@@ -149,7 +149,8 @@ Now that you have successfully created and installed an Agent, you can explore m
- An Agent that [runs on a recurring schedule](./Agent#set-a-cron-job-to-run-flows-periodically)
- An Agent that [executes local processing when a file stored in cloud storage](./Agent#watch-for-flow-outputs-then-save-locally)
- [How to modify the agent code](./Agent#watch-for-files-locally-then-run-flow) to add custom functionality, such as
- - [Adding tags to files](../files/Tags.mdx) to make them easier to find in Ganymede
+ - [Adding tags to files](../files/Tags.mdx) to make captured files easier to find in Ganymede
- Parsing metadata from file contents to determine how files are processed
- - Delivering [multiple files into a single node](../../sdk/markdowns/AgentModelsAndMethods#classes-for-agent-triggered-flows)
-- Incorporating [Agent file utilities](../../sdk/markdowns/AgentModelsAndMethods#file-system-utilities) from the Ganymede SDK
+ - Delivering [multiple files into a single Node](../../sdk/markdowns/AgentModelsAndMethods#classes-for-agent-triggered-flows)
+- Incorporating [Agent utility functions](../../sdk/markdowns/AgentModelsAndMethods) from the Ganymede SDK and Agent SDK
+- Interpreting [Agent log messages](./AgentLogs)
diff --git a/docs/app/flows/FlowEditor.mdx b/docs/app/flows/FlowEditor.mdx
index 44fa6ec9..e20b18b8 100644
--- a/docs/app/flows/FlowEditor.mdx
+++ b/docs/app/flows/FlowEditor.mdx
@@ -89,27 +89,23 @@ To create a new Flow, click the
**Manage
The documentation allows developers to specify instructions for Flow execution; for example, developers could specify the purpose of the Flow and the expected inputs and outputs.
-Initially, the documentation is set to the Flow description given when creating a new Flow.
-
-Flow Documentation can be edited through the "Documentation" tab in the
**Manage **
sidebar in the Flow Editor.
-
-The documentation box accepts [Markdown](https://www.markdownguide.org/basic-syntax/).
+Initially, the documentation is set to the Flow description given when creating a new Flow. Flow Documentation can be edited through the "Documentation" tab in the
**Manage **
sidebar in the Flow Editor. The documentation box accepts [Markdown](https://www.markdownguide.org/basic-syntax/).
Writing the following markdown:
-
+
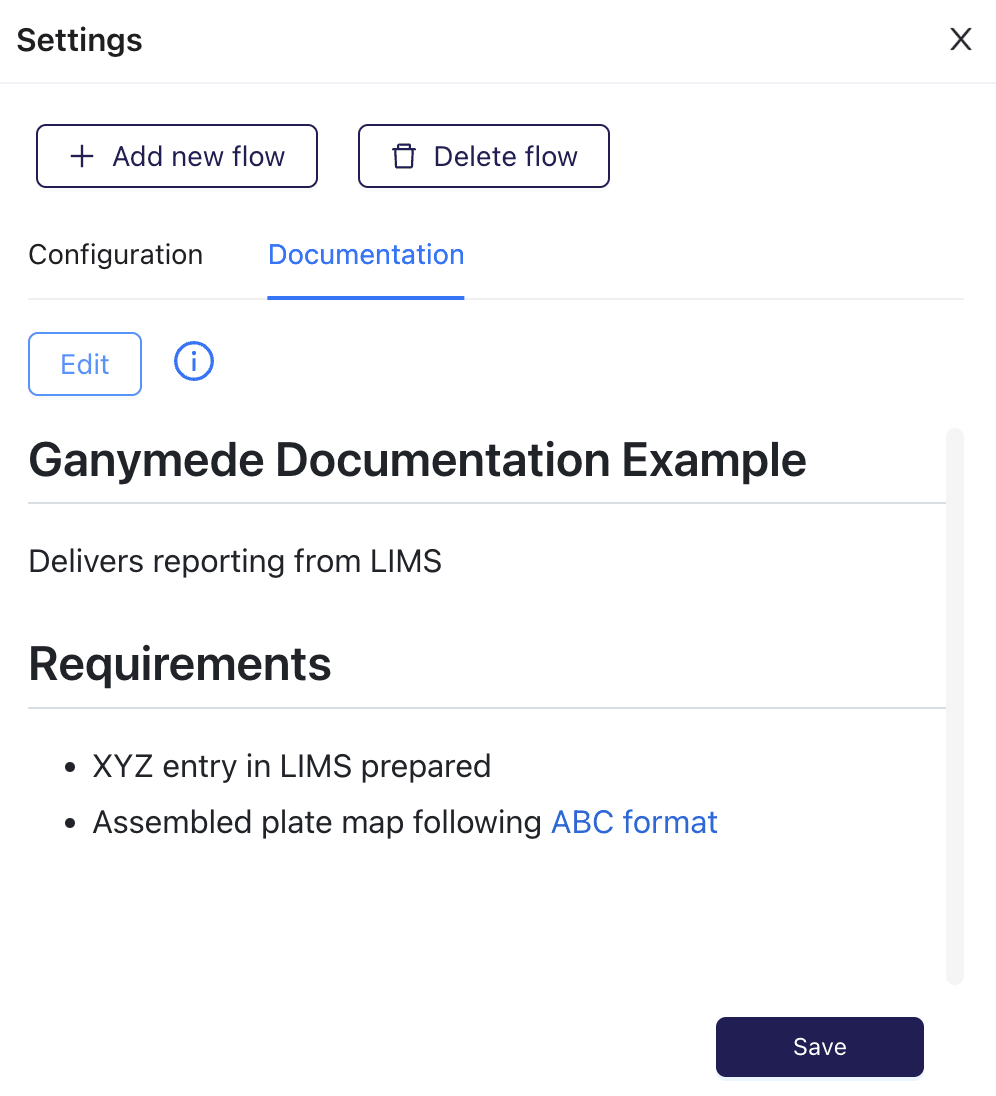
Generates this rendered documentation:
-
+
-The Flow Documentation will also be shown in the Flow View page
+The Flow Documentation is also shown in the Flow View page
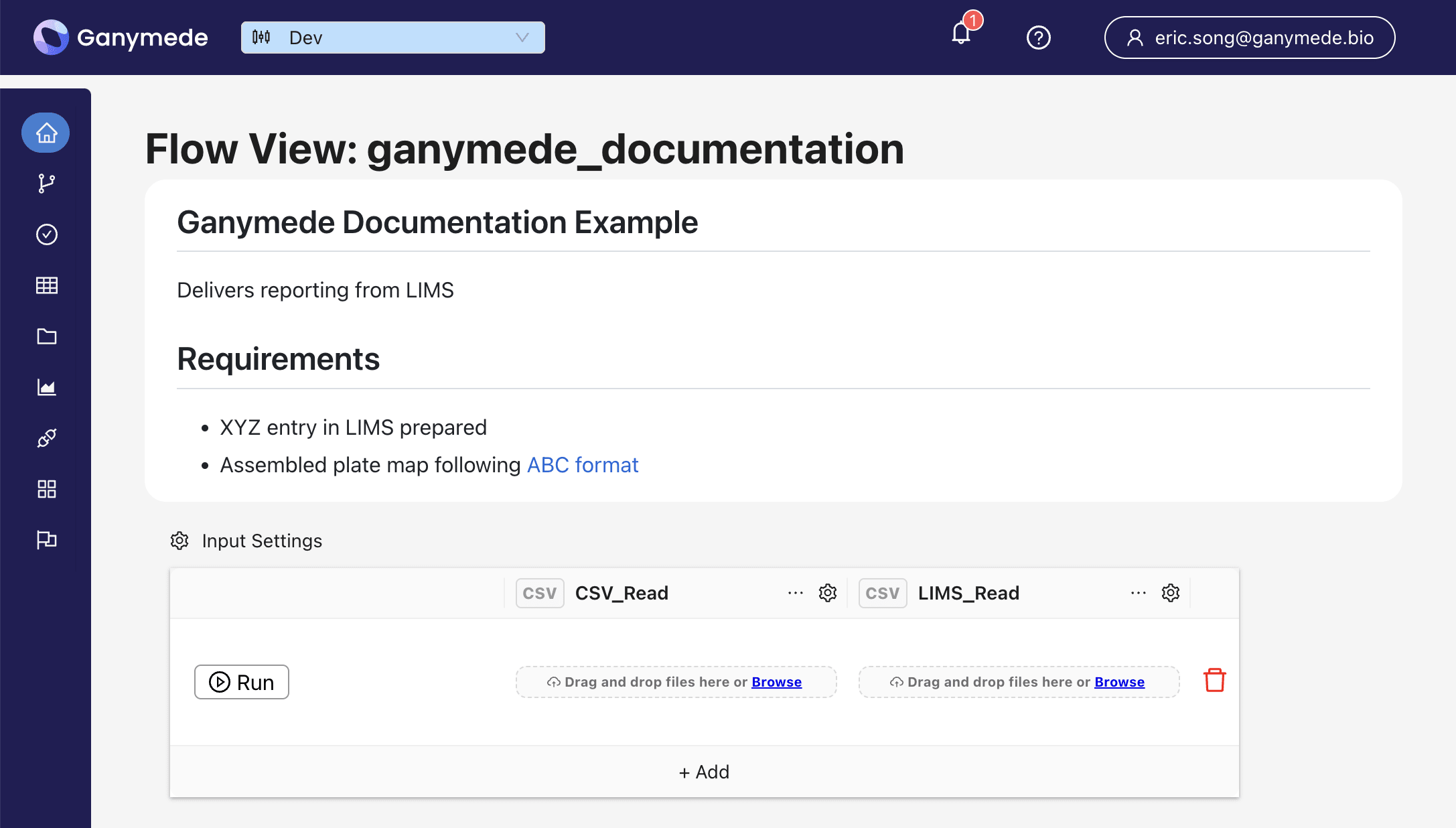
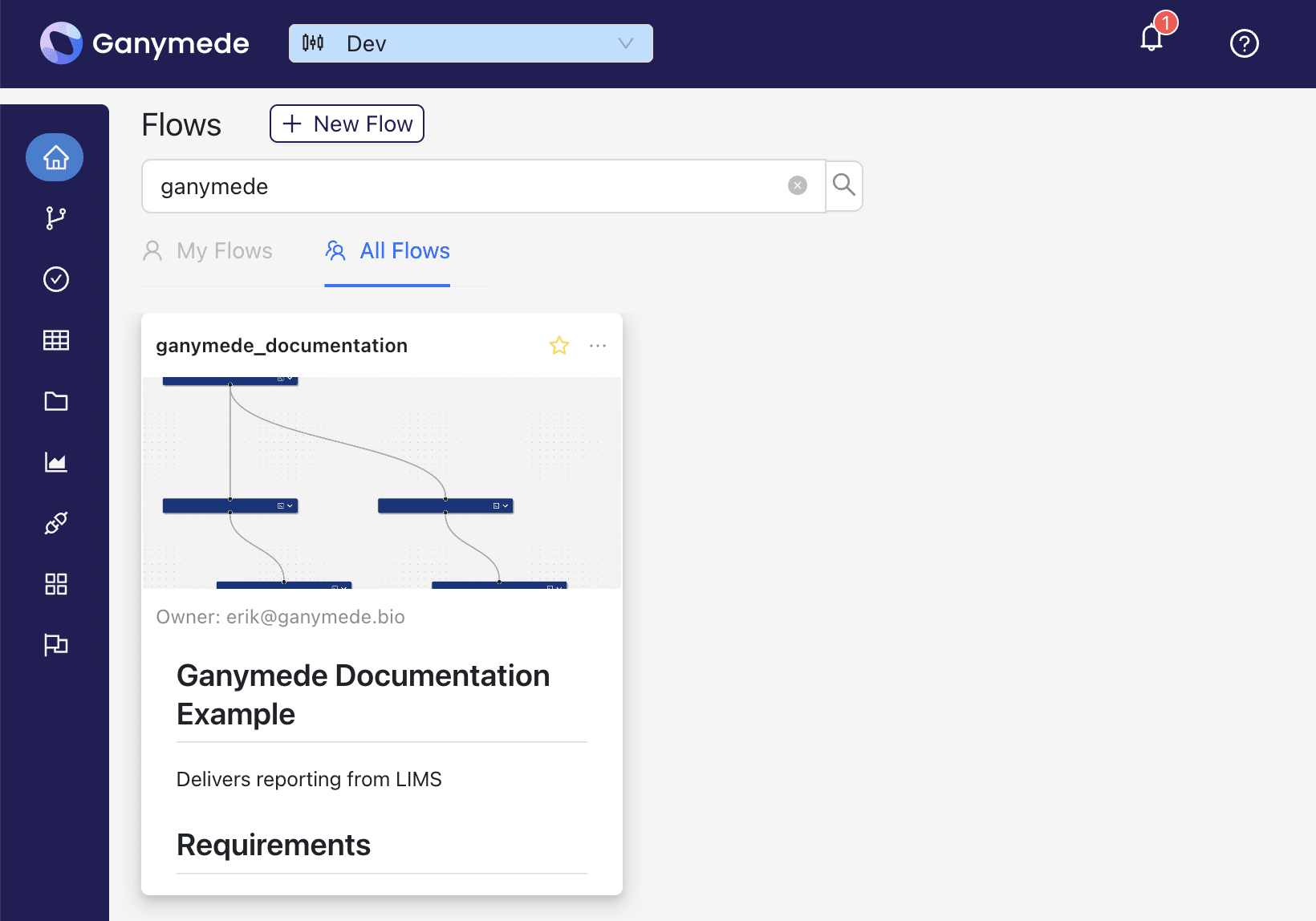
-A smaller preview of the Flow Documentation will also be shown in the Flow Cards of the Home page.
+and a smaller preview of the Flow Documentation is shown in the Flow cards on the Home page.
### Scheduling Flow Execution
diff --git a/docs/app/flows/QuickstartBuildFlow.mdx b/docs/app/flows/QuickstartBuildFlow.mdx
index b490ebf6..82c12a3c 100644
--- a/docs/app/flows/QuickstartBuildFlow.mdx
+++ b/docs/app/flows/QuickstartBuildFlow.mdx
@@ -13,6 +13,7 @@ import {
FolderOutlined,
PlayCircleOutlined,
PlusCircleOutlined,
+ PlusOutlined,
SaveFilled,
SettingOutlined,
TableOutlined,
@@ -30,31 +31,25 @@ This guide walks you through building a Flow for ingesting plate reader absorban
Flows are programs represented as directed graphs comprised of user-editable nodes. For precise definitions of Flows and nodes, check out [the Ganymede Concepts page](../intro/Concepts.mdx).
:::
-### Step 0: Download sample plate reader file
+### Step 1: Download sample plate reader file
Click [here](https://github.com/Ganymede-Bio/website-docusaurus/raw/main/PlateReader.xlsx) to download the plate reader file used for this tutorial.
-### Step 1: Sign into Ganymede
+### Step 2: Sign into Ganymede
Using Google Chrome, sign into Ganymede. The
Sign In
button will be available once authentication is configured for your tenant. This is generally a web address accessible via any web browser with the URL [https://\
.ganymede.bio](#).
You may need to enable pop-ups for Ganymede or disable any ad-blocking software for the ganymede.bio domain.
-

+
**Flow Editor**
button in the sidebar.
-
- -
### Step 3: Create a new Flow
-Click the
-
### Step 3: Create a new Flow
-Click the **Manage **
button in the upper right-hand corner of the header bar and select Add New Flow
from the right sidebar.
+Click the button on the Home page.
-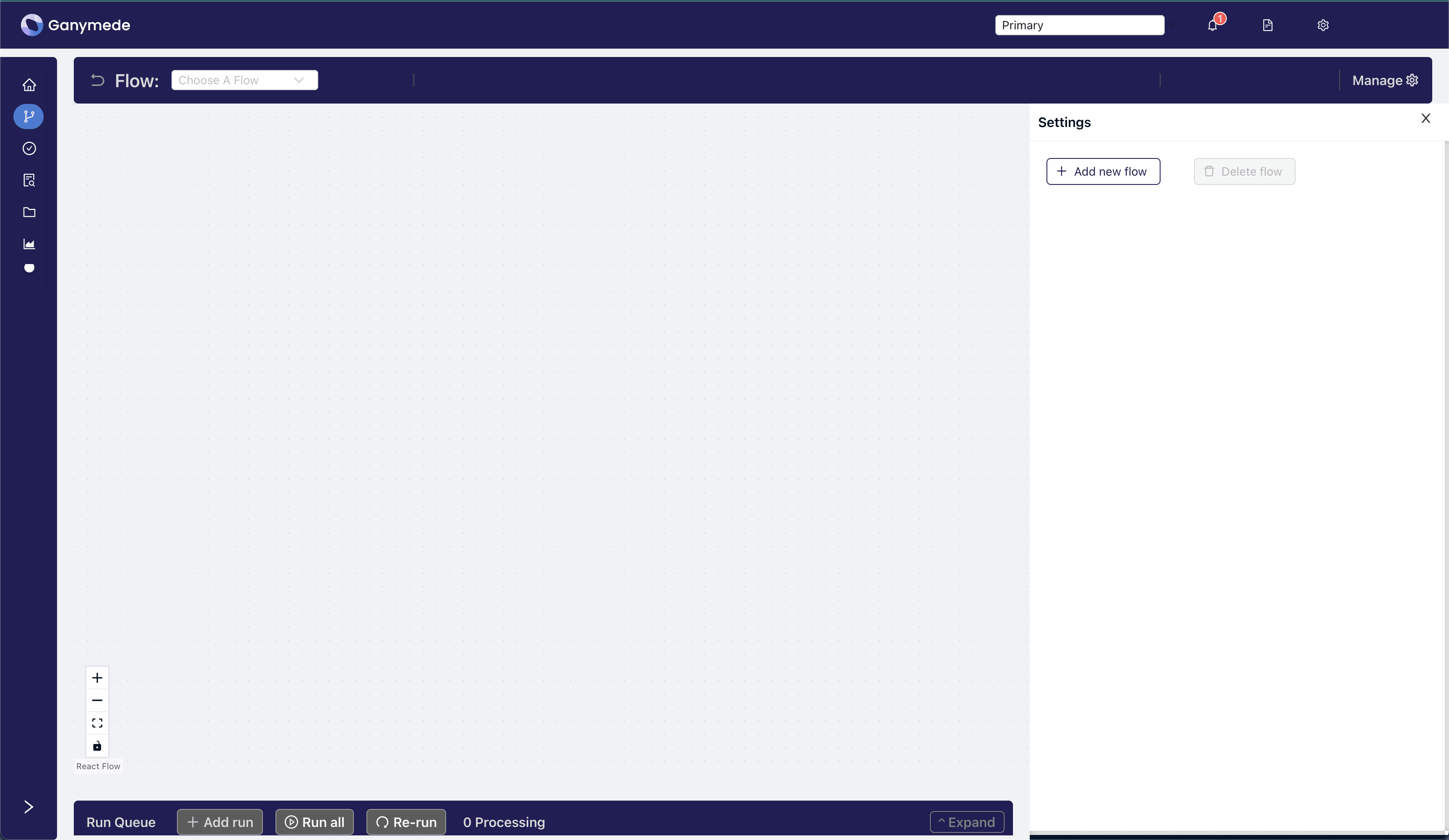 +
+ This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance_Change" as shown below, add a description, and then hit the "Create Flow" button.
@@ -63,7 +58,7 @@ This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance
This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance_Change" as shown below, add a description, and then hit the "Create Flow" button.
@@ -63,7 +58,7 @@ This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance
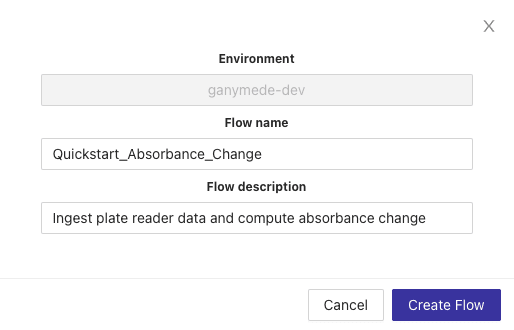
-After a few seconds, your Flow will be created and you'll be redirected to a blank Flow editor page where you can edit your new Flow.
+After a few seconds, your Flow will be created and you'll be redirected to a blank Flow editor page where you can edit your new Flow. Select your Flow from the Choose A Flow dropdown.
### Step 4: Add nodes to ingest and process data
@@ -82,7 +77,7 @@ Click the downward-pointing carat
-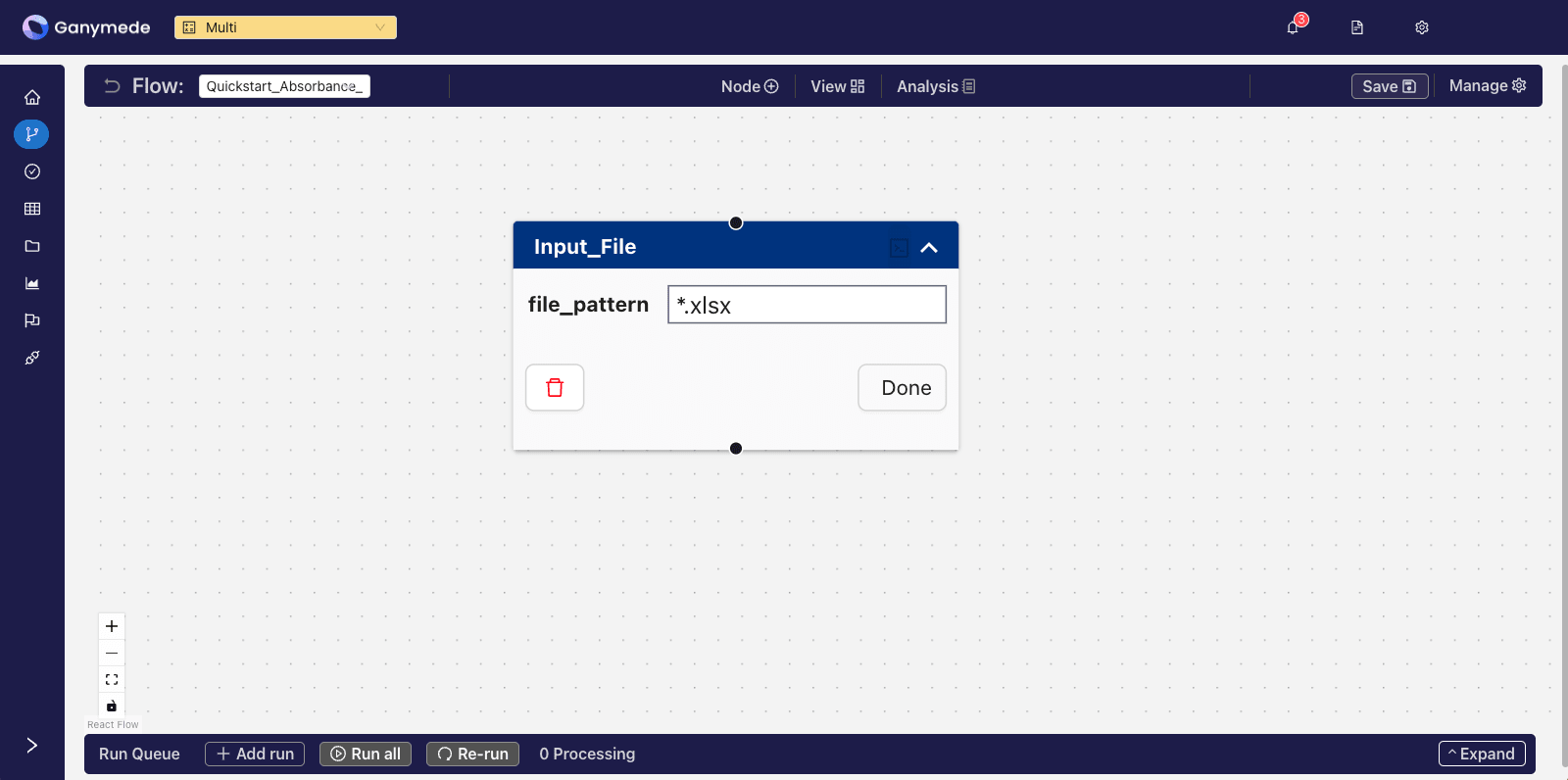 +
+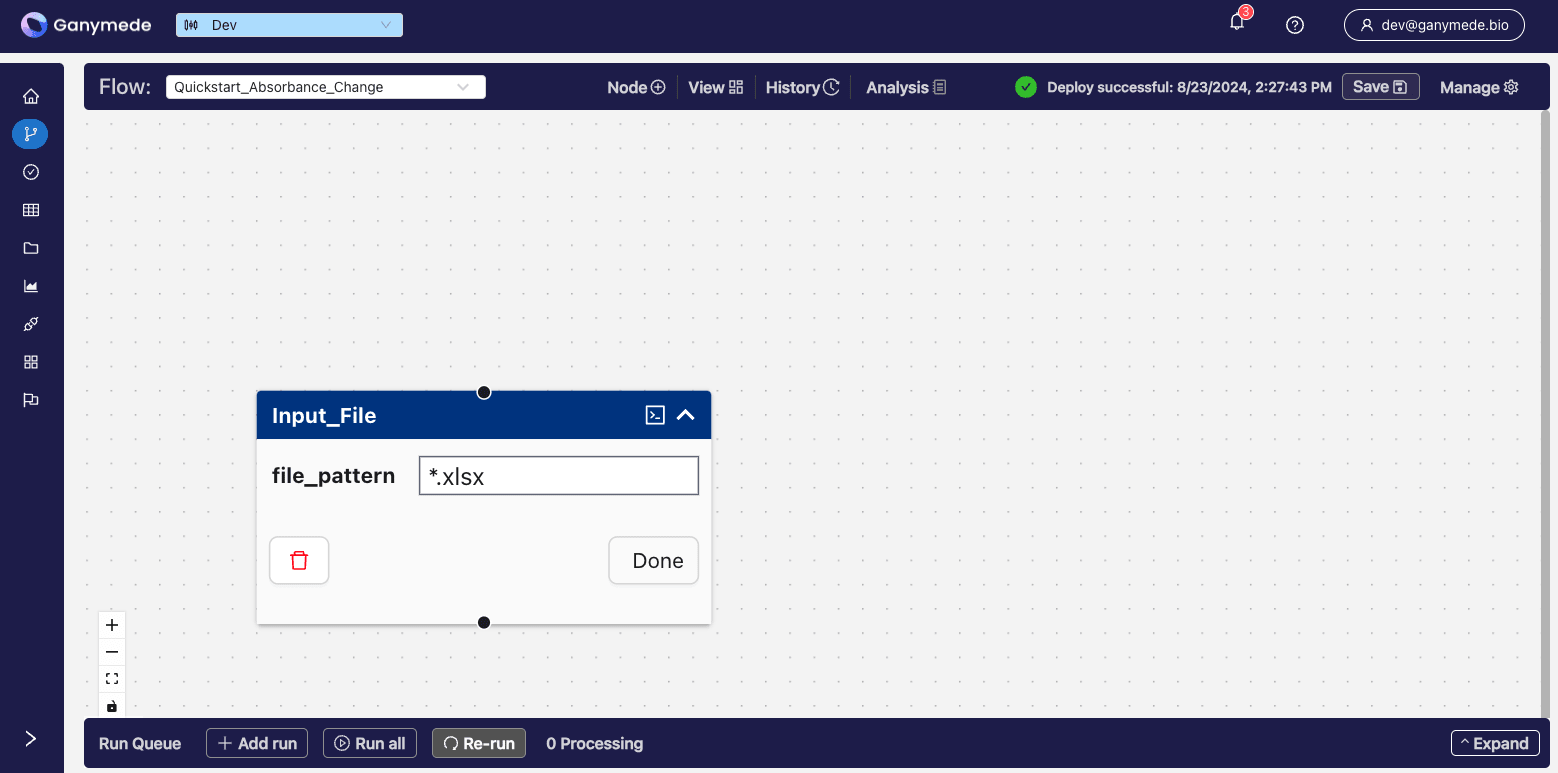
@@ -93,18 +88,17 @@ You can remove extension validation by leaving the file pattern attribute blank.
Click
**Save**
in the header to save this change.
:::tip
-A message will pop up in the upper right when the save is complete. You can also click the
button in the upper right corner to view the Notifications page where you can track the progress of a save.
+A green check box will pop up when the save and deploy are complete. You can also click the
button in the upper right corner to view the Notifications page, where you can track the progress of a save.
:::
-Now you have a one-node Flow that is capable of ingesting Excel files. Let's add a second node to process this incoming raw data.
+Now you have a one-node Flow that is capable of ingesting Excel files. Let's add a second node to process this incoming raw data. Click the
button in the header bar again, and search for the term "Python". This time, add a
**Python**
node to your Flow by clicking the
**Add**
button.
-Click the
button in the header bar again, and search for the term "Python". This time, add a
**Python**
node to your Flow by clicking the
**Add**
button.
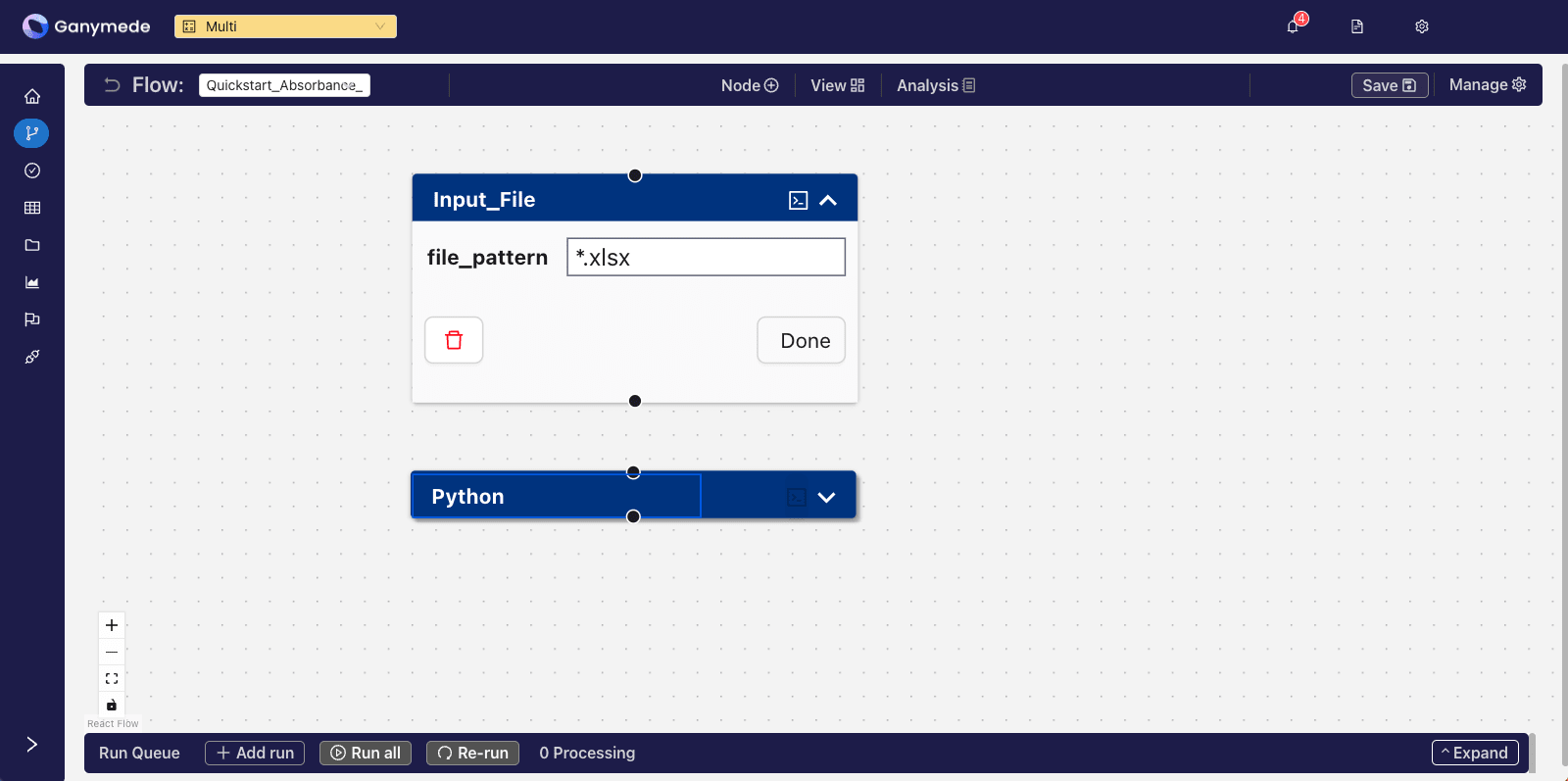
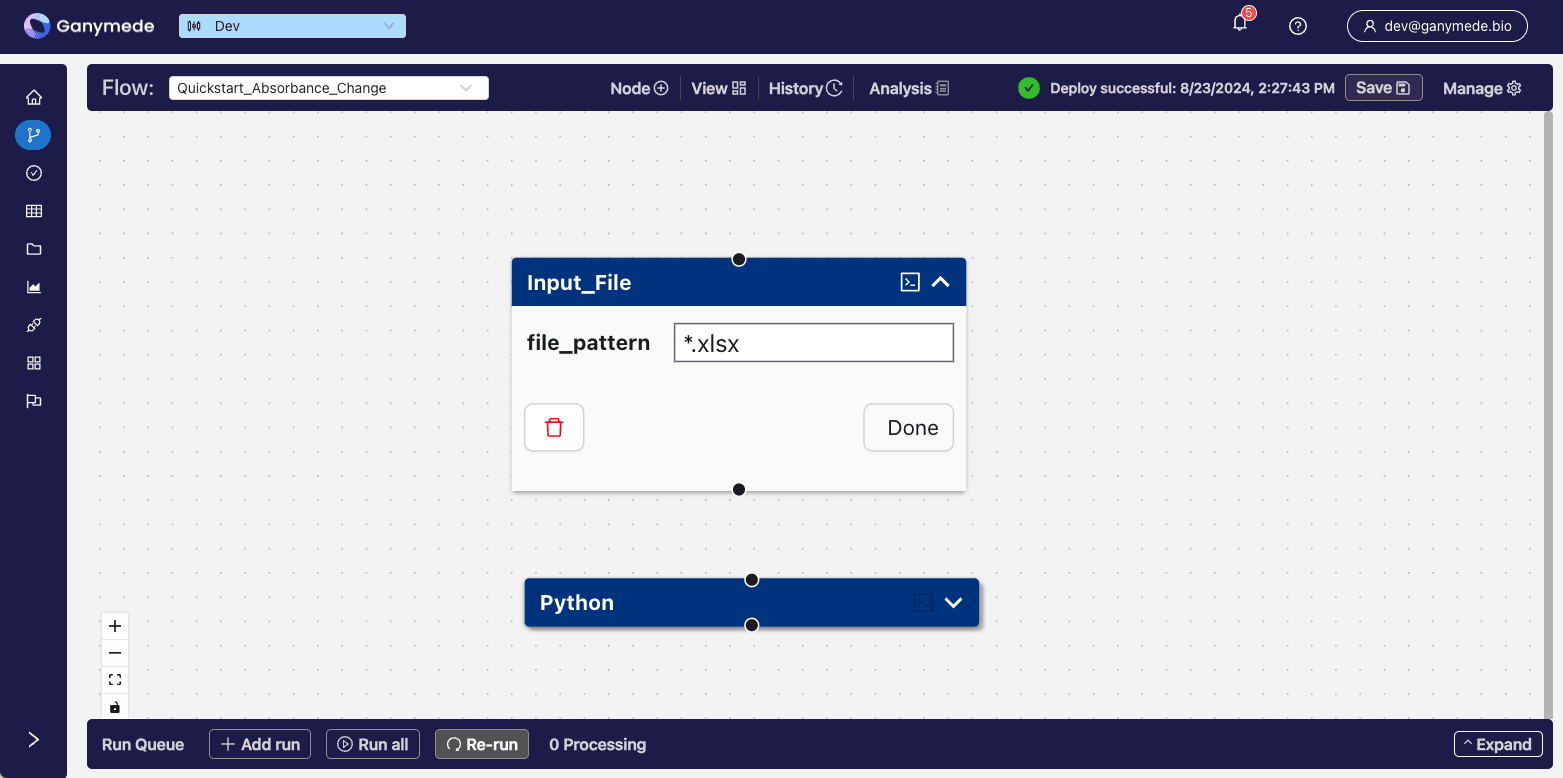
+After adding the Node, the
**Flow Editor**
window should look like this:
-After adding the node, the
**Flow Editor**
window should look like this:
-
-
+
+
-We want data processing to Flow from the
**Input_File**
node to the
**Python**
node. To do this, click on the
**Input_File**
node's bottom attachment point and drag the connection to the top of the
**Python**
node. Then click
**Save**
.
+We want data processing to flow from the
**Input_File**
Node to the
**Python**
Node. To do this, click on the
**Input_File**
node's bottom attachment point and drag the connection to the top of the
**Python**
node. Then click
**Save**
.
### Step 5: Modify the Input File node to store results in a table
@@ -146,7 +140,7 @@ def execute(file_data: Dict[str, bytes], ganymede_context=None) -> NodeReturn:
```
:::info non-tabular data
-Non-tabular data can be stored by returning a dictionary of files to upload in the `files_to_upload` key of the `NodeReturn` object.
+Non-tabular data can be stored by returning a dictionary of files to upload in the `files_to_upload` key of the `NodeReturn` object. Stored files can be retrieved from the [Files page](../files/FileBrowser.mdx) or the [Flow Runs panel](../flows/FlowRuns.mdx).
:::
Run the **User-Defined Function** cell by selecting the cell and then clicking on the
▶️ Run
button in the header, which runs the selected cell. In the "Testing" section below, you can test uploading the original plate reader excel file and passing it into the **User-Defined Function**. This is a great way to debug your code before deploying the Flow.
@@ -154,7 +148,6 @@ Run the **User-Defined Function** cell by selecting the cell and then clicking o
Save and deploy changes made to the Python node by clicking the
button in the toolbar, or by pressing `Cmd+S` (Mac) or `Ctrl+S` (Windows/Linux).
-
:::tip
Below the **User-Defined Function** cell in the notebook, you'll find the **Testing Section**. Here, you can upload a test input file and test the processing performed by the **User-Defined Function** interactively. This is a great way to debug your code before deploying the Flow.
:::
@@ -185,24 +178,37 @@ Next, modify the **User-Defined Function** cell to calculate the absorbance diff
import pandas as pd
from ganymede_sdk.io import NodeReturn
from typing import Union, Dict, List
+from io import BytesIO
+
def execute(
df_sql_result: Union[pd.DataFrame, List[pd.DataFrame]], ganymede_context=None
) -> Union[pd.DataFrame, Dict[str, pd.DataFrame]]:
# remove fields that do not contain well measurements
- df_out = df_sql_result[~df_sql_result['field'].isin(['Cycle Nr.', 'Time [s]', 'Temp. [°C]'])].copy()
-
+ df_out = df_sql_result[
+ ~df_sql_result["field"].isin(["Cycle Nr.", "Time [s]", "Temp. [°C]"])
+ ].copy()
+
# calculate absorbance difference
- df_out['run_diff'] = df_out['run2'] - df_out['run1']
+ df_out["run_diff"] = df_out["run2"] - df_out["run1"]
+
+ # output to CSV file
+ bio = BytesIO()
+ df_out.to_csv(bio, index=False)
+ bio.seek(0)
+ csv_file_output = bio.read()
- return df_out
+ return NodeReturn(
+ tables_to_upload={"quickstart_Python_results": df_out},
+ files_to_upload={"absorbance_diff.csv": csv_file_output},
+ )
```
-This code calculates the difference between the two columns `run1` and `run2`, and stores the result in a new column called `run_diff`.
+This code calculates the difference between the two columns `run1` and `run2`, and stores the result in a new column called `run_diff`. It also outputs a file called absorbance_diff.csv, which contains the results in CSV format.
:::tip
-Near the bottom of the notebook you'll find the **Testing Section**. As with the Input_File node, you can interactively test the SQL query and the **User-Defined Function**.
+Near the bottom of the notebook you'll find the **Testing Section**. As with the Input_File node, you can interactively test the SQL query and the **User-Defined Function**. Note that tables are not generated until the Flow is run (from the Flow Editor, not the notebook), and so the Testing section for this Node will not run successfully unless the Input File Node has been run in a Flow.
:::
After saving the Flow, close the editor notebook associated with the Python node to return to the
**Flow Editor**
view.
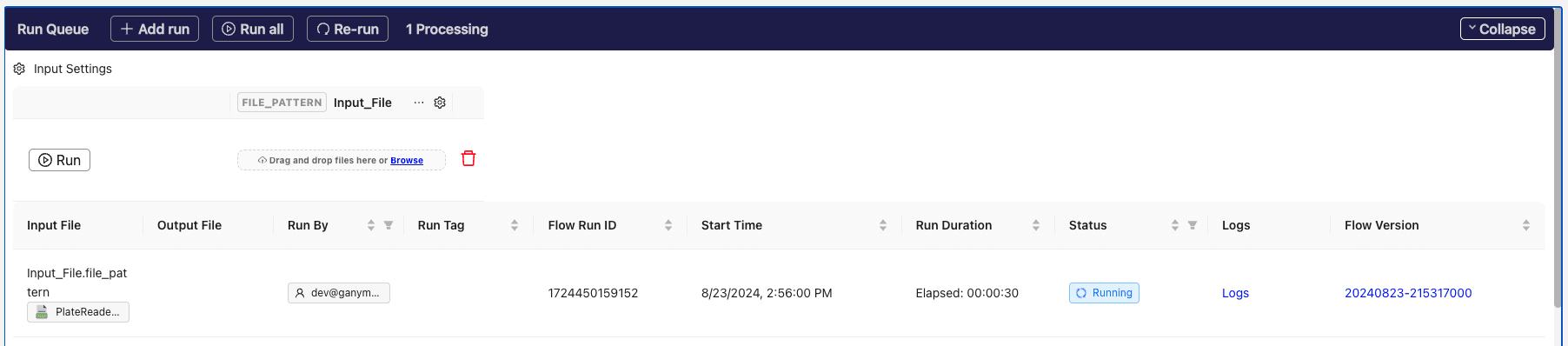
@@ -220,7 +226,7 @@ The associated file ingested into the Flow can be now found in the
button to kick off the Flow. The running Flow run will now be visible below.
-

+
:::tip
You can also click on
**Flow Runs**
in the left sidebar to monitor the current run or browse past runs.
@@ -228,7 +234,7 @@ You can also click on
+

### Step 8: Observe results in Data Explorer
diff --git a/docs/releases/AgentVersionHistory.md b/docs/releases/AgentVersionHistory.md
index 4166930b..cd2d8af4 100644
--- a/docs/releases/AgentVersionHistory.md
+++ b/docs/releases/AgentVersionHistory.md
@@ -6,11 +6,15 @@ displayed_sidebar: releaseSideBar
This page contains a summary of key version updates for the Ganymede [Agent](../app/agents/Agent.mdx). Since the individual instances of Agents (connections) are not updated when the Agent is updated, this page can be helpful for determining the capabilities of specific connections previously installed.
+## 4.9.0
+
+- Ganymede database tables can be accessed using Ganymede SDK
+- File watching is now event-driven rather than polling-based
+
## 4.8.0
- Connection logs are now systematically recorded on method calls to ease debugging
- Connection versions can now be remotely updated for non-major updates
-- Ganymede database tables can be accessed using Ganymede SDK
- Windows Agent Connection names can now have spaces
- Performance improvements made to checking for file existence in Ganymede
@@ -25,9 +29,6 @@ This page contains a summary of key version updates for the Ganymede [Agent](../
## 4.5.0
- Connection configuration can be updated remotely from the Ganymede Cloud. For example, the input path being watched by the Agent can be updated remotely.
-
-## 4.4.12
-
- Added status redundancy check on separate thread to ensure accuracy of connection status and improve connection stability
- Status checks performed over 443 rather that via ICMP
 +
+ 
 +
+ 
 -### Step 3: Download and install the agent
+### Step 3: Download and install the Agent Connection
-If you're not already on the local PC where the agent will be installed, navigate to that PC and sign into your tenant (as described in [step 1](#step-1-sign-into-ganymede)).
+If you're not already on the local PC where the Agent Connection will be installed, navigate to that PC and sign into your tenant (as described in [step 1](#step-1-sign-into-ganymede)).
#### Step 3a: Sync Windows time (if necessary)
@@ -81,7 +81,7 @@ If you choose a pre-existing folder, be aware that the agent will, by default, n
:::
-#### Step 3c: Download the agent
+#### Step 3c: Download the Agent Installer
Once the Agent build is complete, the associated installer will be available for download from the Connections pane.
@@ -96,9 +96,9 @@ After downloading, run the executable to start the installation process. Name th
:::tip
-Be precise when configuring the watch folder path, as errors here can be difficult to troubleshoot. Ensure the input path is correctly specified. For example, if OneDrive is used to back up Desktop files, the path may be "C:\Users\
-### Step 3: Download and install the agent
+### Step 3: Download and install the Agent Connection
-If you're not already on the local PC where the agent will be installed, navigate to that PC and sign into your tenant (as described in [step 1](#step-1-sign-into-ganymede)).
+If you're not already on the local PC where the Agent Connection will be installed, navigate to that PC and sign into your tenant (as described in [step 1](#step-1-sign-into-ganymede)).
#### Step 3a: Sync Windows time (if necessary)
@@ -81,7 +81,7 @@ If you choose a pre-existing folder, be aware that the agent will, by default, n
:::
-#### Step 3c: Download the agent
+#### Step 3c: Download the Agent Installer
Once the Agent build is complete, the associated installer will be available for download from the Connections pane.
@@ -96,9 +96,9 @@ After downloading, run the executable to start the installation process. Name th
:::tip
-Be precise when configuring the watch folder path, as errors here can be difficult to troubleshoot. Ensure the input path is correctly specified. For example, if OneDrive is used to back up Desktop files, the path may be "C:\Users\ @@ -149,7 +149,8 @@ Now that you have successfully created and installed an Agent, you can explore m
- An Agent that [runs on a recurring schedule](./Agent#set-a-cron-job-to-run-flows-periodically)
- An Agent that [executes local processing when a file stored in cloud storage](./Agent#watch-for-flow-outputs-then-save-locally)
- [How to modify the agent code](./Agent#watch-for-files-locally-then-run-flow) to add custom functionality, such as
- - [Adding tags to files](../files/Tags.mdx) to make them easier to find in Ganymede
+ - [Adding tags to files](../files/Tags.mdx) to make captured files easier to find in Ganymede
- Parsing metadata from file contents to determine how files are processed
- - Delivering [multiple files into a single node](../../sdk/markdowns/AgentModelsAndMethods#classes-for-agent-triggered-flows)
-- Incorporating [Agent file utilities](../../sdk/markdowns/AgentModelsAndMethods#file-system-utilities) from the Ganymede SDK
+ - Delivering [multiple files into a single Node](../../sdk/markdowns/AgentModelsAndMethods#classes-for-agent-triggered-flows)
+- Incorporating [Agent utility functions](../../sdk/markdowns/AgentModelsAndMethods) from the Ganymede SDK and Agent SDK
+- Interpreting [Agent log messages](./AgentLogs)
diff --git a/docs/app/flows/FlowEditor.mdx b/docs/app/flows/FlowEditor.mdx
index 44fa6ec9..e20b18b8 100644
--- a/docs/app/flows/FlowEditor.mdx
+++ b/docs/app/flows/FlowEditor.mdx
@@ -89,27 +89,23 @@ To create a new Flow, click the
@@ -149,7 +149,8 @@ Now that you have successfully created and installed an Agent, you can explore m
- An Agent that [runs on a recurring schedule](./Agent#set-a-cron-job-to-run-flows-periodically)
- An Agent that [executes local processing when a file stored in cloud storage](./Agent#watch-for-flow-outputs-then-save-locally)
- [How to modify the agent code](./Agent#watch-for-files-locally-then-run-flow) to add custom functionality, such as
- - [Adding tags to files](../files/Tags.mdx) to make them easier to find in Ganymede
+ - [Adding tags to files](../files/Tags.mdx) to make captured files easier to find in Ganymede
- Parsing metadata from file contents to determine how files are processed
- - Delivering [multiple files into a single node](../../sdk/markdowns/AgentModelsAndMethods#classes-for-agent-triggered-flows)
-- Incorporating [Agent file utilities](../../sdk/markdowns/AgentModelsAndMethods#file-system-utilities) from the Ganymede SDK
+ - Delivering [multiple files into a single Node](../../sdk/markdowns/AgentModelsAndMethods#classes-for-agent-triggered-flows)
+- Incorporating [Agent utility functions](../../sdk/markdowns/AgentModelsAndMethods) from the Ganymede SDK and Agent SDK
+- Interpreting [Agent log messages](./AgentLogs)
diff --git a/docs/app/flows/FlowEditor.mdx b/docs/app/flows/FlowEditor.mdx
index 44fa6ec9..e20b18b8 100644
--- a/docs/app/flows/FlowEditor.mdx
+++ b/docs/app/flows/FlowEditor.mdx
@@ -89,27 +89,23 @@ To create a new Flow, click the 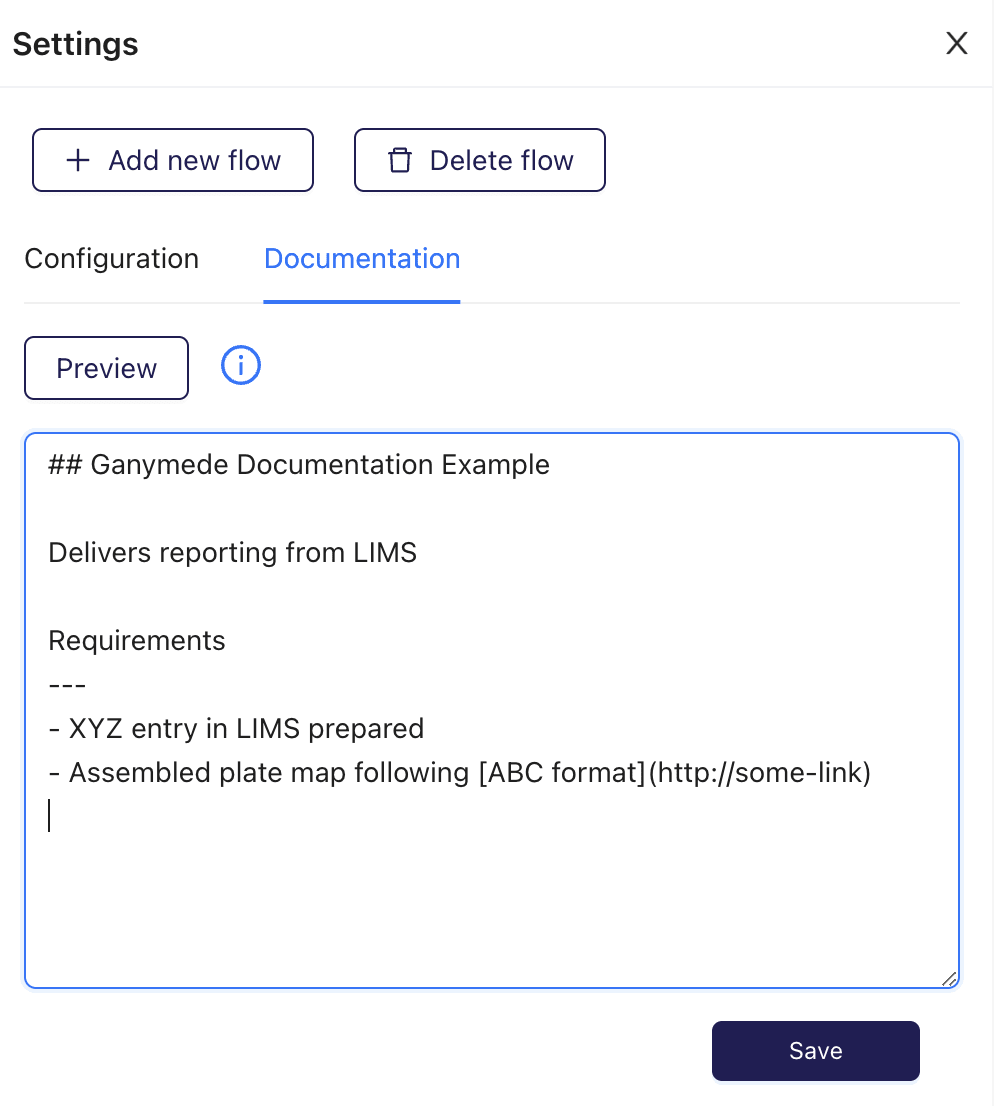
 -A smaller preview of the Flow Documentation will also be shown in the Flow Cards of the Home page.
+and a smaller preview of the Flow Documentation is shown in the Flow cards on the Home page.
-A smaller preview of the Flow Documentation will also be shown in the Flow Cards of the Home page.
+and a smaller preview of the Flow Documentation is shown in the Flow cards on the Home page.
 ### Scheduling Flow Execution
diff --git a/docs/app/flows/QuickstartBuildFlow.mdx b/docs/app/flows/QuickstartBuildFlow.mdx
index b490ebf6..82c12a3c 100644
--- a/docs/app/flows/QuickstartBuildFlow.mdx
+++ b/docs/app/flows/QuickstartBuildFlow.mdx
@@ -13,6 +13,7 @@ import {
FolderOutlined,
PlayCircleOutlined,
PlusCircleOutlined,
+ PlusOutlined,
SaveFilled,
SettingOutlined,
TableOutlined,
@@ -30,31 +31,25 @@ This guide walks you through building a Flow for ingesting plate reader absorban
Flows are programs represented as directed graphs comprised of user-editable nodes. For precise definitions of Flows and nodes, check out [the Ganymede Concepts page](../intro/Concepts.mdx).
:::
-### Step 0: Download sample plate reader file
+### Step 1: Download sample plate reader file
Click [here](https://github.com/Ganymede-Bio/website-docusaurus/raw/main/PlateReader.xlsx) to download the plate reader file used for this tutorial.
-### Step 1: Sign into Ganymede
+### Step 2: Sign into Ganymede
Using Google Chrome, sign into Ganymede. The
### Scheduling Flow Execution
diff --git a/docs/app/flows/QuickstartBuildFlow.mdx b/docs/app/flows/QuickstartBuildFlow.mdx
index b490ebf6..82c12a3c 100644
--- a/docs/app/flows/QuickstartBuildFlow.mdx
+++ b/docs/app/flows/QuickstartBuildFlow.mdx
@@ -13,6 +13,7 @@ import {
FolderOutlined,
PlayCircleOutlined,
PlusCircleOutlined,
+ PlusOutlined,
SaveFilled,
SettingOutlined,
TableOutlined,
@@ -30,31 +31,25 @@ This guide walks you through building a Flow for ingesting plate reader absorban
Flows are programs represented as directed graphs comprised of user-editable nodes. For precise definitions of Flows and nodes, check out [the Ganymede Concepts page](../intro/Concepts.mdx).
:::
-### Step 0: Download sample plate reader file
+### Step 1: Download sample plate reader file
Click [here](https://github.com/Ganymede-Bio/website-docusaurus/raw/main/PlateReader.xlsx) to download the plate reader file used for this tutorial.
-### Step 1: Sign into Ganymede
+### Step 2: Sign into Ganymede
Using Google Chrome, sign into Ganymede. The  +
+
 -
### Step 3: Create a new Flow
-Click the
-
### Step 3: Create a new Flow
-Click the  +
+ This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance_Change" as shown below, add a description, and then hit the "Create Flow" button.
@@ -63,7 +58,7 @@ This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance
This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance_Change" as shown below, add a description, and then hit the "Create Flow" button.
@@ -63,7 +58,7 @@ This exposes a modal for adding a new Flow. Name the Flow "Quickstart_Absorbance

 +
+
 +
+ +
-We want data processing to Flow from the
+
-We want data processing to Flow from the  +
+ :::tip
You can also click on
:::tip
You can also click on  ### Step 8: Observe results in Data Explorer
diff --git a/docs/releases/AgentVersionHistory.md b/docs/releases/AgentVersionHistory.md
index 4166930b..cd2d8af4 100644
--- a/docs/releases/AgentVersionHistory.md
+++ b/docs/releases/AgentVersionHistory.md
@@ -6,11 +6,15 @@ displayed_sidebar: releaseSideBar
This page contains a summary of key version updates for the Ganymede [Agent](../app/agents/Agent.mdx). Since the individual instances of Agents (connections) are not updated when the Agent is updated, this page can be helpful for determining the capabilities of specific connections previously installed.
+## 4.9.0
+
+- Ganymede database tables can be accessed using Ganymede SDK
+- File watching is now event-driven rather than polling-based
+
## 4.8.0
- Connection logs are now systematically recorded on method calls to ease debugging
- Connection versions can now be remotely updated for non-major updates
-- Ganymede database tables can be accessed using Ganymede SDK
- Windows Agent Connection names can now have spaces
- Performance improvements made to checking for file existence in Ganymede
@@ -25,9 +29,6 @@ This page contains a summary of key version updates for the Ganymede [Agent](../
## 4.5.0
- Connection configuration can be updated remotely from the Ganymede Cloud. For example, the input path being watched by the Agent can be updated remotely.
-
-## 4.4.12
-
- Added status redundancy check on separate thread to ensure accuracy of connection status and improve connection stability
- Status checks performed over 443 rather that via ICMP
### Step 8: Observe results in Data Explorer
diff --git a/docs/releases/AgentVersionHistory.md b/docs/releases/AgentVersionHistory.md
index 4166930b..cd2d8af4 100644
--- a/docs/releases/AgentVersionHistory.md
+++ b/docs/releases/AgentVersionHistory.md
@@ -6,11 +6,15 @@ displayed_sidebar: releaseSideBar
This page contains a summary of key version updates for the Ganymede [Agent](../app/agents/Agent.mdx). Since the individual instances of Agents (connections) are not updated when the Agent is updated, this page can be helpful for determining the capabilities of specific connections previously installed.
+## 4.9.0
+
+- Ganymede database tables can be accessed using Ganymede SDK
+- File watching is now event-driven rather than polling-based
+
## 4.8.0
- Connection logs are now systematically recorded on method calls to ease debugging
- Connection versions can now be remotely updated for non-major updates
-- Ganymede database tables can be accessed using Ganymede SDK
- Windows Agent Connection names can now have spaces
- Performance improvements made to checking for file existence in Ganymede
@@ -25,9 +29,6 @@ This page contains a summary of key version updates for the Ganymede [Agent](../
## 4.5.0
- Connection configuration can be updated remotely from the Ganymede Cloud. For example, the input path being watched by the Agent can be updated remotely.
-
-## 4.4.12
-
- Added status redundancy check on separate thread to ensure accuracy of connection status and improve connection stability
- Status checks performed over 443 rather that via ICMP