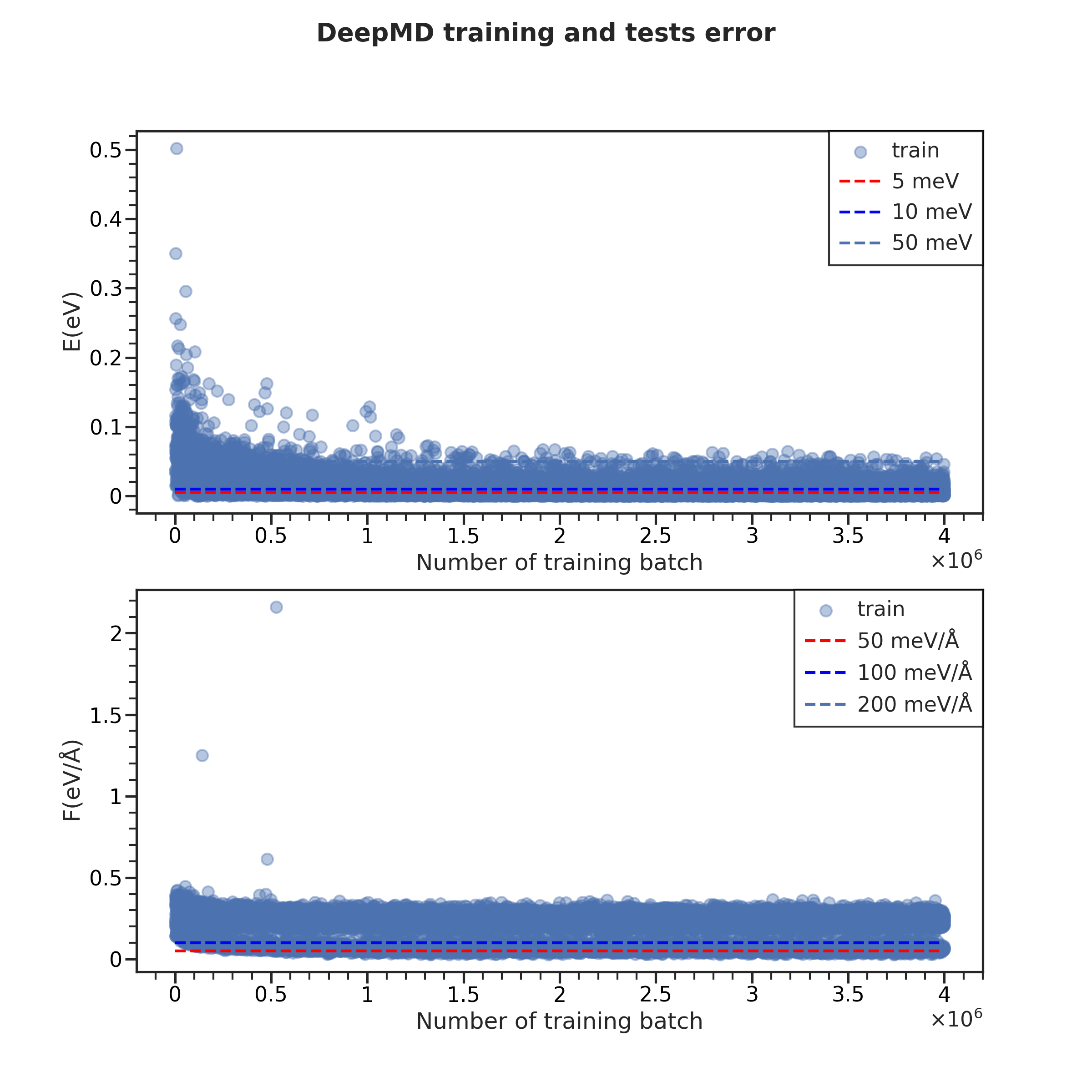
+训练(Training)¶
+导入分析器(DPAnalyzer),这里我们选择训练,即:
from catflow.tesla.dpgen.training import DPTrainingAnalyzer
+从任务初始化分析器实例:
+ana = DPTrainingAnalyzer(t)
+即可利用ana的内置函数进行作图:
fig = ana.plot_lcurve(
+ iteration=28, test=False, style='ticks', context='talk'
+)
+fig.set_size_inches((12,12))
+
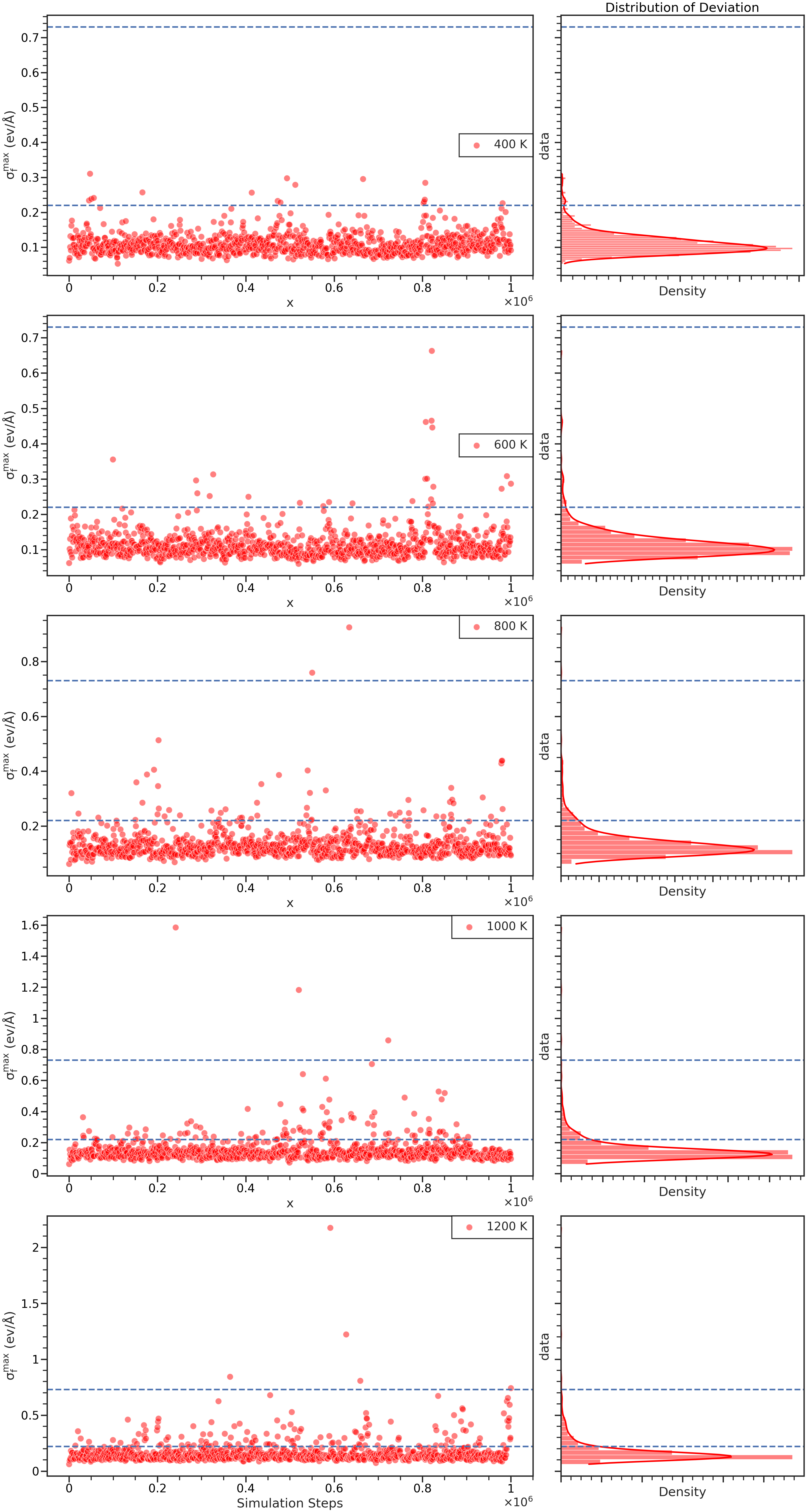
探索(Exploration)¶
+类似地,我们也可以对模型的model deviation分布进行分析:
+from catflow.tesla.dpgen.exploration import DPExplorationAnalyzer
+ana = DPExplorationAnalyzer(t)
+利用分析器自带的方法进行作图:
+fig = ana.plot_single_iteration(
+ iteration=41,
+ temps=[400, 600, 800, 1000, 1200],
+ xlimit=1000000,
+ f_trust_lo=t.param_data['model_devi_f_trust_lo'],
+ f_trust_hi=t.param_data['model_devi_f_trust_hi'],
+ style='ticks',
+ group_by='temps',
+ label_unit='K',
+ context='talk'
+)
+其中:
+-
+
-
+
+iteration对应为所需分析的轮数,默认为最新进行过Exploartion的轮数。
+ -
+
+f_trust_lo和f_trust_hi即对应的最大力偏差上下限设置。
+ -
+
通过
+group_by指定所需作图的参数,对应到param.json中model_devi_jobs中该轮数需要迭代的List,例如:
+
{
+ "template": {
+ "lmp": "lmp/input-meta.lammps",
+ "plm": "lmp/input-meta.plumed"
+ },
+ "sys_idx": [
+ 53
+ ],
+ "traj_freq": 1000,
+ "rev_mat": {
+ "lmp": {
+ "V_NSTEPS": [
+ 1000000
+ ],
+ "V_TEMP": [
+ 400,
+ 600,
+ 800,
+ 1000,
+ 1200
+ ]
+ }
+ },
+ "model_devi_f_trust_lo": 0.23,
+ "model_devi_f_trust_hi": 0.75
+ }
+若指定group_by 参数为 V_TEMP,则根据该轮的热浴温度分组作图,若指定V_TEMP=[400, 600, 800, 1000, 1200],则可由400、600、800、1000、1200K分别对model deviation作图。
-
+
label_unit即group_by参数的单位,例如这里是温度,故为"K"。
+
效果如下:
+