diff --git a/.github/PULL_REQUEST_TEMPLATE.md b/.github/PULL_REQUEST_TEMPLATE.md
index 56cbd186..bb946ef2 100644
--- a/.github/PULL_REQUEST_TEMPLATE.md
+++ b/.github/PULL_REQUEST_TEMPLATE.md
@@ -45,4 +45,4 @@ Please describe the tests that you ran to verify your changes. Provide instructi
- [ ] My changes generate no new warnings
- [ ] I have added tests that prove my fix is effective or that my feature works
- [ ] New and existing unit tests pass locally with my changes
-- [ ] Any dependent changes have been merged and published in downstream modules
\ No newline at end of file
+- [ ] Any dependent changes have been merged and published in downstream modules
diff --git a/.gitignore b/.gitignore
index 26133bb2..fa94582a 100644
--- a/.gitignore
+++ b/.gitignore
@@ -104,7 +104,7 @@ ENV/
# Rope project settings
.ropeproject
-# DS Store
+# DS Store
.DS_Store
# for installation purposes
@@ -114,4 +114,4 @@ install-dev
/catboost_info/
# Data
-shapash/data/telco_customer_churn.csv
\ No newline at end of file
+shapash/data/telco_customer_churn.csv
diff --git a/.pre-commit-config.yaml b/.pre-commit-config.yaml
new file mode 100644
index 00000000..29ae367f
--- /dev/null
+++ b/.pre-commit-config.yaml
@@ -0,0 +1,70 @@
+default_language_version:
+ python: python3
+repos:
+- repo: https://github.com/pre-commit/pre-commit-hooks
+ rev: v3.2.0
+ hooks:
+ - id: check-ast
+ - id: check-byte-order-marker
+ - id: check-case-conflict
+ - id: check-docstring-first
+ - id: check-executables-have-shebangs
+ - id: check-json
+ - id: check-yaml
+ exclude: ^chart/
+ - id: debug-statements
+ - id: end-of-file-fixer
+ exclude: ^(docs/|gdocs/)
+ - id: pretty-format-json
+ args: ['--autofix']
+ exclude: .ipynb
+ - id: trailing-whitespace
+ args: ['--markdown-linebreak-ext=md']
+ exclude: ^(docs/|gdocs/)
+ - id: mixed-line-ending
+ args: ['--fix=lf']
+ exclude: ^(docs/|gdocs/)
+ - id: check-added-large-files
+ args: ['--maxkb=500']
+ - id: no-commit-to-branch
+ args: ['--branch', 'master', '--branch', 'develop']
+- repo: https://github.com/psf/black
+ rev: 21.12b0
+ hooks:
+ - id: black
+ args: [--line-length=120]
+ additional_dependencies: ['click==8.0.4']
+#- repo: https://github.com/pre-commit/mirrors-mypy
+# rev: 'v0.931'
+# hooks:
+# - id: mypy
+# args: [--ignore-missing-imports, --disallow-untyped-defs, --show-error-codes, --no-site-packages]
+# files: src
+# - repo: https://github.com/PyCQA/flake8
+# rev: 6.0.0
+# hooks:
+# - id: flake8
+# exclude: ^tests/
+# args: ['--ignore=E501,D2,D3,D4,D104,D100,D106,D107,W503,D105,E203']
+# additional_dependencies: [ flake8-docstrings, "flake8-bugbear==22.8.23" ]
+- repo: https://github.com/pre-commit/mirrors-isort
+ rev: v5.4.2
+ hooks:
+ - id: isort
+ args: ["--profile", "black", "-l", "120"]
+- repo: https://github.com/asottile/pyupgrade
+ rev: v2.7.2
+ hooks:
+ - id: pyupgrade
+ args: [--py37-plus]
+- repo: https://github.com/asottile/blacken-docs
+ rev: v1.8.0
+ hooks:
+ - id: blacken-docs
+ additional_dependencies: [black==21.12b0]
+- repo: https://github.com/compilerla/conventional-pre-commit
+ rev: v2.1.1
+ hooks:
+ - id: conventional-pre-commit
+ stages: [commit-msg]
+ args: [] # optional: list of Conventional Commits types to allow e.g. [feat, fix, ci, chore, test]
diff --git a/.readthedocs.yml b/.readthedocs.yml
index f44b00d3..0c09bd5e 100644
--- a/.readthedocs.yml
+++ b/.readthedocs.yml
@@ -21,7 +21,7 @@ build:
os: ubuntu-20.04
tools:
python: "3.10"
-
+
# Optionally set the version of Python and requirements required to build your docs
python:
install:
diff --git a/CONTRIBUTING.md b/CONTRIBUTING.md
index 0e7bc96f..7a650ec8 100644
--- a/CONTRIBUTING.md
+++ b/CONTRIBUTING.md
@@ -58,7 +58,7 @@ To contribute to Shapash, you will need to create a personal branch.
```
git checkout -b feature/my-contribution-branch
```
-We recommand to use a convention of naming branch.
+We recommand to use a convention of naming branch.
- **feature/your_feature_name** if you are creating a feature
- **hotfix/your_bug_fix** if you are fixing a bug
@@ -70,7 +70,7 @@ Before committing your modifications, we have some recommendations :
```
pytest
```
-- Try to build Shapash
+- Try to build Shapash
```
python setup.py bdist_wheel
```
@@ -91,7 +91,7 @@ git commit -m ‘fixed a bug’
git push origin feature/my-contribution-branch
```
-Your branch is now available on your remote forked repository, with your changes.
+Your branch is now available on your remote forked repository, with your changes.
Next step is now to create a Pull Request so the Shapash Team can add your changes to the official repository.
@@ -104,7 +104,7 @@ To create one, on the top of your forked repository, you will find a button "Com
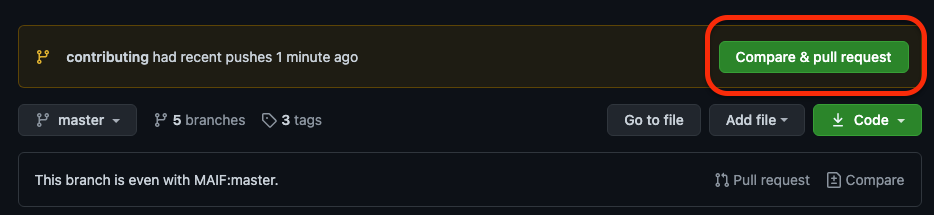 -As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
+As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
On the left side, you will find the official Shapash repository.
@@ -130,4 +130,4 @@ Your pull request is now ready to be submitted. A member of the Shapash team wil
You have contributed to an Open source project, thank you and congratulations ! 🥳
-Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
\ No newline at end of file
+Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
diff --git a/LICENSE b/LICENSE
index 4947287f..f433b1a5 100644
--- a/LICENSE
+++ b/LICENSE
@@ -174,4 +174,4 @@
incurred by, or claims asserted against, such Contributor by reason
of your accepting any such warranty or additional liability.
- END OF TERMS AND CONDITIONS
\ No newline at end of file
+ END OF TERMS AND CONDITIONS
diff --git a/Makefile b/Makefile
index 720b6909..1f65e00e 100644
--- a/Makefile
+++ b/Makefile
@@ -84,4 +84,4 @@ dist: clean ## builds source and wheel package
ls -l dist
install: clean ## install the package to the active Python's site-packages
- python setup.py install
\ No newline at end of file
+ python setup.py install
diff --git a/README.md b/README.md
index 8a159b3a..aee9e717 100644
--- a/README.md
+++ b/README.md
@@ -38,7 +38,7 @@ With Shapash, you can generate a **Webapp** that simplifies the comprehension of
Additionally, Shapash contributes to data science auditing by **presenting valuable information** about any model and data **in a comprehensive report**.
-Shapash is suitable for Regression, Binary Classification, and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
+Shapash is suitable for Regression, Binary Classification and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
> [!NOTE]
> If you want to give us feedback : [Feedback form](https://framaforms.org/shapash-collecting-your-feedback-and-use-cases-1687456776)
@@ -72,9 +72,9 @@ Shapash is suitable for Regression, Binary Classification, and Multiclass proble
| 2.2.x | Dataset Filter
-As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
+As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
On the left side, you will find the official Shapash repository.
@@ -130,4 +130,4 @@ Your pull request is now ready to be submitted. A member of the Shapash team wil
You have contributed to an Open source project, thank you and congratulations ! 🥳
-Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
\ No newline at end of file
+Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
diff --git a/LICENSE b/LICENSE
index 4947287f..f433b1a5 100644
--- a/LICENSE
+++ b/LICENSE
@@ -174,4 +174,4 @@
incurred by, or claims asserted against, such Contributor by reason
of your accepting any such warranty or additional liability.
- END OF TERMS AND CONDITIONS
\ No newline at end of file
+ END OF TERMS AND CONDITIONS
diff --git a/Makefile b/Makefile
index 720b6909..1f65e00e 100644
--- a/Makefile
+++ b/Makefile
@@ -84,4 +84,4 @@ dist: clean ## builds source and wheel package
ls -l dist
install: clean ## install the package to the active Python's site-packages
- python setup.py install
\ No newline at end of file
+ python setup.py install
diff --git a/README.md b/README.md
index 8a159b3a..aee9e717 100644
--- a/README.md
+++ b/README.md
@@ -38,7 +38,7 @@ With Shapash, you can generate a **Webapp** that simplifies the comprehension of
Additionally, Shapash contributes to data science auditing by **presenting valuable information** about any model and data **in a comprehensive report**.
-Shapash is suitable for Regression, Binary Classification, and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
+Shapash is suitable for Regression, Binary Classification and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
> [!NOTE]
> If you want to give us feedback : [Feedback form](https://framaforms.org/shapash-collecting-your-feedback-and-use-cases-1687456776)
@@ -72,9 +72,9 @@ Shapash is suitable for Regression, Binary Classification, and Multiclass proble
| 2.2.x | Dataset Filter
| New tab in the webapp to filter data. And several improvements in the webapp: subtitles, labels, screen adjustments | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/tutorial01-Shapash-Overview-Launch-WebApp.ipynb)
| 2.0.x | Refactoring Shapash
](https://github.com/MAIF/shapash/blob/master/tutorial/tutorial01-Shapash-Overview-Launch-WebApp.ipynb)
| 2.0.x | Refactoring Shapash
| Refactoring attributes of compile methods and init. Refactoring implementation for new backends | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/explainer_and_backend/tuto-expl06-Shapash-custom-backend.ipynb)
| 1.7.x | Variabilize Colors
](https://github.com/MAIF/shapash/blob/master/tutorial/explainer_and_backend/tuto-expl06-Shapash-custom-backend.ipynb)
| 1.7.x | Variabilize Colors
| Giving possibility to have your own colour palette for outputs adapted to your design | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common02-colors.ipynb)
-| 1.6.x | Explainability Quality Metrics
](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common02-colors.ipynb)
-| 1.6.x | Explainability Quality Metrics
[Article](https://towardsdatascience.com/building-confidence-on-explainability-methods-66b9ee575514) | To help increase confidence in explainability methods, you can evaluate the relevance of your explainability using 3 metrics: **Stability**, **Consistency** and **Compacity** | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
-| 1.4.x | Groups of features
](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
-| 1.4.x | Groups of features
[Demo](https://shapash-demo2.ossbymaif.fr/) | You can now regroup features that share common properties together.
This option can be useful if your model has a lot of features. | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
-| 1.3.x | Shapash Report
](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
-| 1.3.x | Shapash Report
[Demo](https://shapash.readthedocs.io/en/latest/report.html) | A standalone HTML report that constitutes a basis of an audit document. | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/generate_report/tuto-shapash-report01.ipynb) |
+| 1.6.x | Explainability Quality Metrics
](https://github.com/MAIF/shapash/blob/master/tutorial/generate_report/tuto-shapash-report01.ipynb) |
+| 1.6.x | Explainability Quality Metrics
[Article](https://towardsdatascience.com/building-confidence-on-explainability-methods-66b9ee575514) | To help increase confidence in explainability methods, you can evaluate the relevance of your explainability using 3 metrics: **Stability**, **Consistency** and **Compacity** | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
+| 1.4.x | Groups of features
](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
+| 1.4.x | Groups of features
[Demo](https://shapash-demo2.ossbymaif.fr/) | You can now regroup features that share common properties together.
This option can be useful if your model has a lot of features. | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
+| 1.3.x | Shapash Report
](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
+| 1.3.x | Shapash Report
[Demo](https://shapash.readthedocs.io/en/latest/report.html) | A standalone HTML report that constitutes a basis of an audit document. | [ ](https://github.com/MAIF/shapash/blob/master/tutorial/generate_report/tuto-shapash-report01.ipynb) |
## 🔥 Features
@@ -83,19 +83,19 @@ Shapash is suitable for Regression, Binary Classification, and Multiclass proble
](https://github.com/MAIF/shapash/blob/master/tutorial/generate_report/tuto-shapash-report01.ipynb) |
## 🔥 Features
@@ -83,19 +83,19 @@ Shapash is suitable for Regression, Binary Classification, and Multiclass proble

 -
-  +
+ 

 -
-  +
+ 

 -
-  +
+ 
@@ -145,13 +145,13 @@ Shapash can use category-encoders object, sklearn ColumnTransformer or simply fe
Shapash is intended to work with Python versions 3.8 to 3.11. Installation can be done with pip:
-```python
+```bash
pip install shapash
```
In order to generate the Shapash Report some extra requirements are needed.
You can install these using the following command :
-```python
+```bash
pip install shapash[report]
```
@@ -167,24 +167,25 @@ The 4 steps to display results:
```python
from shapash import SmartExplainer
+
xpl = SmartExplainer(
- model=regressor,
- features_dict=house_dict, # Optional parameter
- preprocessing=encoder, # Optional: compile step can use inverse_transform method
- postprocessing=postprocess, # Optional: see tutorial postprocessing
+ model=regressor,
+ features_dict=house_dict, # Optional parameter
+ preprocessing=encoder, # Optional: compile step can use inverse_transform method
+ postprocessing=postprocess, # Optional: see tutorial postprocessing
)
```
- Step 2: Compile Dataset, ...
> There 1 mandatory parameter in compile method: Dataset
-
+
```python
xpl.compile(
- x=xtest,
- y_pred=y_pred, # Optional: for your own prediction (by default: model.predict)
- y_target=yTest, # Optional: allows to display True Values vs Predicted Values
- additional_data=xadditional, # Optional: additional dataset of features for Webapp
- additional_features_dict=features_dict_additional, # Optional: dict additional data
+ x=xtest,
+ y_pred=y_pred, # Optional: for your own prediction (by default: model.predict)
+ y_target=yTest, # Optional: allows to display True Values vs Predicted Values
+ additional_data=xadditional, # Optional: additional dataset of features for Webapp
+ additional_features_dict=features_dict_additional, # Optional: dict additional data
)
```
@@ -193,7 +194,7 @@ xpl.compile(
```python
app = xpl.run_app()
-```
+```
[Live Demo Shapash-Monitor](https://shapash-demo.ossbymaif.fr/)
@@ -203,15 +204,15 @@ app = xpl.run_app()
```python
xpl.generate_report(
- output_file='path/to/output/report.html',
- project_info_file='path/to/project_info.yml',
+ output_file="path/to/output/report.html",
+ project_info_file="path/to/project_info.yml",
x_train=xtrain,
y_train=ytrain,
y_test=ytest,
title_story="House prices report",
title_description="""This document is a data science report of the kaggle house prices tutorial project.
It was generated using the Shapash library.""",
- metrics=[{'name': 'MSE', 'path': 'sklearn.metrics.mean_squared_error'}]
+ metrics=[{"name": "MSE", "path": "sklearn.metrics.mean_squared_error"}],
)
```
@@ -220,9 +221,9 @@ xpl.generate_report(
- Step 5: From training to deployment : SmartPredictor Object
> Shapash provides a SmartPredictor object to deploy the summary of local explanation for the operational needs.
It is an object dedicated to deployment, lighter than SmartExplainer with additional consistency checks.
- SmartPredictor can be used with an API or in batch mode. It provides predictions, detailed or summarized local
+ SmartPredictor can be used with an API or in batch mode. It provides predictions, detailed or summarized local
explainability using appropriate wording.
-
+
```python
predictor = xpl.to_smartpredictor()
```
diff --git a/data/house_prices_labels.json b/data/house_prices_labels.json
index cda51b24..1f3e13da 100644
--- a/data/house_prices_labels.json
+++ b/data/house_prices_labels.json
@@ -1,74 +1,74 @@
{
- "MSSubClass": "Building Class",
- "MSZoning": "General zoning classification",
- "LotArea": "Lot size square feet",
- "Street": "Type of road access",
- "LotShape": "General shape of property",
- "LandContour": "Flatness of the property",
- "Utilities": "Type of utilities available",
- "LotConfig": "Lot configuration",
- "LandSlope": "Slope of property",
- "Neighborhood": "Physical locations within Ames city limits",
- "Condition1": "Proximity to various conditions",
- "Condition2": "Proximity to other various conditions",
- "BldgType": "Type of dwelling",
- "HouseStyle": "Style of dwelling",
- "OverallQual": "Overall material and finish of the house",
- "OverallCond": "Overall condition of the house",
- "YearBuilt": "Original construction date",
- "YearRemodAdd": "Remodel date",
- "RoofStyle": "Type of roof",
- "RoofMatl": "Roof material",
- "Exterior1st": "Exterior covering on house",
- "Exterior2nd": "Other exterior covering on house",
- "MasVnrType": "Masonry veneer type",
- "MasVnrArea": "Masonry veneer area in square feet",
- "ExterQual": "Exterior materials' quality",
- "ExterCond": "Exterior materials' condition",
- "Foundation": "Type of foundation",
- "BsmtQual": "Height of the basement",
- "BsmtCond": "General condition of the basement",
- "BsmtExposure": "Refers to walkout or garden level walls",
- "BsmtFinType1": "Rating of basement finished area",
- "BsmtFinSF1": "Type 1 finished square feet",
- "BsmtFinType2": "Rating of basement finished area (if present)",
- "BsmtFinSF2": "Type 2 finished square feet",
- "BsmtUnfSF": "Unfinished square feet of basement area",
- "TotalBsmtSF": "Total square feet of basement area",
- "Heating": "Type of heating",
- "HeatingQC": "Heating quality and condition",
- "CentralAir": "Central air conditioning",
- "Electrical": "Electrical system",
- "1stFlrSF": "First Floor square feet",
- "2ndFlrSF": "Second floor square feet",
- "LowQualFinSF": "Low quality finished square feet",
- "GrLivArea": "Ground living area square feet",
- "BsmtFullBath": "Basement full bathrooms",
- "BsmtHalfBath": "Basement half bathrooms",
- "FullBath": "Full bathrooms above grade",
- "HalfBath": "Half baths above grade",
- "BedroomAbvGr": "Bedrooms above grade",
- "KitchenAbvGr": "Kitchens above grade",
- "KitchenQual": "Kitchen quality",
- "TotRmsAbvGrd": "Total rooms above grade",
- "Functional": "Home functionality",
- "Fireplaces": "Number of fireplaces",
- "GarageType": "Garage location",
- "GarageYrBlt": "Year garage was built",
- "GarageFinish": "Interior finish of the garage?",
- "GarageArea": "Size of garage in square feet",
- "GarageQual": "Garage quality",
- "GarageCond": "Garage condition",
- "PavedDrive": "Paved driveway",
- "WoodDeckSF": "Wood deck area in square feet",
- "OpenPorchSF": "Open porch area in square feet",
- "EnclosedPorch": "Enclosed porch area in square feet",
- "3SsnPorch": "Three season porch area in square feet",
- "ScreenPorch": "Screen porch area in square feet",
- "PoolArea": "Pool area in square feet",
- "MiscVal": "$Value of miscellaneous feature",
- "MoSold": "Month Sold",
- "YrSold": "Year Sold",
- "SaleType": "Type of sale",
- "SaleCondition": "Condition of sale"
-}
\ No newline at end of file
+ "1stFlrSF": "First Floor square feet",
+ "2ndFlrSF": "Second floor square feet",
+ "3SsnPorch": "Three season porch area in square feet",
+ "BedroomAbvGr": "Bedrooms above grade",
+ "BldgType": "Type of dwelling",
+ "BsmtCond": "General condition of the basement",
+ "BsmtExposure": "Refers to walkout or garden level walls",
+ "BsmtFinSF1": "Type 1 finished square feet",
+ "BsmtFinSF2": "Type 2 finished square feet",
+ "BsmtFinType1": "Rating of basement finished area",
+ "BsmtFinType2": "Rating of basement finished area (if present)",
+ "BsmtFullBath": "Basement full bathrooms",
+ "BsmtHalfBath": "Basement half bathrooms",
+ "BsmtQual": "Height of the basement",
+ "BsmtUnfSF": "Unfinished square feet of basement area",
+ "CentralAir": "Central air conditioning",
+ "Condition1": "Proximity to various conditions",
+ "Condition2": "Proximity to other various conditions",
+ "Electrical": "Electrical system",
+ "EnclosedPorch": "Enclosed porch area in square feet",
+ "ExterCond": "Exterior materials' condition",
+ "ExterQual": "Exterior materials' quality",
+ "Exterior1st": "Exterior covering on house",
+ "Exterior2nd": "Other exterior covering on house",
+ "Fireplaces": "Number of fireplaces",
+ "Foundation": "Type of foundation",
+ "FullBath": "Full bathrooms above grade",
+ "Functional": "Home functionality",
+ "GarageArea": "Size of garage in square feet",
+ "GarageCond": "Garage condition",
+ "GarageFinish": "Interior finish of the garage?",
+ "GarageQual": "Garage quality",
+ "GarageType": "Garage location",
+ "GarageYrBlt": "Year garage was built",
+ "GrLivArea": "Ground living area square feet",
+ "HalfBath": "Half baths above grade",
+ "Heating": "Type of heating",
+ "HeatingQC": "Heating quality and condition",
+ "HouseStyle": "Style of dwelling",
+ "KitchenAbvGr": "Kitchens above grade",
+ "KitchenQual": "Kitchen quality",
+ "LandContour": "Flatness of the property",
+ "LandSlope": "Slope of property",
+ "LotArea": "Lot size square feet",
+ "LotConfig": "Lot configuration",
+ "LotShape": "General shape of property",
+ "LowQualFinSF": "Low quality finished square feet",
+ "MSSubClass": "Building Class",
+ "MSZoning": "General zoning classification",
+ "MasVnrArea": "Masonry veneer area in square feet",
+ "MasVnrType": "Masonry veneer type",
+ "MiscVal": "$Value of miscellaneous feature",
+ "MoSold": "Month Sold",
+ "Neighborhood": "Physical locations within Ames city limits",
+ "OpenPorchSF": "Open porch area in square feet",
+ "OverallCond": "Overall condition of the house",
+ "OverallQual": "Overall material and finish of the house",
+ "PavedDrive": "Paved driveway",
+ "PoolArea": "Pool area in square feet",
+ "RoofMatl": "Roof material",
+ "RoofStyle": "Type of roof",
+ "SaleCondition": "Condition of sale",
+ "SaleType": "Type of sale",
+ "ScreenPorch": "Screen porch area in square feet",
+ "Street": "Type of road access",
+ "TotRmsAbvGrd": "Total rooms above grade",
+ "TotalBsmtSF": "Total square feet of basement area",
+ "Utilities": "Type of utilities available",
+ "WoodDeckSF": "Wood deck area in square feet",
+ "YearBuilt": "Original construction date",
+ "YearRemodAdd": "Remodel date",
+ "YrSold": "Year Sold"
+}
diff --git a/data/titaniclabels.json b/data/titaniclabels.json
index 7a0c8e1e..7aff00fd 100644
--- a/data/titaniclabels.json
+++ b/data/titaniclabels.json
@@ -1 +1,13 @@
-{"PassengerID": "PassengerID", "Survival": "Has survived ?", "Pclass": "Ticket class", "Name": "Name, First name", "Sex": "Sex", "Age": "Age", "SibSp": "Relatives such as brother or wife", "Parch": "Relatives like children or parents", "Fare": "Passenger fare", "Embarked": "Port of embarkation", "Title": "Title of passenger"}
\ No newline at end of file
+{
+ "Age": "Age",
+ "Embarked": "Port of embarkation",
+ "Fare": "Passenger fare",
+ "Name": "Name, First name",
+ "Parch": "Relatives like children or parents",
+ "PassengerID": "PassengerID",
+ "Pclass": "Ticket class",
+ "Sex": "Sex",
+ "SibSp": "Relatives such as brother or wife",
+ "Survival": "Has survived ?",
+ "Title": "Title of passenger"
+}
diff --git a/docs/conf.py b/docs/conf.py
index b0f2c98c..6dfb8a60 100644
--- a/docs/conf.py
+++ b/docs/conf.py
@@ -12,15 +12,16 @@
#
import os
import sys
-sys.path.insert(0, '..')
+
+sys.path.insert(0, "..")
import shapash
# -- Project information -----------------------------------------------------
-project = 'Shapash'
-copyright = '2020, Maif'
-author = 'Maif'
+project = "Shapash"
+copyright = "2020, Maif"
+author = "Maif"
# The short X.Y version
version = shapash.__version__
@@ -35,30 +36,30 @@
# extensions coming with Sphinx (named 'sphinx.ext.*') or your custom
# ones.
extensions = [
- 'sphinx.ext.autodoc',
- 'sphinx.ext.viewcode',
- 'sphinx.ext.todo',
- 'sphinx.ext.napoleon',
- 'nbsphinx',
+ "sphinx.ext.autodoc",
+ "sphinx.ext.viewcode",
+ "sphinx.ext.todo",
+ "sphinx.ext.napoleon",
+ "nbsphinx",
]
-nbsphinx_execute = 'never'
-master_doc = 'index'
+nbsphinx_execute = "never"
+master_doc = "index"
# Add any paths that contain templates here, relative to this directory.
-templates_path = ['_templates']
+templates_path = ["_templates"]
# The language for content autogenerated by Sphinx. Refer to documentation
# for a list of supported languages.
#
# This is also used if you do content translation via gettext catalogs.
# Usually you set "language" from the command line for these cases.
-language = 'en'
+language = "en"
# List of patterns, relative to source directory, that match files and
# directories to ignore when looking for source files.
# This pattern also affects html_static_path and html_extra_path.
-exclude_patterns = ['_build', 'Thumbs.db', '.DS_Store']
+exclude_patterns = ["_build", "Thumbs.db", ".DS_Store"]
# -- Options for HTML output -------------------------------------------------
@@ -66,41 +67,35 @@
# The theme to use for HTML and HTML Help pages. See the documentation for
# a list of builtin themes.
#
-html_theme = 'sphinx_material'
-#html_logo = './assets/images/svg/shapash-github.svg'
+html_theme = "sphinx_material"
+# html_logo = './assets/images/svg/shapash-github.svg'
# Material theme options (see theme.conf for more information)
html_theme_options = {
-
# Set the name of the project to appear in the navigation.
- 'nav_title': 'Shapash',
+ "nav_title": "Shapash",
# Set the color and the accent color
- 'color_primary': 'amber',
- 'color_accent': 'deep-orange',
-
+ "color_primary": "amber",
+ "color_accent": "deep-orange",
# Set the repo location to get a badge with stats
- 'repo_url': 'https://github.com/MAIF/shapash',
- 'repo_name': 'shapash',
-
+ "repo_url": "https://github.com/MAIF/shapash",
+ "repo_name": "shapash",
# Icon of the navbar
- 'logo_icon': '',
-
+ "logo_icon": "",
# Visible levels of the global TOC; -1 means unlimited
- 'globaltoc_depth': 3,
+ "globaltoc_depth": 3,
# If False, expand all TOC entries
- 'globaltoc_collapse': True,
+ "globaltoc_collapse": True,
# If True, show hidden TOC entries
- 'globaltoc_includehidden': False,
+ "globaltoc_includehidden": False,
}
-html_sidebars = {
- "**": ["logo-text.html", "globaltoc.html", "localtoc.html", "searchbox.html"]
-}
+html_sidebars = {"**": ["logo-text.html", "globaltoc.html", "localtoc.html", "searchbox.html"]}
# Add any paths that contain custom static files (such as style sheets) here,
# relative to this directory. They are copied after the builtin static files,
# so a file named "default.css" will overwrite the builtin "default.css".
-html_static_path = ['_static']
+html_static_path = ["_static"]
# -- Extension configuration -------------------------------------------------
@@ -112,9 +107,10 @@
# -- Additional html pages -------------------------------------------------
import subprocess
+
# Generates the report example in the documentation
-subprocess.call(['python', '../tutorial/generate_report/shapash_report_example.py'])
-html_extra_path = ['../tutorial/report/output/report.html']
+subprocess.call(["python", "../tutorial/generate_report/shapash_report_example.py"])
+html_extra_path = ["../tutorial/report/output/report.html"]
def setup_tutorials():
@@ -123,33 +119,45 @@ def setup_tutorials():
def _copy_notebooks_and_create_rst(d_path, new_d_path, d_name):
os.makedirs(new_d_path, exist_ok=True)
- list_notebooks = [f for f in os.listdir(d_path) if os.path.splitext(f)[-1] == '.ipynb']
+ list_notebooks = [f for f in os.listdir(d_path) if os.path.splitext(f)[-1] == ".ipynb"]
for notebook_f_name in list_notebooks:
shutil.copyfile(os.path.join(d_path, notebook_f_name), os.path.join(new_d_path, notebook_f_name))
# RST file (see for example docs/overview.rst)
- rst_file = '\n'.join(
- [f'{d_name}', '======================', '', '.. toctree::',
- ' :maxdepth: 1', ' :glob:', '', ' *', '']
+ rst_file = "\n".join(
+ [
+ f"{d_name}",
+ "======================",
+ "",
+ ".. toctree::",
+ " :maxdepth: 1",
+ " :glob:",
+ "",
+ " *",
+ "",
+ ]
)
- with open(os.path.join(new_d_path, 'index.rst'), 'w') as f:
+ with open(os.path.join(new_d_path, "index.rst"), "w") as f:
f.write(rst_file)
docs_path = pathlib.Path(__file__).parent
- tutorials_path = os.path.join(docs_path.parent, 'tutorial')
- tutorials_doc_path = os.path.join(docs_path, 'tutorials')
+ tutorials_path = os.path.join(docs_path.parent, "tutorial")
+ tutorials_doc_path = os.path.join(docs_path, "tutorials")
# Create a directory in shapash/docs/tutorials for each directory of shapash/tutorial
# And copy each notebook file in it
- list_dir = [d for d in os.listdir(tutorials_path) if os.path.isdir(os.path.join(tutorials_path, d))
- and not d.startswith('.')]
+ list_dir = [
+ d
+ for d in os.listdir(tutorials_path)
+ if os.path.isdir(os.path.join(tutorials_path, d)) and not d.startswith(".")
+ ]
for d_name in list_dir:
d_path = os.path.join(tutorials_path, d_name)
new_d_path = os.path.join(tutorials_doc_path, d_name)
_copy_notebooks_and_create_rst(d_path, new_d_path, d_name)
# Also copying all the overview tutorials (shapash/tutorial/shapash-overview-in-jupyter.ipynb for example)
- _copy_notebooks_and_create_rst(tutorials_path, tutorials_doc_path, 'overview')
+ _copy_notebooks_and_create_rst(tutorials_path, tutorials_doc_path, "overview")
setup_tutorials()
diff --git a/requirements.dev.txt b/requirements.dev.txt
index 9dfe603e..6e89a7b2 100644
--- a/requirements.dev.txt
+++ b/requirements.dev.txt
@@ -25,7 +25,7 @@ nbsphinx==0.8.8
sphinx_material==0.0.35
pytest>=6.2.5
pytest-cov>=2.8.1
-scikit-learn>=1.0.1
+scikit-learn>=1.0.1,<1.4
xgboost>=1.0.0
nbformat>4.2.0
numba>=0.53.1
diff --git a/setup.py b/setup.py
index 11b1c462..76df371c 100644
--- a/setup.py
+++ b/setup.py
@@ -1,108 +1,121 @@
#!/usr/bin/env python
-# -*- coding: utf-8 -*-
"""The setup script."""
import os
+
from setuptools import setup
here = os.path.abspath(os.path.dirname(__file__))
-with open('README.md', encoding='utf8') as readme_file:
+with open("README.md", encoding="utf8") as readme_file:
long_description = readme_file.read()
# Load the package's __version__.py module as a dictionary.
version_d: dict = {}
-with open(os.path.join(here, 'shapash', "__version__.py")) as f:
+with open(os.path.join(here, "shapash", "__version__.py")) as f:
exec(f.read(), version_d)
requirements = [
- 'plotly>=5.0.0',
- 'matplotlib>=3.2.0',
- 'numpy>1.18.0',
- 'pandas>1.0.2',
- 'shap>=0.38.1',
- 'Flask<2.3.0',
- 'dash>=2.3.1',
- 'dash-bootstrap-components>=1.1.0',
- 'dash-core-components>=2.0.0',
- 'dash-daq>=0.5.0',
- 'dash-html-components>=2.0.0',
- 'dash-renderer==1.8.3',
- 'dash-table>=5.0.0',
- 'nbformat>4.2.0',
- 'numba>=0.53.1',
- 'scikit-learn>=1.0.1',
- 'category_encoders>=2.6.0',
- 'scipy>=0.19.1',
+ "plotly>=5.0.0",
+ "matplotlib>=3.2.0",
+ "numpy>1.18.0",
+ "pandas>1.0.2",
+ "shap>=0.38.1",
+ "Flask<2.3.0",
+ "dash>=2.3.1",
+ "dash-bootstrap-components>=1.1.0",
+ "dash-core-components>=2.0.0",
+ "dash-daq>=0.5.0",
+ "dash-html-components>=2.0.0",
+ "dash-renderer==1.8.3",
+ "dash-table>=5.0.0",
+ "nbformat>4.2.0",
+ "numba>=0.53.1",
+ "scikit-learn>=1.0.1,<1.4",
+ "category_encoders>=2.6.0",
+ "scipy>=0.19.1",
]
extras = dict()
# This list should be identical to the list in shapash/report/__init__.py
-extras['report'] = [
- 'nbconvert>=6.0.7',
- 'papermill>=2.0.0',
- 'jupyter-client>=7.4.0',
- 'seaborn==0.12.2',
- 'notebook',
- 'Jinja2>=2.11.0',
- 'phik'
+extras["report"] = [
+ "nbconvert>=6.0.7",
+ "papermill>=2.0.0",
+ "jupyter-client>=7.4.0",
+ "seaborn==0.12.2",
+ "notebook",
+ "Jinja2>=2.11.0",
+ "phik",
]
-extras['xgboost'] = ['xgboost>=1.0.0']
-extras['lightgbm'] = ['lightgbm>=2.3.0']
-extras['catboost'] = ['catboost>=1.0.1']
-extras['lime'] = ['lime>=0.2.0.0']
+extras["xgboost"] = ["xgboost>=1.0.0"]
+extras["lightgbm"] = ["lightgbm>=2.3.0"]
+extras["catboost"] = ["catboost>=1.0.1"]
+extras["lime"] = ["lime>=0.2.0.0"]
-setup_requirements = ['pytest-runner', ]
+setup_requirements = [
+ "pytest-runner",
+]
-test_requirements = ['pytest', ]
+test_requirements = [
+ "pytest",
+]
setup(
name="shapash",
- version=version_d['__version__'],
- python_requires='>3.7, <3.12',
- url='https://github.com/MAIF/shapash',
+ version=version_d["__version__"],
+ python_requires=">3.7, <3.12",
+ url="https://github.com/MAIF/shapash",
author="Yann Golhen, Sebastien Bidault, Yann Lagre, Maxime Gendre",
author_email="yann.golhen@maif.fr",
description="Shapash is a Python library which aims to make machine learning interpretable and understandable by everyone.",
long_description=long_description,
- long_description_content_type='text/markdown',
+ long_description_content_type="text/markdown",
classifiers=[
- 'Programming Language :: Python :: 3',
- 'Programming Language :: Python :: 3.8',
- 'Programming Language :: Python :: 3.9',
- 'Programming Language :: Python :: 3.10',
- 'Programming Language :: Python :: 3.11',
+ "Programming Language :: Python :: 3",
+ "Programming Language :: Python :: 3.8",
+ "Programming Language :: Python :: 3.9",
+ "Programming Language :: Python :: 3.10",
+ "Programming Language :: Python :: 3.11",
"License :: OSI Approved :: Apache Software License",
"Operating System :: OS Independent",
],
install_requires=requirements,
extras_require=extras,
license="Apache Software License 2.0",
- keywords='shapash',
+ keywords="shapash",
package_dir={
- 'shapash': 'shapash',
- 'shapash.data': 'shapash/data',
- 'shapash.decomposition': 'shapash/decomposition',
- 'shapash.explainer': 'shapash/explainer',
- 'shapash.backend': 'shapash/backend',
- 'shapash.manipulation': 'shapash/manipulation',
- 'shapash.report': 'shapash/report',
- 'shapash.utils': 'shapash/utils',
- 'shapash.webapp': 'shapash/webapp',
- 'shapash.webapp.utils': 'shapash/webapp/utils',
- 'shapash.style': 'shapash/style',
+ "shapash": "shapash",
+ "shapash.data": "shapash/data",
+ "shapash.decomposition": "shapash/decomposition",
+ "shapash.explainer": "shapash/explainer",
+ "shapash.backend": "shapash/backend",
+ "shapash.manipulation": "shapash/manipulation",
+ "shapash.report": "shapash/report",
+ "shapash.utils": "shapash/utils",
+ "shapash.webapp": "shapash/webapp",
+ "shapash.webapp.utils": "shapash/webapp/utils",
+ "shapash.style": "shapash/style",
},
- packages=['shapash', 'shapash.data', 'shapash.decomposition',
- 'shapash.explainer', 'shapash.backend', 'shapash.manipulation',
- 'shapash.utils', 'shapash.webapp', 'shapash.webapp.utils',
- 'shapash.report', 'shapash.style'],
- data_files=[('style', ['shapash/style/colors.json'])],
+ packages=[
+ "shapash",
+ "shapash.data",
+ "shapash.decomposition",
+ "shapash.explainer",
+ "shapash.backend",
+ "shapash.manipulation",
+ "shapash.utils",
+ "shapash.webapp",
+ "shapash.webapp.utils",
+ "shapash.report",
+ "shapash.style",
+ ],
+ data_files=[("style", ["shapash/style/colors.json"])],
include_package_data=True,
setup_requires=setup_requirements,
- test_suite='tests',
+ test_suite="tests",
tests_require=test_requirements,
zip_safe=False,
)
diff --git a/shapash/__init__.py b/shapash/__init__.py
index 52cbbfb2..59117772 100644
--- a/shapash/__init__.py
+++ b/shapash/__init__.py
@@ -1,8 +1,10 @@
-
"""Top-level package."""
-__author__ = """Yann Golhen, Yann Lagré, Sebastien Bidault, Maxime Gendre, Thomas Bouche, Johann Martin, Guillaume Vignal"""
-__email__ = 'yann.golhen@maif.fr, yann.lagre@maif.fr, sebabstien.bidault.marketing@maif.fr, thomas.bouche@maif.fr, guillaume.vignal@maif.fr'
+__author__ = (
+ """Yann Golhen, Yann Lagré, Sebastien Bidault, Maxime Gendre, Thomas Bouche, Johann Martin, Guillaume Vignal"""
+)
+__email__ = "yann.golhen@maif.fr, yann.lagre@maif.fr, sebabstien.bidault.marketing@maif.fr, thomas.bouche@maif.fr, guillaume.vignal@maif.fr"
-from .__version__ import __version__
from shapash.explainer.smart_explainer import SmartExplainer
+
+from .__version__ import __version__
diff --git a/shapash/backend/__init__.py b/shapash/backend/__init__.py
index b9d262ab..0627fb74 100644
--- a/shapash/backend/__init__.py
+++ b/shapash/backend/__init__.py
@@ -1,9 +1,9 @@
-import sys
import inspect
+import sys
from .base_backend import BaseBackend
-from .shap_backend import ShapBackend
from .lime_backend import LimeBackend
+from .shap_backend import ShapBackend
def get_backend_cls_from_name(name):
@@ -14,10 +14,10 @@ def get_backend_cls_from_name(name):
cls
for _, cls in inspect.getmembers(sys.modules[__name__])
if (
- inspect.isclass(cls)
- and issubclass(cls, BaseBackend)
- and cls.name.lower() == name.lower()
- and cls.name.lower() != 'base'
+ inspect.isclass(cls)
+ and issubclass(cls, BaseBackend)
+ and cls.name.lower() == name.lower()
+ and cls.name.lower() != "base"
)
]
diff --git a/shapash/backend/base_backend.py b/shapash/backend/base_backend.py
index cd7ef5b8..6e3699a3 100644
--- a/shapash/backend/base_backend.py
+++ b/shapash/backend/base_backend.py
@@ -1,9 +1,10 @@
from abc import ABC, abstractmethod
from typing import Any, List, Optional, Union
-import pandas as pd
+
import numpy as np
+import pandas as pd
-from shapash.utils.check import check_model, check_contribution_object
+from shapash.utils.check import check_contribution_object, check_model
from shapash.utils.transform import adapt_contributions, get_preprocessing_mapping
from shapash.utils.utils import choose_state
@@ -19,13 +20,13 @@ class BaseBackend(ABC):
# `column_aggregation` defines a way to aggregate local contributions.
# Default is sum, possible values are 'sum' or 'first'.
# It allows to compute (column-wise) aggregation of local contributions.
- column_aggregation = 'sum'
+ column_aggregation = "sum"
# `name` defines the string name of the backend allowing to identify and
# construct the backend from it.
- name = 'base'
+ name = "base"
support_groups = True
- supported_cases = ['classification', 'regression']
+ supported_cases = ["classification", "regression"]
def __init__(self, model: Any, preprocessing: Optional[Any] = None):
"""Create a backend instance using a given implementation.
@@ -43,7 +44,7 @@ def __init__(self, model: Any, preprocessing: Optional[Any] = None):
self.state = None
self._case, self._classes = check_model(model)
if self._case not in self.supported_cases:
- raise ValueError(f'Model not supported by the backend as it does not cover {self._case} case')
+ raise ValueError(f"Model not supported by the backend as it does not cover {self._case} case")
@abstractmethod
def run_explainer(self, x: pd.DataFrame) -> dict:
@@ -53,10 +54,7 @@ def run_explainer(self, x: pd.DataFrame) -> dict:
)
def get_local_contributions(
- self,
- x: pd.DataFrame,
- explain_data: Any,
- subset: Optional[List[int]] = None
+ self, x: pd.DataFrame, explain_data: Any, subset: Optional[List[int]] = None
) -> Union[pd.DataFrame, List[pd.DataFrame]]:
"""Get local contributions using the explainer data computed in the `run_explainer`
method.
@@ -81,24 +79,21 @@ def get_local_contributions(
The local contributions computed by the backend.
"""
assert isinstance(explain_data, dict), "The _run_explainer method should return a dict"
- if 'contributions' not in explain_data.keys():
+ if "contributions" not in explain_data.keys():
raise ValueError(
- 'The _run_explainer method should return a dict'
- ' with at least `contributions` key containing '
- 'the local contributions'
+ "The _run_explainer method should return a dict"
+ " with at least `contributions` key containing "
+ "the local contributions"
)
- local_contributions = explain_data['contributions']
+ local_contributions = explain_data["contributions"]
if subset is not None:
local_contributions = local_contributions.loc[subset]
local_contributions = self.format_and_aggregate_local_contributions(x, local_contributions)
return local_contributions
def get_global_features_importance(
- self,
- contributions: pd.DataFrame,
- explain_data: Optional[dict] = None,
- subset: Optional[List[int]] = None
+ self, contributions: pd.DataFrame, explain_data: Optional[dict] = None, subset: Optional[List[int]] = None
) -> Union[pd.Series, List[pd.Series]]:
"""Get global contributions using the explainer data computed in the `run_explainer`
method.
@@ -126,9 +121,9 @@ def get_global_features_importance(
return state.compute_features_import(contributions)
def format_and_aggregate_local_contributions(
- self,
- x: pd.DataFrame,
- contributions: Union[pd.DataFrame, np.array, List[pd.DataFrame], List[np.array]],
+ self,
+ x: pd.DataFrame,
+ contributions: Union[pd.DataFrame, np.array, List[pd.DataFrame], List[np.array]],
) -> Union[pd.DataFrame, List[pd.DataFrame]]:
"""
This function allows to format and aggregate contributions in the right format
@@ -153,7 +148,8 @@ def format_and_aggregate_local_contributions(
check_contribution_object(self._case, self._classes, contributions)
contributions = self.state.validate_contributions(contributions, x)
contributions_cols = (
- contributions.columns.to_list() if isinstance(contributions, pd.DataFrame)
+ contributions.columns.to_list()
+ if isinstance(contributions, pd.DataFrame)
else contributions[0].columns.to_list()
)
if _needs_preprocessing(contributions_cols, x, self.preprocessing):
@@ -161,8 +157,7 @@ def format_and_aggregate_local_contributions(
return contributions
def _apply_preprocessing(
- self,
- contributions: Union[pd.DataFrame, List[pd.DataFrame]]
+ self, contributions: Union[pd.DataFrame, List[pd.DataFrame]]
) -> Union[pd.DataFrame, List[pd.DataFrame]]:
"""
Reconstruct contributions for original features, taken into account a preprocessing.
@@ -179,9 +174,7 @@ def _apply_preprocessing(
"""
if self.preprocessing:
return self.state.inverse_transform_contributions(
- contributions,
- self.preprocessing,
- agg_columns=self.column_aggregation
+ contributions, self.preprocessing, agg_columns=self.column_aggregation
)
else:
return contributions
diff --git a/shapash/backend/lime_backend.py b/shapash/backend/lime_backend.py
index ada25113..3830ba21 100644
--- a/shapash/backend/lime_backend.py
+++ b/shapash/backend/lime_backend.py
@@ -1,10 +1,11 @@
try:
from lime import lime_tabular
+
is_lime_available = True
except ImportError:
is_lime_available = False
-from typing import Any, Optional, List, Union
+from typing import Any, List, Optional, Union
import pandas as pd
@@ -12,12 +13,12 @@
class LimeBackend(BaseBackend):
- column_aggregation = 'sum'
- name = 'lime'
+ column_aggregation = "sum"
+ name = "lime"
support_groups = False
def __init__(self, model, preprocessing=None, data=None, **kwargs):
- super(LimeBackend, self).__init__(model, preprocessing)
+ super().__init__(model, preprocessing)
self.explainer = None
self.data = data
@@ -36,11 +37,7 @@ def run_explainer(self, x: pd.DataFrame):
dict containing local contributions
"""
data = self.data if self.data is not None else x
- explainer = lime_tabular.LimeTabularExplainer(
- data.values,
- feature_names=x.columns,
- mode=self._case
- )
+ explainer = lime_tabular.LimeTabularExplainer(data.values, feature_names=x.columns, mode=self._case)
lime_contrib = []
for i in x.index:
@@ -49,8 +46,7 @@ def run_explainer(self, x: pd.DataFrame):
if num_classes <= 2:
exp = explainer.explain_instance(x.loc[i], self.model.predict_proba, num_features=x.shape[1])
- lime_contrib.append(
- dict([[_transform_name(var_name[0], x), var_name[1]] for var_name in exp.as_list()]))
+ lime_contrib.append({_transform_name(var_name[0], x): var_name[1] for var_name in exp.as_list()})
elif num_classes > 2:
contribution = []
@@ -59,11 +55,11 @@ def run_explainer(self, x: pd.DataFrame):
df_contrib = pd.DataFrame()
for i in x.index:
exp = explainer.explain_instance(
- x.loc[i], self.model.predict_proba, top_labels=num_classes,
- num_features=x.shape[1])
+ x.loc[i], self.model.predict_proba, top_labels=num_classes, num_features=x.shape[1]
+ )
list_contrib.append(
- dict([[_transform_name(var_name[0], x), var_name[1]] for var_name in
- exp.as_list(j)]))
+ {_transform_name(var_name[0], x): var_name[1] for var_name in exp.as_list(j)}
+ )
df_contrib = pd.DataFrame(list_contrib)
df_contrib = df_contrib[list(x.columns)]
contribution.append(df_contrib.values)
@@ -71,8 +67,7 @@ def run_explainer(self, x: pd.DataFrame):
else:
exp = explainer.explain_instance(x.loc[i], self.model.predict, num_features=x.shape[1])
- lime_contrib.append(
- dict([[_transform_name(var_name[0], x), var_name[1]] for var_name in exp.as_list()]))
+ lime_contrib.append({_transform_name(var_name[0], x): var_name[1] for var_name in exp.as_list()})
contributions = pd.DataFrame(lime_contrib, index=x.index)
contributions = contributions[list(x.columns)]
@@ -83,9 +78,8 @@ def run_explainer(self, x: pd.DataFrame):
def _transform_name(var_name, x_df):
- """Function for transform name of LIME contribution shape to a comprehensive name
- """
+ """Function for transform name of LIME contribution shape to a comprehensive name"""
for colname in list(x_df.columns):
- if f' {colname} ' in f' {var_name} ':
+ if f" {colname} " in f" {var_name} ":
col_rename = colname
return col_rename
diff --git a/shapash/backend/shap_backend.py b/shapash/backend/shap_backend.py
index 4771758c..716a304a 100644
--- a/shapash/backend/shap_backend.py
+++ b/shapash/backend/shap_backend.py
@@ -1,5 +1,5 @@
-import pandas as pd
import numpy as np
+import pandas as pd
import shap
from shapash.backend.base_backend import BaseBackend
@@ -8,18 +8,18 @@
class ShapBackend(BaseBackend):
# When grouping features contributions together, Shap uses the sum of the contributions
# of the features that belong to the group
- column_aggregation = 'sum'
- name = 'shap'
+ column_aggregation = "sum"
+ name = "shap"
def __init__(self, model, preprocessing=None, masker=None, explainer_args=None, explainer_compute_args=None):
- super(ShapBackend, self).__init__(model, preprocessing)
+ super().__init__(model, preprocessing)
self.masker = masker
self.explainer_args = explainer_args if explainer_args else {}
self.explainer_compute_args = explainer_compute_args if explainer_compute_args else {}
if self.explainer_args:
if "explainer" in self.explainer_args.keys():
- shap_parameters = {k: v for k, v in self.explainer_args.items() if k != 'explainer'}
+ shap_parameters = {k: v for k, v in self.explainer_args.items() if k != "explainer"}
self.explainer = self.explainer_args["explainer"](**shap_parameters)
else:
self.explainer = shap.Explainer(**self.explainer_args)
@@ -31,15 +31,14 @@ def __init__(self, model, preprocessing=None, masker=None, explainer_args=None,
elif shap.explainers.Additive.supports_model_with_masker(model, self.masker):
self.explainer = shap.Explainer(model=model, masker=self.masker)
# otherwise use a model agnostic method
- elif hasattr(model, 'predict_proba'):
+ elif hasattr(model, "predict_proba"):
self.explainer = shap.Explainer(model=model.predict_proba, masker=self.masker)
- elif hasattr(model, 'predict'):
+ elif hasattr(model, "predict"):
self.explainer = shap.Explainer(model=model.predict, masker=self.masker)
# if we get here then we don't know how to handle what was given to us
else:
raise ValueError("The model is not recognized by Shapash! Model: " + str(model))
-
def run_explainer(self, x: pd.DataFrame) -> dict:
"""
Computes and returns local contributions using Shap explainer
@@ -78,8 +77,10 @@ def get_shap_interaction_values(x_df, explainer):
Shap interaction values for each sample as an array of shape (# samples x # features x # features).
"""
if not isinstance(explainer, shap.TreeExplainer):
- raise ValueError(f"Explainer type ({type(explainer)}) is not a TreeExplainer. "
- f"Shap interaction values can only be computed for TreeExplainer types")
+ raise ValueError(
+ f"Explainer type ({type(explainer)}) is not a TreeExplainer. "
+ f"Shap interaction values can only be computed for TreeExplainer types"
+ )
shap_interaction_values = explainer.shap_interaction_values(x_df)
diff --git a/shapash/data/data_loader.py b/shapash/data/data_loader.py
index bf91ca20..e1ad6699 100644
--- a/shapash/data/data_loader.py
+++ b/shapash/data/data_loader.py
@@ -1,12 +1,13 @@
"""
Data loader module
"""
+import json
import os
from pathlib import Path
-import json
-import pandas as pd
-from urllib.request import urlopen
from urllib.error import URLError
+from urllib.request import urlopen
+
+import pandas as pd
def _find_file(data_path, github_data_url, filename):
@@ -29,7 +30,7 @@ def _find_file(data_path, github_data_url, filename):
"""
file = os.path.join(data_path, filename)
if os.path.isfile(file) is False:
- file = github_data_url+filename
+ file = github_data_url + filename
try:
urlopen(file)
except URLError:
@@ -63,42 +64,42 @@ def data_loading(dataset):
If exist, columns labels dictionnary associated to the dataset.
"""
data_path = str(Path(__file__).parents[2] / "data")
- if dataset == 'house_prices':
- github_data_url = 'https://github.com/MAIF/shapash/raw/master/data/'
+ if dataset == "house_prices":
+ github_data_url = "https://github.com/MAIF/shapash/raw/master/data/"
data_house_prices_path = _find_file(data_path, github_data_url, "house_prices_dataset.csv")
dict_house_prices_path = _find_file(data_path, github_data_url, "house_prices_labels.json")
- data = pd.read_csv(data_house_prices_path, header=0, index_col=0, engine='python')
+ data = pd.read_csv(data_house_prices_path, header=0, index_col=0, engine="python")
if github_data_url in dict_house_prices_path:
with urlopen(dict_house_prices_path) as openfile:
dic = json.load(openfile)
else:
- with open(dict_house_prices_path, 'r') as openfile:
+ with open(dict_house_prices_path) as openfile:
dic = json.load(openfile)
return data, dic
- elif dataset == 'titanic':
- github_data_url = 'https://github.com/MAIF/shapash/raw/master/data/'
+ elif dataset == "titanic":
+ github_data_url = "https://github.com/MAIF/shapash/raw/master/data/"
data_titanic_path = _find_file(data_path, github_data_url, "titanicdata.csv")
- dict_titanic_path = _find_file(data_path, github_data_url, 'titaniclabels.json')
- data = pd.read_csv(data_titanic_path, header=0, index_col=0, engine='python')
+ dict_titanic_path = _find_file(data_path, github_data_url, "titaniclabels.json")
+ data = pd.read_csv(data_titanic_path, header=0, index_col=0, engine="python")
if github_data_url in dict_titanic_path:
with urlopen(dict_titanic_path) as openfile:
dic = json.load(openfile)
else:
- with open(dict_titanic_path, 'r') as openfile:
+ with open(dict_titanic_path) as openfile:
dic = json.load(openfile)
return data, dic
- elif dataset == 'telco_customer_churn':
- github_data_url = 'https://github.com/IBM/telco-customer-churn-on-icp4d/raw/master/data/'
+ elif dataset == "telco_customer_churn":
+ github_data_url = "https://github.com/IBM/telco-customer-churn-on-icp4d/raw/master/data/"
data_telco_path = _find_file(data_path, github_data_url, "Telco-Customer-Churn.csv")
- data = pd.read_csv(data_telco_path, header=0, index_col=0, engine='python')
+ data = pd.read_csv(data_telco_path, header=0, index_col=0, engine="python")
return data
- elif dataset == 'us_car_accident':
- github_data_url = 'https://github.com/MAIF/shapash/raw/master/data/'
+ elif dataset == "us_car_accident":
+ github_data_url = "https://github.com/MAIF/shapash/raw/master/data/"
data_accidents_path = _find_file(data_path, github_data_url, "US_Accidents_extract.csv")
- data = pd.read_csv(data_accidents_path, header=0, engine='python')
+ data = pd.read_csv(data_accidents_path, header=0, engine="python")
return data
else:
diff --git a/shapash/decomposition/contributions.py b/shapash/decomposition/contributions.py
index 9820de2f..79f21409 100644
--- a/shapash/decomposition/contributions.py
+++ b/shapash/decomposition/contributions.py
@@ -2,14 +2,15 @@
Contributions
"""
-import pandas as pd
import numpy as np
-from shapash.utils.transform import preprocessing_tolist
-from shapash.utils.transform import check_transformers
+import pandas as pd
+
from shapash.utils.category_encoder_backend import calc_inv_contrib_ce
from shapash.utils.columntransformer_backend import calc_inv_contrib_ct
+from shapash.utils.transform import check_transformers, preprocessing_tolist
-def inverse_transform_contributions(contributions, preprocessing=None, agg_columns='sum'):
+
+def inverse_transform_contributions(contributions, preprocessing=None, agg_columns="sum"):
"""
Reverse contribution giving a preprocessing.
@@ -38,12 +39,12 @@ def inverse_transform_contributions(contributions, preprocessing=None, agg_colum
"""
if not isinstance(contributions, pd.DataFrame):
- raise Exception('Shap values must be a pandas dataframe.')
+ raise Exception("Shap values must be a pandas dataframe.")
if preprocessing is None:
return contributions
else:
- #Transform preprocessing into a list
+ # Transform preprocessing into a list
list_encoding = preprocessing_tolist(preprocessing)
# check supported inverse
@@ -59,6 +60,7 @@ def inverse_transform_contributions(contributions, preprocessing=None, agg_colum
x_contrib_invers = calc_inv_contrib_ce(x_contrib_invers, encoding, agg_columns)
return x_contrib_invers
+
def rank_contributions(s_df, x_df):
"""
Function to sort contributions and input features
@@ -85,14 +87,15 @@ def rank_contributions(s_df, x_df):
sorted_contrib = np.take_along_axis(s_df.values, argsort, axis=1)
sorted_features = np.take_along_axis(x_df.values, argsort, axis=1)
- contrib_col = ['contribution_' + str(i) for i in range(s_df.shape[1])]
- col = ['feature_' + str(i) for i in range(s_df.shape[1])]
+ contrib_col = ["contribution_" + str(i) for i in range(s_df.shape[1])]
+ col = ["feature_" + str(i) for i in range(s_df.shape[1])]
s_dict = pd.DataFrame(data=argsort, columns=col, index=x_df.index)
s_ord = pd.DataFrame(data=sorted_contrib, columns=contrib_col, index=x_df.index)
x_ord = pd.DataFrame(data=sorted_features, columns=col, index=x_df.index)
return [s_ord, x_ord, s_dict]
+
def assign_contributions(ranked):
"""
Turn a list of results into a dict.
@@ -114,11 +117,7 @@ def assign_contributions(ranked):
"""
if len(ranked) != 3:
raise ValueError(
- 'Expected lenght : 3, observed lenght : {},'
- 'please check the outputs of rank_contributions.'.format(len(ranked))
+ "Expected lenght : 3, observed lenght : {},"
+ "please check the outputs of rank_contributions.".format(len(ranked))
)
- return {
- 'contrib_sorted': ranked[0],
- 'x_sorted': ranked[1],
- 'var_dict': ranked[2]
- }
+ return {"contrib_sorted": ranked[0], "x_sorted": ranked[1], "var_dict": ranked[2]}
diff --git a/shapash/explainer/consistency.py b/shapash/explainer/consistency.py
index 0dd56cb3..631287c9 100644
--- a/shapash/explainer/consistency.py
+++ b/shapash/explainer/consistency.py
@@ -1,20 +1,20 @@
-from category_encoders import OrdinalEncoder
import copy
import itertools
+
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
+from category_encoders import OrdinalEncoder
from plotly import graph_objs as go
from plotly.offline import plot
from plotly.subplots import make_subplots
from sklearn.manifold import MDS
from shapash import SmartExplainer
-from shapash.style.style_utils import colors_loading, select_palette, define_style
-
+from shapash.style.style_utils import colors_loading, define_style, select_palette
-class Consistency():
+class Consistency:
def __init__(self):
self._palette_name = list(colors_loading().keys())[0]
self._style_dict = define_style(select_palette(colors_loading(), self._palette_name))
@@ -27,9 +27,8 @@ def tuning_colorscale(self, values):
values whose quantiles must be calculated
"""
desc_df = values.describe(percentiles=np.arange(0.1, 1, 0.1).tolist())
- min_pred, max_init = list(desc_df.loc[['min', 'max']].values)
- desc_pct_df = (desc_df.loc[~desc_df.index.isin(['count', 'mean', 'std'])] - min_pred) / \
- (max_init - min_pred)
+ min_pred, max_init = list(desc_df.loc[["min", "max"]].values)
+ desc_pct_df = (desc_df.loc[~desc_df.index.isin(["count", "mean", "std"])] - min_pred) / (max_init - min_pred)
color_scale = list(map(list, (zip(desc_pct_df.values.flatten(), self._style_dict["init_contrib_colorscale"]))))
return color_scale
@@ -59,7 +58,7 @@ def compile(self, contributions, x=None, preprocessing=None):
self.x = x
self.preprocessing = preprocessing
if not isinstance(contributions, dict):
- raise ValueError('Contributions must be a dictionary')
+ raise ValueError("Contributions must be a dictionary")
self.methods = list(contributions.keys())
self.weights = list(contributions.values())
@@ -77,15 +76,15 @@ def check_consistency_contributions(self, weights):
List of contributions from different methods
"""
if weights[0].ndim == 1:
- raise ValueError('Multiple datapoints are required to compute the metric')
+ raise ValueError("Multiple datapoints are required to compute the metric")
if not all(isinstance(x, pd.DataFrame) for x in weights):
- raise ValueError('Contributions must be pandas DataFrames')
+ raise ValueError("Contributions must be pandas DataFrames")
if not all(x.shape == weights[0].shape for x in weights):
- raise ValueError('Contributions must be of same shape')
+ raise ValueError("Contributions must be of same shape")
if not all(x.columns.tolist() == weights[0].columns.tolist() for x in weights):
- raise ValueError('Columns names are different between contributions')
+ raise ValueError("Columns names are different between contributions")
if not all(x.index.tolist() == weights[0].index.tolist() for x in weights):
- raise ValueError('Index names are different between contributions')
+ raise ValueError("Index names are different between contributions")
def consistency_plot(self, selection=None, max_features=20):
"""
@@ -113,16 +112,17 @@ def consistency_plot(self, selection=None, max_features=20):
weights = [weight.values for weight in self.weights]
elif isinstance(selection, list):
if len(selection) == 1:
- raise ValueError('Selection must include multiple points')
+ raise ValueError("Selection must include multiple points")
else:
weights = [weight.values[selection] for weight in self.weights]
else:
- raise ValueError('Parameter selection must be a list')
+ raise ValueError("Parameter selection must be a list")
all_comparisons, mean_distances = self.calculate_all_distances(self.methods, weights)
- method_1, method_2, l2, index, backend_name_1, backend_name_2 = \
- self.find_examples(mean_distances, all_comparisons, weights)
+ method_1, method_2, l2, index, backend_name_1, backend_name_2 = self.find_examples(
+ mean_distances, all_comparisons, weights
+ )
self.plot_comparison(mean_distances)
self.plot_examples(method_1, method_2, l2, index, backend_name_1, backend_name_2, max_features)
@@ -156,7 +156,12 @@ def calculate_all_distances(self, methods, weights):
l2_dist = self.calculate_pairwise_distances(weights, index_i, index_j)
# Populate the (n choose 2)x4 array
pairwise_comparison = np.column_stack(
- (np.repeat(index_i, len(l2_dist)), np.repeat(index_j, len(l2_dist)), np.arange(len(l2_dist)), l2_dist,)
+ (
+ np.repeat(index_i, len(l2_dist)),
+ np.repeat(index_j, len(l2_dist)),
+ np.arange(len(l2_dist)),
+ l2_dist,
+ )
)
all_comparisons = np.concatenate((all_comparisons, pairwise_comparison), axis=0)
@@ -249,7 +254,9 @@ def find_examples(self, mean_distances, all_comparisons, weights):
l2 = []
# Evenly split the scale of L2 distances (from min to max excluding 0)
- for i in np.linspace(start=mean_distances[mean_distances > 0].min().min(), stop=mean_distances.max().max(), num=5):
+ for i in np.linspace(
+ start=mean_distances[mean_distances > 0].min().min(), stop=mean_distances.max().max(), num=5
+ ):
# For each split, find the closest existing L2 distance
closest_l2 = all_comparisons[:, -1][np.abs(all_comparisons[:, -1] - i).argmin()]
# Return the row that contains this L2 distance
@@ -298,18 +305,25 @@ def plot_comparison(self, mean_distances):
mean_distances : DataFrame
DataFrame storing all pairwise distances between methods
"""
- font = {"color": '#{:02x}{:02x}{:02x}'.format(50, 50, 50)}
+ font = {"color": "#{:02x}{:02x}{:02x}".format(50, 50, 50)}
fig, ax = plt.subplots(ncols=1, figsize=(10, 6))
- ax.text(x=0.5, y=1.04, s="Consistency of explanations:", fontsize=24, ha="center", transform=fig.transFigure, **font)
- ax.text(x=0.5, y=0.98, s="How similar are explanations from different methods?",
- fontsize=18, ha="center", transform=fig.transFigure, **font)
-
- ax.set_title(
- "Average distances between the explanations", fontsize=14, pad=-60
+ ax.text(
+ x=0.5, y=1.04, s="Consistency of explanations:", fontsize=24, ha="center", transform=fig.transFigure, **font
+ )
+ ax.text(
+ x=0.5,
+ y=0.98,
+ s="How similar are explanations from different methods?",
+ fontsize=18,
+ ha="center",
+ transform=fig.transFigure,
+ **font,
)
+ ax.set_title("Average distances between the explanations", fontsize=14, pad=-60)
+
coords = self.calculate_coords(mean_distances)
ax.scatter(coords[:, 0], coords[:, 1], marker="o")
@@ -331,9 +345,9 @@ def plot_comparison(self, mean_distances):
)
# set gray background
- ax.set_facecolor('#F5F5F2')
+ ax.set_facecolor("#F5F5F2")
# draw solid white grid lines
- ax.grid(color='w', linestyle='solid')
+ ax.grid(color="w", linestyle="solid")
lim = (coords.min().min(), coords.max().max())
margin = 0.1 * (lim[1] - lim[0])
@@ -395,8 +409,8 @@ def plot_examples(self, method_1, method_2, l2, index, backend_name_1, backend_n
figure
"""
y = np.arange(method_1[0].shape[0])
- fig, axes = plt.subplots(ncols=len(l2), figsize=(3*len(l2), 4))
- fig.subplots_adjust(wspace=.3, top=.8)
+ fig, axes = plt.subplots(ncols=len(l2), figsize=(3 * len(l2), 4))
+ fig.subplots_adjust(wspace=0.3, top=0.8)
if len(l2) == 1:
axes = np.array([axes])
fig.suptitle("Examples of explanations' comparisons for various distances (L2 norm)")
@@ -411,27 +425,36 @@ def plot_examples(self, method_1, method_2, l2, index, backend_name_1, backend_n
idx = np.flip(i.argsort())
i, j = i[idx], j[idx]
- axes[n].barh(y, i, label='method 1', left=0, color='#{:02x}{:02x}{:02x}'.format(255, 166, 17))
- axes[n].barh(y, j, label='method 2', left=np.abs(np.max(i)) + np.abs(np.min(j)) + np.max(i)/3,
- color='#{:02x}{:02x}{:02x}'.format(117, 152, 189)) # /3 to add space
+ axes[n].barh(y, i, label="method 1", left=0, color="#{:02x}{:02x}{:02x}".format(255, 166, 17))
+ axes[n].barh(
+ y,
+ j,
+ label="method 2",
+ left=np.abs(np.max(i)) + np.abs(np.min(j)) + np.max(i) / 3,
+ color="#{:02x}{:02x}{:02x}".format(117, 152, 189),
+ ) # /3 to add space
# set gray background
- axes[n].set_facecolor('#F5F5F2')
+ axes[n].set_facecolor("#F5F5F2")
# draw solid white grid lines
- axes[n].grid(color='w', linestyle='solid')
+ axes[n].grid(color="w", linestyle="solid")
- axes[n].set(title="%s: %s" %
- (self.index.name if self.index.name is not None else "Id", l) + "\n$d_{L2}$ = " + str(round(k, 2)))
+ axes[n].set(
+ title="{}: {}".format(self.index.name if self.index.name is not None else "Id", l)
+ + "\n$d_{L2}$ = "
+ + str(round(k, 2))
+ )
axes[n].set_xlabel("Contributions")
axes[n].set_ylabel(f"Top {max_features} features")
- axes[n].set_xticks([0, np.abs(np.max(i)) + np.abs(np.min(j)) + np.max(i)/3])
+ axes[n].set_xticks([0, np.abs(np.max(i)) + np.abs(np.min(j)) + np.max(i) / 3])
axes[n].set_xticklabels([m, o])
axes[n].set_yticks([])
return fig
- def pairwise_consistency_plot(self, methods, selection=None,
- max_features=10, max_points=100, file_name=None, auto_open=False):
+ def pairwise_consistency_plot(
+ self, methods, selection=None, max_features=10, max_points=100, file_name=None, auto_open=False
+ ):
"""The Pairwise_Consistency_plot compares the difference of 2 explainability methods across each feature and each data point,
and plots the distribution of those differences.
@@ -463,11 +486,11 @@ def pairwise_consistency_plot(self, methods, selection=None,
figure
"""
if self.x is None:
- raise ValueError('x must be defined in the compile to display the plot')
+ raise ValueError("x must be defined in the compile to display the plot")
if not isinstance(self.x, pd.DataFrame):
- raise ValueError('x must be a pandas DataFrame')
+ raise ValueError("x must be a pandas DataFrame")
if len(methods) != 2:
- raise ValueError('Choose 2 methods among methods of the contributions')
+ raise ValueError("Choose 2 methods among methods of the contributions")
# Select contributions of input methods
pair_indices = [self.methods.index(x) for x in methods]
@@ -480,12 +503,12 @@ def pairwise_consistency_plot(self, methods, selection=None,
x = self.x.iloc[ind_max_points]
elif isinstance(selection, list):
if len(selection) == 1:
- raise ValueError('Selection must include multiple points')
+ raise ValueError("Selection must include multiple points")
else:
weights = [weight.iloc[selection] for weight in pair_weights]
x = self.x.iloc[selection]
else:
- raise ValueError('Parameter selection must be a list')
+ raise ValueError("Parameter selection must be a list")
# Remove constant columns
const_cols = x.loc[:, x.apply(pd.Series.nunique) == 1]
@@ -526,15 +549,17 @@ def plot_pairwise_consistency(self, weights, x, top_features, methods, file_name
if isinstance(self.preprocessing, OrdinalEncoder):
encoder = self.preprocessing
else:
- categorical_features = [col for col in x.columns if x[col].dtype == 'object']
- encoder = OrdinalEncoder(cols=categorical_features,
- handle_unknown='ignore',
- return_df=True).fit(x)
+ categorical_features = [col for col in x.columns if x[col].dtype == "object"]
+ encoder = OrdinalEncoder(cols=categorical_features, handle_unknown="ignore", return_df=True).fit(x)
x = encoder.transform(x)
- xaxis_title = "Difference of contributions between the 2 methods" \
- + f"
{methods[0]} - {methods[1]}"
- yaxis_title = "Top features
(Ordered by mean of absolute contributions)"
+ xaxis_title = (
+ "Difference of contributions between the 2 methods"
+ + f"
{methods[0]} - {methods[1]}"
+ )
+ yaxis_title = (
+ "Top features
(Ordered by mean of absolute contributions)"
+ )
fig = make_subplots(specs=[[{"secondary_y": True}]])
@@ -551,11 +576,15 @@ def plot_pairwise_consistency(self, weights, x, top_features, methods, file_name
inverse_mapping = {v: k for k, v in mapping.to_dict().items()}
feature_value = x[c].map(inverse_mapping)
- hv_text = [f"Feature value: {i}
{methods[0]}: {j}
{methods[1]}: {k}
Diff: {l}"
- for i, j, k, l in zip(feature_value if switch else x[c].round(3),
- weights[0][c].round(2),
- weights[1][c].round(2),
- (weights[0][c] - weights[1][c]).round(2))]
+ hv_text = [
+ f"Feature value: {i}
{methods[0]}: {j}
{methods[1]}: {k}
Diff: {l}"
+ for i, j, k, l in zip(
+ feature_value if switch else x[c].round(3),
+ weights[0][c].round(2),
+ weights[1][c].round(2),
+ (weights[0][c] - weights[1][c]).round(2),
+ )
+ ]
fig.add_trace(
go.Violin(
@@ -565,25 +594,23 @@ def plot_pairwise_consistency(self, weights, x, top_features, methods, file_name
fillcolor="rgba(255, 0, 0, 0.1)",
line={"color": "black", "width": 0.5},
showlegend=False,
- ), secondary_y=False
+ ),
+ secondary_y=False,
)
fig.add_trace(
go.Scatter(
x=(weights[0][c] - weights[1][c]).values,
- y=len(x)*[i] + np.random.normal(0, 0.1, len(x)),
- mode='markers',
- marker={"color": x[c].values,
- "colorscale": self.tuning_colorscale(x[c]),
- "opacity": 0.7},
+ y=len(x) * [i] + np.random.normal(0, 0.1, len(x)),
+ mode="markers",
+ marker={"color": x[c].values, "colorscale": self.tuning_colorscale(x[c]), "opacity": 0.7},
name=c,
- text=len(x)*[c],
+ text=len(x) * [c],
hovertext=hv_text,
- hovertemplate="%{text}
" +
- "%{hovertext}
" +
- "",
+ hovertemplate="%{text}
" + "%{hovertext}
" + "",
showlegend=False,
- ), secondary_y=True
+ ),
+ secondary_y=True,
)
# Dummy invisible plot to add the color scale
@@ -595,15 +622,17 @@ def plot_pairwise_consistency(self, weights, x, top_features, methods, file_name
size=1,
color=[x.min(), x.max()],
colorscale=self.tuning_colorscale(pd.Series(np.linspace(x.min().min(), x.max().max(), 10))),
- colorbar=dict(thickness=20,
- lenmode="pixels",
- len=400,
- yanchor="top",
- y=1.1,
- ypad=20,
- title="Feature values",
- tickvals=[x.min().min(), x.max().max()],
- ticktext=["Low", "High"]),
+ colorbar=dict(
+ thickness=20,
+ lenmode="pixels",
+ len=400,
+ yanchor="top",
+ y=1.1,
+ ypad=20,
+ title="Feature values",
+ tickvals=[x.min().min(), x.max().max()],
+ ticktext=["Low", "High"],
+ ),
showscale=True,
),
hoverinfo="none",
@@ -612,12 +641,14 @@ def plot_pairwise_consistency(self, weights, x, top_features, methods, file_name
fig.add_trace(colorbar_trace)
- self._update_pairwise_consistency_fig(fig=fig,
- top_features=top_features,
- xaxis_title=xaxis_title,
- yaxis_title=yaxis_title,
- file_name=file_name,
- auto_open=auto_open)
+ self._update_pairwise_consistency_fig(
+ fig=fig,
+ top_features=top_features,
+ xaxis_title=xaxis_title,
+ yaxis_title=yaxis_title,
+ file_name=file_name,
+ auto_open=auto_open,
+ )
return fig
@@ -645,19 +676,21 @@ def _update_pairwise_consistency_fig(self, fig, top_features, xaxis_title, yaxis
dict_t = copy.deepcopy(self._style_dict["dict_title_stability"])
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_xaxis['text'] = xaxis_title
- dict_yaxis['text'] = yaxis_title
- dict_t['text'] = title
+ dict_xaxis["text"] = xaxis_title
+ dict_yaxis["text"] = yaxis_title
+ dict_t["text"] = title
fig.layout.yaxis.update(showticklabels=True)
fig.layout.yaxis2.update(showticklabels=False)
- fig.update_layout(template="none",
- title=dict_t,
- xaxis_title=dict_xaxis,
- yaxis_title=dict_yaxis,
- yaxis=dict(range=[-0.7, len(top_features)-0.3]),
- yaxis2=dict(range=[-0.7, len(top_features)-0.3]),
- height=max(500, 40 * len(top_features)))
+ fig.update_layout(
+ template="none",
+ title=dict_t,
+ xaxis_title=dict_xaxis,
+ yaxis_title=dict_yaxis,
+ yaxis=dict(range=[-0.7, len(top_features) - 0.3]),
+ yaxis2=dict(range=[-0.7, len(top_features) - 0.3]),
+ height=max(500, 40 * len(top_features)),
+ )
fig.update_yaxes(automargin=True, zeroline=False)
fig.update_xaxes(automargin=True)
diff --git a/shapash/explainer/multi_decorator.py b/shapash/explainer/multi_decorator.py
index 5a0d1663..f734d10f 100644
--- a/shapash/explainer/multi_decorator.py
+++ b/shapash/explainer/multi_decorator.py
@@ -14,9 +14,11 @@ def __init__(self, member):
self.member = member
def __getattr__(self, item):
- if item in [x for x in dir(SmartState) if not x.startswith('__')]:
+ if item in [x for x in dir(SmartState) if not x.startswith("__")]:
+
def wrapper(*args, **kwargs):
return self.delegate(item, *args, **kwargs)
+
return wrapper
else:
return self.__getattribute__(item)
@@ -73,8 +75,7 @@ def check_args(self, args, name):
"""
if not args:
raise ValueError(
- '{} is applied without arguments,'
- 'please check that you have specified contributions.'.format(name)
+ "{} is applied without arguments," "please check that you have specified contributions.".format(name)
)
def check_method(self, method, name):
@@ -94,9 +95,7 @@ def check_method(self, method, name):
Raise if not callable.
"""
if not callable(method):
- raise ValueError(
- '{} is not an allowed function, please check for any typo'.format(name)
- )
+ raise ValueError(f"{name} is not an allowed function, please check for any typo")
def check_first_arg(self, arg, name):
"""
@@ -116,8 +115,8 @@ def check_first_arg(self, arg, name):
"""
if not isinstance(arg, list):
raise ValueError(
- '{} is not applied to a list of contributions,'
- 'please check that you are dealing with a multi-class problem.'.format(name)
+ "{} is not applied to a list of contributions,"
+ "please check that you are dealing with a multi-class problem.".format(name)
)
def assign_contributions(self, ranked):
@@ -139,7 +138,7 @@ def assign_contributions(self, ranked):
ValueError
The output of a single call to rank_contributions should always be of length three.
"""
- dicts = self.delegate('assign_contributions', ranked)
+ dicts = self.delegate("assign_contributions", ranked)
keys = list(dicts[0].keys())
return {key: [d[key] for d in dicts] for key in keys}
@@ -160,7 +159,7 @@ def check_contributions(self, contributions, x_init, features_names=True):
Bool
True if all inputs share same shape and index with the prediction set.
"""
- bools = self.delegate('check_contributions', contributions, x_init, features_names)
+ bools = self.delegate("check_contributions", contributions, x_init, features_names)
return all(bools)
def combine_masks(self, masks):
@@ -178,7 +177,7 @@ def combine_masks(self, masks):
Combination of all masks.
"""
transposed_masks = list(map(list, zip(*masks)))
- return self.delegate('combine_masks', transposed_masks)
+ return self.delegate("combine_masks", transposed_masks)
def compute_masked_contributions(self, s_contrib, masks):
"""
@@ -198,7 +197,7 @@ def compute_masked_contributions(self, s_contrib, masks):
List of masked contributions (pandas.Series).
"""
arg_tup = list(zip(s_contrib, masks))
- return self.delegate('compute_masked_contributions', arg_tup)
+ return self.delegate("compute_masked_contributions", arg_tup)
def summarize(self, s_contribs, var_dicts, xs_sorted, masks, columns_dict, features_dict):
"""
@@ -225,7 +224,7 @@ def summarize(self, s_contribs, var_dicts, xs_sorted, masks, columns_dict, featu
Result of the summarize step
"""
arg_tup = list(zip(s_contribs, var_dicts, xs_sorted, masks))
- return self.delegate('summarize', arg_tup, columns_dict, features_dict)
+ return self.delegate("summarize", arg_tup, columns_dict, features_dict)
def compute_features_import(self, contributions):
"""
@@ -243,7 +242,7 @@ def compute_features_import(self, contributions):
list
list of features importance pandas.series
"""
- return self.delegate('compute_features_import', contributions)
+ return self.delegate("compute_features_import", contributions)
def compute_grouped_contributions(self, contributions, features_groups):
"""
@@ -260,4 +259,4 @@ def compute_grouped_contributions(self, contributions, features_groups):
-------
pd.DataFrame
"""
- return self.delegate('compute_grouped_contributions', contributions, features_groups)
+ return self.delegate("compute_grouped_contributions", contributions, features_groups)
diff --git a/shapash/explainer/smart_explainer.py b/shapash/explainer/smart_explainer.py
index fc085281..5ea92fb7 100644
--- a/shapash/explainer/smart_explainer.py
+++ b/shapash/explainer/smart_explainer.py
@@ -1,29 +1,38 @@
"""
Smart explainer module
"""
-import logging
import copy
-import tempfile
+import logging
import shutil
+import tempfile
+
import numpy as np
import pandas as pd
-from shapash.webapp.smart_app import SmartApp
+
+import shapash.explainer.smart_predictor
from shapash.backend import BaseBackend, get_backend_cls_from_name
-from shapash.utils.io import save_pickle
-from shapash.utils.io import load_pickle
-from shapash.utils.transform import inverse_transform, apply_postprocessing, handle_categorical_missing
-from shapash.utils.utils import get_host_name
-from shapash.utils.threading import CustomThread
-from shapash.utils.check import check_model, check_label_dict, check_y, check_postprocessing, check_features_name, check_additional_data
from shapash.backend.shap_backend import get_shap_interaction_values
from shapash.manipulation.select_lines import keep_right_contributions
-from shapash.report import check_report_requirements
from shapash.manipulation.summarize import create_grouped_features_values
-from .smart_plotter import SmartPlotter
-import shapash.explainer.smart_predictor
-from shapash.utils.model import predict_proba, predict, predict_error
-from shapash.utils.explanation_metrics import find_neighbors, shap_neighbors, get_min_nb_features, get_distance
+from shapash.report import check_report_requirements
from shapash.style.style_utils import colors_loading, select_palette
+from shapash.utils.check import (
+ check_additional_data,
+ check_features_name,
+ check_label_dict,
+ check_model,
+ check_postprocessing,
+ check_y,
+)
+from shapash.utils.explanation_metrics import find_neighbors, get_distance, get_min_nb_features, shap_neighbors
+from shapash.utils.io import load_pickle, save_pickle
+from shapash.utils.model import predict, predict_error, predict_proba
+from shapash.utils.threading import CustomThread
+from shapash.utils.transform import apply_postprocessing, handle_categorical_missing, inverse_transform
+from shapash.utils.utils import get_host_name
+from shapash.webapp.smart_app import SmartApp
+
+from .smart_plotter import SmartPlotter
logging.basicConfig(level=logging.INFO)
@@ -117,7 +126,7 @@ class SmartExplainer:
It gives, for each line, the list of most important features values regarding the local
decomposition. These values can only be understood with respect to data['var_dict']
backend_name:
- backend name if backend passed is a string
+ backend name if backend passed is a string
x_encoded: pandas.DataFrame
preprocessed dataset used by the model to perform the prediction.
x_init: pandas.DataFrame
@@ -168,18 +177,18 @@ class SmartExplainer:
"""
def __init__(
- self,
- model,
- backend='shap',
- preprocessing=None,
- postprocessing=None,
- features_groups=None,
- features_dict=None,
- label_dict=None,
- title_story: str = None,
- palette_name=None,
- colors_dict=None,
- **backend_kwargs
+ self,
+ model,
+ backend="shap",
+ preprocessing=None,
+ postprocessing=None,
+ features_groups=None,
+ features_dict=None,
+ label_dict=None,
+ title_story: str = None,
+ palette_name=None,
+ colors_dict=None,
+ **backend_kwargs,
):
if features_dict is not None and not isinstance(features_dict, dict):
raise ValueError(
@@ -203,16 +212,15 @@ def __init__(
if backend.preprocessing is None and self.preprocessing is not None:
self.backend.preprocessing = self.preprocessing
else:
- raise NotImplementedError(f'Unknown backend : {backend}')
+ raise NotImplementedError(f"Unknown backend : {backend}")
self.backend_kwargs = backend_kwargs
self.features_dict = dict() if features_dict is None else copy.deepcopy(features_dict)
self.label_dict = label_dict
self.plot = SmartPlotter(self)
- self.title_story = title_story if title_story is not None else ''
- self.palette_name = palette_name if palette_name else 'default'
- self.colors_dict = copy.deepcopy(
- select_palette(colors_loading(), self.palette_name))
+ self.title_story = title_story if title_story is not None else ""
+ self.palette_name = palette_name if palette_name else "default"
+ self.colors_dict = copy.deepcopy(select_palette(colors_loading(), self.palette_name))
if colors_dict is not None:
self.colors_dict.update(colors_dict)
self.plot.define_style_attributes(colors_dict=self.colors_dict)
@@ -231,13 +239,9 @@ def __init__(
self.explain_data = None
self.features_imp = None
- def compile(self,
- x,
- contributions=None,
- y_pred=None,
- y_target=None,
- additional_data=None,
- additional_features_dict=None):
+ def compile(
+ self, x, contributions=None, y_pred=None, y_target=None, additional_data=None, additional_features_dict=None
+ ):
"""
The compile method is the first step to understand model and
prediction. It performs the sorting of contributions, the reverse
@@ -281,7 +285,8 @@ def compile(self,
if isinstance(self.backend_name, str):
backend_cls = get_backend_cls_from_name(self.backend_name)
self.backend = backend_cls(
- model=self.model, preprocessing=self.preprocessing, masker=x, **self.backend_kwargs)
+ model=self.model, preprocessing=self.preprocessing, masker=x, **self.backend_kwargs
+ )
self.x_encoded = handle_categorical_missing(x)
x_init = inverse_transform(self.x_encoded, self.preprocessing)
self.x_init = handle_categorical_missing(x_init)
@@ -297,27 +302,22 @@ def compile(self,
self.inv_features_dict = {v: k for k, v in self.features_dict.items()}
self._apply_all_postprocessing_modifications()
- self.data = self.state.assign_contributions(
- self.state.rank_contributions(
- self.contributions,
- self.x_init
- )
- )
+ self.data = self.state.assign_contributions(self.state.rank_contributions(self.contributions, self.x_init))
self.features_desc = dict(self.x_init.nunique())
if self.features_groups is not None:
self._compile_features_groups(self.features_groups)
- self.additional_features_dict = dict() if additional_features_dict is None else self._compile_additional_features_dict(additional_features_dict)
+ self.additional_features_dict = (
+ dict()
+ if additional_features_dict is None
+ else self._compile_additional_features_dict(additional_features_dict)
+ )
self.additional_data = self._compile_additional_data(additional_data)
- def _get_contributions_from_backend_or_user(self,
- x,
- contributions):
+ def _get_contributions_from_backend_or_user(self, x, contributions):
# Computing contributions using backend
if contributions is None:
self.explain_data = self.backend.run_explainer(x=x)
- self.contributions = self.backend.get_local_contributions(
- x=x,
- explain_data=self.explain_data)
+ self.contributions = self.backend.get_local_contributions(x=x, explain_data=self.explain_data)
else:
self.explain_data = contributions
self.contributions = self.backend.format_and_aggregate_local_contributions(
@@ -335,38 +335,31 @@ def _apply_all_postprocessing_modifications(self):
self.x_contrib_plot = copy.deepcopy(self.x_init)
self.x_init = self.apply_postprocessing(postprocessing)
- def _compile_features_groups(self,
- features_groups):
+ def _compile_features_groups(self, features_groups):
"""
Performs required computations for groups of features.
"""
if self.backend.support_groups is False:
- raise AssertionError(
- f'Selected backend ({self.backend.name}) '
- f'does not support groups of features.'
- )
+ raise AssertionError(f"Selected backend ({self.backend.name}) " f"does not support groups of features.")
# Compute contributions for groups of features
- self.contributions_groups = self.state.compute_grouped_contributions(
- self.contributions, features_groups)
+ self.contributions_groups = self.state.compute_grouped_contributions(self.contributions, features_groups)
self.features_imp_groups = None
# Update features dict with groups names
self._update_features_dict_with_groups(features_groups=features_groups)
# Compute t-sne projections for groups of features
self.x_init_groups = create_grouped_features_values(
- x_init=self.x_init, x_encoded=self.x_encoded,
+ x_init=self.x_init,
+ x_encoded=self.x_encoded,
preprocessing=self.preprocessing,
features_groups=self.features_groups,
features_dict=self.features_dict,
- how='dict_of_values')
+ how="dict_of_values",
+ )
# Compute data attribute for groups of features
self.data_groups = self.state.assign_contributions(
- self.state.rank_contributions(
- self.contributions_groups,
- self.x_init_groups
- )
+ self.state.rank_contributions(self.contributions_groups, self.x_init_groups)
)
- self.columns_dict_groups = {
- i: col for i, col in enumerate(self.x_init_groups.columns)}
+ self.columns_dict_groups = {i: col for i, col in enumerate(self.x_init_groups.columns)}
def _compile_additional_features_dict(self, additional_features_dict):
"""
@@ -396,27 +389,26 @@ def _compile_additional_data(self, additional_data):
self.additional_features_dict[feature] = feature
return additional_data
- def define_style(self,
- palette_name=None,
- colors_dict=None):
+ def define_style(self, palette_name=None, colors_dict=None):
if palette_name is None and colors_dict is None:
raise ValueError("At least one of palette_name or colors_dict parameters must be defined")
new_palette_name = palette_name or self.palette_name
- new_colors_dict = copy.deepcopy(
- select_palette(colors_loading(), new_palette_name))
+ new_colors_dict = copy.deepcopy(select_palette(colors_loading(), new_palette_name))
if colors_dict is not None:
new_colors_dict.update(colors_dict)
self.colors_dict.update(new_colors_dict)
self.plot.define_style_attributes(colors_dict=self.colors_dict)
- def add(self,
- y_pred=None,
- y_target=None,
- label_dict=None,
- features_dict=None,
- title_story: str = None,
- additional_data=None,
- additional_features_dict=None):
+ def add(
+ self,
+ y_pred=None,
+ y_target=None,
+ label_dict=None,
+ features_dict=None,
+ title_story: str = None,
+ additional_data=None,
+ additional_features_dict=None,
+ ):
"""
add method allows the user to add a label_dict, features_dict
or y_pred without compiling again (and it can last a few moments).
@@ -449,11 +441,11 @@ def add(self,
"""
if y_pred is not None:
self.y_pred = check_y(self.x_init, y_pred, y_name="y_pred")
- if hasattr(self, 'y_target'):
+ if hasattr(self, "y_target"):
self.prediction_error = predict_error(self.y_target, self.y_pred, self._case)
if y_target is not None:
self.y_target = check_y(self.x_init, y_target, y_name="y_target")
- if hasattr(self, 'y_pred'):
+ if hasattr(self, "y_pred"):
self.prediction_error = predict_error(self.y_target, self.y_pred, self._case)
if label_dict is not None:
if isinstance(label_dict, dict) is False:
@@ -503,7 +495,7 @@ def get_interaction_values(self, n_samples_max=None, selection=None):
if selection:
x = x.loc[selection]
- if hasattr(self, 'x_interaction'):
+ if hasattr(self, "x_interaction"):
if self.x_interaction.equals(x[:n_samples_max]):
return self.interaction_values
@@ -528,8 +520,7 @@ def check_postprocessing_modif_strings(self, postprocessing=None):
if postprocessing is not None:
for key in postprocessing.keys():
dict_postprocess = postprocessing[key]
- if dict_postprocess['type'] in {'prefix', 'suffix'} \
- and pd.api.types.is_numeric_dtype(self.x_init[key]):
+ if dict_postprocess["type"] in {"prefix", "suffix"} and pd.api.types.is_numeric_dtype(self.x_init[key]):
modif = True
return modif
@@ -593,7 +584,7 @@ def check_features_dict(self):
Check the features_dict and add the necessary keys if all the
input X columns are not present
"""
- for feature in (set(list(self.columns_dict.values())) - set(list(self.features_dict))):
+ for feature in set(list(self.columns_dict.values())) - set(list(self.features_dict)):
self.features_dict[feature] = feature
def _update_features_dict_with_groups(self, features_groups):
@@ -636,22 +627,22 @@ def check_label_name(self, label, origin=None):
"""
if origin is None:
if label in self._classes:
- origin = 'code'
+ origin = "code"
elif self.label_dict is not None and label in self.label_dict.values():
- origin = 'value'
+ origin = "value"
elif isinstance(label, int) and label in range(-1, len(self._classes)):
- origin = 'num'
+ origin = "num"
try:
- if origin == 'num':
+ if origin == "num":
label_num = label
label_code = self._classes[label]
label_value = self.label_dict[label_code] if self.label_dict else label_code
- elif origin == 'code':
+ elif origin == "code":
label_code = label
label_num = self._classes.index(label)
label_value = self.label_dict[label_code] if self.label_dict else label_code
- elif origin == 'value':
+ elif origin == "value":
label_code = self.inv_label_dict[label]
label_num = self._classes.index(label_code)
label_value = label
@@ -698,19 +689,15 @@ def check_attributes(self, attribute):
if not hasattr(self, attribute):
raise ValueError(
"""
- attribute {0} isn't an attribute of the explainer precised.
- """.format(attribute))
+ attribute {} isn't an attribute of the explainer precised.
+ """.format(
+ attribute
+ )
+ )
return self.__dict__[attribute]
- def filter(
- self,
- features_to_hide=None,
- threshold=None,
- positive=None,
- max_contrib=None,
- display_groups=None
- ):
+ def filter(self, features_to_hide=None, threshold=None, positive=None, max_contrib=None, display_groups=None):
"""
The filter method is an important method which allows to summarize the local explainability
by using the user defined parameters which correspond to its use case.
@@ -738,40 +725,27 @@ def filter(
data = self.data_groups
else:
data = self.data
- mask = [self.state.init_mask(data['contrib_sorted'], True)]
+ mask = [self.state.init_mask(data["contrib_sorted"], True)]
if features_to_hide:
mask.append(
self.state.hide_contributions(
- data['var_dict'],
- features_list=self.check_features_name(features_to_hide, use_groups=display_groups)
+ data["var_dict"],
+ features_list=self.check_features_name(features_to_hide, use_groups=display_groups),
)
)
if threshold:
- mask.append(
- self.state.cap_contributions(
- data['contrib_sorted'],
- threshold=threshold
- )
- )
+ mask.append(self.state.cap_contributions(data["contrib_sorted"], threshold=threshold))
if positive is not None:
- mask.append(
- self.state.sign_contributions(
- data['contrib_sorted'],
- positive=positive
- )
- )
+ mask.append(self.state.sign_contributions(data["contrib_sorted"], positive=positive))
self.mask = self.state.combine_masks(mask)
if max_contrib:
self.mask = self.state.cutoff_contributions(self.mask, max_contrib=max_contrib)
- self.masked_contributions = self.state.compute_masked_contributions(
- data['contrib_sorted'],
- self.mask
- )
+ self.masked_contributions = self.state.compute_masked_contributions(data["contrib_sorted"], self.mask)
self.mask_params = {
- 'features_to_hide': features_to_hide,
- 'threshold': threshold,
- 'positive': positive,
- 'max_contrib': max_contrib
+ "features_to_hide": features_to_hide,
+ "threshold": threshold,
+ "positive": positive,
+ "max_contrib": max_contrib,
}
def save(self, path):
@@ -788,7 +762,7 @@ def save(self, path):
--------
>>> xpl.save('path_to_pkl/xpl.pkl')
"""
- if hasattr(self, 'smartapp'):
+ if hasattr(self, "smartapp"):
self.smartapp = None
save_pickle(self, path)
@@ -812,9 +786,7 @@ def load(cls, path):
smart_explainer.__dict__.update(xpl.__dict__)
return smart_explainer
else:
- raise ValueError(
- "File is not a SmartExplainer object"
- )
+ raise ValueError("File is not a SmartExplainer object")
def predict_proba(self):
"""
@@ -827,17 +799,11 @@ def predict(self):
The predict method computes the model output for each x_encoded row and stores it in y_pred attribute
"""
self.y_pred = predict(self.model, self.x_encoded)
- if hasattr(self, 'y_target'):
+ if hasattr(self, "y_target"):
self.prediction_error = predict_error(self.y_target, self.y_pred, self._case)
def to_pandas(
- self,
- features_to_hide=None,
- threshold=None,
- positive=None,
- max_contrib=None,
- proba=False,
- use_groups=None
+ self, features_to_hide=None, threshold=None, positive=None, max_contrib=None, proba=False, use_groups=None
):
"""
The to_pandas method allows to export the summary of local explainability.
@@ -888,41 +854,41 @@ def to_pandas(
# Classification: y_pred is needed
if self.y_pred is None:
- raise ValueError(
- "You have to specify y_pred argument. Please use add() or compile() method"
- )
+ raise ValueError("You have to specify y_pred argument. Please use add() or compile() method")
# Apply filter method if necessary
- if all(var is None for var in [features_to_hide, threshold, positive, max_contrib]) \
- and hasattr(self, 'mask_params') \
- and (
+ if (
+ all(var is None for var in [features_to_hide, threshold, positive, max_contrib])
+ and hasattr(self, "mask_params")
+ and (
# if the already computed mask does not have the right shape (this can happen when
# we use groups of features once and then use method without groups)
- (isinstance(data['contrib_sorted'], pd.DataFrame)
- and len(data["contrib_sorted"].columns) == len(self.mask.columns))
- or
- (isinstance(data['contrib_sorted'], list)
- and len(data["contrib_sorted"][0].columns) == len(self.mask[0].columns))
- ):
- print('to_pandas params: ' + str(self.mask_params))
+ (
+ isinstance(data["contrib_sorted"], pd.DataFrame)
+ and len(data["contrib_sorted"].columns) == len(self.mask.columns)
+ )
+ or (
+ isinstance(data["contrib_sorted"], list)
+ and len(data["contrib_sorted"][0].columns) == len(self.mask[0].columns)
+ )
+ )
+ ):
+ print("to_pandas params: " + str(self.mask_params))
else:
- self.filter(features_to_hide=features_to_hide,
- threshold=threshold,
- positive=positive,
- max_contrib=max_contrib,
- display_groups=use_groups)
+ self.filter(
+ features_to_hide=features_to_hide,
+ threshold=threshold,
+ positive=positive,
+ max_contrib=max_contrib,
+ display_groups=use_groups,
+ )
if use_groups:
columns_dict = {i: col for i, col in enumerate(self.x_init_groups.columns)}
else:
columns_dict = self.columns_dict
# Summarize information
- data['summary'] = self.state.summarize(
- data['contrib_sorted'],
- data['var_dict'],
- data['x_sorted'],
- self.mask,
- columns_dict,
- self.features_dict
+ data["summary"] = self.state.summarize(
+ data["contrib_sorted"], data["var_dict"], data["x_sorted"], self.mask, columns_dict, self.features_dict
)
# Matching with y_pred
if proba:
@@ -931,9 +897,9 @@ def to_pandas(
else:
proba_values = None
- y_pred, summary = keep_right_contributions(self.y_pred, data['summary'],
- self._case, self._classes,
- self.label_dict, proba_values)
+ y_pred, summary = keep_right_contributions(
+ self.y_pred, data["summary"], self._case, self._classes, self.label_dict, proba_values
+ )
return pd.concat([y_pred, summary], axis=1)
@@ -955,9 +921,7 @@ def compute_features_import(self, force=False):
index of the serie = contributions.columns
"""
self.features_imp = self.backend.get_global_features_importance(
- contributions=self.contributions,
- explain_data=self.explain_data,
- subset=None
+ contributions=self.contributions, explain_data=self.explain_data, subset=None
)
if self.features_groups is not None and self.features_imp_groups is None:
@@ -996,7 +960,11 @@ def compute_features_stability(self, selection):
variability = np.zeros((numb_expl, self.x_init.shape[1]))
# For each instance (+ neighbors), compute explanation
for i in range(numb_expl):
- (_, variability[i, :], amplitude[i, :],) = shap_neighbors(all_neighbors[i], self.x_encoded, self.contributions, self._case)
+ (
+ _,
+ variability[i, :],
+ amplitude[i, :],
+ ) = shap_neighbors(all_neighbors[i], self.x_encoded, self.contributions, self._case)
self.features_stability = {"variability": variability, "amplitude": amplitude}
def compute_features_compacity(self, selection, distance, nb_features):
@@ -1027,7 +995,7 @@ def compute_features_compacity(self, selection, distance, nb_features):
def init_app(self, settings: dict = None):
"""
Simple init of SmartApp in case of host smartapp by another way
-
+
Parameters
----------
settings : dict (default: None)
@@ -1039,7 +1007,9 @@ def init_app(self, settings: dict = None):
self.predict()
self.smartapp = SmartApp(self, settings)
- def run_app(self, port: int = None, host: str = None, title_story: str = None, settings: dict = None) -> CustomThread:
+ def run_app(
+ self, port: int = None, host: str = None, title_story: str = None, settings: dict = None
+ ) -> CustomThread:
"""
run_app method launches the interpretability web app associated with the shapash object.
run_app method can be used directly in a Jupyter notebook
@@ -1075,7 +1045,7 @@ def run_app(self, port: int = None, host: str = None, title_story: str = None, s
self.title_story = title_story
if self.y_pred is None:
self.predict()
- if hasattr(self, '_case'):
+ if hasattr(self, "_case"):
self.smartapp = SmartApp(self, settings)
if host is None:
host = "0.0.0.0"
@@ -1083,7 +1053,8 @@ def run_app(self, port: int = None, host: str = None, title_story: str = None, s
port = 8050
host_name = get_host_name()
server_instance = CustomThread(
- target=lambda: self.smartapp.app.run_server(debug=False, host=host, port=port))
+ target=lambda: self.smartapp.app.run_server(debug=False, host=host, port=port)
+ )
if host_name is None:
host_name = host
elif host != "0.0.0.0":
@@ -1134,18 +1105,22 @@ def to_smartpredictor(self):
self.features_types = {features: str(self.x_init[features].dtypes) for features in self.x_init.columns}
- listattributes = ["features_dict", "model", "columns_dict", "backend", "features_types",
- "label_dict", "preprocessing", "postprocessing", "features_groups"]
+ listattributes = [
+ "features_dict",
+ "model",
+ "columns_dict",
+ "backend",
+ "features_types",
+ "label_dict",
+ "preprocessing",
+ "postprocessing",
+ "features_groups",
+ ]
params_smartpredictor = [self.check_attributes(attribute) for attribute in listattributes]
if not hasattr(self, "mask_params"):
- self.mask_params = {
- "features_to_hide": None,
- "threshold": None,
- "positive": None,
- "max_contrib": None
- }
+ self.mask_params = {"features_to_hide": None, "threshold": None, "positive": None, "max_contrib": None}
params_smartpredictor.append(self.mask_params)
return shapash.explainer.smart_predictor.SmartPredictor(*params_smartpredictor)
@@ -1179,18 +1154,20 @@ def check_x_y_attributes(self, x_str, y_str):
params_checkypred.append(None)
return params_checkypred
- def generate_report(self,
- output_file,
- project_info_file,
- x_train=None,
- y_train=None,
- y_test=None,
- title_story=None,
- title_description=None,
- metrics=None,
- working_dir=None,
- notebook_path=None,
- kernel_name=None):
+ def generate_report(
+ self,
+ output_file,
+ project_info_file,
+ x_train=None,
+ y_train=None,
+ y_test=None,
+ title_story=None,
+ title_description=None,
+ metrics=None,
+ working_dir=None,
+ notebook_path=None,
+ kernel_name=None,
+ ):
"""
This method will generate an HTML report containing different information about the project.
It analyzes the data and the model used in order to provide interesting
@@ -1265,9 +1242,11 @@ def generate_report(self,
working_dir = tempfile.mkdtemp()
rm_working_dir = True
- if not hasattr(self, 'model'):
- raise AssertionError("Explainer object was not compiled. Please compile the explainer "
- "object using .compile(...) method before generating the report.")
+ if not hasattr(self, "model"):
+ raise AssertionError(
+ "Explainer object was not compiled. Please compile the explainer "
+ "object using .compile(...) method before generating the report."
+ )
try:
execute_report(
@@ -1283,7 +1262,7 @@ def generate_report(self,
metrics=metrics,
),
notebook_path=notebook_path,
- kernel_name=kernel_name
+ kernel_name=kernel_name,
)
export_and_save_report(working_dir=working_dir, output_file=output_file)
diff --git a/shapash/explainer/smart_plotter.py b/shapash/explainer/smart_plotter.py
index 0eaacb74..bbf46a1b 100644
--- a/shapash/explainer/smart_plotter.py
+++ b/shapash/explainer/smart_plotter.py
@@ -1,26 +1,34 @@
"""
Smart plotter module
"""
-import warnings
-from numbers import Number
-import random
import copy
import math
+import random
+import warnings
+from numbers import Number
+
import numpy as np
-from scipy.optimize import fsolve
-import scipy.cluster.hierarchy as sch
import pandas as pd
-from plotly import graph_objs as go
import plotly.express as px
-from plotly.subplots import make_subplots
+import scipy.cluster.hierarchy as sch
+from plotly import graph_objs as go
from plotly.offline import plot
+from plotly.subplots import make_subplots
+from scipy.optimize import fsolve
+
from shapash.manipulation.select_lines import select_lines
-from shapash.manipulation.summarize import compute_features_import, project_feature_values_1d, compute_corr
-from shapash.utils.utils import add_line_break, truncate_str, compute_digit_number, add_text, \
- maximum_difference_sort_value, compute_sorted_variables_interactions_list_indices, \
- compute_top_correlations_features
+from shapash.manipulation.summarize import compute_corr, compute_features_import, project_feature_values_1d
+from shapash.style.style_utils import colors_loading, define_style, select_palette
+from shapash.utils.utils import (
+ add_line_break,
+ add_text,
+ compute_digit_number,
+ compute_sorted_variables_interactions_list_indices,
+ compute_top_correlations_features,
+ maximum_difference_sort_value,
+ truncate_str,
+)
from shapash.webapp.utils.utils import round_to_k
-from shapash.style.style_utils import colors_loading, select_palette, define_style
class SmartPlotter:
@@ -37,8 +45,7 @@ class SmartPlotter:
>>> xpl.plot.my_plot_method(param=value)
"""
- def __init__(self,
- explainer):
+ def __init__(self, explainer):
self.explainer = explainer
self._palette_name = list(colors_loading().keys())[0]
self._style_dict = define_style(select_palette(colors_loading(), self._palette_name))
@@ -46,8 +53,7 @@ def __init__(self,
self.last_stability_selection = False
self.last_compacity_selection = False
- def define_style_attributes(self,
- colors_dict):
+ def define_style_attributes(self, colors_dict):
"""
define_style_attributes allows shapash user to change the color of plot
Parameters
@@ -60,8 +66,7 @@ def define_style_attributes(self,
if hasattr(self, "pred_colorscale"):
delattr(self, "pred_colorscale")
- def tuning_colorscale(self,
- values):
+ def tuning_colorscale(self, values):
"""
adapts the color scale to the distribution of points
Parameters
@@ -70,11 +75,9 @@ def tuning_colorscale(self,
values whose quantiles must be calculated
"""
desc_df = values.describe(percentiles=np.arange(0.1, 1, 0.1).tolist())
- min_pred, max_init = list(desc_df.loc[['min', 'max']].values)
- desc_pct_df = (desc_df.loc[~desc_df.index.isin(['count', 'mean', 'std'])] - min_pred) / \
- (max_init - min_pred)
- color_scale = list(map(list, (zip(desc_pct_df.values.flatten(),
- self._style_dict["init_contrib_colorscale"]))))
+ min_pred, max_init = list(desc_df.loc[["min", "max"]].values)
+ desc_pct_df = (desc_df.loc[~desc_df.index.isin(["count", "mean", "std"])] - min_pred) / (max_init - min_pred)
+ color_scale = list(map(list, (zip(desc_pct_df.values.flatten(), self._style_dict["init_contrib_colorscale"]))))
return color_scale
def tuning_round_digit(self):
@@ -83,23 +86,25 @@ def tuning_round_digit(self):
"""
quantile = [0.25, 0.75]
desc_df = self.explainer.y_pred.describe(percentiles=quantile)
- perc1, perc2 = list(desc_df.loc[[str(int(p * 100)) + '%' for p in quantile]].values)
+ perc1, perc2 = list(desc_df.loc[[str(int(p * 100)) + "%" for p in quantile]].values)
p_diff = perc2 - perc1
self.round_digit = compute_digit_number(p_diff)
- def _update_contributions_fig(self,
- fig,
- feature_name,
- pred,
- proba_values,
- col_modality,
- col_scale,
- addnote,
- subtitle,
- width,
- height,
- file_name,
- auto_open):
+ def _update_contributions_fig(
+ self,
+ fig,
+ feature_name,
+ pred,
+ proba_values,
+ col_modality,
+ col_scale,
+ addnote,
+ subtitle,
+ width,
+ height,
+ file_name,
+ auto_open,
+ ):
"""
Function used by both violin and scatter methods for contributions plots in order to
update the layout of the (already) created plotly figure.
@@ -134,7 +139,7 @@ def _update_contributions_fig(self,
title = f"{truncate_str(feature_name)} - Feature Contribution"
# Add subtitle and / or addnote
if subtitle or addnote:
- #title += f"
{add_text([subtitle, addnote], sep=' - ')}"
+ # title += f"
{add_text([subtitle, addnote], sep=' - ')}"
if subtitle and addnote:
title += "
" + subtitle + " - " + addnote + ""
elif subtitle:
@@ -144,47 +149,43 @@ def _update_contributions_fig(self,
dict_t = copy.deepcopy(self._style_dict["dict_title"])
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_t['text'] = title
- dict_xaxis['text'] = truncate_str(feature_name, 110)
- dict_yaxis['text'] = 'Contribution'
+ dict_t["text"] = title
+ dict_xaxis["text"] = truncate_str(feature_name, 110)
+ dict_yaxis["text"] = "Contribution"
if self.explainer._case == "regression":
colorpoints = pred
- colorbar_title = 'Predicted'
+ colorbar_title = "Predicted"
elif self.explainer._case == "classification":
colorpoints = proba_values
- colorbar_title = 'Predicted Proba'
+ colorbar_title = "Predicted Proba"
if colorpoints is not None:
fig.data[-1].marker.color = colorpoints.values.flatten()
- fig.data[-1].marker.coloraxis = 'coloraxis'
+ fig.data[-1].marker.coloraxis = "coloraxis"
fig.layout.coloraxis.colorscale = col_scale
- fig.layout.coloraxis.colorbar = {'title': {'text': colorbar_title}}
-
- elif fig.data[0].type != 'violin':
- if self.explainer._case == 'classification' and pred is not None:
- fig.data[-1].marker.color = pred.iloc[:, 0].apply(lambda
- x: self._style_dict["violin_area_classif"][1] if x == col_modality else
- self._style_dict["violin_area_classif"][0])
+ fig.layout.coloraxis.colorbar = {"title": {"text": colorbar_title}}
+
+ elif fig.data[0].type != "violin":
+ if self.explainer._case == "classification" and pred is not None:
+ fig.data[-1].marker.color = pred.iloc[:, 0].apply(
+ lambda x: self._style_dict["violin_area_classif"][1]
+ if x == col_modality
+ else self._style_dict["violin_area_classif"][0]
+ )
else:
fig.data[-1].marker.color = self._style_dict["violin_default"]
- fig.update_traces(
- marker={
- 'size': 10,
- 'opacity': 0.8,
- 'line': {'width': 0.8, 'color': 'white'}
- }
- )
+ fig.update_traces(marker={"size": 10, "opacity": 0.8, "line": {"width": 0.8, "color": "white"}})
fig.update_layout(
- template='none',
+ template="none",
title=dict_t,
width=width,
height=height,
xaxis_title=dict_xaxis,
yaxis_title=dict_yaxis,
- hovermode='closest'
+ hovermode="closest",
)
fig.update_yaxes(automargin=True)
@@ -192,22 +193,24 @@ def _update_contributions_fig(self,
if file_name:
plot(fig, filename=file_name, auto_open=auto_open)
- def plot_scatter(self,
- feature_values,
- contributions,
- feature_name,
- pred=None,
- proba_values=None,
- col_modality=None,
- col_scale=None,
- metadata=None,
- addnote=None,
- subtitle=None,
- width=900,
- height=600,
- file_name=None,
- auto_open=False,
- zoom=False):
+ def plot_scatter(
+ self,
+ feature_values,
+ contributions,
+ feature_name,
+ pred=None,
+ proba_values=None,
+ col_modality=None,
+ col_scale=None,
+ metadata=None,
+ addnote=None,
+ subtitle=None,
+ width=900,
+ height=600,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
Scatter plot of one feature contribution across the prediction set.
Parameters
@@ -245,95 +248,101 @@ def plot_scatter(self,
fig = go.Figure()
# add break line to X label if necessary
- max_len_by_row = max([
- round(50 / self.explainer.features_desc[feature_values.columns.values[0]]), 8])
+ max_len_by_row = max([round(50 / self.explainer.features_desc[feature_values.columns.values[0]]), 8])
feature_values.iloc[:, 0] = feature_values.iloc[:, 0].apply(
- add_line_break, args=(max_len_by_row, 120,))
+ add_line_break,
+ args=(
+ max_len_by_row,
+ 120,
+ ),
+ )
if pred is not None:
- hv_text = [f"Id: {x}
Predict: {y}" for x, y in
- zip(feature_values.index, pred.values.flatten())]
+ hv_text = [f"Id: {x}
Predict: {y}" for x, y in zip(feature_values.index, pred.values.flatten())]
else:
hv_text = [f"Id: {x}" for x in feature_values.index]
if metadata:
- metadata = {k: [round_to_k(x, 3) if isinstance(x, Number) else x for x in v]
- for k, v in metadata.items()}
- text_groups_features = np.swap = np.array(
- [col_values for col_values in metadata.values()])
+ metadata = {k: [round_to_k(x, 3) if isinstance(x, Number) else x for x in v] for k, v in metadata.items()}
+ text_groups_features = np.swap = np.array([col_values for col_values in metadata.values()])
text_groups_features = np.swapaxes(text_groups_features, 0, 1)
text_groups_features_keys = list(metadata.keys())
- hovertemplate = '%{hovertext}
' + \
- 'Contribution: %{y:.4f}
' + \
- '
'.join([
- '{}: %{{text[{}]}}'.format(text_groups_features_keys[i], i)
- for i in range(len(text_groups_features_keys))
- ]) + ''
+ hovertemplate = (
+ "%{hovertext}
"
+ + "Contribution: %{y:.4f}
"
+ + "
".join(
+ [
+ "{}: %{{text[{}]}}".format(text_groups_features_keys[i], i)
+ for i in range(len(text_groups_features_keys))
+ ]
+ )
+ + ""
+ )
else:
- hovertemplate = '%{hovertext}
' +\
- f'{feature_name}: ' +\
- '%{customdata[0]}
Contribution: %{y:.4f}'
+ hovertemplate = (
+ "%{hovertext}
"
+ + f"{feature_name}: "
+ + "%{customdata[0]}
Contribution: %{y:.4f}"
+ )
text_groups_features = None
fig.add_scatter(
x=feature_values.values.flatten(),
y=contributions.values.flatten(),
- mode='markers',
+ mode="markers",
hovertext=hv_text,
hovertemplate=hovertemplate,
text=text_groups_features,
)
# To change ticktext when the x label size is upper than 10 and zoom is False
if (type(feature_values.values.flatten()[0]) == str) & (not zoom):
- feature_val = [x.replace('
', '') for x in feature_values.values.flatten()]
- feature_val = [
- x.replace(x[3: len(x)-3], '...') if len(x) > 10 else x for x in feature_val]
+ feature_val = [x.replace("
", "") for x in feature_values.values.flatten()]
+ feature_val = [x.replace(x[3 : len(x) - 3], "...") if len(x) > 10 else x for x in feature_val]
fig.update_xaxes(
- tickangle=45,
- ticktext=feature_val,
- tickvals=feature_values.values.flatten(),
- tickmode="array",
- dtick=1
+ tickangle=45, ticktext=feature_val, tickvals=feature_values.values.flatten(), tickmode="array", dtick=1
)
# Customdata contains the values and index of feature_values.
# The values are used in the hovertext and the indexes are used for
# the interactions between the graphics.
- customdata = np.stack((feature_values.values.flatten(),
- feature_values.index.values), axis=-1)
+ customdata = np.stack((feature_values.values.flatten(), feature_values.index.values), axis=-1)
fig.update_traces(customdata=customdata, hovertemplate=hovertemplate)
- self._update_contributions_fig(fig=fig,
- feature_name=feature_name,
- pred=pred,
- proba_values=proba_values,
- col_modality=col_modality,
- col_scale=col_scale,
- addnote=addnote,
- subtitle=subtitle,
- width=width,
- height=height,
- file_name=file_name,
- auto_open=auto_open)
+ self._update_contributions_fig(
+ fig=fig,
+ feature_name=feature_name,
+ pred=pred,
+ proba_values=proba_values,
+ col_modality=col_modality,
+ col_scale=col_scale,
+ addnote=addnote,
+ subtitle=subtitle,
+ width=width,
+ height=height,
+ file_name=file_name,
+ auto_open=auto_open,
+ )
return fig
- def plot_violin(self,
- feature_values,
- contributions,
- feature_name,
- pred=None,
- proba_values=None,
- col_modality=None,
- col_scale=None,
- addnote=None,
- subtitle=None,
- width=900,
- height=600,
- file_name=None,
- auto_open=False,
- zoom=False):
+ def plot_violin(
+ self,
+ feature_values,
+ contributions,
+ feature_name,
+ pred=None,
+ proba_values=None,
+ col_modality=None,
+ col_scale=None,
+ addnote=None,
+ subtitle=None,
+ width=900,
+ height=600,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
Violin plot of one feature contribution across the prediction set.
Parameters
@@ -374,112 +383,129 @@ def plot_violin(self,
jitter_param = 0.075
if pred is not None:
- hv_text = [f"Id: {x}
Predict: {y}" for x, y in zip(
- feature_values.index, pred.values.flatten())]
+ hv_text = [f"Id: {x}
Predict: {y}" for x, y in zip(feature_values.index, pred.values.flatten())]
else:
hv_text = [f"Id: {x}" for x in feature_values.index]
- hv_text_df = pd.DataFrame(hv_text, columns=['text'], index=feature_values.index)
- hv_temp = f'{feature_name} :
' + '%{customdata[0]}
Contribution: %{y:.4f}'
+ hv_text_df = pd.DataFrame(hv_text, columns=["text"], index=feature_values.index)
+ hv_temp = f"{feature_name} :
" + "%{customdata[0]}
Contribution: %{y:.4f}"
# add break line to X label
- max_len_by_row = max([round(
- 50 / self.explainer.features_desc[feature_values.columns.values[0]]), 8])
+ max_len_by_row = max([round(50 / self.explainer.features_desc[feature_values.columns.values[0]]), 8])
feature_values.iloc[:, 0] = feature_values.iloc[:, 0].apply(
- add_line_break, args=(max_len_by_row, 120,))
+ add_line_break,
+ args=(
+ max_len_by_row,
+ 120,
+ ),
+ )
uniq_l = list(pd.unique(feature_values.values.flatten()))
uniq_l.sort()
for i in uniq_l:
- if pred is not None and self.explainer._case == 'classification':
- contribution_neg = contributions.loc[(pred.iloc[:, 0] != col_modality) &
- (feature_values.iloc[:, 0] == i)].values.flatten()
+ if pred is not None and self.explainer._case == "classification":
+ contribution_neg = contributions.loc[
+ (pred.iloc[:, 0] != col_modality) & (feature_values.iloc[:, 0] == i)
+ ].values.flatten()
# Check if contribution is not empty
if len(contribution_neg) != 0:
- fig.add_trace(go.Violin(x=feature_values.loc[(pred.iloc[:, 0] != col_modality) &
- (feature_values.iloc[:, 0] == i)].values.flatten(),
- y=contribution_neg,
- points=points_param,
- pointpos=-0.1,
- side='negative',
- line_color=self._style_dict["violin_area_classif"][0],
- showlegend=False,
- jitter=jitter_param,
- meanline_visible=True,
- hovertext=hv_text_df.loc[(pred.iloc[:, 0] != col_modality) &
- (feature_values.iloc[:, 0] == i)].values.flatten()
- ))
-
- contribution_pos = contributions.loc[(pred.iloc[:, 0] == col_modality) &
- (feature_values.iloc[:, 0] == i)].values.flatten()
+ fig.add_trace(
+ go.Violin(
+ x=feature_values.loc[
+ (pred.iloc[:, 0] != col_modality) & (feature_values.iloc[:, 0] == i)
+ ].values.flatten(),
+ y=contribution_neg,
+ points=points_param,
+ pointpos=-0.1,
+ side="negative",
+ line_color=self._style_dict["violin_area_classif"][0],
+ showlegend=False,
+ jitter=jitter_param,
+ meanline_visible=True,
+ hovertext=hv_text_df.loc[
+ (pred.iloc[:, 0] != col_modality) & (feature_values.iloc[:, 0] == i)
+ ].values.flatten(),
+ )
+ )
+
+ contribution_pos = contributions.loc[
+ (pred.iloc[:, 0] == col_modality) & (feature_values.iloc[:, 0] == i)
+ ].values.flatten()
if len(contribution_pos) != 0:
- fig.add_trace(go.Violin(x=feature_values.loc[(pred.iloc[:, 0] == col_modality) &
- (feature_values.iloc[:, 0] == i)].values.flatten(),
- y=contribution_pos,
- points=points_param,
- pointpos=0.1,
- side='positive',
- line_color=self._style_dict["violin_area_classif"][1],
- showlegend=False,
- jitter=jitter_param,
- meanline_visible=True,
- scalemode='count',
- hovertext=hv_text_df.loc[(pred.iloc[:, 0] == col_modality) &
- (feature_values.iloc[:, 0] == i)].values.flatten()
- ))
+ fig.add_trace(
+ go.Violin(
+ x=feature_values.loc[
+ (pred.iloc[:, 0] == col_modality) & (feature_values.iloc[:, 0] == i)
+ ].values.flatten(),
+ y=contribution_pos,
+ points=points_param,
+ pointpos=0.1,
+ side="positive",
+ line_color=self._style_dict["violin_area_classif"][1],
+ showlegend=False,
+ jitter=jitter_param,
+ meanline_visible=True,
+ scalemode="count",
+ hovertext=hv_text_df.loc[
+ (pred.iloc[:, 0] == col_modality) & (feature_values.iloc[:, 0] == i)
+ ].values.flatten(),
+ )
+ )
else:
feature = feature_values.loc[feature_values.iloc[:, 0] == i].values.flatten()
- fig.add_trace(go.Violin(x=feature,
- y=contributions.loc[feature_values.iloc[:, 0] == i].values.flatten(),
- line_color=self._style_dict["violin_default"],
- showlegend=False,
- meanline_visible=True,
- scalemode='count',
- hovertext=hv_text_df.loc[feature_values.iloc[:, 0] == i].values.flatten()
- ))
+ fig.add_trace(
+ go.Violin(
+ x=feature,
+ y=contributions.loc[feature_values.iloc[:, 0] == i].values.flatten(),
+ line_color=self._style_dict["violin_default"],
+ showlegend=False,
+ meanline_visible=True,
+ scalemode="count",
+ hovertext=hv_text_df.loc[feature_values.iloc[:, 0] == i].values.flatten(),
+ )
+ )
if pred is None:
fig.data[-1].points = points_param
fig.data[-1].pointpos = 0
fig.data[-1].jitter = jitter_param
- colorpoints = pred if self.explainer._case == "regression" else proba_values if \
- self.explainer._case == 'classification' else None
+ colorpoints = (
+ pred
+ if self.explainer._case == "regression"
+ else proba_values
+ if self.explainer._case == "classification"
+ else None
+ )
- hovertemplate = '%{hovertext}
' + hv_temp
+ hovertemplate = "%{hovertext}
" + hv_temp
feature = feature_values.values.flatten()
- customdata = np.stack((feature_values.values.flatten(),
- contributions.index.values), axis=-1)
+ customdata = np.stack((feature_values.values.flatten(), contributions.index.values), axis=-1)
if colorpoints is not None:
- fig.add_trace(go.Scatter(
- x=feature_values.values.flatten(),
- y=contributions.values.flatten(),
- mode='markers',
- showlegend=False,
- hovertext=hv_text,
- hovertemplate=hovertemplate
- ))
+ fig.add_trace(
+ go.Scatter(
+ x=feature_values.values.flatten(),
+ y=contributions.values.flatten(),
+ mode="markers",
+ showlegend=False,
+ hovertext=hv_text,
+ hovertemplate=hovertemplate,
+ )
+ )
- fig.update_layout(
- violingap=0.05,
- violingroupgap=0,
- violinmode='overlay',
- xaxis_type='category'
- )
+ fig.update_layout(violingap=0.05, violingroupgap=0, violinmode="overlay", xaxis_type="category")
# To change ticktext when the x label size is upper than 10 and zoom is False
if (type(feature[0]) == str) & (not zoom):
- feature_val = [x.replace('
', '') for x in np.unique(
- feature_values.values.flatten())]
- feature_val = [
- x.replace(x[3: len(x)-3], '...') if len(x) > 10 else x for x in feature_val]
+ feature_val = [x.replace("
", "") for x in np.unique(feature_values.values.flatten())]
+ feature_val = [x.replace(x[3 : len(x) - 3], "...") if len(x) > 10 else x for x in feature_val]
fig.update_xaxes(
tickangle=45,
ticktext=feature_val,
tickvals=np.unique(feature_values.values.flatten()),
tickmode="array",
dtick=1,
- range=[-0.6, len(uniq_l) - 0.4]
+ range=[-0.6, len(uniq_l) - 0.4],
)
else:
fig.update_xaxes(range=[-0.6, len(uniq_l) - 0.4])
@@ -487,32 +513,36 @@ def plot_violin(self,
# Update customdata and hovertemplate
fig.update_traces(customdata=customdata, hovertemplate=hovertemplate)
- self._update_contributions_fig(fig=fig,
- feature_name=feature_name,
- pred=pred,
- proba_values=proba_values,
- col_modality=col_modality,
- col_scale=col_scale,
- addnote=addnote,
- subtitle=subtitle,
- width=width,
- height=height,
- file_name=file_name,
- auto_open=auto_open)
+ self._update_contributions_fig(
+ fig=fig,
+ feature_name=feature_name,
+ pred=pred,
+ proba_values=proba_values,
+ col_modality=col_modality,
+ col_scale=col_scale,
+ addnote=addnote,
+ subtitle=subtitle,
+ width=width,
+ height=height,
+ file_name=file_name,
+ auto_open=auto_open,
+ )
return fig
- def plot_features_import(self,
- feature_imp1,
- feature_imp2=None,
- title='Features Importance',
- addnote=None,
- subtitle=None,
- width=900,
- height=500,
- file_name=None,
- auto_open=False,
- zoom=False):
+ def plot_features_import(
+ self,
+ feature_imp1,
+ feature_imp2=None,
+ title="Features Importance",
+ addnote=None,
+ subtitle=None,
+ width=900,
+ height=500,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
Plot features importance computed with the prediction set.
Parameters
@@ -552,16 +582,16 @@ def plot_features_import(self,
topmargin = topmargin + 15
dict_t.update(text=title)
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
- dict_xaxis.update(text='Mean absolute Contribution')
+ dict_xaxis.update(text="Mean absolute Contribution")
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
dict_yaxis.update(text=None)
dict_style_bar1 = self._style_dict["dict_featimp_colors"][1]
dict_style_bar2 = self._style_dict["dict_featimp_colors"][2]
- dict_yaxis['text'] = None
+ dict_yaxis["text"] = None
# Change bar color for groups of features
marker_color = [
- self._style_dict['featureimp_groups'][0]
+ self._style_dict["featureimp_groups"][0]
if (
self.explainer.features_groups is not None
and self.explainer.inv_features_dict.get(f.replace("", "").replace("", ""))
@@ -572,48 +602,42 @@ def plot_features_import(self,
]
layout = go.Layout(
- barmode='group',
- template='none',
+ barmode="group",
+ template="none",
autosize=False,
width=width,
height=height,
title=dict_t,
xaxis_title=dict_xaxis,
yaxis_title=dict_yaxis,
- hovermode='closest',
- margin={
- 'l': 160,
- 'r': 0,
- 't': topmargin,
- 'b': 50
- }
+ hovermode="closest",
+ margin={"l": 160, "r": 0, "t": topmargin, "b": 50},
)
# To change ticktext when the x label size is upper than 30 and zoom is False
if (type(feature_imp1.index[0]) == str) & (not zoom):
# change index to abc...abc if its length is upper than 30
- index_val = [
- y.replace(y[24: len(y)-3], '...') if len(y) > 30 else y for y in feature_imp1.index]
+ index_val = [y.replace(y[24 : len(y) - 3], "...") if len(y) > 30 else y for y in feature_imp1.index]
else:
index_val = feature_imp1.index
bar1 = go.Bar(
x=feature_imp1.round(4),
y=feature_imp1.index,
- orientation='h',
- name='Global',
+ orientation="h",
+ name="Global",
marker=dict_style_bar1,
marker_color=marker_color,
- hovertemplate='Feature: %{customdata}
Contribution: %{x:.4f}',
- customdata=feature_imp1.index
+ hovertemplate="Feature: %{customdata}
Contribution: %{x:.4f}",
+ customdata=feature_imp1.index,
)
if feature_imp2 is not None:
bar2 = go.Bar(
x=feature_imp2.round(4),
y=feature_imp2.index,
- orientation='h',
- name='Subset',
+ orientation="h",
+ name="Subset",
marker=dict_style_bar2,
- hovertemplate='Feature: %{customdata}
Contribution: %{x:.4f}',
- customdata=feature_imp2.index
+ hovertemplate="Feature: %{customdata}
Contribution: %{x:.4f}",
+ customdata=feature_imp2.index,
)
data = [bar2, bar1]
else:
@@ -621,28 +645,26 @@ def plot_features_import(self,
fig = go.Figure(data=data, layout=layout)
# Update ticktext
- fig.update_yaxes(ticktext=index_val,
- tickvals=feature_imp1.index,
- tickmode="array",
- dtick=1)
+ fig.update_yaxes(ticktext=index_val, tickvals=feature_imp1.index, tickmode="array", dtick=1)
fig.update_yaxes(automargin=True)
if file_name:
plot(fig, filename=file_name, auto_open=auto_open)
return fig
- def plot_bar_chart(self,
- index_value,
- var_dict,
- x_val,
- contrib,
- yaxis_max_label=12,
- subtitle=None,
- width=900,
- height=550,
- file_name=None,
- auto_open=False,
- zoom=False
- ):
+ def plot_bar_chart(
+ self,
+ index_value,
+ var_dict,
+ x_val,
+ contrib,
+ yaxis_max_label=12,
+ subtitle=None,
+ width=900,
+ height=550,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
Plotly bar plot of local explainers
Parameters
@@ -686,95 +708,102 @@ def plot_bar_chart(self,
if subtitle:
title += "
" + subtitle + ""
topmargin += 15
- dict_t['text'] = title
- dict_xaxis['text'] = 'Contribution'
- dict_yaxis['text'] = None
+ dict_t["text"] = title
+ dict_xaxis["text"] = "Contribution"
+ dict_yaxis["text"] = None
layout = go.Layout(
- barmode='group',
- template='none',
+ barmode="group",
+ template="none",
width=width,
height=height,
title=dict_t,
xaxis_title=dict_xaxis,
yaxis_title=dict_yaxis,
- yaxis_type='category',
- hovermode='closest',
- margin={
- 'l': 150,
- 'r': 20,
- 't': topmargin,
- 'b': 70
- }
+ yaxis_type="category",
+ hovermode="closest",
+ margin={"l": 150, "r": 20, "t": topmargin, "b": 70},
)
bars = []
for num, expl in enumerate(list(zip(var_dict, x_val, contrib))):
group_name = None
- if expl[1] == '':
- ylabel = '{}'.format(expl[0])
- hoverlabel = '{}'.format(expl[0])
+ if expl[1] == "":
+ ylabel = "{}".format(expl[0])
+ hoverlabel = "{}".format(expl[0])
else:
# If bar is a group of features, hovertext includes the values of the features of the group
# And color changes
- if (self.explainer.features_groups is not None
- and self.explainer.inv_features_dict.get(expl[0]) in self.explainer.features_groups.keys()
- and len(index_value) > 0):
+ if (
+ self.explainer.features_groups is not None
+ and self.explainer.inv_features_dict.get(expl[0]) in self.explainer.features_groups.keys()
+ and len(index_value) > 0
+ ):
group_name = self.explainer.inv_features_dict.get(expl[0])
- feat_groups_values = self.explainer.x_init[self.explainer.features_groups[group_name]]\
- .loc[index_value[0]]
- hoverlabel = '
'.join([
- '{} :{}'.format(add_line_break(self.explainer.features_dict.get(f_name, f_name),
- 40, maxlen=120),
- add_line_break(f_value, 40, maxlen=160))
- for f_name, f_value in feat_groups_values.to_dict().items()
- ])
+ feat_groups_values = self.explainer.x_init[self.explainer.features_groups[group_name]].loc[
+ index_value[0]
+ ]
+ hoverlabel = "
".join(
+ [
+ "{} :{}".format(
+ add_line_break(self.explainer.features_dict.get(f_name, f_name), 40, maxlen=120),
+ add_line_break(f_value, 40, maxlen=160),
+ )
+ for f_name, f_value in feat_groups_values.to_dict().items()
+ ]
+ )
else:
- hoverlabel = '{} :
{}'.format(
- add_line_break(expl[0], 40, maxlen=120),
- add_line_break(expl[1], 40, maxlen=160))
+ hoverlabel = "{} :
{}".format(
+ add_line_break(expl[0], 40, maxlen=120), add_line_break(expl[1], 40, maxlen=160)
+ )
trunc_value = truncate_str(expl[0], 45)
if not zoom:
- # Truncate value if length is upper than 30
- trunc_new_value = trunc_value.replace(
- trunc_value[24: len(trunc_value)-3], '...') if len(trunc_value) > 30 else trunc_value
+ # Truncate value if length is upper than 30
+ trunc_new_value = (
+ trunc_value.replace(trunc_value[24 : len(trunc_value) - 3], "...")
+ if len(trunc_value) > 30
+ else trunc_value
+ )
else:
trunc_new_value = trunc_value
if len(contrib) <= yaxis_max_label and (
- self.explainer.features_groups is None
- # We don't want to display label values for t-sne projected values of groups of features.
- or (
- self.explainer.features_groups is not None
- and self.explainer.inv_features_dict.get(expl[0])
- not in self.explainer.features_groups.keys()
- )
+ self.explainer.features_groups is None
+ # We don't want to display label values for t-sne projected values of groups of features.
+ or (
+ self.explainer.features_groups is not None
+ and self.explainer.inv_features_dict.get(expl[0])
+ not in self.explainer.features_groups.keys()
+ )
):
# ylabel is based on trunc_new_value
- ylabel = '{} :
{}'.format(
- trunc_new_value, truncate_str(expl[1], 45))
+ ylabel = "{} :
{}".format(trunc_new_value, truncate_str(expl[1], 45))
else:
- ylabel = ('{}'.format(trunc_new_value))
+ ylabel = f"{trunc_new_value}"
contrib_value = expl[2]
# colors
if contrib_value >= 0:
- color = 1 if expl[1] != '' else 0
+ color = 1 if expl[1] != "" else 0
else:
- color = -1 if expl[1] != '' else -2
+ color = -1 if expl[1] != "" else -2
# If the bar is a group of features we modify the color
if group_name is not None:
- bar_color = self._style_dict["featureimp_groups"][0] if color == 1 else self._style_dict["featureimp_groups"][1]
+ bar_color = (
+ self._style_dict["featureimp_groups"][0]
+ if color == 1
+ else self._style_dict["featureimp_groups"][1]
+ )
else:
- bar_color = dict_local_plot_colors[color]['color']
+ bar_color = dict_local_plot_colors[color]["color"]
barobj = go.Bar(
x=[contrib_value],
y=[ylabel],
customdata=[hoverlabel],
- orientation='h',
+ orientation="h",
marker=dict_local_plot_colors[color],
marker_color=bar_color,
showlegend=False,
- hovertemplate='%{customdata}
Contribution: %{x:.4f}'
+ hovertemplate="%{customdata}
Contribution: %{x:.4f}",
)
bars.append([color, contrib_value, num, barobj])
@@ -790,19 +819,17 @@ def plot_bar_chart(self,
else:
fig = go.Figure()
fig.update_layout(
- xaxis = { "visible": False },
- yaxis = { "visible": False },
- annotations = [
+ xaxis={"visible": False},
+ yaxis={"visible": False},
+ annotations=[
{
"text": "Select a valid single sample to display
Local Explanation plot.",
"xref": "paper",
"yref": "paper",
"showarrow": False,
- "font": {
- "size": 14
- }
+ "font": {"size": 14},
}
- ]
+ ],
)
return fig
@@ -855,7 +882,7 @@ def apply_mask_one_line(self, line, var_dict, x_val, contrib, label=None):
Masked input lists.
"""
mask = np.array([True] * len(contrib))
- if hasattr(self.explainer, 'mask'):
+ if hasattr(self.explainer, "mask"):
if isinstance(self.explainer.mask, list):
mask = self.explainer.mask[label].loc[line[0], :].values
else:
@@ -887,14 +914,14 @@ def check_masked_contributions(self, line, var_dict, x_val, contrib, label=None)
numpy arrays
Input arrays updated with masked contributions.
"""
- if hasattr(self.explainer, 'masked_contributions'):
+ if hasattr(self.explainer, "masked_contributions"):
if isinstance(self.explainer.masked_contributions, list):
ext_contrib = self.explainer.masked_contributions[label].loc[line[0], :].values
else:
ext_contrib = self.explainer.masked_contributions.loc[line[0], :].values
- ext_var_dict = ['Hidden Negative Contributions', 'Hidden Positive Contributions']
- ext_x = ['', '']
+ ext_var_dict = ["Hidden Negative Contributions", "Hidden Positive Contributions"]
+ ext_x = ["", ""]
ext_contrib = ext_contrib.tolist()
exclusion = np.where(np.array(ext_contrib) == 0)[0].tolist()
@@ -910,9 +937,7 @@ def check_masked_contributions(self, line, var_dict, x_val, contrib, label=None)
return var_dict, x_val, contrib
- def local_pred(self,
- index,
- label=None):
+ def local_pred(self, index, label=None):
"""
compute a local pred to display in local_plot
Parameters
@@ -925,7 +950,7 @@ def local_pred(self,
float: Predict or predict_proba value
"""
if self.explainer._case == "classification":
- if hasattr(self.explainer.model, 'predict_proba'):
+ if hasattr(self.explainer.model, "predict_proba"):
if not hasattr(self.explainer, "proba_values"):
self.explainer.predict_proba()
value = self.explainer.proba_values.iloc[:, [label]].loc[index].values[0]
@@ -942,20 +967,22 @@ def local_pred(self,
return value
- def local_plot(self,
- index=None,
- row_num=None,
- query=None,
- label=None,
- show_masked=True,
- show_predict=True,
- display_groups=None,
- yaxis_max_label=12,
- width=900,
- height=550,
- file_name=None,
- auto_open=False,
- zoom=False):
+ def local_plot(
+ self,
+ index=None,
+ row_num=None,
+ query=None,
+ label=None,
+ show_masked=True,
+ show_predict=True,
+ display_groups=None,
+ yaxis_max_label=12,
+ width=900,
+ height=550,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
The local_plot method is used to display the local contributions of
an individual in the dataset.
@@ -1028,7 +1055,7 @@ def local_plot(self,
if len(line) != 1:
if len(line) > 1:
- raise ValueError('Only one line/observation must match the condition')
+ raise ValueError("Only one line/observation must match the condition")
contrib = []
x_val = []
var_dict = []
@@ -1038,10 +1065,14 @@ def local_plot(self,
if (
not hasattr(self.explainer, "mask_params") # If the filter method has not been called yet
# Or if the already computed mask was not updated with current display_groups parameter
- or (isinstance(data["contrib_sorted"], pd.DataFrame)
- and len(data["contrib_sorted"].columns) != len(self.explainer.mask.columns))
- or (isinstance(data["contrib_sorted"], list)
- and len(data["contrib_sorted"][0].columns) != len(self.explainer.mask[0].columns))
+ or (
+ isinstance(data["contrib_sorted"], pd.DataFrame)
+ and len(data["contrib_sorted"].columns) != len(self.explainer.mask.columns)
+ )
+ or (
+ isinstance(data["contrib_sorted"], list)
+ and len(data["contrib_sorted"][0].columns) != len(self.explainer.mask[0].columns)
+ )
):
self.explainer.filter(max_contrib=20, display_groups=display_groups)
@@ -1051,9 +1082,9 @@ def local_plot(self,
label_num, _, label_value = self.explainer.check_label_name(label)
- contrib = data['contrib_sorted'][label_num]
- x_val = data['x_sorted'][label_num]
- var_dict = data['var_dict'][label_num]
+ contrib = data["contrib_sorted"][label_num]
+ x_val = data["x_sorted"][label_num]
+ var_dict = data["var_dict"][label_num]
if show_predict is True:
pred = self.local_pred(line[0], label_num)
@@ -1063,9 +1094,9 @@ def local_plot(self,
subtitle = f"Response: {label_value} - Proba: {pred:.4f}"
elif self.explainer._case == "regression":
- contrib = data['contrib_sorted']
- x_val = data['x_sorted']
- var_dict = data['var_dict']
+ contrib = data["contrib_sorted"]
+ x_val = data["x_sorted"]
+ var_dict = data["var_dict"]
label_num = None
if show_predict is True:
pred_value = self.local_pred(line[0])
@@ -1085,13 +1116,15 @@ def local_plot(self,
else:
var_dict = [self.explainer.features_dict[self.explainer.columns_dict[x]] for x in var_dict]
if show_masked:
- var_dict, x_val, contrib = self.check_masked_contributions(line, var_dict, x_val, contrib, label=label_num)
+ var_dict, x_val, contrib = self.check_masked_contributions(
+ line, var_dict, x_val, contrib, label=label_num
+ )
# Filtering all negative or positive contrib if specify in mask
exclusion = []
- if hasattr(self.explainer, 'mask_params'):
- if self.explainer.mask_params['positive'] is True:
+ if hasattr(self.explainer, "mask_params"):
+ if self.explainer.mask_params["positive"] is True:
exclusion = np.where(np.array(contrib) < 0)[0].tolist()
- elif self.explainer.mask_params['positive'] is False:
+ elif self.explainer.mask_params["positive"] is False:
exclusion = np.where(np.array(contrib) > 0)[0].tolist()
exclusion.sort(reverse=True)
for expl in exclusion:
@@ -1099,22 +1132,25 @@ def local_plot(self,
del x_val[expl]
del contrib[expl]
- fig = self.plot_bar_chart(line, var_dict, x_val, contrib, yaxis_max_label, subtitle, width, height, file_name,
- auto_open, zoom)
+ fig = self.plot_bar_chart(
+ line, var_dict, x_val, contrib, yaxis_max_label, subtitle, width, height, file_name, auto_open, zoom
+ )
return fig
- def contribution_plot(self,
- col,
- selection=None,
- label=-1,
- violin_maxf=10,
- max_points=2000,
- proba=True,
- width=900,
- height=600,
- file_name=None,
- auto_open=False,
- zoom=False):
+ def contribution_plot(
+ self,
+ col,
+ selection=None,
+ label=-1,
+ violin_maxf=10,
+ max_points=2000,
+ proba=True,
+ width=900,
+ height=600,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
contribution_plot method diplays a Plotly scatter or violin plot of a selected feature.
It represents the contribution of the selected feature to the predicted value.
@@ -1166,8 +1202,8 @@ def contribution_plot(self,
label_num, _, label_value = self.explainer.check_label_name(label)
if not isinstance(col, (str, int)):
- raise ValueError('parameter col must be string or int.')
- if hasattr(self.explainer, 'inv_features_dict'):
+ raise ValueError("parameter col must be string or int.")
+ if hasattr(self.explainer, "inv_features_dict"):
col = self.explainer.inv_features_dict.get(col, col)
col_is_group = self.explainer.features_groups and col in self.explainer.features_groups.keys()
@@ -1229,19 +1265,27 @@ def contribution_plot(self,
feature_values = self.explainer.x_init.loc[list_ind, col_name]
if col_is_group:
- feature_values = project_feature_values_1d(feature_values, col, self.explainer.x_init,
- self.explainer.x_encoded, self.explainer.preprocessing,
- features_dict=self.explainer.features_dict)
+ feature_values = project_feature_values_1d(
+ feature_values,
+ col,
+ self.explainer.x_init,
+ self.explainer.x_encoded,
+ self.explainer.preprocessing,
+ features_dict=self.explainer.features_dict,
+ )
contrib = subcontrib.loc[list_ind, col].to_frame()
if self.explainer.features_imp is None:
self.explainer.compute_features_import()
- features_imp = self.explainer.features_imp if isinstance(self.explainer.features_imp, pd.Series) \
+ features_imp = (
+ self.explainer.features_imp
+ if isinstance(self.explainer.features_imp, pd.Series)
else self.explainer.features_imp[0]
- top_features_of_group = features_imp.loc[self.explainer.features_groups[col]] \
- .sort_values(ascending=False)[:4].index # Displaying top 4 features
+ )
+ top_features_of_group = (
+ features_imp.loc[self.explainer.features_groups[col]].sort_values(ascending=False)[:4].index
+ ) # Displaying top 4 features
metadata = {
- self.explainer.features_dict[f_name]: self.explainer.x_init[f_name]
- for f_name in top_features_of_group
+ self.explainer.features_dict[f_name]: self.explainer.x_init[f_name] for f_name in top_features_of_group
}
text_group = "Features values were projected on the x axis using t-SNE"
# if group don't show addnote, if not, it's too long
@@ -1257,11 +1301,11 @@ def contribution_plot(self,
if self.explainer.y_pred is not None:
y_pred = self.explainer.y_pred.loc[list_ind]
# Add labels if exist
- if self.explainer._case == 'classification' and self.explainer.label_dict is not None:
+ if self.explainer._case == "classification" and self.explainer.label_dict is not None:
y_pred = y_pred.applymap(lambda x: self.explainer.label_dict[x])
col_value = self.explainer.label_dict[col_value]
# round predict
- elif self.explainer._case == 'regression':
+ elif self.explainer._case == "regression":
if self.round_digit is None:
self.tuning_round_digit()
y_pred = y_pred.applymap(lambda x: round(x, self.round_digit))
@@ -1270,30 +1314,57 @@ def contribution_plot(self,
# selecting the best plot : Scatter, Violin?
if col_value_count > violin_maxf:
- fig = self.plot_scatter(feature_values, contrib, col_label, y_pred,
- proba_values, col_value, col_scale, metadata,
- addnote, subtitle, width, height, file_name,
- auto_open, zoom)
+ fig = self.plot_scatter(
+ feature_values,
+ contrib,
+ col_label,
+ y_pred,
+ proba_values,
+ col_value,
+ col_scale,
+ metadata,
+ addnote,
+ subtitle,
+ width,
+ height,
+ file_name,
+ auto_open,
+ zoom,
+ )
else:
- fig = self.plot_violin(feature_values, contrib, col_label, y_pred,
- proba_values, col_value, col_scale, addnote,
- subtitle, width, height, file_name, auto_open,
- zoom)
+ fig = self.plot_violin(
+ feature_values,
+ contrib,
+ col_label,
+ y_pred,
+ proba_values,
+ col_value,
+ col_scale,
+ addnote,
+ subtitle,
+ width,
+ height,
+ file_name,
+ auto_open,
+ zoom,
+ )
return fig
- def features_importance(self,
- max_features=20,
- selection=None,
- label=-1,
- group_name=None,
- display_groups=True,
- force=False,
- width=900,
- height=500,
- file_name=None,
- auto_open=False,
- zoom=False):
+ def features_importance(
+ self,
+ max_features=20,
+ selection=None,
+ label=-1,
+ group_name=None,
+ display_groups=True,
+ force=False,
+ width=900,
+ height=500,
+ file_name=None,
+ auto_open=False,
+ zoom=False,
+ ):
"""
features_importance display a plotly features importance plot.
in Multiclass Case, this features_importance focus on a label value.
@@ -1344,14 +1415,16 @@ def features_importance(self,
"""
self.explainer.compute_features_import(force=force)
subtitle = None
- title = 'Features Importance'
+ title = "Features Importance"
display_groups = self.explainer.features_groups is not None and display_groups
if display_groups:
if group_name: # Case where we have groups of features and we want to display only features inside a group
if group_name not in self.explainer.features_groups.keys():
- raise ValueError(f"group_name parameter : {group_name} is not in features_groups keys. "
- f"Possible values are : {list(self.explainer.features_groups.keys())}")
- title += f' - {truncate_str(self.explainer.features_dict.get(group_name), 20)}'
+ raise ValueError(
+ f"group_name parameter : {group_name} is not in features_groups keys. "
+ f"Possible values are : {list(self.explainer.features_groups.keys())}"
+ )
+ title += f" - {truncate_str(self.explainer.features_dict.get(group_name), 20)}"
if isinstance(self.explainer.features_imp, list):
features_importance = [
label_feat_imp.loc[label_feat_imp.index.isin(self.explainer.features_groups[group_name])]
@@ -1375,9 +1448,7 @@ def features_importance(self,
global_feat_imp = features_importance[label_num].tail(max_features)
if selection is not None:
subset_feat_imp = self.explainer.backend.get_global_features_importance(
- contributions=contributions[label_num],
- explain_data=self.explainer.explain_data,
- subset=selection
+ contributions=contributions[label_num], explain_data=self.explainer.explain_data, subset=selection
)
else:
subset_feat_imp = None
@@ -1387,13 +1458,11 @@ def features_importance(self,
global_feat_imp = features_importance.tail(max_features)
if selection is not None:
subset_feat_imp = self.explainer.backend.get_global_features_importance(
- contributions=contributions,
- explain_data=self.explainer.explain_data,
- subset=selection
+ contributions=contributions, explain_data=self.explainer.explain_data, subset=selection
)
else:
subset_feat_imp = None
- addnote = ''
+ addnote = ""
if subset_feat_imp is not None:
subset_feat_imp = subset_feat_imp.reindex(global_feat_imp.index)
subset_feat_imp.index = subset_feat_imp.index.map(self.explainer.features_dict)
@@ -1401,45 +1470,47 @@ def features_importance(self,
raise ValueError("selection argument doesn't return any row")
subset_len = len(selection)
total_len = self.explainer.x_init.shape[0]
- addnote = add_text([addnote,
- f"Subset length: {subset_len} ({int(np.round(100 * subset_len / total_len))}%)"],
- sep=" - ")
+ addnote = add_text(
+ [addnote, f"Subset length: {subset_len} ({int(np.round(100 * subset_len / total_len))}%)"], sep=" - "
+ )
if self.explainer.x_init.shape[1] >= max_features:
- addnote = add_text([addnote,
- f"Total number of features: {int(self.explainer.x_init.shape[1])}"],
- sep=" - ")
+ addnote = add_text([addnote, f"Total number of features: {int(self.explainer.x_init.shape[1])}"], sep=" - ")
global_feat_imp.index = global_feat_imp.index.map(self.explainer.features_dict)
if display_groups:
# Bold font for groups of features
global_feat_imp.index = [
- '' + str(f)
+ "" + str(f)
if self.explainer.inv_features_dict.get(f) in self.explainer.features_groups.keys()
- else str(f) for f in global_feat_imp.index
+ else str(f)
+ for f in global_feat_imp.index
]
if subset_feat_imp is not None:
subset_feat_imp.index = [
- '' + str(f)
+ "" + str(f)
if self.explainer.inv_features_dict.get(f) in self.explainer.features_groups.keys()
- else str(f) for f in subset_feat_imp.index
+ else str(f)
+ for f in subset_feat_imp.index
]
- fig = self.plot_features_import(global_feat_imp, subset_feat_imp,
- title, addnote, subtitle, width,
- height, file_name, auto_open, zoom)
+ fig = self.plot_features_import(
+ global_feat_imp, subset_feat_imp, title, addnote, subtitle, width, height, file_name, auto_open, zoom
+ )
return fig
- def plot_line_comparison(self,
- index,
- feature_values,
- contributions,
- predictions=None,
- dict_features=None,
- subtitle=None,
- width=900,
- height=550,
- file_name=None,
- auto_open=False):
+ def plot_line_comparison(
+ self,
+ index,
+ feature_values,
+ contributions,
+ predictions=None,
+ dict_features=None,
+ subtitle=None,
+ width=900,
+ height=550,
+ file_name=None,
+ auto_open=False,
+ ):
"""
Plotly plot for comparisons. Displays
the contributions of several individuals. One line represents
@@ -1479,39 +1550,36 @@ def plot_line_comparison(self,
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
if len(index) == 0:
- warnings.warn('No individuals matched', UserWarning)
- dict_t['text'] = "Compare plot - No Matching Reference Entry"
+ warnings.warn("No individuals matched", UserWarning)
+ dict_t["text"] = "Compare plot - No Matching Reference Entry"
elif len(index) < 2:
- warnings.warn('Comparison needs at least 2 individuals', UserWarning)
- dict_t['text'] = "Compare plot - index : " + ' ; '.join(['' + str(id) + '' for id in index])
+ warnings.warn("Comparison needs at least 2 individuals", UserWarning)
+ dict_t["text"] = "Compare plot - index : " + " ; ".join(["" + str(id) + "" for id in index])
else:
- dict_t['text'] = "Compare plot - index : " + ' ; '.join(['' + str(id) + '' for id in index])
+ dict_t["text"] = "Compare plot - index : " + " ; ".join(["" + str(id) + "" for id in index])
- dict_xaxis['text'] = "Contributions"
+ dict_xaxis["text"] = "Contributions"
- dict_yaxis['text'] = None
+ dict_yaxis["text"] = None
if subtitle is not None:
topmargin += 15 * height / 275
- dict_t['text'] = truncate_str(dict_t['text'], 120) \
+ dict_t["text"] = (
+ truncate_str(dict_t["text"], 120)
+ f"
{truncate_str(subtitle, 200)}"
+ )
layout = go.Layout(
- template='none',
+ template="none",
title=dict_t,
xaxis_title=dict_xaxis,
yaxis_title=dict_yaxis,
- yaxis_type='category',
+ yaxis_type="category",
width=width,
height=height,
- hovermode='closest',
+ hovermode="closest",
legend=dict(x=1, y=1),
- margin={
- 'l': 150,
- 'r': 20,
- 't': topmargin,
- 'b': 70
- }
+ margin={"l": 150, "r": 20, "t": topmargin, "b": 70},
)
iteration_list = list(zip(contributions, feature_values))
@@ -1527,23 +1595,26 @@ def plot_line_comparison(self,
for contrib, feat in iteration_list:
x_i.append(contrib[i])
- features.append('' + str(feat) + '')
+ features.append("" + str(feat) + "")
pred_x_val = x_val[dict_features[feat]]
- x_hover.append(f"Id: {add_line_break(id_i, 40, 160)}"
- + f"
{add_line_break(feat, 40, 160)}
"
- + f"Contribution: {contrib[i]:.4f}
Value: "
- + str(add_line_break(pred_x_val, 40, 160)))
-
- lines.append(go.Scatter(
- x=x_i,
- y=features,
- mode='lines+markers',
- showlegend=True,
- name=f"Id: {index[i]}",
- hoverinfo="text",
- hovertext=x_hover,
- marker={'color': dic_color[i % len(dic_color)]}
- )
+ x_hover.append(
+ f"Id: {add_line_break(id_i, 40, 160)}"
+ + f"
{add_line_break(feat, 40, 160)}
"
+ + f"Contribution: {contrib[i]:.4f}
Value: "
+ + str(add_line_break(pred_x_val, 40, 160))
+ )
+
+ lines.append(
+ go.Scatter(
+ x=x_i,
+ y=features,
+ mode="lines+markers",
+ showlegend=True,
+ name=f"Id: {index[i]}",
+ hoverinfo="text",
+ hovertext=x_hover,
+ marker={"color": dic_color[i % len(dic_color)]},
+ )
)
fig = go.Figure(data=lines, layout=layout)
@@ -1555,16 +1626,18 @@ def plot_line_comparison(self,
return fig
- def compare_plot(self,
- index=None,
- row_num=None,
- label=None,
- max_features=20,
- width=900,
- height=550,
- show_predict=True,
- file_name=None,
- auto_open=True):
+ def compare_plot(
+ self,
+ index=None,
+ row_num=None,
+ label=None,
+ max_features=20,
+ width=900,
+ height=550,
+ show_predict=True,
+ file_name=None,
+ auto_open=True,
+ ):
"""
Plotly comparison plot of several individuals' contributions. Plots contributions feature by feature.
Allows to see the differences of contributions between two or more individuals,
@@ -1603,9 +1676,7 @@ def compare_plot(self,
"""
# Checking input is okay
if sum(arg is not None for arg in [row_num, index]) != 1:
- raise ValueError(
- "You have to specify just one of these arguments: index, row_num"
- )
+ raise ValueError("You have to specify just one of these arguments: index, row_num")
# Getting indexes in a list
line_reference = []
if index is not None:
@@ -1614,16 +1685,18 @@ def compare_plot(self,
line_reference.append(ident)
elif row_num is not None:
- line_reference = [self.explainer.x_init.index.values[row_nb_reference]
- for row_nb_reference in row_num
- if self.explainer.x_init.index.values[row_nb_reference] in self.explainer.x_init.index]
+ line_reference = [
+ self.explainer.x_init.index.values[row_nb_reference]
+ for row_nb_reference in row_num
+ if self.explainer.x_init.index.values[row_nb_reference] in self.explainer.x_init.index
+ ]
subtitle = ""
if len(line_reference) < 1:
- raise ValueError('No matching entry for index')
+ raise ValueError("No matching entry for index")
# Classification case
- if self.explainer._case == 'classification':
+ if self.explainer._case == "classification":
if label is None:
label = -1
@@ -1632,19 +1705,23 @@ def compare_plot(self,
if show_predict:
preds = [self.local_pred(line, label_num) for line in line_reference]
- subtitle = f"Response: {label_value} - " \
- + "Probas: " \
- + ' ; '.join([str(id) + ': ' + str(round(proba, 2)) + ''
- for proba, id in zip(preds, line_reference)])
+ subtitle = (
+ f"Response: {label_value} - "
+ + "Probas: "
+ + " ; ".join(
+ [str(id) + ": " + str(round(proba, 2)) + "" for proba, id in zip(preds, line_reference)]
+ )
+ )
# Regression case
- elif self.explainer._case == 'regression':
+ elif self.explainer._case == "regression":
contrib = self.explainer.contributions
if show_predict:
preds = [self.local_pred(line) for line in line_reference]
- subtitle = "Predictions: " + ' ; '.join([str(id) + ': ' + str(round(pred, 2)) + ''
- for id, pred in zip(line_reference, preds)])
+ subtitle = "Predictions: " + " ; ".join(
+ [str(id) + ": " + str(round(pred, 2)) + "" for id, pred in zip(line_reference, preds)]
+ )
new_contrib = list()
for ident in line_reference:
@@ -1653,7 +1730,7 @@ def compare_plot(self,
# Well labels if available
feature_values = [0] * len(contrib.columns)
- if hasattr(self.explainer, 'columns_dict'):
+ if hasattr(self.explainer, "columns_dict"):
for i, name in enumerate(contrib.columns):
feature_name = self.explainer.features_dict[name]
feature_values[i] = feature_name
@@ -1667,21 +1744,22 @@ def compare_plot(self,
iteration_list = iteration_list[::-1]
new_contrib, feature_values = list(zip(*iteration_list))
- fig = self.plot_line_comparison(line_reference, feature_values, new_contrib,
- predictions=preds, dict_features=dict_features,
- width=width, height=height, subtitle=subtitle,
- file_name=file_name, auto_open=auto_open)
+ fig = self.plot_line_comparison(
+ line_reference,
+ feature_values,
+ new_contrib,
+ predictions=preds,
+ dict_features=dict_features,
+ width=width,
+ height=height,
+ subtitle=subtitle,
+ file_name=file_name,
+ auto_open=auto_open,
+ )
return fig
- def _plot_interactions_scatter(self,
- x_name,
- y_name,
- col_name,
- x_values,
- y_values,
- col_values,
- col_scale):
+ def _plot_interactions_scatter(self, x_name, y_name, col_name, x_values, y_values, col_values, col_scale):
"""
Function used to generate a scatter plot figure for the interactions plots.
Parameters
@@ -1706,32 +1784,38 @@ def _plot_interactions_scatter(self,
"""
# add break line to X label if necessary
max_len_by_row = max([round(50 / self.explainer.features_desc[x_values.columns.values[0]]), 8])
- x_values.iloc[:, 0] = x_values.iloc[:, 0].apply(add_line_break, args=(max_len_by_row, 120,))
+ x_values.iloc[:, 0] = x_values.iloc[:, 0].apply(
+ add_line_break,
+ args=(
+ max_len_by_row,
+ 120,
+ ),
+ )
- data_df = pd.DataFrame({
- x_name: x_values.values.flatten(),
- y_name: y_values.values.flatten(),
- col_name: col_values.values.flatten()
- })
+ data_df = pd.DataFrame(
+ {
+ x_name: x_values.values.flatten(),
+ y_name: y_values.values.flatten(),
+ col_name: col_values.values.flatten(),
+ }
+ )
if isinstance(col_values.values.flatten()[0], str):
- fig = px.scatter(data_df, x=x_name, y=y_name, color=col_name,
- color_discrete_sequence=self._style_dict["interactions_discrete_colors"])
+ fig = px.scatter(
+ data_df,
+ x=x_name,
+ y=y_name,
+ color=col_name,
+ color_discrete_sequence=self._style_dict["interactions_discrete_colors"],
+ )
else:
fig = px.scatter(data_df, x=x_name, y=y_name, color=col_name, color_continuous_scale=col_scale)
- fig.update_traces(mode='markers')
+ fig.update_traces(mode="markers")
return fig
- def _plot_interactions_violin(self,
- x_name,
- y_name,
- col_name,
- x_values,
- y_values,
- col_values,
- col_scale):
+ def _plot_interactions_violin(self, x_name, y_name, col_name, x_values, y_values, col_values, col_scale):
"""
Function used to generate a violin plot figure for the interactions plots.
Parameters
@@ -1759,47 +1843,54 @@ def _plot_interactions_violin(self,
# add break line to X label
max_len_by_row = max([round(50 / self.explainer.features_desc[x_values.columns.values[0]]), 8])
- x_values.iloc[:, 0] = x_values.iloc[:, 0].apply(add_line_break, args=(max_len_by_row, 120,))
+ x_values.iloc[:, 0] = x_values.iloc[:, 0].apply(
+ add_line_break,
+ args=(
+ max_len_by_row,
+ 120,
+ ),
+ )
uniq_l = list(pd.unique(x_values.values.flatten()))
uniq_l.sort()
for i in uniq_l:
- fig.add_trace(go.Violin(x=x_values.loc[x_values.iloc[:, 0] == i].values.flatten(),
- y=y_values.loc[x_values.iloc[:, 0] == i].values.flatten(),
- line_color=self._style_dict["violin_default"],
- showlegend=False,
- meanline_visible=True,
- scalemode='count',
- ))
- scatter_fig = self._plot_interactions_scatter(x_name=x_name, y_name=y_name, col_name=col_name,
- x_values=x_values, y_values=y_values, col_values=col_values,
- col_scale=col_scale)
+ fig.add_trace(
+ go.Violin(
+ x=x_values.loc[x_values.iloc[:, 0] == i].values.flatten(),
+ y=y_values.loc[x_values.iloc[:, 0] == i].values.flatten(),
+ line_color=self._style_dict["violin_default"],
+ showlegend=False,
+ meanline_visible=True,
+ scalemode="count",
+ )
+ )
+ scatter_fig = self._plot_interactions_scatter(
+ x_name=x_name,
+ y_name=y_name,
+ col_name=col_name,
+ x_values=x_values,
+ y_values=y_values,
+ col_values=col_values,
+ col_scale=col_scale,
+ )
for trace in scatter_fig.data:
fig.add_trace(trace)
fig.update_layout(
autosize=False,
- hovermode='closest',
+ hovermode="closest",
violingap=0.05,
violingroupgap=0,
- violinmode='overlay',
- xaxis_type='category'
+ violinmode="overlay",
+ xaxis_type="category",
)
fig.update_xaxes(range=[-0.6, len(uniq_l) - 0.4])
return fig
- def _update_interactions_fig(self,
- fig,
- col_name1,
- col_name2,
- addnote,
- width,
- height,
- file_name,
- auto_open):
+ def _update_interactions_fig(self, fig, col_name1, col_name2, addnote, width, height, file_name, auto_open):
"""
Function used for the interactions plot to update the layout of the plotly figure.
Parameters
@@ -1823,7 +1914,7 @@ def _update_interactions_fig(self,
go.Figure
"""
- if fig.data[-1]['showlegend'] is False: # Case where col2 is not categorical
+ if fig.data[-1]["showlegend"] is False: # Case where col2 is not categorical
fig.layout.coloraxis.colorscale = self._style_dict["interactions_col_scale"]
else:
fig.update_layout(legend=dict(title=dict(text=col_name2)))
@@ -1832,30 +1923,24 @@ def _update_interactions_fig(self,
if addnote:
title += f"
{add_text([addnote], sep=' - ')}"
dict_t = copy.deepcopy(self._style_dict["dict_title"])
- dict_t['text'] = title
+ dict_t["text"] = title
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
- dict_xaxis['text'] = truncate_str(col_name1, 110)
+ dict_xaxis["text"] = truncate_str(col_name1, 110)
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_yaxis['text'] = 'Shap interaction value'
+ dict_yaxis["text"] = "Shap interaction value"
- fig.update_traces(
- marker={
- 'size': 8,
- 'opacity': 0.8,
- 'line': {'width': 0.8, 'color': 'white'}
- }
- )
+ fig.update_traces(marker={"size": 8, "opacity": 0.8, "line": {"width": 0.8, "color": "white"}})
fig.update_layout(
- coloraxis=dict(colorbar={'title': {'text': col_name2}}),
+ coloraxis=dict(colorbar={"title": {"text": col_name2}}),
yaxis_title=dict_yaxis,
title=dict_t,
- template='none',
+ template="none",
width=width,
height=height,
xaxis_title=dict_xaxis,
- hovermode='closest'
+ hovermode="closest",
)
fig.update_yaxes(automargin=True)
@@ -1888,7 +1973,7 @@ def _select_indices_interactions_plot(self, selection, max_points):
addnote = None
if selection is None:
# interaction_selection attribute is used to store already computed indices of interaction_values
- if hasattr(self, 'interaction_selection'):
+ if hasattr(self, "interaction_selection"):
list_ind = self.interaction_selection
elif self.explainer.x_init.shape[0] <= max_points:
list_ind = self.explainer.x_init.index.tolist()
@@ -1899,28 +1984,30 @@ def _select_indices_interactions_plot(self, selection, max_points):
if len(selection) <= max_points:
list_ind = selection
addnote = "Length of user-defined Subset : "
- elif hasattr(self, 'interaction_selection'):
+ elif hasattr(self, "interaction_selection"):
if set(selection).issubset(set(self.interaction_selection)):
list_ind = self.interaction_selection
else:
list_ind = random.sample(selection, max_points)
addnote = "Length of random Subset : "
else:
- ValueError('parameter selection must be a list')
+ ValueError("parameter selection must be a list")
self.interaction_selection = list_ind
return list_ind, addnote
- def interactions_plot(self,
- col1,
- col2,
- selection=None,
- violin_maxf=10,
- max_points=500,
- width=900,
- height=600,
- file_name=None,
- auto_open=False):
+ def interactions_plot(
+ self,
+ col1,
+ col2,
+ selection=None,
+ violin_maxf=10,
+ max_points=500,
+ width=900,
+ height=600,
+ file_name=None,
+ auto_open=False,
+ ):
"""
Diplays a Plotly scatter plot or violin plot of two selected features and their combined
contributions for each of their values.
@@ -1959,7 +2046,7 @@ def interactions_plot(self,
"""
if not (isinstance(col1, (str, int)) or isinstance(col2, (str, int))):
- raise ValueError('parameters col1 and col2 must be string or int.')
+ raise ValueError("parameters col1 and col2 must be string or int.")
col_id1 = self.explainer.check_features_name([col1])[0]
col_name1 = self.explainer.columns_dict[col_id1]
@@ -1972,9 +2059,10 @@ def interactions_plot(self,
list_ind, addnote = self._select_indices_interactions_plot(selection=selection, max_points=max_points)
if addnote is not None:
- addnote = add_text([addnote,
- f"{len(list_ind)} ({int(np.round(100 * len(list_ind) / self.explainer.x_init.shape[0]))}%)"],
- sep='')
+ addnote = add_text(
+ [addnote, f"{len(list_ind)} ({int(np.round(100 * len(list_ind) / self.explainer.x_init.shape[0]))}%)"],
+ sep="",
+ )
# Subset
if self.explainer.postprocessing_modifications:
@@ -1992,22 +2080,22 @@ def interactions_plot(self,
if col_value_count1 > violin_maxf:
fig = self._plot_interactions_scatter(
x_name=col_name1,
- y_name='Shap interaction value',
+ y_name="Shap interaction value",
col_name=col_name2,
x_values=feature_values1,
y_values=pd.DataFrame(interaction_values, index=feature_values1.index),
col_values=feature_values2,
- col_scale=self._style_dict["interactions_col_scale"]
+ col_scale=self._style_dict["interactions_col_scale"],
)
else:
fig = self._plot_interactions_violin(
x_name=col_name1,
- y_name='Shap interaction value',
+ y_name="Shap interaction value",
col_name=col_name2,
x_values=feature_values1,
y_values=pd.DataFrame(interaction_values, index=feature_values1.index),
col_values=feature_values2,
- col_scale=self._style_dict["interactions_col_scale"]
+ col_scale=self._style_dict["interactions_col_scale"],
)
self._update_interactions_fig(
@@ -2018,20 +2106,22 @@ def interactions_plot(self,
width=width,
height=height,
file_name=file_name,
- auto_open=auto_open
+ auto_open=auto_open,
)
return fig
- def top_interactions_plot(self,
- nb_top_interactions=5,
- selection=None,
- violin_maxf=10,
- max_points=500,
- width=900,
- height=600,
- file_name=None,
- auto_open=False):
+ def top_interactions_plot(
+ self,
+ nb_top_interactions=5,
+ selection=None,
+ violin_maxf=10,
+ max_points=500,
+ width=900,
+ height=600,
+ file_name=None,
+ auto_open=False,
+ ):
"""
Displays a dynamic plot with the `nb_top_interactions` most important interactions existing
between two variables.
@@ -2088,7 +2178,7 @@ def top_interactions_plot(self,
width=width,
height=height,
file_name=None,
- auto_open=False
+ auto_open=False,
)
# The number of traces of each figure is stored
@@ -2103,7 +2193,7 @@ def generate_title_dict(col_name1, col_name2, addnote):
if addnote:
title += f"
{add_text([addnote], sep=' - ')}"
dict_t = copy.deepcopy(self._style_dict["dict_title"])
- dict_t.update({'text': title, 'y': 0.88, 'x': 0.5, 'xanchor': 'center', 'yanchor': 'top'})
+ dict_t.update({"text": title, "y": 0.88, "x": 0.5, "xanchor": "center", "yanchor": "top"})
return dict_t
fig.layout.coloraxis.colorscale = self._style_dict["interactions_col_scale"]
@@ -2113,33 +2203,60 @@ def generate_title_dict(col_name1, col_name2, addnote):
updatemenus=[
dict(
active=0,
- buttons=list([
- dict(label=f"{self.explainer.columns_dict[i]} - {self.explainer.columns_dict[j]}",
- method="update",
- args=[{"visible": [True if i == id_trace else False
- for i, x in enumerate(interactions_indices_traces_mapping)
- for _ in range(x)]},
- {'xaxis': {'title': {**{'text': self.explainer.columns_dict[i]}, **self._style_dict["dict_xaxis"]}},
- 'legend': {'title': {'text': self.explainer.columns_dict[j]}},
- 'coloraxis': {'colorbar': {'title': {'text': self.explainer.columns_dict[j]}},
- 'colorscale': fig.layout.coloraxis.colorscale},
- 'title': generate_title_dict(self.explainer.columns_dict[i],
- self.explainer.columns_dict[j], addnote)},
- ])
- for id_trace, (i, j) in enumerate(indices_to_plot)
- ]),
+ buttons=list(
+ [
+ dict(
+ label=f"{self.explainer.columns_dict[i]} - {self.explainer.columns_dict[j]}",
+ method="update",
+ args=[
+ {
+ "visible": [
+ True if i == id_trace else False
+ for i, x in enumerate(interactions_indices_traces_mapping)
+ for _ in range(x)
+ ]
+ },
+ {
+ "xaxis": {
+ "title": {
+ **{"text": self.explainer.columns_dict[i]},
+ **self._style_dict["dict_xaxis"],
+ }
+ },
+ "legend": {"title": {"text": self.explainer.columns_dict[j]}},
+ "coloraxis": {
+ "colorbar": {"title": {"text": self.explainer.columns_dict[j]}},
+ "colorscale": fig.layout.coloraxis.colorscale,
+ },
+ "title": generate_title_dict(
+ self.explainer.columns_dict[i], self.explainer.columns_dict[j], addnote
+ ),
+ },
+ ],
+ )
+ for id_trace, (i, j) in enumerate(indices_to_plot)
+ ]
+ ),
direction="down",
pad={"r": 10, "t": 10},
showactive=True,
x=0.37,
xanchor="left",
y=1.25,
- yanchor="top"
- )],
+ yanchor="top",
+ )
+ ],
annotations=[
- dict(text=f"Sorted top {len(indices_to_plot)} SHAP interaction Variables :",
- x=0, xref="paper", y=1.2, yref="paper", align="left", showarrow=False)
- ]
+ dict(
+ text=f"Sorted top {len(indices_to_plot)} SHAP interaction Variables :",
+ x=0,
+ xref="paper",
+ y=1.2,
+ yref="paper",
+ align="left",
+ showarrow=False,
+ )
+ ],
)
self._update_interactions_fig(
@@ -2150,16 +2267,10 @@ def generate_title_dict(col_name1, col_name2, addnote):
width=width,
height=height,
file_name=None,
- auto_open=False
+ auto_open=False,
)
- fig.update_layout(
- title={
- 'y': 0.88,
- 'x': 0.5,
- 'xanchor': 'center',
- 'yanchor': 'top'}
- )
+ fig.update_layout(title={"y": 0.88, "x": 0.5, "xanchor": "center", "yanchor": "top"})
if file_name:
plot(fig, filename=file_name, auto_open=auto_open)
@@ -2167,18 +2278,18 @@ def generate_title_dict(col_name1, col_name2, addnote):
return fig
def correlations(
- self,
- df=None,
- max_features=20,
- features_to_hide=None,
- facet_col=None,
- how='phik',
- width=900,
- height=500,
- degree=2.5,
- decimals=2,
- file_name=None,
- auto_open=False
+ self,
+ df=None,
+ max_features=20,
+ features_to_hide=None,
+ facet_col=None,
+ how="phik",
+ width=900,
+ height=500,
+ degree=2.5,
+ decimals=2,
+ file_name=None,
+ auto_open=False,
):
"""
Correlations matrix heatmap plot.
@@ -2220,8 +2331,8 @@ def correlations(
def cluster_corr(corr, degree, inplace=False):
"""
- Rearranges the correlation matrix, corr, so that groups of highly
- correlated variables are next to eachother
+ Rearranges the correlation matrix, corr, so that groups of highly
+ correlated variables are next to eachother
Parameters
----------
@@ -2242,10 +2353,10 @@ def cluster_corr(corr, degree, inplace=False):
if corr.shape[0] < 2:
return corr
- pairwise_distances = sch.distance.pdist(corr**degree)
- linkage = sch.linkage(pairwise_distances, method='complete')
- cluster_distance_threshold = pairwise_distances.max()/2
- idx_to_cluster_array = sch.fcluster(linkage, cluster_distance_threshold, criterion='distance')
+ pairwise_distances = sch.distance.pdist(corr ** degree)
+ linkage = sch.linkage(pairwise_distances, method="complete")
+ cluster_distance_threshold = pairwise_distances.max() / 2
+ idx_to_cluster_array = sch.fcluster(linkage, cluster_distance_threshold, criterion="distance")
idx = np.argsort(idx_to_cluster_array)
if not inplace:
@@ -2267,17 +2378,18 @@ def cluster_corr(corr, degree, inplace=False):
features_to_hide += [facet_col]
# We use phik by default as it is a convenient method for numeric and categorical data
- if how == 'phik':
+ if how == "phik":
try:
from phik import phik_matrix
- compute_method = 'phik'
+
+ compute_method = "phik"
except (ImportError, ModuleNotFoundError):
warnings.warn('Cannot compute phik correlations. Install phik using "pip install phik".', UserWarning)
compute_method = "pearson"
else:
compute_method = how
- hovertemplate = '%{text}
Correlation: %{z}'
+ hovertemplate = "%{text}
Correlation: %{z}"
list_features = []
if facet_col:
@@ -2286,7 +2398,7 @@ def cluster_corr(corr, degree, inplace=False):
rows=1,
cols=df[facet_col].nunique(),
subplot_titles=[t + " correlation" for t in facet_col_values],
- horizontal_spacing=0.15
+ horizontal_spacing=0.15,
)
# Used for the Shapash report to get train then test set
for i, col_v in enumerate(facet_col_values):
@@ -2303,12 +2415,20 @@ def cluster_corr(corr, degree, inplace=False):
z=corr.loc[list_features, list_features].round(decimals).values,
x=list_features,
y=list_features,
- coloraxis='coloraxis',
- text=[[f'Feature 1: {self.explainer.features_dict.get(y, y)}
'
- f'Feature 2: {self.explainer.features_dict.get(x, x)}' for x in list_features]
- for y in list_features],
+ coloraxis="coloraxis",
+ text=[
+ [
+ f"Feature 1: {self.explainer.features_dict.get(y, y)}
"
+ f"Feature 2: {self.explainer.features_dict.get(x, x)}"
+ for x in list_features
+ ]
+ for y in list_features
+ ],
hovertemplate=hovertemplate,
- ), row=1, col=i+1)
+ ),
+ row=1,
+ col=i + 1,
+ )
else:
corr = compute_corr(df.drop(features_to_hide, axis=1), compute_method)
@@ -2316,30 +2436,37 @@ def cluster_corr(corr, degree, inplace=False):
corr = cluster_corr(corr.loc[top_features, top_features], degree=degree)
list_features = [col for col in corr.columns if col in top_features]
- fig = go.Figure(go.Heatmap(
- z=corr.loc[list_features, list_features].round(decimals).values,
- x=list_features,
- y=list_features,
- coloraxis='coloraxis',
- text=[[f'Feature 1: {self.explainer.features_dict.get(y, y)}
'
- f'Feature 2: {self.explainer.features_dict.get(x, x)}' for x in list_features]
- for y in list_features],
- hovertemplate=hovertemplate,
- ))
-
- title = f'Correlation ({compute_method})'
+ fig = go.Figure(
+ go.Heatmap(
+ z=corr.loc[list_features, list_features].round(decimals).values,
+ x=list_features,
+ y=list_features,
+ coloraxis="coloraxis",
+ text=[
+ [
+ f"Feature 1: {self.explainer.features_dict.get(y, y)}
"
+ f"Feature 2: {self.explainer.features_dict.get(x, x)}"
+ for x in list_features
+ ]
+ for y in list_features
+ ],
+ hovertemplate=hovertemplate,
+ )
+ )
+
+ title = f"Correlation ({compute_method})"
if len(list_features) < len(df.drop(features_to_hide, axis=1).columns):
subtitle = f"Top {len(list_features)} correlations"
title += f"
{subtitle}"
dict_t = copy.deepcopy(self._style_dict["dict_title"])
- dict_t['text'] = title
+ dict_t["text"] = title
fig.update_layout(
- coloraxis=dict(colorscale=['rgb(255, 255, 255)'] + self._style_dict["init_contrib_colorscale"][5:-1]),
+ coloraxis=dict(colorscale=["rgb(255, 255, 255)"] + self._style_dict["init_contrib_colorscale"][5:-1]),
showlegend=True,
title=dict_t,
width=width,
- height=height
+ height=height,
)
fig.update_yaxes(automargin=True)
@@ -2370,51 +2497,51 @@ def plot_amplitude_vs_stability(self, mean_variability, mean_amplitude, column_n
-------
go.Figure
"""
- xaxis_title = "Variability of the Normalized Local Contribution Values" \
- + "
(standard deviation / mean)"
+ xaxis_title = (
+ "Variability of the Normalized Local Contribution Values"
+ + "
(standard deviation / mean)"
+ )
yaxis_title = "Importance
(Average contributions)"
col_scale = self.tuning_colorscale(pd.DataFrame(mean_amplitude))
- hv_text = [f"Feature: {col}
Importance: {y}
Variability: {x}"
- for col, x, y in zip(column_names, mean_variability, mean_amplitude)]
- hovertemplate = "%{hovertext}" + ''
+ hv_text = [
+ f"Feature: {col}
Importance: {y}
Variability: {x}"
+ for col, x, y in zip(column_names, mean_variability, mean_amplitude)
+ ]
+ hovertemplate = "%{hovertext}" + ""
fig = go.Figure()
fig.add_scatter(
x=mean_variability,
y=mean_amplitude,
showlegend=False,
- mode='markers',
+ mode="markers",
marker={
- 'color': mean_amplitude,
- 'size': 10,
- 'opacity': 0.8,
- 'line': {'width': 0.8, 'color': 'white'},
- 'colorscale': col_scale
+ "color": mean_amplitude,
+ "size": 10,
+ "opacity": 0.8,
+ "line": {"width": 0.8, "color": "white"},
+ "colorscale": col_scale,
},
hovertext=hv_text,
- hovertemplate=hovertemplate
+ hovertemplate=hovertemplate,
)
- fig.update_xaxes(range=[np.min(np.append(mean_variability, [0.15])) - 0.03,
- np.max(mean_variability) + 0.03])
+ fig.update_xaxes(range=[np.min(np.append(mean_variability, [0.15])) - 0.03, np.max(mean_variability) + 0.03])
- self._update_stability_fig(fig=fig,
- x_barlen=len(mean_amplitude),
- y_bar=[0, mean_amplitude.max()],
- xaxis_title=xaxis_title,
- yaxis_title=yaxis_title,
- file_name=file_name,
- auto_open=auto_open)
+ self._update_stability_fig(
+ fig=fig,
+ x_barlen=len(mean_amplitude),
+ y_bar=[0, mean_amplitude.max()],
+ xaxis_title=xaxis_title,
+ yaxis_title=yaxis_title,
+ file_name=file_name,
+ auto_open=auto_open,
+ )
return fig
- def plot_stability_distribution(self,
- variability,
- plot_type,
- mean_amplitude,
- dataset,
- column_names,
- file_name,
- auto_open):
+ def plot_stability_distribution(
+ self, variability, plot_type, mean_amplitude, dataset, column_names, file_name, auto_open
+ ):
"""
Intermediate function used to display the stability plot when plot_type is "boxplot" or
"violin"
@@ -2490,13 +2617,15 @@ def plot_stability_distribution(self,
size=1,
color=[mean_amplitude.min(), mean_amplitude.max()],
colorscale=col_scale,
- colorbar=dict(thickness=20,
- lenmode="pixels",
- len=300,
- yanchor="top",
- y=1,
- ypad=60,
- title="Importance
(Average contributions)"),
+ colorbar=dict(
+ thickness=20,
+ lenmode="pixels",
+ len=300,
+ yanchor="top",
+ y=1,
+ ypad=60,
+ title="Importance
(Average contributions)",
+ ),
showscale=True,
),
hoverinfo="none",
@@ -2509,13 +2638,15 @@ def plot_stability_distribution(self,
height=height_value,
)
- self._update_stability_fig(fig=fig,
- x_barlen=len(mean_amplitude),
- y_bar=column_names,
- xaxis_title=xaxis_title,
- yaxis_title=yaxis_title,
- file_name=file_name,
- auto_open=auto_open)
+ self._update_stability_fig(
+ fig=fig,
+ x_barlen=len(mean_amplitude),
+ y_bar=column_names,
+ xaxis_title=xaxis_title,
+ yaxis_title=yaxis_title,
+ file_name=file_name,
+ auto_open=auto_open,
+ )
return fig
@@ -2548,10 +2679,10 @@ def _update_stability_fig(self, fig, x_barlen, y_bar, xaxis_title, yaxis_title,
dict_t = copy.deepcopy(self._style_dict["dict_title_stability"])
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_xaxis['text'] = xaxis_title
- dict_yaxis['text'] = yaxis_title
+ dict_xaxis["text"] = xaxis_title
+ dict_yaxis["text"] = yaxis_title
dict_stability_bar_colors = copy.deepcopy(self._style_dict["dict_stability_bar_colors"])
- dict_t['text'] = title
+ dict_t["text"] = title
fig.add_trace(
go.Scatter(
@@ -2576,12 +2707,12 @@ def _update_stability_fig(self, fig, x_barlen, y_bar, xaxis_title, yaxis_title,
)
fig.update_layout(
- template='none',
+ template="none",
title=dict_t,
xaxis_title=dict_xaxis,
yaxis_title=dict_yaxis,
coloraxis_showscale=False,
- hovermode='closest'
+ hovermode="closest",
)
fig.update_yaxes(automargin=True)
@@ -2590,11 +2721,7 @@ def _update_stability_fig(self, fig, x_barlen, y_bar, xaxis_title, yaxis_title,
if file_name:
plot(fig, filename=file_name, auto_open=auto_open)
- def local_neighbors_plot(self,
- index,
- max_features=10,
- file_name=None,
- auto_open=False):
+ def local_neighbors_plot(self, index, max_features=10, file_name=None, auto_open=False):
"""
The Local_neighbors_plot has the main objective of increasing confidence \
in interpreting the contribution values of a selected instance.
@@ -2637,7 +2764,9 @@ def local_neighbors_plot(self,
self.explainer.compute_features_stability([index])
column_names = np.array([self.explainer.features_dict.get(x) for x in self.explainer.x_init.columns])
- def ordinal(n): return "%d%s" % (n, "tsnrhtdd"[(math.floor(n / 10) % 10 != 1) * (n % 10 < 4) * n % 10:: 4])
+
+ def ordinal(n):
+ return "%d%s" % (n, "tsnrhtdd"[(math.floor(n / 10) % 10 != 1) * (n % 10 < 4) * n % 10 :: 4])
# Compute explanations for instance and neighbors
g = self.explainer.local_neighbors["norm_shap"]
@@ -2659,32 +2788,41 @@ def ordinal(n): return "%d%s" % (n, "tsnrhtdd"[(math.floor(n / 10) % 10 != 1) *
if max_features is not None:
g_df = g_df[:max_features]
- fig = go.Figure(data=[go.Bar(name=g_df.iloc[::-1, ::-1].columns[i],
- y=g_df.iloc[::-1, ::-1].index.tolist(),
- x=g_df.iloc[::-1, ::-1].iloc[:, i],
- marker_color=self._style_dict["dict_stability_bar_colors"][1] if i == g_df.shape[1]-1
- else self._style_dict["dict_stability_bar_colors"][0],
- orientation='h',
- opacity=np.clip(0.2+i*(1-0.2)/(g_df.shape[1]-1), 0.2, 1)
- if g_df.shape[1] > 1 else 1) for i in range(g_df.shape[1])])
+ fig = go.Figure(
+ data=[
+ go.Bar(
+ name=g_df.iloc[::-1, ::-1].columns[i],
+ y=g_df.iloc[::-1, ::-1].index.tolist(),
+ x=g_df.iloc[::-1, ::-1].iloc[:, i],
+ marker_color=self._style_dict["dict_stability_bar_colors"][1]
+ if i == g_df.shape[1] - 1
+ else self._style_dict["dict_stability_bar_colors"][0],
+ orientation="h",
+ opacity=np.clip(0.2 + i * (1 - 0.2) / (g_df.shape[1] - 1), 0.2, 1) if g_df.shape[1] > 1 else 1,
+ )
+ for i in range(g_df.shape[1])
+ ]
+ )
title = f"Comparing local explanations in a neighborhood - Id: {index}"
title += "
How similar are explanations for closeby neighbours?"
dict_t = copy.deepcopy(self._style_dict["dict_title_stability"])
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_xaxis['text'] = "Normalized contribution values"
- dict_yaxis['text'] = ""
- dict_t['text'] = title
- fig.update_layout(template="none",
- title=dict_t,
- xaxis_title=dict_xaxis,
- yaxis_title=dict_yaxis,
- hovermode='closest',
- barmode="group",
- height=max(500, 11*g_df.shape[0]*g_df.shape[1]),
- legend={"traceorder": "reversed"},
- xaxis={"side": "bottom"})
+ dict_xaxis["text"] = "Normalized contribution values"
+ dict_yaxis["text"] = ""
+ dict_t["text"] = title
+ fig.update_layout(
+ template="none",
+ title=dict_t,
+ xaxis_title=dict_xaxis,
+ yaxis_title=dict_yaxis,
+ hovermode="closest",
+ barmode="group",
+ height=max(500, 11 * g_df.shape[0] * g_df.shape[1]),
+ legend={"traceorder": "reversed"},
+ xaxis={"side": "bottom"},
+ )
fig.update_yaxes(automargin=True)
fig.update_xaxes(automargin=True)
@@ -2694,14 +2832,16 @@ def ordinal(n): return "%d%s" % (n, "tsnrhtdd"[(math.floor(n / 10) % 10 != 1) *
return fig
- def stability_plot(self,
- selection=None,
- max_points=500,
- force=False,
- max_features=10,
- distribution='none',
- file_name=None,
- auto_open=False):
+ def stability_plot(
+ self,
+ selection=None,
+ max_points=500,
+ force=False,
+ max_features=10,
+ distribution="none",
+ file_name=None,
+ auto_open=False,
+ ):
"""
The Stability_plot has the main objective of increasing confidence in contribution values, \
and helping determine if we can trust an explanation.
@@ -2766,14 +2906,16 @@ def stability_plot(self,
self.last_stability_selection = False
elif isinstance(selection, list):
if len(selection) == 1:
- raise ValueError('Selection must include multiple points')
+ raise ValueError("Selection must include multiple points")
if len(selection) > max_points:
- print(f"Size of selection is bigger than max_points (default: {max_points}).\
- Computation time might be affected")
+ print(
+ f"Size of selection is bigger than max_points (default: {max_points}).\
+ Computation time might be affected"
+ )
self.explainer.compute_features_stability(selection)
self.last_stability_selection = True
else:
- raise ValueError('Parameter selection must be a list')
+ raise ValueError("Parameter selection must be a list")
column_names = np.array([self.explainer.features_dict.get(x) for x in self.explainer.x_init.columns])
@@ -2784,7 +2926,7 @@ def stability_plot(self,
mean_amplitude = amplitude.mean(axis=0)
# Plot 1 : only show average variability on y-axis
- if distribution not in ['boxplot', 'violin']:
+ if distribution not in ["boxplot", "violin"]:
fig = self.plot_amplitude_vs_stability(mean_variability, mean_amplitude, column_names, file_name, auto_open)
# Plot 2 : Show distribution of variability
@@ -2800,19 +2942,15 @@ def stability_plot(self,
dataset = self.explainer.x_init.iloc[:, keep]
column_names = column_names[keep]
- fig = self.plot_stability_distribution(variability, distribution, mean_amplitude, dataset,
- column_names, file_name, auto_open)
+ fig = self.plot_stability_distribution(
+ variability, distribution, mean_amplitude, dataset, column_names, file_name, auto_open
+ )
return fig
- def compacity_plot(self,
- selection=None,
- max_points=2000,
- force=False,
- approx=0.9,
- nb_features=5,
- file_name=None,
- auto_open=False):
+ def compacity_plot(
+ self, selection=None, max_points=2000, force=False, approx=0.9, nb_features=5, file_name=None, auto_open=False
+ ):
"""
The Compacity_plot has the main objective of determining if a small subset of features \
can be extracted to provide a simpler explanation of the model. \
@@ -2861,12 +2999,14 @@ def compacity_plot(self,
self.last_compacity_selection = False
elif isinstance(selection, list):
if len(selection) > max_points:
- print(f"Size of selection is bigger than max_points (default: {max_points}).\
- Computation time might be affected")
+ print(
+ f"Size of selection is bigger than max_points (default: {max_points}).\
+ Computation time might be affected"
+ )
self.explainer.compute_features_compacity(selection, 1 - approx, nb_features)
self.last_compacity_selection = True
else:
- raise ValueError('Parameter selection must be a list')
+ raise ValueError("Parameter selection must be a list")
features_needed = self.explainer.features_compacity["features_needed"]
distance_reached = self.explainer.features_compacity["distance_reached"]
@@ -2876,14 +3016,12 @@ def compacity_plot(self,
rows=1,
cols=2,
subplot_titles=[
- "Number of features required
to explain "
- + str(round(100 * approx))
- + "% of the model's output",
+ "Number of features required
to explain " + str(round(100 * approx)) + "% of the model's output",
"Percentage of the model output
explained by the "
+ str(nb_features)
+ " most important
features per instance",
],
- horizontal_spacing=0.2
+ horizontal_spacing=0.2,
)
# Used as titles in make_subplots are considered annotations
@@ -2908,8 +3046,8 @@ def compacity_plot(self,
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_xaxis['text'] = "Number of selected features"
- dict_yaxis['text'] = "Cumulative distribution over
dataset's instances (%)"
+ dict_xaxis["text"] = "Number of selected features"
+ dict_yaxis["text"] = "Cumulative distribution over
dataset's instances (%)"
fig.update_xaxes(title=dict_xaxis, row=1, col=1)
fig.update_yaxes(title=dict_yaxis, row=1, col=1)
@@ -2921,7 +3059,9 @@ def compacity_plot(self,
histnorm="percent",
cumulative={"enabled": True, "direction": "decreasing"},
name="",
- hovertemplate="Top " + str(nb_features) + " features explain at least "
+ hovertemplate="Top "
+ + str(nb_features)
+ + " features explain at least "
+ "%{x:.0f}"
+ "%
of the model for %{y:.1f}% of the instances",
marker_color=self._style_dict["dict_compacity_bar_colors"][0],
@@ -2932,8 +3072,8 @@ def compacity_plot(self,
dict_xaxis2 = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis2 = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_xaxis2['text'] = "Percentage of model output
explained (%)"
- dict_yaxis2['text'] = "Cumulative distribution over
dataset's instances (%)"
+ dict_xaxis2["text"] = "Percentage of model output
explained (%)"
+ dict_yaxis2["text"] = "Cumulative distribution over
dataset's instances (%)"
fig.update_xaxes(title=dict_xaxis2, row=1, col=2)
fig.update_yaxes(title=dict_yaxis2, row=1, col=2)
@@ -2941,13 +3081,13 @@ def compacity_plot(self,
title = "Compacity of explanations:"
title += "
How many variables are enough to produce accurate explanations?"
dict_t = copy.deepcopy(self._style_dict["dict_title_stability"])
- dict_t['text'] = title
+ dict_t["text"] = title
fig.update_layout(
template="none",
title=dict_t,
title_y=0.8,
- hovermode='closest',
+ hovermode="closest",
margin={"t": 150},
showlegend=False,
)
@@ -2957,15 +3097,16 @@ def compacity_plot(self,
return fig
- def scatter_plot_prediction(self,
- selection=None,
- label=-1,
- max_points=2000,
- width=900,
- height=600,
- file_name=None,
- auto_open=False,
- ):
+ def scatter_plot_prediction(
+ self,
+ selection=None,
+ label=-1,
+ max_points=2000,
+ width=900,
+ height=600,
+ file_name=None,
+ auto_open=False,
+ ):
"""
scatter_plot_prediction displays a Plotly scatter or violin plot of predictions in comparison to the target variable.
This plot represents Trues Values versus Predicted Values.
@@ -3016,26 +3157,20 @@ def scatter_plot_prediction(self,
dict_t = copy.deepcopy(self._style_dict["dict_title"])
dict_xaxis = copy.deepcopy(self._style_dict["dict_xaxis"])
dict_yaxis = copy.deepcopy(self._style_dict["dict_yaxis"])
- dict_t['text'] = title
- dict_xaxis['text'] = truncate_str('True Values', 110)
- dict_yaxis['text'] = 'Predicted Values'
-
- fig.update_traces(
- marker={
- 'size': 10,
- 'opacity': 0.8,
- 'line': {'width': 0.8, 'color': 'white'}
- }
- )
+ dict_t["text"] = title
+ dict_xaxis["text"] = truncate_str("True Values", 110)
+ dict_yaxis["text"] = "Predicted Values"
+
+ fig.update_traces(marker={"size": 10, "opacity": 0.8, "line": {"width": 0.8, "color": "white"}})
fig.update_layout(
- template='none',
+ template="none",
title=dict_t,
width=width,
height=height,
xaxis_title=dict_xaxis,
yaxis_title=dict_yaxis,
- hovermode='closest'
+ hovermode="closest",
)
fig.update_yaxes(automargin=True)
@@ -3046,27 +3181,26 @@ def scatter_plot_prediction(self,
else:
fig = go.Figure()
fig.update_layout(
- xaxis = { "visible": False },
- yaxis = { "visible": False },
- annotations = [
- {
- "text": "Provide the y_target argument in the compile() method to display this plot.",
- "xref": "paper",
- "yref": "paper",
- "showarrow": False,
- "font": {
- "size": 14
+ xaxis={"visible": False},
+ yaxis={"visible": False},
+ annotations=[
+ {
+ "text": "Provide the y_target argument in the compile() method to display this plot.",
+ "xref": "paper",
+ "yref": "paper",
+ "showarrow": False,
+ "font": {"size": 14},
}
- }
- ]
- )
+ ],
+ )
return fig
- def _prediction_classification_plot(self,
- list_ind,
- label=-1,
- ):
+ def _prediction_classification_plot(
+ self,
+ list_ind,
+ label=-1,
+ ):
"""
_prediction_classification_plot displays a Plotly violin plot of predictions in comparison to the target variable.
This plot represents Trues Values versus Predicted Values.
@@ -3094,82 +3228,107 @@ def _prediction_classification_plot(self,
self.explainer.predict()
# Assign proba values of the target
df_proba_target = self.explainer.proba_values.copy()
- df_proba_target['proba_target'] = df_proba_target.iloc[:, label_num]
- proba_values = df_proba_target[['proba_target']]
+ df_proba_target["proba_target"] = df_proba_target.iloc[:, label_num]
+ proba_values = df_proba_target[["proba_target"]]
# Proba subset:
proba_values = proba_values.loc[list_ind, :]
target = self.explainer.y_target.loc[list_ind, :]
y_pred = self.explainer.y_pred.loc[list_ind, :]
- df_pred = pd.concat([proba_values.reset_index(), y_pred.reset_index(drop=True),
- target.reset_index(drop=True)], axis=1)
+ df_pred = pd.concat(
+ [proba_values.reset_index(), y_pred.reset_index(drop=True), target.reset_index(drop=True)], axis=1
+ )
df_pred.set_index(df_pred.columns[0], inplace=True)
df_pred.columns = ["proba_values", "predict_class", "target"]
- df_pred['wrong_predict'] = 1
- df_pred.loc[(df_pred['predict_class'] == df_pred['target']), 'wrong_predict'] = 0
+ df_pred["wrong_predict"] = 1
+ df_pred.loc[(df_pred["predict_class"] == df_pred["target"]), "wrong_predict"] = 0
subtitle = f"Response: {label_value}"
# Plot distribution
- fig.add_trace(go.Violin(
- x=df_pred['target'].values.flatten(),
- y=df_pred['proba_values'].values.flatten(),
- points=False,
- legendgroup='M', scalegroup='M', name='Correct Prediction',
- line_color=self._style_dict["violin_area_classif"][1],
- pointpos=-0.1,
- showlegend=False,
- jitter=0.075,
- meanline_visible=True,
- spanmode="hard",
- customdata=df_pred['proba_values'].index.values,
- scalemode='count',
- ))
+ fig.add_trace(
+ go.Violin(
+ x=df_pred["target"].values.flatten(),
+ y=df_pred["proba_values"].values.flatten(),
+ points=False,
+ legendgroup="M",
+ scalegroup="M",
+ name="Correct Prediction",
+ line_color=self._style_dict["violin_area_classif"][1],
+ pointpos=-0.1,
+ showlegend=False,
+ jitter=0.075,
+ meanline_visible=True,
+ spanmode="hard",
+ customdata=df_pred["proba_values"].index.values,
+ scalemode="count",
+ )
+ )
# Plot points depending if wrong or correct prediction
- df_correct_predict = df_pred[(df_pred['wrong_predict'] == 0)]
- df_wrong_predict = df_pred[(df_pred['wrong_predict'] == 1)]
- hv_text_correct_predict = [f"Id: {x}
Predicted Values: {y:.3f}
Predicted class: {w}
True Values: {z}
" for
- x, y, w, z in zip(df_correct_predict.index, df_correct_predict.proba_values.values.round(3).flatten(),
- df_correct_predict.predict_class.values.flatten(), df_correct_predict.target.values.flatten())]
- hv_text_wrong_predict = [f"Id: {x}
Predicted Values: {y:.3f}
Predicted class: {w}
True Values: {z}
" for
- x, y, w, z in zip(df_wrong_predict.index, df_wrong_predict.proba_values.values.round(3).flatten(),
- df_wrong_predict.predict_class.values.flatten(), df_wrong_predict.target.values.flatten())]
-
- fig.add_trace(go.Scatter(
- x=df_correct_predict['target'].values.flatten() + np.random.normal(0, 0.02, len(df_correct_predict)),
- y=df_correct_predict['proba_values'].values.flatten(),
- mode='markers',
- marker_color=self._style_dict["prediction_plot"][0],
- showlegend=True,
- name="Correct Prediction",
- hovertext=hv_text_correct_predict,
- hovertemplate='%{hovertext}
',
- customdata=df_correct_predict['proba_values'].index.values,
- ))
-
- fig.add_trace(go.Scatter(
- x=df_wrong_predict['target'].values.flatten() + np.random.normal(0, 0.02, len(df_wrong_predict)),
- y=df_wrong_predict['proba_values'].values.flatten(),
- mode='markers',
- marker_color=self._style_dict["prediction_plot"][1],
- showlegend=True,
- name="Wrong Prediction",
- hovertext=hv_text_wrong_predict,
- hovertemplate='%{hovertext}
',
- customdata=df_wrong_predict['proba_values'].index.values,
- ))
+ df_correct_predict = df_pred[(df_pred["wrong_predict"] == 0)]
+ df_wrong_predict = df_pred[(df_pred["wrong_predict"] == 1)]
+ hv_text_correct_predict = [
+ f"Id: {x}
Predicted Values: {y:.3f}
Predicted class: {w}
True Values: {z}
"
+ for x, y, w, z in zip(
+ df_correct_predict.index,
+ df_correct_predict.proba_values.values.round(3).flatten(),
+ df_correct_predict.predict_class.values.flatten(),
+ df_correct_predict.target.values.flatten(),
+ )
+ ]
+ hv_text_wrong_predict = [
+ f"Id: {x}
Predicted Values: {y:.3f}
Predicted class: {w}
True Values: {z}
"
+ for x, y, w, z in zip(
+ df_wrong_predict.index,
+ df_wrong_predict.proba_values.values.round(3).flatten(),
+ df_wrong_predict.predict_class.values.flatten(),
+ df_wrong_predict.target.values.flatten(),
+ )
+ ]
+
+ fig.add_trace(
+ go.Scatter(
+ x=df_correct_predict["target"].values.flatten() + np.random.normal(0, 0.02, len(df_correct_predict)),
+ y=df_correct_predict["proba_values"].values.flatten(),
+ mode="markers",
+ marker_color=self._style_dict["prediction_plot"][0],
+ showlegend=True,
+ name="Correct Prediction",
+ hovertext=hv_text_correct_predict,
+ hovertemplate="%{hovertext}
",
+ customdata=df_correct_predict["proba_values"].index.values,
+ )
+ )
- fig.update_layout(violingap=0, violinmode='overlay')
+ fig.add_trace(
+ go.Scatter(
+ x=df_wrong_predict["target"].values.flatten() + np.random.normal(0, 0.02, len(df_wrong_predict)),
+ y=df_wrong_predict["proba_values"].values.flatten(),
+ mode="markers",
+ marker_color=self._style_dict["prediction_plot"][1],
+ showlegend=True,
+ name="Wrong Prediction",
+ hovertext=hv_text_wrong_predict,
+ hovertemplate="%{hovertext}
",
+ customdata=df_wrong_predict["proba_values"].index.values,
+ )
+ )
+
+ fig.update_layout(violingap=0, violinmode="overlay")
if self.explainer.label_dict is not None:
- fig.update_xaxes(tickmode='array', tickvals=list(df_pred['target'].unique()),
- ticktext=list(df_pred['target'].apply(lambda x: self.explainer.label_dict[x]).unique()))
+ fig.update_xaxes(
+ tickmode="array",
+ tickvals=list(df_pred["target"].unique()),
+ ticktext=list(df_pred["target"].apply(lambda x: self.explainer.label_dict[x]).unique()),
+ )
if self.explainer.label_dict is None:
- fig.update_xaxes(tickvals=sorted(list(df_pred['target'].unique())))
+ fig.update_xaxes(tickvals=sorted(list(df_pred["target"].unique())))
return fig, subtitle
- def _prediction_regression_plot(self,
- list_ind,
- ):
+ def _prediction_regression_plot(
+ self,
+ list_ind,
+ ):
"""
_prediction_regression_plot displays a Plotly scatter plot of predictions in comparison to the target variable.
This plot represents Trues Values versus Predicted Values.
@@ -3195,10 +3354,13 @@ def _prediction_regression_plot(self,
else:
subtitle = "Prediction Error = abs(True Values - Predicted Values) / True Values"
df_equal_bins = prediction_error.describe(percentiles=np.arange(0.1, 1, 0.1).tolist())
- equal_bins = df_equal_bins.loc[~df_equal_bins.index.isin(['count', 'mean', 'std'])].values
+ equal_bins = df_equal_bins.loc[~df_equal_bins.index.isin(["count", "mean", "std"])].values
equal_bins = np.unique(equal_bins)
- self.pred_colorscale = self.tuning_colorscale(pd.DataFrame(pd.cut([val[0] for val in prediction_error.values],
- bins=[i for i in equal_bins], labels=False)))
+ self.pred_colorscale = self.tuning_colorscale(
+ pd.DataFrame(
+ pd.cut([val[0] for val in prediction_error.values], bins=[i for i in equal_bins], labels=False)
+ )
+ )
col_scale = self.pred_colorscale
y_pred = self.explainer.y_pred.loc[list_ind]
@@ -3209,36 +3371,59 @@ def _prediction_regression_plot(self,
self.tuning_round_digit()
y_pred = y_pred.applymap(lambda x: round(x, self.round_digit))
- hv_text = [f"Id: {x}
True Values: {y:,.2f}
Predicted Values: {z:,.2f}
Prediction Error: {w:,.2f}" for x, y, z, w in
- zip(y_target.index, y_target.values.flatten(), y_pred.values.flatten(), prediction_error.flatten())]
+ hv_text = [
+ f"Id: {x}
True Values: {y:,.2f}
Predicted Values: {z:,.2f}
Prediction Error: {w:,.2f}"
+ for x, y, z, w in zip(
+ y_target.index, y_target.values.flatten(), y_pred.values.flatten(), prediction_error.flatten()
+ )
+ ]
fig.add_scatter(
x=y_target.values.flatten(),
y=y_pred.values.flatten(),
- mode='markers',
+ mode="markers",
hovertext=hv_text,
- hovertemplate='%{hovertext}
',
- customdata=y_pred.index.values
+ hovertemplate="%{hovertext}
",
+ customdata=y_pred.index.values,
)
- colorpoints = pd.cut([val[0] for val in prediction_error], bins=[i for i in equal_bins], labels=False)/10
- colorbar_title = 'Prediction Error'
+ colorpoints = pd.cut([val[0] for val in prediction_error], bins=[i for i in equal_bins], labels=False) / 10
+ colorbar_title = "Prediction Error"
fig.data[-1].marker.color = colorpoints.flatten()
- fig.data[-1].marker.coloraxis = 'coloraxis'
+ fig.data[-1].marker.coloraxis = "coloraxis"
fig.layout.coloraxis.colorscale = col_scale
- fig.layout.coloraxis.colorbar = {'title': {'text': colorbar_title}, "tickvals": [col_scale[0][0], col_scale[-1][0]-0.15],
- "ticktext":[float('{:0.3f}'.format(equal_bins[0])), float('{:0.3f}'.format(equal_bins[-2]))], "tickformat": ".2s",
- "yanchor": "top", "y": 1.1}
- range_axis = [min(min(y_target.values.flatten()), min(y_pred.values.flatten())), max(max(y_target.values.flatten()),
- max(y_pred.values.flatten()))]
+ fig.layout.coloraxis.colorbar = {
+ "title": {"text": colorbar_title},
+ "tickvals": [col_scale[0][0], col_scale[-1][0] - 0.15],
+ "ticktext": [float("{:0.3f}".format(equal_bins[0])), float("{:0.3f}".format(equal_bins[-2]))],
+ "tickformat": ".2s",
+ "yanchor": "top",
+ "y": 1.1,
+ }
+ range_axis = [
+ min(min(y_target.values.flatten()), min(y_pred.values.flatten())),
+ max(max(y_target.values.flatten()), max(y_pred.values.flatten())),
+ ]
fig.update_xaxes(range=range_axis)
fig.update_yaxes(range=range_axis)
- fig.update_layout(shapes=[{'type': 'line', 'yref': 'y domain', 'xref': 'x domain', 'y0': 0, 'y1': 1,
- 'x0': 0, 'x1': 1, 'line': dict(color="grey", width=1, dash="dot")}])
+ fig.update_layout(
+ shapes=[
+ {
+ "type": "line",
+ "yref": "y domain",
+ "xref": "x domain",
+ "y0": 0,
+ "y1": 1,
+ "x0": 0,
+ "x1": 1,
+ "line": dict(color="grey", width=1, dash="dot"),
+ }
+ ]
+ )
return fig, subtitle
- def _subset_sampling(self, selection = None, max_points = 2000):
+ def _subset_sampling(self, selection=None, max_points=2000):
"""
Subset sampling for plots and create addnote for subtitle
@@ -3263,8 +3448,7 @@ def _subset_sampling(self, selection = None, max_points = 2000):
addnote = None
else:
random.seed(79)
- list_ind = random.sample(
- self.explainer.x_init.index.tolist(), max_points)
+ list_ind = random.sample(self.explainer.x_init.index.tolist(), max_points)
addnote = "Length of random Subset: "
elif isinstance(selection, list):
if len(selection) <= max_points:
@@ -3275,10 +3459,11 @@ def _subset_sampling(self, selection = None, max_points = 2000):
list_ind = random.sample(selection, max_points)
addnote = "Length of random Subset: "
else:
- raise ValueError('parameter selection must be a list')
+ raise ValueError("parameter selection must be a list")
if addnote is not None:
- addnote = add_text([addnote,
- f"{len(list_ind)} ({int(np.round(100 * len(list_ind) / self.explainer.x_init.shape[0]))}%)"],
- sep='')
+ addnote = add_text(
+ [addnote, f"{len(list_ind)} ({int(np.round(100 * len(list_ind) / self.explainer.x_init.shape[0]))}%)"],
+ sep="",
+ )
return list_ind, addnote
diff --git a/shapash/explainer/smart_predictor.py b/shapash/explainer/smart_predictor.py
index b655ec77..053f39c6 100644
--- a/shapash/explainer/smart_predictor.py
+++ b/shapash/explainer/smart_predictor.py
@@ -1,30 +1,41 @@
"""
Smart predictor module
"""
-from shapash.utils.check import check_consistency_model_features, check_consistency_model_label
-from shapash.utils.check import check_model, check_preprocessing, check_preprocessing_options
-from shapash.utils.check import check_label_dict, check_mask_params, check_y, check_contribution_object,\
- check_features_name
+import copy
+
import pandas as pd
-from shapash.utils.transform import adapt_contributions
+
+import shapash.explainer.smart_explainer
+from shapash.decomposition.contributions import assign_contributions, rank_contributions
+from shapash.manipulation.filters import (
+ cap_contributions,
+ combine_masks,
+ cutoff_contributions,
+ hide_contributions,
+ sign_contributions,
+)
+from shapash.manipulation.mask import compute_masked_contributions, init_mask
from shapash.manipulation.select_lines import keep_right_contributions
-from shapash.utils.model import predict_proba
-from shapash.utils.io import save_pickle
-from shapash.utils.transform import apply_preprocessing, apply_postprocessing, preprocessing_tolist
-from shapash.manipulation.filters import hide_contributions
-from shapash.manipulation.filters import cap_contributions
-from shapash.manipulation.filters import sign_contributions
-from shapash.manipulation.filters import cutoff_contributions
-from shapash.manipulation.filters import combine_masks
-from shapash.manipulation.mask import init_mask
-from shapash.manipulation.mask import compute_masked_contributions
-from shapash.manipulation.summarize import summarize, create_grouped_features_values, group_contributions
-from shapash.decomposition.contributions import rank_contributions, assign_contributions
+from shapash.manipulation.summarize import create_grouped_features_values, group_contributions, summarize
+from shapash.utils.check import (
+ check_consistency_model_features,
+ check_consistency_model_label,
+ check_contribution_object,
+ check_features_name,
+ check_label_dict,
+ check_mask_params,
+ check_model,
+ check_preprocessing,
+ check_preprocessing_options,
+ check_y,
+)
from shapash.utils.columntransformer_backend import columntransformer
-import copy
-import shapash.explainer.smart_explainer
+from shapash.utils.io import save_pickle
+from shapash.utils.model import predict_proba
+from shapash.utils.transform import adapt_contributions, apply_postprocessing, apply_preprocessing, preprocessing_tolist
+
-class SmartPredictor :
+class SmartPredictor:
"""
The SmartPredictor class is an object lighter than SmartExplainer Object with
additionnal consistency checks.
@@ -93,17 +104,19 @@ class SmartPredictor :
SmartExplainer instance to point to.
"""
- def __init__(self, features_dict, model,
- columns_dict, backend, features_types,
- label_dict=None, preprocessing=None,
- postprocessing=None,
- features_groups=None,
- mask_params = {"features_to_hide": None,
- "threshold": None,
- "positive": None,
- "max_contrib": None
- }
- ):
+ def __init__(
+ self,
+ features_dict,
+ model,
+ columns_dict,
+ backend,
+ features_types,
+ label_dict=None,
+ preprocessing=None,
+ postprocessing=None,
+ features_groups=None,
+ mask_params={"features_to_hide": None, "threshold": None, "positive": None, "max_contrib": None},
+ ):
params_dict = [features_dict, features_types, label_dict, columns_dict, postprocessing]
@@ -111,8 +124,10 @@ def __init__(self, features_dict, model,
if params is not None and isinstance(params, dict) == False:
raise ValueError(
"""
- {0} must be a dict.
- """.format(str(params))
+ {} must be a dict.
+ """.format(
+ str(params)
+ )
)
self.model = model
@@ -130,9 +145,17 @@ def __init__(self, features_dict, model,
self.postprocessing = postprocessing
self.features_groups = features_groups
list_preprocessing = preprocessing_tolist(self.preprocessing)
- check_consistency_model_features(self.features_dict, self.model, self.columns_dict,
- self.features_types, self.mask_params, self.preprocessing,
- self.postprocessing, list_preprocessing, self.features_groups)
+ check_consistency_model_features(
+ self.features_dict,
+ self.model,
+ self.columns_dict,
+ self.features_types,
+ self.mask_params,
+ self.preprocessing,
+ self.postprocessing,
+ list_preprocessing,
+ self.features_groups,
+ )
check_consistency_model_label(self.columns_dict, self.label_dict)
self._drop_option = check_preprocessing_options(columns_dict, features_dict, preprocessing, list_preprocessing)
@@ -200,25 +223,24 @@ def add_input(self, x=None, ypred=None, contributions=None):
x = self.check_dataset_features(self.check_dataset_type(x))
self.data = self.clean_data(x)
self.data["x_postprocessed"] = self.apply_postprocessing()
- try :
+ try:
self.data["x_preprocessed"] = self.apply_preprocessing()
- except BaseException :
+ except BaseException:
raise ValueError(
"""
Preprocessing has failed. The preprocessing specified or the dataset doesn't match.
"""
)
else:
- if not hasattr(self,"data"):
- raise ValueError ("No dataset x specified.")
+ if not hasattr(self, "data"):
+ raise ValueError("No dataset x specified.")
if ypred is not None:
self.data["ypred_init"] = self.check_ypred(ypred)
if contributions is not None:
self.data["ypred"], self.data["contributions"] = self.compute_contributions(
- contributions=contributions,
- use_groups=False
+ contributions=contributions, use_groups=False
)
else:
self.data["ypred"], self.data["contributions"] = self.compute_contributions(use_groups=False)
@@ -232,18 +254,18 @@ def _add_groups_input(self):
and stores it in data_groups attribute
"""
self.data_groups = dict()
- self.data_groups['x_postprocessed'] = create_grouped_features_values(x_init=self.data["x_postprocessed"],
- x_encoded=self.data["x_preprocessed"],
- preprocessing=self.preprocessing,
- features_groups=self.features_groups,
- features_dict=self.features_dict,
- how='dict_of_values')
- self.data_groups['ypred'] = self.data["ypred"]
- self.data_groups['contributions'] = group_contributions(
- contributions=self.data['contributions'],
- features_groups=self.features_groups
+ self.data_groups["x_postprocessed"] = create_grouped_features_values(
+ x_init=self.data["x_postprocessed"],
+ x_encoded=self.data["x_preprocessed"],
+ preprocessing=self.preprocessing,
+ features_groups=self.features_groups,
+ features_dict=self.features_dict,
+ how="dict_of_values",
+ )
+ self.data_groups["ypred"] = self.data["ypred"]
+ self.data_groups["contributions"] = group_contributions(
+ contributions=self.data["contributions"], features_groups=self.features_groups
)
-
def check_dataset_type(self, x=None):
"""
@@ -266,7 +288,7 @@ def check_dataset_type(self, x=None):
x must be a dict or a pandas.DataFrame.
"""
)
- else :
+ else:
x = self.convert_dict_dataset(x)
return x
@@ -286,9 +308,11 @@ def convert_dict_dataset(self, x):
"""
if type(x) == dict:
if not all([column in self.features_types.keys() for column in x.keys()]):
- raise ValueError("""
+ raise ValueError(
+ """
All features from dataset x must be in the features_types dict initialized.
- """)
+ """
+ )
try:
x = pd.DataFrame.from_dict(x, orient="index").T
for feature, type_feature in self.features_types.items():
@@ -320,10 +344,12 @@ def check_dataset_features(self, x):
assert all(column in self.features_types.keys() for column in x.columns)
if not all([str(x[feature].dtypes) == self.features_types[feature] for feature in x.columns]):
- raise ValueError("""
+ raise ValueError(
+ """
Types of features in x doesn't match with the expected one in features_types.
x input must be initial dataset without preprocessing applied.
- """)
+ """
+ )
return x
def check_ypred(self, ypred=None):
@@ -385,13 +411,14 @@ def clean_data(self, x):
-------
dict of data stored
"""
- return {"x" : x,
- "ypred_init": None,
- "ypred" : None,
- "contributions" : None,
- "x_preprocessed": None,
- "x_postprocessed": None
- }
+ return {
+ "x": x,
+ "ypred_init": None,
+ "ypred": None,
+ "contributions": None,
+ "x_preprocessed": None,
+ "x_postprocessed": None,
+ }
def predict_proba(self):
"""
@@ -446,19 +473,17 @@ def compute_contributions(self, contributions=None, use_groups=None):
if contributions is None:
explain_data = self.backend.run_explainer(x=self.data["x_preprocessed"])
contributions = self.backend.get_local_contributions(
- explain_data=explain_data,
- x=self.data["x_preprocessed"]
+ explain_data=explain_data, x=self.data["x_preprocessed"]
)
else:
contributions = self.backend.format_and_aggregate_local_contributions(
- x=self.data["x_preprocessed"],
- contributions=contributions
+ x=self.data["x_preprocessed"], contributions=contributions
)
self.check_contributions(contributions)
proba_values = self.predict_proba() if self._case == "classification" else None
- y_pred, match_contrib = keep_right_contributions(self.data["ypred_init"], contributions,
- self._case, self._classes,
- self.label_dict, proba_values)
+ y_pred, match_contrib = keep_right_contributions(
+ self.data["ypred_init"], contributions, self._case, self._classes, self.label_dict, proba_values
+ )
if use_groups:
match_contrib = group_contributions(match_contrib, features_groups=self.features_groups)
@@ -523,35 +548,22 @@ def filter(self):
The filter method is an important method which allows to summarize the local explainability
by using the user defined mask_params parameters which correspond to its use case.
"""
- mask = [init_mask(self.summary['contrib_sorted'], True)]
+ mask = [init_mask(self.summary["contrib_sorted"], True)]
if self.mask_params["features_to_hide"] is not None:
mask.append(
hide_contributions(
- self.summary['var_dict'],
- features_list=self.check_features_name(self.mask_params["features_to_hide"])
+ self.summary["var_dict"],
+ features_list=self.check_features_name(self.mask_params["features_to_hide"]),
)
)
if self.mask_params["threshold"] is not None:
- mask.append(
- cap_contributions(
- self.summary['contrib_sorted'],
- threshold=self.mask_params["threshold"]
- )
- )
+ mask.append(cap_contributions(self.summary["contrib_sorted"], threshold=self.mask_params["threshold"]))
if self.mask_params["positive"] is not None:
- mask.append(
- sign_contributions(
- self.summary['contrib_sorted'],
- positive=self.mask_params["positive"]
- )
- )
+ mask.append(sign_contributions(self.summary["contrib_sorted"], positive=self.mask_params["positive"]))
self.mask = combine_masks(mask)
if self.mask_params["max_contrib"] is not None:
self.mask = cutoff_contributions(mask=self.mask, k=self.mask_params["max_contrib"])
- self.masked_contributions = compute_masked_contributions(
- self.summary['contrib_sorted'],
- self.mask
- )
+ self.masked_contributions = compute_masked_contributions(self.summary["contrib_sorted"], self.mask)
def summarize(self, use_groups=None):
"""
@@ -580,7 +592,7 @@ def summarize(self, use_groups=None):
--------
>>> summary_df = predictor.summarize()
>>> summary_df
- pred proba feature_1 value_1 contribution_1 feature_2 value_2 contribution_2
+ pred proba feature_1 value_1 contribution_1 feature_2 value_2 contribution_2
0 0 0.756416 Sex 1.0 0.322308 Pclass 3.0 0.155069
1 3 0.628911 Sex 2.0 0.585475 Pclass 1.0 0.370504
2 0 0.543308 Sex 2.0 -0.486667 Pclass 3.0 0.255072
@@ -588,7 +600,7 @@ def summarize(self, use_groups=None):
>>> predictor.modify_mask(max_contrib=1)
>>> summary_df = predictor.summarize()
>>> summary_df
- pred proba feature_1 value_1 contribution_1
+ pred proba feature_1 value_1 contribution_1
0 0 0.756416 Sex 1.0 0.322308
1 3 0.628911 Sex 2.0 0.585475
2 0 0.543308 Sex 2.0 -0.486667
@@ -605,8 +617,9 @@ def summarize(self, use_groups=None):
data = self.data
if self._drop_option is not None:
- columns_to_keep = [x for x in self._drop_option["columns_dict_op"].values()
- if x in data["x_postprocessed"].columns]
+ columns_to_keep = [
+ x for x in self._drop_option["columns_dict_op"].values() if x in data["x_postprocessed"].columns
+ ]
if use_groups:
columns_to_keep += list(self.features_groups.keys())
x_preprocessed = data["x_postprocessed"][columns_to_keep]
@@ -616,33 +629,24 @@ def summarize(self, use_groups=None):
columns_dict = {i: col for i, col in enumerate(x_preprocessed.columns)}
features_dict = {k: v for k, v in self.features_dict.items() if k in x_preprocessed.columns}
- self.summary = assign_contributions(
- rank_contributions(
- data["contributions"],
- x_preprocessed
- )
- )
+ self.summary = assign_contributions(rank_contributions(data["contributions"], x_preprocessed))
# Apply filter method with mask_params attributes parameters
self.filter()
# Summarize information
- data['summary'] = summarize(self.summary['contrib_sorted'],
- self.summary['var_dict'],
- self.summary['x_sorted'],
- self.mask,
- columns_dict,
- features_dict)
+ data["summary"] = summarize(
+ self.summary["contrib_sorted"],
+ self.summary["var_dict"],
+ self.summary["x_sorted"],
+ self.mask,
+ columns_dict,
+ features_dict,
+ )
# Matching with y_pred
- return pd.concat([data["ypred"], data['summary']], axis=1)
-
- def modify_mask(
- self,
- features_to_hide=None,
- threshold=None,
- positive=None,
- max_contrib=None
- ):
+ return pd.concat([data["ypred"], data["summary"]], axis=1)
+
+ def modify_mask(self, features_to_hide=None, threshold=None, positive=None, max_contrib=None):
"""
This method allows the users to modify the mask_params values.
Each parameter is optional, modify_mask method modifies only the values specified in parameters.
@@ -666,17 +670,19 @@ def modify_mask(
>>> predictor.modify_mask(max_contrib=1)
>>> summary_df = predictor.summarize()
>>> summary_df
- pred proba feature_1 value_1 contribution_1
+ pred proba feature_1 value_1 contribution_1
0 0 0.756416 Sex 1.0 0.322308
1 3 0.628911 Sex 2.0 0.585475
2 0 0.543308 Sex 2.0 -0.486667
"""
- Attributes = {"features_to_hide": features_to_hide,
- "threshold": threshold,
- "positive": positive,
- "max_contrib": max_contrib}
- for label, attribute in Attributes.items() :
+ Attributes = {
+ "features_to_hide": features_to_hide,
+ "threshold": threshold,
+ "positive": positive,
+ "max_contrib": max_contrib,
+ }
+ for label, attribute in Attributes.items():
if attribute is not None:
self.mask_params[label] = attribute
@@ -704,11 +710,12 @@ def predict(self):
x must be specified in an add_input method to apply predict.
"""
)
- if hasattr(self.model, 'predict'):
+ if hasattr(self.model, "predict"):
self.data["ypred_init"] = pd.DataFrame(
self.model.predict(self.data["x_preprocessed"]),
- columns=['ypred'],
- index=self.data["x_preprocessed"].index)
+ columns=["ypred"],
+ index=self.data["x_preprocessed"].index,
+ )
else:
raise ValueError("model has no predict method")
@@ -777,7 +784,7 @@ def to_smartexplainer(self):
postprocessing=self.postprocessing,
features_groups=self.features_groups,
features_dict=copy.deepcopy(self.features_dict),
- label_dict=copy.deepcopy(self.label_dict)
+ label_dict=copy.deepcopy(self.label_dict),
)
xpl.compile(x=copy.deepcopy(self.data["x_preprocessed"]), y_pred=copy.deepcopy(self.data["ypred_init"]))
return xpl
diff --git a/shapash/explainer/smart_state.py b/shapash/explainer/smart_state.py
index 910461fa..b9e78364 100644
--- a/shapash/explainer/smart_state.py
+++ b/shapash/explainer/smart_state.py
@@ -3,16 +3,21 @@
"""
import numpy as np
import pandas as pd
-from shapash.decomposition.contributions import inverse_transform_contributions
-from shapash.decomposition.contributions import rank_contributions, assign_contributions
-from shapash.manipulation.filters import hide_contributions
-from shapash.manipulation.filters import cap_contributions
-from shapash.manipulation.filters import sign_contributions
-from shapash.manipulation.filters import cutoff_contributions
-from shapash.manipulation.filters import combine_masks
-from shapash.manipulation.mask import compute_masked_contributions
-from shapash.manipulation.mask import init_mask
-from shapash.manipulation.summarize import summarize, compute_features_import, group_contributions
+
+from shapash.decomposition.contributions import (
+ assign_contributions,
+ inverse_transform_contributions,
+ rank_contributions,
+)
+from shapash.manipulation.filters import (
+ cap_contributions,
+ combine_masks,
+ cutoff_contributions,
+ hide_contributions,
+ sign_contributions,
+)
+from shapash.manipulation.mask import compute_masked_contributions, init_mask
+from shapash.manipulation.summarize import compute_features_import, group_contributions, summarize
class SmartState:
@@ -38,19 +43,13 @@ def validate_contributions(self, contributions, x_init):
Local contributions on the original feature space (no encoding).
"""
if not isinstance(contributions, (np.ndarray, pd.DataFrame)):
- raise ValueError(
- 'Type of contributions must be pd.DataFrame or np.ndarray'
- )
+ raise ValueError("Type of contributions must be pd.DataFrame or np.ndarray")
if isinstance(contributions, np.ndarray):
- return pd.DataFrame(
- contributions,
- columns=x_init.columns,
- index=x_init.index
- )
+ return pd.DataFrame(contributions, columns=x_init.columns, index=x_init.index)
else:
return contributions
- def inverse_transform_contributions(self, contributions, preprocessing, agg_columns='sum'):
+ def inverse_transform_contributions(self, contributions, preprocessing, agg_columns="sum"):
"""
Compute local contributions in the original feature space, despite category encoding.
diff --git a/shapash/manipulation/filters.py b/shapash/manipulation/filters.py
index 965007a3..6d959558 100644
--- a/shapash/manipulation/filters.py
+++ b/shapash/manipulation/filters.py
@@ -93,11 +93,7 @@ def cutoff_contributions_old(dataframe, max_contrib):
"""
mask = np.full_like(dataframe, False).astype(bool)
mask[:, :max_contrib] = True
- return pd.DataFrame(
- mask,
- columns=dataframe.columns,
- index=dataframe.index
- )
+ return pd.DataFrame(mask, columns=dataframe.columns, index=dataframe.index)
def cutoff_contributions(mask, k=10):
@@ -117,7 +113,7 @@ def cutoff_contributions(mask, k=10):
pd.Dataframe
Mask where only the k-top contributions are considered.
"""
- return mask.replace(False, np.nan).cumsum(axis=1).isin(range(1, k+1))
+ return mask.replace(False, np.nan).cumsum(axis=1).isin(range(1, k + 1))
def combine_masks(masks_list):
@@ -138,13 +134,11 @@ def combine_masks(masks_list):
"""
if len(set(map(lambda x: x.shape, masks_list))) != 1:
- raise ValueError('Masks must have same dimensions.')
+ raise ValueError("Masks must have same dimensions.")
masks_cube = np.dstack(masks_list)
mask_final = np.min(masks_cube, axis=2)
return pd.DataFrame(
- mask_final,
- columns=['contrib_{}'.format(i+1) for i in range(mask_final.shape[1])],
- index=masks_list[0].index
+ mask_final, columns=["contrib_{}".format(i + 1) for i in range(mask_final.shape[1])], index=masks_list[0].index
)
diff --git a/shapash/manipulation/mask.py b/shapash/manipulation/mask.py
index ebd38457..6dd65062 100644
--- a/shapash/manipulation/mask.py
+++ b/shapash/manipulation/mask.py
@@ -22,15 +22,9 @@ def compute_masked_contributions(s_contrib, mask):
pd.DataFrame
Sum of contributions of hidden features.
"""
- colname = ['masked_neg', 'masked_pos']
- hidden_neg = np.sum(
- ma.array(s_contrib, mask=np.max(np.dstack([mask, (s_contrib > 0)]), axis=2)),
- axis=1
- )
- hidden_pos = np.sum(
- ma.array(s_contrib, mask=np.max(np.dstack([mask, (s_contrib < 0)]), axis=2)),
- axis=1
- )
+ colname = ["masked_neg", "masked_pos"]
+ hidden_neg = np.sum(ma.array(s_contrib, mask=np.max(np.dstack([mask, (s_contrib > 0)]), axis=2)), axis=1)
+ hidden_pos = np.sum(ma.array(s_contrib, mask=np.max(np.dstack([mask, (s_contrib < 0)]), axis=2)), axis=1)
hidden_contrib = np.array([hidden_neg, hidden_pos])
return pd.DataFrame(hidden_contrib.T, columns=colname, index=s_contrib.index)
@@ -56,8 +50,4 @@ def init_mask(s_contrib, value=True):
else:
mask = np.zeros(s_contrib.shape, dtype=bool)
- return pd.DataFrame(
- mask,
- columns=s_contrib.columns,
- index=s_contrib.index
- )
+ return pd.DataFrame(mask, columns=s_contrib.columns, index=s_contrib.index)
diff --git a/shapash/manipulation/select_lines.py b/shapash/manipulation/select_lines.py
index 47ede894..9f2e00c2 100644
--- a/shapash/manipulation/select_lines.py
+++ b/shapash/manipulation/select_lines.py
@@ -4,6 +4,7 @@
import pandas as pd
from pandas.core.common import flatten
+
def select_lines(dataframe, condition=None):
"""
Select lines of a pandas.DataFrame based
@@ -24,6 +25,7 @@ def select_lines(dataframe, condition=None):
else:
return []
+
def keep_right_contributions(y_pred, contributions, _case, _classes, label_dict, proba_values=None):
"""
Keep the right contributions/summary for the right ypred.
@@ -48,18 +50,20 @@ def keep_right_contributions(y_pred, contributions, _case, _classes, label_dict,
if _case == "classification":
complete_sum = [list(x) for x in list(zip(*[df.values.tolist() for df in contributions]))]
indexclas = [_classes.index(x) for x in list(flatten(y_pred.values))]
- summary = pd.DataFrame([summar[ind]
- for ind, summar in zip(indexclas, complete_sum)],
- columns=contributions[0].columns,
- index=contributions[0].index,
- dtype=object)
+ summary = pd.DataFrame(
+ [summar[ind] for ind, summar in zip(indexclas, complete_sum)],
+ columns=contributions[0].columns,
+ index=contributions[0].index,
+ dtype=object,
+ )
if label_dict is not None:
y_pred = y_pred.applymap(lambda x: label_dict[x])
if proba_values is not None:
- y_proba = pd.DataFrame([proba[ind]
- for ind, proba in zip(indexclas, proba_values.values)],
- columns=['proba'],
- index=y_pred.index)
+ y_proba = pd.DataFrame(
+ [proba[ind] for ind, proba in zip(indexclas, proba_values.values)],
+ columns=["proba"],
+ index=y_pred.index,
+ )
y_pred = pd.concat([y_pred, y_proba], axis=1)
else:
diff --git a/shapash/manipulation/summarize.py b/shapash/manipulation/summarize.py
index db1fdc5e..835b2a1b 100644
--- a/shapash/manipulation/summarize.py
+++ b/shapash/manipulation/summarize.py
@@ -2,10 +2,12 @@
Summarize Module
"""
import warnings
+
import numpy as np
import pandas as pd
from pandas.core.common import flatten
from sklearn.manifold import TSNE
+
from shapash.utils.transform import get_features_transform_mapping
@@ -29,17 +31,14 @@ def summarize_el(dataframe, mask, prefix):
Result of the summarize step
"""
matrix = dataframe.where(mask.to_numpy()).values.tolist()
- summarized_matrix = [[x for x in l if str(x) != 'nan'] for l in matrix]
+ summarized_matrix = [[x for x in l if str(x) != "nan"] for l in matrix]
# Padding to create pd.DataFrame
max_length = max(len(l) for l in summarized_matrix)
for elem in summarized_matrix:
elem.extend([np.nan] * (max_length - len(elem)))
# Create DataFrame
col_list = [prefix + str(x + 1) for x in list(range(max_length))]
- df_summarized_matrix = pd.DataFrame(summarized_matrix,
- index=list(dataframe.index),
- columns=col_list,
- dtype=object)
+ df_summarized_matrix = pd.DataFrame(summarized_matrix, index=list(dataframe.index), columns=col_list, dtype=object)
return df_summarized_matrix
@@ -64,6 +63,7 @@ def compute_features_import(dataframe):
tot = feat_imp.sum()
return feat_imp / tot
+
def summarize(s_contrib, var_dict, x_sorted, mask, columns_dict, features_dict):
"""
Compute the summarized contributions of features.
@@ -88,10 +88,11 @@ def summarize(s_contrib, var_dict, x_sorted, mask, columns_dict, features_dict):
pd.DataFrame
Result of the summarize step
"""
- contrib_sum = summarize_el(s_contrib, mask, 'contribution_')
- var_dict_sum = summarize_el(var_dict, mask, 'feature_').applymap(
- lambda x: features_dict[columns_dict[x]] if not np.isnan(x) else x)
- x_sorted_sum = summarize_el(x_sorted, mask, 'value_')
+ contrib_sum = summarize_el(s_contrib, mask, "contribution_")
+ var_dict_sum = summarize_el(var_dict, mask, "feature_").applymap(
+ lambda x: features_dict[columns_dict[x]] if not np.isnan(x) else x
+ )
+ x_sorted_sum = summarize_el(x_sorted, mask, "value_")
# Concatenate pd.DataFrame
summary = pd.concat([contrib_sum, var_dict_sum, x_sorted_sum], axis=1)
@@ -130,7 +131,7 @@ def group_contributions(contributions, features_groups):
return new_contributions
-def project_feature_values_1d(feature_values, col, x_init, x_encoded, preprocessing, features_dict, how='tsne'):
+def project_feature_values_1d(feature_values, col, x_init, x_encoded, preprocessing, features_dict, how="tsne"):
"""
Project feature values of a group of features in 1 dimension.
If feature_values contains categorical features, use preprocessing to get
@@ -167,19 +168,20 @@ def project_feature_values_1d(feature_values, col, x_init, x_encoded, preprocess
col_names_in_xinit.extend(encoding_mapping.get(c, [c]))
feature_values = x_encoded.loc[feature_values.index, col_names_in_xinit]
# Project in 1D the feature values
- if how == 'tsne':
+ if how == "tsne":
try:
feature_values_proj_1d = TSNE(n_components=1, random_state=1).fit_transform(feature_values)
feature_values = pd.Series(feature_values_proj_1d[:, 0], name=col, index=feature_values.index)
except Exception as e:
- warnings.warn(f'Could not project group features values : {e}', UserWarning)
+ warnings.warn(f"Could not project group features values : {e}", UserWarning)
feature_values = pd.Series(feature_values.iloc[:, 0], name=col, index=feature_values.index)
- elif how == 'dict_of_values':
+ elif how == "dict_of_values":
feature_values.columns = [features_dict.get(x, x) for x in feature_values.columns]
- feature_values = pd.Series(feature_values.apply(lambda x: x.to_dict(), axis=1), name=col,
- index=feature_values.index)
+ feature_values = pd.Series(
+ feature_values.apply(lambda x: x.to_dict(), axis=1), name=col, index=feature_values.index
+ )
else:
- raise NotImplementedError(f'Unknown method : {how}')
+ raise NotImplementedError(f"Unknown method : {how}")
return feature_values
@@ -200,22 +202,18 @@ def compute_corr(df, compute_method):
"""
# Remove user warnings (when not enough values to compute correlation).
warnings.filterwarnings("ignore")
- if compute_method == 'phik':
+ if compute_method == "phik":
from phik import phik_matrix
+
return phik_matrix(df, verbose=False)
- elif compute_method == 'pearson':
+ elif compute_method == "pearson":
return df.corr()
else:
- raise NotImplementedError(f'Not implemented correlation method : {compute_method}')
+ raise NotImplementedError(f"Not implemented correlation method : {compute_method}")
def create_grouped_features_values(
- x_init,
- x_encoded,
- preprocessing,
- features_groups,
- features_dict,
- how='tsne'
+ x_init, x_encoded, preprocessing, features_groups, features_dict, how="tsne"
) -> pd.DataFrame:
"""
Compute projections of groups of features using t-sne.
@@ -243,7 +241,7 @@ def create_grouped_features_values(
df = x_init.copy()
for group in features_groups.keys():
if not isinstance(features_groups[group], list):
- raise ValueError(f'features_groups[{group}] should be a list of features')
+ raise ValueError(f"features_groups[{group}] should be a list of features")
features_values = x_init[features_groups[group]]
df[group] = project_feature_values_1d(
features_values,
@@ -252,7 +250,7 @@ def create_grouped_features_values(
x_encoded=x_encoded,
preprocessing=preprocessing,
features_dict=features_dict,
- how=how
+ how=how,
)
for f in features_groups[group]:
if f in df.columns:
diff --git a/shapash/report/__init__.py b/shapash/report/__init__.py
index 09af927f..bb3718f3 100644
--- a/shapash/report/__init__.py
+++ b/shapash/report/__init__.py
@@ -1,14 +1,7 @@
import importlib
# This list should be identical to the list in setup.py
-report_requirements = [
- 'nbconvert==6.0.7',
- 'papermill',
- 'matplotlib',
- 'seaborn',
- 'notebook',
- 'Jinja2'
-]
+report_requirements = ["nbconvert==6.0.7", "papermill", "matplotlib", "seaborn", "notebook", "Jinja2"]
def check_report_requirements():
@@ -17,9 +10,11 @@ def check_report_requirements():
This function should be called before executing the report.
"""
for req in report_requirements:
- pkg = req.split('=')[0]
+ pkg = req.split("=")[0]
try:
importlib.import_module(pkg.lower())
except (ModuleNotFoundError, ImportError):
- raise ModuleNotFoundError(f"The following package is necessary to generate the Shapash Report : {pkg}. "
- f"Try 'pip install shapash[report]' to install all required packages.")
\ No newline at end of file
+ raise ModuleNotFoundError(
+ f"The following package is necessary to generate the Shapash Report : {pkg}. "
+ f"Try 'pip install shapash[report]' to install all required packages."
+ )
diff --git a/shapash/report/common.py b/shapash/report/common.py
index bdbe922f..99abf0ee 100644
--- a/shapash/report/common.py
+++ b/shapash/report/common.py
@@ -1,16 +1,17 @@
-from typing import Union, Optional
+import os
from enum import Enum
from numbers import Number
+from typing import Optional, Union
import pandas as pd
-import os
-from pandas.api.types import is_string_dtype, is_numeric_dtype, is_bool_dtype
+from pandas.api.types import is_bool_dtype, is_numeric_dtype, is_string_dtype
class VarType(Enum):
"""
Helper class to indicate the type of a variable.
"""
+
TYPE_CAT = "Categorical"
TYPE_NUM = "Numeric"
TYPE_UNSUPPORTED = "Unsupported"
@@ -36,7 +37,7 @@ def series_dtype(s: pd.Series) -> VarType:
return VarType.TYPE_CAT
elif is_string_dtype(s):
return VarType.TYPE_CAT
- elif s.dtype.name == 'object':
+ elif s.dtype.name == "object":
return VarType.TYPE_CAT
elif is_numeric_dtype(s):
if numeric_is_continuous(s):
@@ -114,9 +115,7 @@ def get_callable(path: str):
try:
import_module(mod)
except Exception as e:
- raise ImportError(
- f"Encountered error: `{e}` when loading module '{path}'"
- ) from e
+ raise ImportError(f"Encountered error: `{e}` when loading module '{path}'") from e
obj = getattr(obj, part)
if isinstance(obj, type):
obj_type: type = obj
@@ -149,7 +148,7 @@ def load_saved_df(path: str) -> Union[pd.DataFrame, None]:
return None
-def display_value(value: float, thousands_separator: str = ',', decimal_separator: str = '.') -> str:
+def display_value(value: float, thousands_separator: str = ",", decimal_separator: str = ".") -> str:
"""
Display a value as a string with specific format.
@@ -172,8 +171,8 @@ def display_value(value: float, thousands_separator: str = ',', decimal_separato
'1,255,000'
"""
- value_str = '{:,}'.format(value).replace(',', '/thousands/').replace('.', '/decimal/')
- return value_str.replace('/thousands/', thousands_separator).replace('/decimal/', decimal_separator)
+ value_str = f"{value:,}".replace(",", "/thousands/").replace(".", "/decimal/")
+ return value_str.replace("/thousands/", thousands_separator).replace("/decimal/", decimal_separator)
def replace_dict_values(obj: dict, replace_fn: callable, *args) -> dict:
diff --git a/shapash/report/data_analysis.py b/shapash/report/data_analysis.py
index 51751b1d..671ad863 100644
--- a/shapash/report/data_analysis.py
+++ b/shapash/report/data_analysis.py
@@ -25,19 +25,19 @@ def perform_global_dataframe_analysis(df: Optional[pd.DataFrame]) -> dict:
return dict()
missing_values = df.isna().sum().sum()
global_d = {
- 'number of features': len(df.columns),
- 'number of observations': df.shape[0],
- 'missing values': missing_values,
- '% missing values': missing_values / (df.shape[0] * df.shape[1]),
+ "number of features": len(df.columns),
+ "number of observations": df.shape[0],
+ "missing values": missing_values,
+ "% missing values": missing_values / (df.shape[0] * df.shape[1]),
}
for stat in global_d.keys():
- if stat == 'number of observations':
+ if stat == "number of observations":
global_d[stat] = int(global_d[stat]) # Keeping the exact number
elif isinstance(global_d[stat], float):
global_d[stat] = round_to_k(global_d[stat], 3)
- replace_dict_values(global_d, display_value, ',', '.')
+ replace_dict_values(global_d, display_value, ",", ".")
return global_d
@@ -64,20 +64,15 @@ def perform_univariate_dataframe_analysis(df: Optional[pd.DataFrame], col_types:
d = df.describe().to_dict()
for col in df.columns:
if col_types[col] == VarType.TYPE_CAT:
- d[col] = {
- 'distinct values': df[col].nunique(),
- 'missing values': df[col].isna().sum()
- }
+ d[col] = {"distinct values": df[col].nunique(), "missing values": df[col].isna().sum()}
for col in d.keys():
for stat in d[col].keys():
- if stat in ['count', 'distinct values']:
+ if stat in ["count", "distinct values"]:
d[col][stat] = int(d[col][stat]) # Keeping the exact number here
elif isinstance(d[col][stat], float):
d[col][stat] = round_to_k(d[col][stat], 3) # Rounding to 3 important figures
- replace_dict_values(d, display_value, ',', '.')
+ replace_dict_values(d, display_value, ",", ".")
return d
-
-
diff --git a/shapash/report/generation.py b/shapash/report/generation.py
index e8114328..32853330 100644
--- a/shapash/report/generation.py
+++ b/shapash/report/generation.py
@@ -1,25 +1,26 @@
"""
Report generation helper module.
"""
-from typing import Optional, Union
import os
+from typing import Optional, Union
+
import pandas as pd
-from nbconvert import HTMLExporter
import papermill as pm
+from nbconvert import HTMLExporter
from shapash.utils.utils import get_project_root
def execute_report(
- working_dir: str,
- explainer: object,
- project_info_file: str,
- x_train: Optional[pd.DataFrame] = None,
- y_train: Optional[pd.DataFrame] = None,
- y_test: Optional[Union[pd.Series, pd.DataFrame]] = None,
- config: Optional[dict] = None,
- notebook_path: Optional[str] = None,
- kernel_name: Optional[str] = None
+ working_dir: str,
+ explainer: object,
+ project_info_file: str,
+ x_train: Optional[pd.DataFrame] = None,
+ y_train: Optional[pd.DataFrame] = None,
+ y_test: Optional[Union[pd.Series, pd.DataFrame]] = None,
+ config: Optional[dict] = None,
+ notebook_path: Optional[str] = None,
+ kernel_name: Optional[str] = None,
):
"""
Executes the base_report.ipynb notebook and saves the results in working_dir.
@@ -50,26 +51,22 @@ def execute_report(
"""
if config is None:
config = {}
- explainer.save(path=os.path.join(working_dir, 'smart_explainer.pickle'))
+ explainer.save(path=os.path.join(working_dir, "smart_explainer.pickle"))
if x_train is not None:
- x_train.to_csv(os.path.join(working_dir, 'x_train.csv'))
+ x_train.to_csv(os.path.join(working_dir, "x_train.csv"))
if y_train is not None:
- y_train.to_csv(os.path.join(working_dir, 'y_train.csv'))
+ y_train.to_csv(os.path.join(working_dir, "y_train.csv"))
if y_test is not None:
- y_test.to_csv(os.path.join(working_dir, 'y_test.csv'))
+ y_test.to_csv(os.path.join(working_dir, "y_test.csv"))
root_path = get_project_root()
- if notebook_path is None or notebook_path == '':
- notebook_path = os.path.join(root_path, 'shapash', 'report', 'base_report.ipynb')
+ if notebook_path is None or notebook_path == "":
+ notebook_path = os.path.join(root_path, "shapash", "report", "base_report.ipynb")
pm.execute_notebook(
notebook_path,
- os.path.join(working_dir, 'base_report.ipynb'),
- parameters=dict(
- dir_path=working_dir,
- project_info_file=project_info_file,
- config=config
- ),
- kernel_name=kernel_name
+ os.path.join(working_dir, "base_report.ipynb"),
+ parameters=dict(dir_path=working_dir, project_info_file=project_info_file, config=config),
+ kernel_name=kernel_name,
)
@@ -85,10 +82,13 @@ def export_and_save_report(working_dir: str, output_file: str):
Path to the html file that will be created.
"""
- exporter = HTMLExporter(exclude_input=True,
- extra_template_basedirs=[os.path.join(get_project_root(), 'shapash', 'report', 'template')],
- template_name='custom', exclude_anchor_links=True)
- (body, resources) = exporter.from_filename(filename=os.path.join(working_dir, 'base_report.ipynb'))
+ exporter = HTMLExporter(
+ exclude_input=True,
+ extra_template_basedirs=[os.path.join(get_project_root(), "shapash", "report", "template")],
+ template_name="custom",
+ exclude_anchor_links=True,
+ )
+ (body, resources) = exporter.from_filename(filename=os.path.join(working_dir, "base_report.ipynb"))
with open(output_file, "w") as file:
file.write(body)
diff --git a/shapash/report/html/double_table.html b/shapash/report/html/double_table.html
index 11f032ee..a4acd513 100644
--- a/shapash/report/html/double_table.html
+++ b/shapash/report/html/double_table.html
@@ -9,4 +9,4 @@
{% include "table_two_columns.html" %}
{% endwith %}
-
\ No newline at end of file
+
diff --git a/shapash/report/html/dropdown.html b/shapash/report/html/dropdown.html
index 05f773c3..201da706 100644
--- a/shapash/report/html/dropdown.html
+++ b/shapash/report/html/dropdown.html
@@ -16,4 +16,4 @@
{% endfor %}
-
\ No newline at end of file
+
diff --git a/shapash/report/html/explainability.html b/shapash/report/html/explainability.html
index 90b0976f..6ef1bb66 100644
--- a/shapash/report/html/explainability.html
+++ b/shapash/report/html/explainability.html
@@ -27,4 +27,4 @@ {{ col['name'] }} - {{ col['type'] }}
{% endfor %}
-{% endfor %}
\ No newline at end of file
+{% endfor %}
diff --git a/shapash/report/html/table_two_columns.html b/shapash/report/html/table_two_columns.html
index 8402242e..962641da 100644
--- a/shapash/report/html/table_two_columns.html
+++ b/shapash/report/html/table_two_columns.html
@@ -16,4 +16,4 @@
{% endfor %}
-
\ No newline at end of file
+
diff --git a/shapash/report/html/univariate.html b/shapash/report/html/univariate.html
index 42fc2118..b29da33f 100644
--- a/shapash/report/html/univariate.html
+++ b/shapash/report/html/univariate.html
@@ -15,4 +15,4 @@ {{ col['name'] }} - {{ col['type'] }}
{{ col['image'] }}
-{% endfor %}
\ No newline at end of file
+{% endfor %}
diff --git a/shapash/report/plots.py b/shapash/report/plots.py
index 420dfdd7..e611a33c 100644
--- a/shapash/report/plots.py
+++ b/shapash/report/plots.py
@@ -1,21 +1,18 @@
-from typing import Union, Optional
-import pandas as pd
+from typing import Optional, Union
+
+import matplotlib.pyplot as plt
import numpy as np
+import pandas as pd
import seaborn as sns
-import matplotlib.pyplot as plt
from matplotlib.colors import LinearSegmentedColormap
-from shapash.utils.utils import truncate_str
from shapash.report.common import VarType
-from shapash.style.style_utils import get_pyplot_color, get_palette
+from shapash.style.style_utils import get_palette, get_pyplot_color
+from shapash.utils.utils import truncate_str
def generate_fig_univariate(
- df_all: pd.DataFrame,
- col: str,
- hue: str,
- type: VarType,
- colors_dict: Optional[dict] = None
+ df_all: pd.DataFrame, col: str, hue: str, type: VarType, colors_dict: Optional[dict] = None
) -> plt.Figure:
"""
Returns a matplotlib figure containing the distribution of any kind of feature
@@ -55,10 +52,7 @@ def generate_fig_univariate(
def generate_fig_univariate_continuous(
- df_all: pd.DataFrame,
- col: str,
- hue: str,
- colors_dict: Optional[dict] = None
+ df_all: pd.DataFrame, col: str, hue: str, colors_dict: Optional[dict] = None
) -> plt.Figure:
"""
Returns a matplotlib figure containing the distribution of a continuous feature.
@@ -78,9 +72,16 @@ def generate_fig_univariate_continuous(
-------
matplotlib.pyplot.Figure
"""
- colors_dict = colors_dict or get_palette('default')
- g = sns.displot(df_all, x=col, hue=hue, kind="kde", fill=True, common_norm=False,
- palette=get_pyplot_color(colors=colors_dict['report_feature_distribution']))
+ colors_dict = colors_dict or get_palette("default")
+ g = sns.displot(
+ df_all,
+ x=col,
+ hue=hue,
+ kind="kde",
+ fill=True,
+ common_norm=False,
+ palette=get_pyplot_color(colors=colors_dict["report_feature_distribution"]),
+ )
g.set_xticklabels(rotation=30)
fig = g.fig
@@ -92,11 +93,7 @@ def generate_fig_univariate_continuous(
def generate_fig_univariate_categorical(
- df_all: pd.DataFrame,
- col: str,
- hue: str,
- nb_cat_max: int = 7,
- colors_dict: Optional[dict] = None
+ df_all: pd.DataFrame, col: str, hue: str, nb_cat_max: int = 7, colors_dict: Optional[dict] = None
) -> plt.Figure:
"""
Returns a matplotlib figure containing the distribution of a categorical feature.
@@ -124,40 +121,50 @@ def generate_fig_univariate_categorical(
-------
matplotlib.pyplot.Figure
"""
- colors_dict = colors_dict or get_palette('default')
- df_cat = df_all.groupby([col, hue]).agg({col: 'count'})\
- .rename(columns={col: "count"}).reset_index()
- df_cat['Percent'] = df_cat['count'] * 100 / df_cat.groupby(hue)['count'].transform('sum')
+ colors_dict = colors_dict or get_palette("default")
+ df_cat = df_all.groupby([col, hue]).agg({col: "count"}).rename(columns={col: "count"}).reset_index()
+ df_cat["Percent"] = df_cat["count"] * 100 / df_cat.groupby(hue)["count"].transform("sum")
if pd.api.types.is_numeric_dtype(df_cat[col].dtype):
df_cat = df_cat.sort_values(col, ascending=True)
df_cat[col] = df_cat[col].astype(str)
- nb_cat = df_cat.groupby([col]).agg({'count': 'sum'}).reset_index()[col].nunique()
+ nb_cat = df_cat.groupby([col]).agg({"count": "sum"}).reset_index()[col].nunique()
if nb_cat > nb_cat_max:
df_cat = _merge_small_categories(df_cat=df_cat, col=col, hue=hue, nb_cat_max=nb_cat_max)
fig, ax = plt.subplots(figsize=(7, 4))
- sns.barplot(data=df_cat, x='Percent', y=col, hue=hue,
- palette=get_pyplot_color(colors=colors_dict['report_feature_distribution']), ax=ax)
+ sns.barplot(
+ data=df_cat,
+ x="Percent",
+ y=col,
+ hue=hue,
+ palette=get_pyplot_color(colors=colors_dict["report_feature_distribution"]),
+ ax=ax,
+ )
for p in ax.patches:
- ax.annotate("{:.1f}%".format(np.nan_to_num(p.get_width(), nan=0)),
- xy=(p.get_width(), p.get_y() + p.get_height() / 2),
- xytext=(5, 0), textcoords='offset points', ha="left", va="center")
+ ax.annotate(
+ "{:.1f}%".format(np.nan_to_num(p.get_width(), nan=0)),
+ xy=(p.get_width(), p.get_y() + p.get_height() / 2),
+ xytext=(5, 0),
+ textcoords="offset points",
+ ha="left",
+ va="center",
+ )
# Shrink current axis by 20%
box = ax.get_position()
ax.set_position([box.x0, box.y0, box.width * 0.8, box.height])
# Put a legend to the right of the current axis
- ax.legend(loc='center left', bbox_to_anchor=(1, 0.5))
+ ax.legend(loc="center left", bbox_to_anchor=(1, 0.5))
# Removes plot borders
- ax.spines['top'].set_visible(False)
- ax.spines['right'].set_visible(False)
+ ax.spines["top"].set_visible(False)
+ ax.spines["right"].set_visible(False)
new_labels = [truncate_str(i.get_text(), maxlen=45) for i in ax.yaxis.get_ticklabels()]
ax.yaxis.set_ticklabels(new_labels)
@@ -165,23 +172,22 @@ def generate_fig_univariate_categorical(
return fig
-def _merge_small_categories(df_cat: pd.DataFrame, col: str, hue: str, nb_cat_max: int) -> pd.DataFrame:
+def _merge_small_categories(df_cat: pd.DataFrame, col: str, hue: str, nb_cat_max: int) -> pd.DataFrame:
"""
Merges categories of column 'col' of df_cat into 'Other' category so that
the number of categories is less than nb_cat_max.
"""
- df_cat_sum_hue = df_cat.groupby([col]).agg({'count': 'sum'}).reset_index()
- list_cat_to_merge = df_cat_sum_hue.sort_values('count', ascending=False)[col].to_list()[nb_cat_max - 1:]
- df_cat_other = df_cat.loc[df_cat[col].isin(list_cat_to_merge)] \
- .groupby(hue, as_index=False)[["count", "Percent"]].sum()
+ df_cat_sum_hue = df_cat.groupby([col]).agg({"count": "sum"}).reset_index()
+ list_cat_to_merge = df_cat_sum_hue.sort_values("count", ascending=False)[col].to_list()[nb_cat_max - 1 :]
+ df_cat_other = (
+ df_cat.loc[df_cat[col].isin(list_cat_to_merge)].groupby(hue, as_index=False)[["count", "Percent"]].sum()
+ )
df_cat_other[col] = "Other"
- return pd.concat([df_cat.loc[~df_cat[col].isin(list_cat_to_merge)],df_cat_other], axis=0)
+ return pd.concat([df_cat.loc[~df_cat[col].isin(list_cat_to_merge)], df_cat_other], axis=0)
def generate_confusion_matrix_plot(
- y_true: Union[np.array, list],
- y_pred: Union[np.array, list],
- colors_dict: Optional[dict] = None
+ y_true: Union[np.array, list], y_pred: Union[np.array, list], colors_dict: Optional[dict] = None
) -> plt.Figure:
"""
Returns a matplotlib figure containing a confusion matrix that is computed using y_true and
@@ -199,11 +205,11 @@ def generate_confusion_matrix_plot(
-------
matplotlib.pyplot.Figure
"""
- colors_dict = colors_dict or get_palette('default')
- col_scale = get_pyplot_color(colors=colors_dict['report_confusion_matrix'])
- cmap_gradient = LinearSegmentedColormap.from_list('col_corr', col_scale, N=100)
+ colors_dict = colors_dict or get_palette("default")
+ col_scale = get_pyplot_color(colors=colors_dict["report_confusion_matrix"])
+ cmap_gradient = LinearSegmentedColormap.from_list("col_corr", col_scale, N=100)
- df_cm = pd.crosstab(y_true, y_pred, rownames=['Actual'], colnames=['Predicted'])
+ df_cm = pd.crosstab(y_true, y_pred, rownames=["Actual"], colnames=["Predicted"])
fig, ax = plt.subplots(figsize=(7, 4))
- sns.heatmap(df_cm, ax=ax, annot=True, cmap=cmap_gradient, fmt='g')
+ sns.heatmap(df_cm, ax=ax, annot=True, cmap=cmap_gradient, fmt="g")
return fig
diff --git a/shapash/report/project_report.py b/shapash/report/project_report.py
index 149a71a2..3c578d01 100644
--- a/shapash/report/project_report.py
+++ b/shapash/report/project_report.py
@@ -1,28 +1,34 @@
-from typing import Optional, Union, Tuple
import logging
-import sys
import os
-from numbers import Number
+import sys
from datetime import date
+from numbers import Number
+from typing import Optional, Tuple, Union
+
import jinja2
-import pandas as pd
import numpy as np
+import pandas as pd
import plotly
-from shapash.utils.transform import inverse_transform, apply_postprocessing, handle_categorical_missing
from shapash import SmartExplainer
+from shapash.report.common import VarType, compute_col_types, display_value, get_callable, series_dtype
+from shapash.report.data_analysis import perform_global_dataframe_analysis, perform_univariate_dataframe_analysis
+from shapash.report.plots import generate_confusion_matrix_plot, generate_fig_univariate
+from shapash.report.visualisation import (
+ convert_fig_to_html,
+ print_css_style,
+ print_html,
+ print_javascript_misc,
+ print_md,
+)
from shapash.utils.io import load_yml
+from shapash.utils.transform import apply_postprocessing, handle_categorical_missing, inverse_transform
from shapash.utils.utils import get_project_root, truncate_str
-from shapash.report.visualisation import print_md, print_html, print_css_style, convert_fig_to_html, \
- print_javascript_misc
-from shapash.report.data_analysis import perform_global_dataframe_analysis, perform_univariate_dataframe_analysis
-from shapash.report.plots import generate_fig_univariate, generate_confusion_matrix_plot
-from shapash.report.common import series_dtype, get_callable, compute_col_types, VarType, display_value
from shapash.webapp.utils.utils import round_to_k
logging.basicConfig(level=logging.INFO)
-template_loader = jinja2.FileSystemLoader(searchpath=os.path.join(get_project_root(), 'shapash', 'report', 'html'))
+template_loader = jinja2.FileSystemLoader(searchpath=os.path.join(get_project_root(), "shapash", "report", "html"))
template_env = jinja2.Environment(loader=template_loader)
@@ -60,13 +66,13 @@ class ProjectReport:
"""
def __init__(
- self,
- explainer: SmartExplainer,
- project_info_file: str,
- x_train: Optional[pd.DataFrame] = None,
- y_train: Optional[pd.DataFrame] = None,
- y_test: Optional[pd.DataFrame] = None,
- config: Optional[dict] = None
+ self,
+ explainer: SmartExplainer,
+ project_info_file: str,
+ x_train: Optional[pd.DataFrame] = None,
+ y_train: Optional[pd.DataFrame] = None,
+ y_test: Optional[pd.DataFrame] = None,
+ config: Optional[dict] = None,
):
self.explainer = explainer
self.metadata = load_yml(path=project_info_file)
@@ -87,35 +93,34 @@ def __init__(
self.y_pred = np.array(self.explainer.y_pred.T)[0]
else:
self.y_pred = self.explainer.model.predict(self.explainer.x_encoded)
- self.y_test, target_name_test = self._get_values_and_name(y_test, 'target')
- self.y_train, target_name_train = self._get_values_and_name(y_train, 'target')
+ self.y_test, target_name_test = self._get_values_and_name(y_test, "target")
+ self.y_train, target_name_train = self._get_values_and_name(y_train, "target")
self.target_name = target_name_train or target_name_test
- if 'title_story' in self.config.keys():
- self.title_story = config['title_story']
- elif self.explainer.title_story != '':
+ if "title_story" in self.config.keys():
+ self.title_story = config["title_story"]
+ elif self.explainer.title_story != "":
self.title_story = self.explainer.title_story
else:
- self.title_story = 'Shapash report'
- self.title_description = self.config['title_description'] if 'title_description' in self.config.keys() else ''
+ self.title_story = "Shapash report"
+ self.title_description = self.config["title_description"] if "title_description" in self.config.keys() else ""
print_css_style()
print_javascript_misc()
- if 'metrics' in self.config.keys():
- if not isinstance(self.config['metrics'], list) or not isinstance(self.config['metrics'][0], dict):
+ if "metrics" in self.config.keys():
+ if not isinstance(self.config["metrics"], list) or not isinstance(self.config["metrics"][0], dict):
raise ValueError("The metrics parameter expects a list of dict.")
- for metric in self.config['metrics']:
+ for metric in self.config["metrics"]:
for key in metric:
- if key not in ['path', 'name', 'use_proba_values']:
+ if key not in ["path", "name", "use_proba_values"]:
raise ValueError(f"Unknown key : {key}. Key should be in ['path', 'name', 'use_proba_values']")
- if key == 'use_proba_values' and not isinstance(metric['use_proba_values'], bool):
+ if key == "use_proba_values" and not isinstance(metric["use_proba_values"], bool):
raise ValueError('"use_proba_values" metric key expects a boolean value.')
@staticmethod
def _get_values_and_name(
- y: Optional[Union[pd.DataFrame, pd.Series, list]],
- default_name: str
+ y: Optional[Union[pd.DataFrame, pd.Series, list]], default_name: str
) -> Union[Tuple[list, str], Tuple[None, None]]:
"""
Extracts vales and column name from a Pandas Series, DataFrame, or assign a default
@@ -170,20 +175,25 @@ def _create_train_test_df(test: Optional[pd.DataFrame], train: Optional[pd.DataF
The concatenation of train and test as a dataframe containing train and test values with
a new 'data_train_test' column allowing to distinguish the values.
"""
- if (test is not None and 'data_train_test' in test.columns) or \
- (train is not None and 'data_train_test' in train.columns):
+ if (test is not None and "data_train_test" in test.columns) or (
+ train is not None and "data_train_test" in train.columns
+ ):
raise ValueError('"data_train_test" column must be renamed as it is used in ProjectReport')
if test is None and train is None:
return None
- return pd.concat([test.assign(data_train_test="test") if test is not None else None,
- train.assign(data_train_test="train") if train is not None else None]).reset_index(drop=True)
+ return pd.concat(
+ [
+ test.assign(data_train_test="test") if test is not None else None,
+ train.assign(data_train_test="train") if train is not None else None,
+ ]
+ ).reset_index(drop=True)
def display_title_description(self):
"""
Displays title of the report and its description if defined.
"""
print_html(f"""{self.title_story} """)
- if self.title_description != '':
+ if self.title_description != "":
print_html(f'{self.title_description}
')
def display_project_information(self):
@@ -193,11 +203,11 @@ def display_project_information(self):
for section in self.metadata.keys():
print_md(f"## {section.title()}")
for k, v in self.metadata[section].items():
- if k.lower() == 'date' and v.lower() == 'auto':
+ if k.lower() == "date" and v.lower() == "auto":
print_md(f"**{k.title()}** : {date.today()}")
else:
print_md(f"**{k.title()}** : {v}")
- print_md('---')
+ print_md("---")
def display_model_analysis(self):
"""
@@ -209,29 +219,37 @@ def display_model_analysis(self):
print_md(f"**Library :** {self.explainer.model.__class__.__module__}")
for name, module in sorted(sys.modules.items()):
- if hasattr(module, '__version__') \
- and self.explainer.model.__class__.__module__.split('.')[0] == module.__name__:
+ if (
+ hasattr(module, "__version__")
+ and self.explainer.model.__class__.__module__.split(".")[0] == module.__name__
+ ):
print_md(f"**Library version :** {module.__version__}")
print_md("**Model parameters :** ")
model_params = self.explainer.model.__dict__
table_template = template_env.get_template("double_table.html")
- print_html(table_template.render(
- columns1=["Parameter key", "Parameter value"],
- rows1=[{"name": truncate_str(str(k), 50), "value": truncate_str(str(v), 300)}
- for k, v in list(model_params.items())[:len(model_params)//2:]], # Getting half of the parameters
- columns2=["Parameter key", "Parameter value"],
- rows2=[{"name": truncate_str(str(k), 50), "value": truncate_str(str(v), 300)}
- for k, v in list(model_params.items())[len(model_params)//2:]] # Getting 2nd half of the parameters
- ))
- print_md('---')
+ print_html(
+ table_template.render(
+ columns1=["Parameter key", "Parameter value"],
+ rows1=[
+ {"name": truncate_str(str(k), 50), "value": truncate_str(str(v), 300)}
+ for k, v in list(model_params.items())[: len(model_params) // 2 :]
+ ], # Getting half of the parameters
+ columns2=["Parameter key", "Parameter value"],
+ rows2=[
+ {"name": truncate_str(str(k), 50), "value": truncate_str(str(v), 300)}
+ for k, v in list(model_params.items())[len(model_params) // 2 :]
+ ], # Getting 2nd half of the parameters
+ )
+ )
+ print_md("---")
def display_dataset_analysis(
- self,
- global_analysis: bool = True,
- univariate_analysis: bool = True,
- target_analysis: bool = True,
- multivariate_analysis: bool = True
+ self,
+ global_analysis: bool = True,
+ univariate_analysis: bool = True,
+ target_analysis: bool = True,
+ multivariate_analysis: bool = True,
):
"""
This method performs and displays an exploration of the data given.
@@ -262,14 +280,16 @@ def display_dataset_analysis(
col_splitter="data_train_test",
split_values=["test", "train"],
names=["Prediction dataset", "Training dataset"],
- group_id='univariate'
+ group_id="univariate",
)
if target_analysis:
df_target = self._create_train_test_df(
- test=pd.DataFrame({self.target_name: self.y_test},
- index=range(len(self.y_test))) if self.y_test is not None else None,
- train=pd.DataFrame({self.target_name: self.y_train},
- index=range(len(self.y_train))) if self.y_train is not None else None
+ test=pd.DataFrame({self.target_name: self.y_test}, index=range(len(self.y_test)))
+ if self.y_test is not None
+ else None,
+ train=pd.DataFrame({self.target_name: self.y_train}, index=range(len(self.y_train)))
+ if self.y_train is not None
+ else None,
)
if df_target is not None:
if target_analysis:
@@ -279,77 +299,78 @@ def display_dataset_analysis(
col_splitter="data_train_test",
split_values=["test", "train"],
names=["Prediction dataset", "Training dataset"],
- group_id='target'
+ group_id="target",
)
if multivariate_analysis:
print_md("### Multivariate analysis")
fig_corr = self.explainer.plot.correlations(
self.df_train_test,
- facet_col='data_train_test',
+ facet_col="data_train_test",
max_features=20,
- width=900 if len(self.df_train_test['data_train_test'].unique()) > 1 else 500,
+ width=900 if len(self.df_train_test["data_train_test"].unique()) > 1 else 500,
height=500,
)
print_html(plotly.io.to_html(fig_corr))
- print_md('---')
+ print_md("---")
def _display_dataset_analysis_global(self):
- df_stats_global = self._stats_to_table(test_stats=perform_global_dataframe_analysis(self.x_init),
- train_stats=perform_global_dataframe_analysis(self.x_train_pre),
- names=["Prediction dataset", "Training dataset"])
+ df_stats_global = self._stats_to_table(
+ test_stats=perform_global_dataframe_analysis(self.x_init),
+ train_stats=perform_global_dataframe_analysis(self.x_train_pre),
+ names=["Prediction dataset", "Training dataset"],
+ )
print_html(df_stats_global.to_html(classes="greyGridTable"))
def _perform_and_display_analysis_univariate(
- self,
- df: pd.DataFrame,
- col_splitter: str,
- split_values: list,
- names: list,
- group_id: str
+ self, df: pd.DataFrame, col_splitter: str, split_values: list, names: list, group_id: str
):
col_types = compute_col_types(df)
n_splits = df[col_splitter].nunique()
inv_columns_dict = {v: k for k, v in self.explainer.columns_dict.items()}
- test_stats_univariate = perform_univariate_dataframe_analysis(df.loc[df[col_splitter] == split_values[0]],
- col_types=col_types)
+ test_stats_univariate = perform_univariate_dataframe_analysis(
+ df.loc[df[col_splitter] == split_values[0]], col_types=col_types
+ )
if n_splits > 1:
- train_stats_univariate = perform_univariate_dataframe_analysis(df.loc[df[col_splitter] == split_values[1]],
- col_types=col_types)
+ train_stats_univariate = perform_univariate_dataframe_analysis(
+ df.loc[df[col_splitter] == split_values[1]], col_types=col_types
+ )
univariate_template = template_env.get_template("univariate.html")
univariate_features_desc = list()
- list_cols_labels = [self.explainer.features_dict.get(col, col)
- for col in df.drop(col_splitter, axis=1).columns.to_list()]
+ list_cols_labels = [
+ self.explainer.features_dict.get(col, col) for col in df.drop(col_splitter, axis=1).columns.to_list()
+ ]
for col_label in sorted(list_cols_labels):
col = self.explainer.inv_features_dict.get(col_label, col_label)
- fig = generate_fig_univariate(df_all=df, col=col, hue=col_splitter, type=col_types[col],
- colors_dict=self.explainer.colors_dict)
+ fig = generate_fig_univariate(
+ df_all=df, col=col, hue=col_splitter, type=col_types[col], colors_dict=self.explainer.colors_dict
+ )
df_col_stats = self._stats_to_table(
test_stats=test_stats_univariate[col],
train_stats=train_stats_univariate[col] if n_splits > 1 else None,
- names=names
+ names=names,
)
- univariate_features_desc.append({
- 'feature_index': int(inv_columns_dict.get(col, 0)),
- 'name': col,
- 'type': str(series_dtype(df[col])),
- 'description': col_label,
- 'table': df_col_stats.to_html(classes="greyGridTable"),
- 'image': convert_fig_to_html(fig)
- })
+ univariate_features_desc.append(
+ {
+ "feature_index": int(inv_columns_dict.get(col, 0)),
+ "name": col,
+ "type": str(series_dtype(df[col])),
+ "description": col_label,
+ "table": df_col_stats.to_html(classes="greyGridTable"),
+ "image": convert_fig_to_html(fig),
+ }
+ )
print_html(univariate_template.render(features=univariate_features_desc, groupId=group_id))
@staticmethod
- def _stats_to_table(test_stats: dict,
- names: list,
- train_stats: Optional[dict] = None,
- ) -> pd.DataFrame:
+ def _stats_to_table(
+ test_stats: dict,
+ names: list,
+ train_stats: Optional[dict] = None,
+ ) -> pd.DataFrame:
if train_stats is not None:
- return pd.DataFrame({
- names[1]: pd.Series(train_stats),
- names[0]: pd.Series(test_stats)
- })
+ return pd.DataFrame({names[1]: pd.Series(train_stats), names[0]: pd.Series(test_stats)})
else:
return pd.DataFrame({names[0]: pd.Series(test_stats)})
@@ -364,30 +385,34 @@ def display_model_explainability(self):
multiclass = True if (self.explainer._classes and len(self.explainer._classes) > 2) else False
c_list = self.explainer._classes if multiclass else [1] # list just used for multiclass
for index_label, label in enumerate(c_list): # Iterating over all labels in multiclass case
- label_value = self.explainer.check_label_name(label)[2] if multiclass else ''
+ label_value = self.explainer.check_label_name(label)[2] if multiclass else ""
fig_features_importance = self.explainer.plot.features_importance(label=label)
explain_contrib_data = list()
- list_cols_labels = [self.explainer.features_dict.get(col, col)
- for col in self.col_names]
+ list_cols_labels = [self.explainer.features_dict.get(col, col) for col in self.col_names]
for feature_label in sorted(list_cols_labels):
feature = self.explainer.inv_features_dict.get(feature_label, feature_label)
fig = self.explainer.plot.contribution_plot(feature, label=label, max_points=200)
- explain_contrib_data.append({
- 'feature_index': int(inv_columns_dict[feature]),
- 'name': feature,
- 'description': self.explainer.features_dict[feature],
- 'plot': plotly.io.to_html(fig, include_plotlyjs=False, full_html=False)
- })
- explain_data.append({
- 'index': index_label,
- 'name': label_value,
- 'feature_importance_plot': plotly.io.to_html(fig_features_importance, include_plotlyjs=False,
- full_html=False),
- 'features': explain_contrib_data
- })
+ explain_contrib_data.append(
+ {
+ "feature_index": int(inv_columns_dict[feature]),
+ "name": feature,
+ "description": self.explainer.features_dict[feature],
+ "plot": plotly.io.to_html(fig, include_plotlyjs=False, full_html=False),
+ }
+ )
+ explain_data.append(
+ {
+ "index": index_label,
+ "name": label_value,
+ "feature_importance_plot": plotly.io.to_html(
+ fig_features_importance, include_plotlyjs=False, full_html=False
+ ),
+ "features": explain_contrib_data,
+ }
+ )
print_html(explainability_template.render(labels=explain_data))
- print_md('---')
+ print_md("---")
def display_model_performance(self):
"""
@@ -418,39 +443,48 @@ def display_model_performance(self):
return
print_md("### Univariate analysis of target variable")
- df = pd.concat([pd.DataFrame({self.target_name: self.y_pred}).assign(_dataset="pred"),
- pd.DataFrame({self.target_name: self.y_test}).assign(_dataset="true")
- if self.y_test is not None else None]).reset_index(drop=True)
+ df = pd.concat(
+ [
+ pd.DataFrame({self.target_name: self.y_pred}).assign(_dataset="pred"),
+ pd.DataFrame({self.target_name: self.y_test}).assign(_dataset="true")
+ if self.y_test is not None
+ else None,
+ ]
+ ).reset_index(drop=True)
self._perform_and_display_analysis_univariate(
df=df,
col_splitter="_dataset",
split_values=["pred", "true"],
names=["Prediction values", "True values"],
- group_id='target-distribution'
+ group_id="target-distribution",
)
- if 'metrics' not in self.config.keys():
+ if "metrics" not in self.config.keys():
logging.info("No 'metrics' key found in report config dict. Skipping model performance part.")
return
print_md("### Metrics")
- for metric in self.config['metrics']:
- if 'name' not in metric.keys():
- metric['name'] = metric['path']
+ for metric in self.config["metrics"]:
+ if "name" not in metric.keys():
+ metric["name"] = metric["path"]
- if metric['path'] in ['confusion_matrix', 'sklearn.metrics.confusion_matrix'] or \
- metric['name'] == 'confusion_matrix':
+ if (
+ metric["path"] in ["confusion_matrix", "sklearn.metrics.confusion_matrix"]
+ or metric["name"] == "confusion_matrix"
+ ):
print_md(f"**{metric['name']} :**")
- print_html(convert_fig_to_html(generate_confusion_matrix_plot(
- y_true=self.y_test,
- y_pred=self.y_pred,
- colors_dict=self.explainer.colors_dict
- )))
+ print_html(
+ convert_fig_to_html(
+ generate_confusion_matrix_plot(
+ y_true=self.y_test, y_pred=self.y_pred, colors_dict=self.explainer.colors_dict
+ )
+ )
+ )
else:
try:
- metric_fn = get_callable(path=metric['path'])
+ metric_fn = get_callable(path=metric["path"])
# Look if we should use proba values instead of predicted values
- if 'use_proba_values' in metric.keys() and metric['use_proba_values'] is True:
+ if "use_proba_values" in metric.keys() and metric["use_proba_values"] is True:
y_pred = self.explainer.proba_values
else:
y_pred = self.y_pred
@@ -468,6 +502,8 @@ def display_model_performance(self):
print_md(f"**{metric['name']} :**")
print_html(f"{res}")
else:
- logging.info(f"Could not compute following metric : {metric['path']}. \n"
- f"Result of type {res} cannot be displayed")
- print_md('---')
+ logging.info(
+ f"Could not compute following metric : {metric['path']}. \n"
+ f"Result of type {res} cannot be displayed"
+ )
+ print_md("---")
diff --git a/shapash/report/template/custom/conf.json b/shapash/report/template/custom/conf.json
index 76a9c7c5..a5974f9f 100644
--- a/shapash/report/template/custom/conf.json
+++ b/shapash/report/template/custom/conf.json
@@ -1,12 +1,12 @@
{
- "base_template": "classic",
- "mimetypes": {
- "text/html": true
- },
- "preprocessors": {
- "100-pygments": {
- "type": "nbconvert.preprocessors.CSSHTMLHeaderPreprocessor",
- "enabled": true
- }
+ "base_template": "classic",
+ "mimetypes": {
+ "text/html": true
+ },
+ "preprocessors": {
+ "100-pygments": {
+ "enabled": true,
+ "type": "nbconvert.preprocessors.CSSHTMLHeaderPreprocessor"
}
-}
\ No newline at end of file
+ }
+}
diff --git a/shapash/report/visualisation.py b/shapash/report/visualisation.py
index b82d0d8d..66a11b2a 100644
--- a/shapash/report/visualisation.py
+++ b/shapash/report/visualisation.py
@@ -1,7 +1,6 @@
-from IPython.display import display, Markdown, Latex, HTML
import matplotlib.pyplot as plt
-
import pandas as pd
+from IPython.display import HTML, Latex, Markdown, display
def print_md(text: str):
@@ -26,7 +25,8 @@ def print_html(text: str):
def print_css_style():
- print_html("""
+ print_html(
+ """
- """)
+ """
+ )
def print_javascript_misc():
- print_html("""
+ print_html(
+ """
- """)
+ """
+ )
def convert_fig_to_html(fig):
- """ Convert Matplotlib figure 'fig' into a ![]() tag for HTML use using base64 encoding. """
- import io
+ """Convert Matplotlib figure 'fig' into a
tag for HTML use using base64 encoding. """
- import io
+ """Convert Matplotlib figure 'fig' into a ![]() tag for HTML use using base64 encoding."""
import base64
+ import io
+
s = io.BytesIO()
- fig.savefig(s, format='png', bbox_inches="tight")
+ fig.savefig(s, format="png", bbox_inches="tight")
plt.close()
s = base64.b64encode(s.getvalue()).decode("utf-8").replace("\n", "")
return '
tag for HTML use using base64 encoding."""
import base64
+ import io
+
s = io.BytesIO()
- fig.savefig(s, format='png', bbox_inches="tight")
+ fig.savefig(s, format="png", bbox_inches="tight")
plt.close()
s = base64.b64encode(s.getvalue()).decode("utf-8").replace("\n", "")
return ' ' % s
@@ -136,4 +140,3 @@ def html_str_df_and_image(df: pd.DataFrame, fig: plt.Figure) -> str:
def print_figure(fig):
print_html(convert_fig_to_html(fig))
-
diff --git a/shapash/style/colors.json b/shapash/style/colors.json
index 513d9922..32999b6e 100644
--- a/shapash/style/colors.json
+++ b/shapash/style/colors.json
@@ -1,200 +1,200 @@
{
- "default": {
- "title_color": "rgb(50, 50, 50)",
- "axis_color": "rgb(50, 50, 50)",
- "featureimp_bar": {
- "1": "rgba(244, 192, 0, 1.0)",
- "2": "rgba(52, 55, 54, 0.7)"
- },
- "featureimp_line": "rgba(52, 55, 54, 0.8)",
- "featureimp_groups": {
- "0": "rgb(245, 133, 24)",
- "1": "rgb(49, 99, 149)"
- },
- "contrib_colorscale": [
- "rgb(52, 55, 54)",
- "rgb(74, 99, 138)",
- "rgb(116, 153, 214)",
- "rgb(162, 188, 213)",
- "rgb(212, 234, 242)",
- "rgb(235, 216, 134)",
- "rgb(255, 204, 83)",
- "rgb(244, 192, 0)",
- "rgb(255, 166, 17)",
- "rgb(255, 123, 38)",
- "rgb(255, 77, 7)"
- ],
- "violin_area_classif": {
- "0": "rgba(117, 152, 189, 0.9)",
- "1": "rgba(255, 166, 17, 0.9)"
- },
- "violin_default": "rgba(117, 152, 189, 0.9)",
- "localplot_bar": {
- "1": "rgba(244, 192, 0, 1.0)",
- "-1": "rgba(74, 99, 138, 0.7)",
- "0": "rgba(113, 101, 59, 1.0)",
- "-2": "rgba(52, 55, 54, 0.7)"
- },
- "localplot_line": {
- "1": "rgba(52, 55, 54, 0.8)",
- "-1": "rgba(27, 28, 28, 1.0)",
- "0": "rgba(52, 55, 54, 0.8)",
- "-2": "rgba(27, 28, 28, 1.0)"
- },
- "compare_plot": [
- "rgba(244, 192, 0, 1.0)",
- "rgba(74, 99, 138, 0.7)",
- "rgba(113, 101, 59, 1.0)",
- "rgba(183, 58, 56, 0.9)",
- "rgba(255, 123, 38, 1.0)",
- "rgba(0, 21, 179, 0.97)",
- "rgba(116, 1, 179, 0.9)"
- ],
- "interaction_scale": [
- "rgb(175, 169, 157)",
- "rgb(255, 255, 255)",
- "rgb(255, 77, 7)"
- ],
- "interaction_discrete": [
- "rgb(133, 92, 117)",
- "rgb(217, 175, 107)",
- "rgb(175, 100, 88)",
- "rgb(115, 111, 76)",
- "rgb(82, 106, 131)",
- "rgb(98, 83, 119)",
- "rgb(104, 133, 92)",
- "rgb(156, 156, 94)",
- "rgb(160, 97, 119)",
- "rgb(140, 120, 93)",
- "rgb(124, 124, 124)"
- ],
- "stability_bar": {
- "1": "rgba(255, 166, 17, 0.9)",
- "0": "rgba(117, 152, 189, 0.9)"
- },
- "compacity_bar": {
- "1": "rgba(255, 166, 17, 0.9)",
- "0": "rgba(117, 152, 189, 0.9)"
- },
- "webapp_button": {
- "0": "rgb(244, 192, 0)",
- "1": "rgb(113, 101, 59)"
- },
- "webapp_bkg": "rgb(52,55,54)",
- "webapp_title": "rgb(244, 192, 0)",
- "report_feature_distribution": {
- "train": "rgba(74, 99, 128, 0.7)",
- "test": "rgba(244, 192, 0, 1)",
- "true": "rgba(74, 99, 138, 0.7)",
- "pred": "rgba(244, 192, 0, 1)"
- },
- "report_confusion_matrix": [
- "rgb(255, 255, 255)",
- "rgb(244, 192, 0)",
- "rgb(255, 166, 17)"
- ],
- "prediction_plot": {
- "1": "rgba(52, 55, 54, 0.9)",
- "0": "rgba(255, 77, 7, 0.9)"
- }
- },
- "blues": {
- "title_color": "rgb(50, 50, 50)",
- "axis_color": "rgb(50, 50, 50)",
- "featureimp_bar": {
- "1": "rgba(0, 154, 203, 1)",
- "2": "rgba(223, 103, 0, 0.8)"
- },
- "featureimp_line": "rgba(52, 55, 54, 0.8)",
- "featureimp_groups": {
- "0": "rgb(10, 204, 143)",
- "1": "rgb(176, 140, 104)"
- },
- "contrib_colorscale": [
- "rgb(168, 84, 0)",
- "rgb(204, 102, 0)",
- "rgb(245, 122, 0)",
- "rgb(240, 150, 67)",
- "rgb(240, 195, 162)",
- "rgb(237, 235, 232)",
- "rgb(161, 221, 254)",
- "rgb(103, 208, 255)",
- "rgb(0, 154, 203)",
- "rgb(0, 98, 128)",
- "rgb(0, 70, 92)"
- ],
- "violin_area_classif": {
- "0": "rgba(223, 103, 0, 0.9)",
- "1": "rgba(0, 154, 203, 0.9)"
- },
- "violin_default": "rgba(0, 98, 128, 0.7)",
- "localplot_bar": {
- "1": "rgba(0, 154, 203, 1)",
- "-1": "rgba(235, 149, 72, 0.9)",
- "0": "rgba(106, 178, 204, 0.8)",
- "-2": "rgba(217, 116, 0, 0.5)"
- },
- "localplot_line": {
- "1": "rgba(52, 55, 54, 0.8)",
- "-1": "rgba(27, 28, 28, 1.0)",
- "0": "rgba(52, 55, 54, 0.8)",
- "-2": "rgba(27, 28, 28, 1.0)"
- },
- "compare_plot": [
- "rgba(0, 154, 203, 1.0)",
- "rgba(223, 103, 0, 0.8)",
- "rgba(11, 60, 217, 1.0)",
- "rgba(204, 50, 10, 0.9)",
- "rgba(11, 217, 152, 1.0)",
- "rgba(129, 64, 0, 0.9)",
- "rgba(0, 98, 128, 0.9)"
- ],
- "interaction_scale": [
- "rgb(175, 169, 157)",
- "rgb(255, 255, 255)",
- "rgb(0, 98, 128)"
- ],
- "interaction_discrete": [
- "rgb(95, 70, 144)",
- "rgb(29, 105, 150)",
- "rgb(56, 166, 165)",
- "rgb(15, 133, 84)",
- "rgb(115, 175, 72)",
- "rgb(237, 173, 8)",
- "rgb(225, 124, 5)",
- "rgb(204, 80, 62)",
- "rgb(148, 52, 110)",
- "rgb(111, 64, 112)",
- "rgb(102, 102, 102)"
- ],
- "stability_bar": {
- "1": "rgba(0, 154, 203, 0.9)",
- "0": "rgba(230, 111, 0, 0.9)"
- },
- "compacity_bar": {
- "1": "rgba(0, 154, 203, 0.9)",
- "0": "rgba(230, 111, 0, 0.9)"
- },
- "webapp_button": {
- "0": "rgb(0, 154, 203)",
- "1": "rgb(50, 90, 117)"
- },
- "webapp_bkg": "rgb(52,55,54)",
- "webapp_title": "rgb(0, 154, 203)",
- "report_feature_distribution": {
- "train": "rgba(0, 154, 203, 0.9)",
- "test": "rgba(230, 111, 0, 0.9)",
- "true": "rgba(0, 154, 203, 0.9)",
- "pred": "rgba(230, 111, 0, 0.9)"
- },
- "report_confusion_matrix": [
- "rgb(255, 255, 255)",
- "rgb(244, 192, 0)",
- "rgb(255, 166, 17)"
- ],
- "prediction_plot": {
- "1": "rgba(168, 84, 0, 0.9)",
- "0": "rgba(0, 70, 92, 0.9)"
- }
- }
-}
\ No newline at end of file
+ "blues": {
+ "axis_color": "rgb(50, 50, 50)",
+ "compacity_bar": {
+ "0": "rgba(230, 111, 0, 0.9)",
+ "1": "rgba(0, 154, 203, 0.9)"
+ },
+ "compare_plot": [
+ "rgba(0, 154, 203, 1.0)",
+ "rgba(223, 103, 0, 0.8)",
+ "rgba(11, 60, 217, 1.0)",
+ "rgba(204, 50, 10, 0.9)",
+ "rgba(11, 217, 152, 1.0)",
+ "rgba(129, 64, 0, 0.9)",
+ "rgba(0, 98, 128, 0.9)"
+ ],
+ "contrib_colorscale": [
+ "rgb(168, 84, 0)",
+ "rgb(204, 102, 0)",
+ "rgb(245, 122, 0)",
+ "rgb(240, 150, 67)",
+ "rgb(240, 195, 162)",
+ "rgb(237, 235, 232)",
+ "rgb(161, 221, 254)",
+ "rgb(103, 208, 255)",
+ "rgb(0, 154, 203)",
+ "rgb(0, 98, 128)",
+ "rgb(0, 70, 92)"
+ ],
+ "featureimp_bar": {
+ "1": "rgba(0, 154, 203, 1)",
+ "2": "rgba(223, 103, 0, 0.8)"
+ },
+ "featureimp_groups": {
+ "0": "rgb(10, 204, 143)",
+ "1": "rgb(176, 140, 104)"
+ },
+ "featureimp_line": "rgba(52, 55, 54, 0.8)",
+ "interaction_discrete": [
+ "rgb(95, 70, 144)",
+ "rgb(29, 105, 150)",
+ "rgb(56, 166, 165)",
+ "rgb(15, 133, 84)",
+ "rgb(115, 175, 72)",
+ "rgb(237, 173, 8)",
+ "rgb(225, 124, 5)",
+ "rgb(204, 80, 62)",
+ "rgb(148, 52, 110)",
+ "rgb(111, 64, 112)",
+ "rgb(102, 102, 102)"
+ ],
+ "interaction_scale": [
+ "rgb(175, 169, 157)",
+ "rgb(255, 255, 255)",
+ "rgb(0, 98, 128)"
+ ],
+ "localplot_bar": {
+ "-1": "rgba(235, 149, 72, 0.9)",
+ "-2": "rgba(217, 116, 0, 0.5)",
+ "0": "rgba(106, 178, 204, 0.8)",
+ "1": "rgba(0, 154, 203, 1)"
+ },
+ "localplot_line": {
+ "-1": "rgba(27, 28, 28, 1.0)",
+ "-2": "rgba(27, 28, 28, 1.0)",
+ "0": "rgba(52, 55, 54, 0.8)",
+ "1": "rgba(52, 55, 54, 0.8)"
+ },
+ "prediction_plot": {
+ "0": "rgba(0, 70, 92, 0.9)",
+ "1": "rgba(168, 84, 0, 0.9)"
+ },
+ "report_confusion_matrix": [
+ "rgb(255, 255, 255)",
+ "rgb(244, 192, 0)",
+ "rgb(255, 166, 17)"
+ ],
+ "report_feature_distribution": {
+ "pred": "rgba(230, 111, 0, 0.9)",
+ "test": "rgba(230, 111, 0, 0.9)",
+ "train": "rgba(0, 154, 203, 0.9)",
+ "true": "rgba(0, 154, 203, 0.9)"
+ },
+ "stability_bar": {
+ "0": "rgba(230, 111, 0, 0.9)",
+ "1": "rgba(0, 154, 203, 0.9)"
+ },
+ "title_color": "rgb(50, 50, 50)",
+ "violin_area_classif": {
+ "0": "rgba(223, 103, 0, 0.9)",
+ "1": "rgba(0, 154, 203, 0.9)"
+ },
+ "violin_default": "rgba(0, 98, 128, 0.7)",
+ "webapp_bkg": "rgb(52,55,54)",
+ "webapp_button": {
+ "0": "rgb(0, 154, 203)",
+ "1": "rgb(50, 90, 117)"
+ },
+ "webapp_title": "rgb(0, 154, 203)"
+ },
+ "default": {
+ "axis_color": "rgb(50, 50, 50)",
+ "compacity_bar": {
+ "0": "rgba(117, 152, 189, 0.9)",
+ "1": "rgba(255, 166, 17, 0.9)"
+ },
+ "compare_plot": [
+ "rgba(244, 192, 0, 1.0)",
+ "rgba(74, 99, 138, 0.7)",
+ "rgba(113, 101, 59, 1.0)",
+ "rgba(183, 58, 56, 0.9)",
+ "rgba(255, 123, 38, 1.0)",
+ "rgba(0, 21, 179, 0.97)",
+ "rgba(116, 1, 179, 0.9)"
+ ],
+ "contrib_colorscale": [
+ "rgb(52, 55, 54)",
+ "rgb(74, 99, 138)",
+ "rgb(116, 153, 214)",
+ "rgb(162, 188, 213)",
+ "rgb(212, 234, 242)",
+ "rgb(235, 216, 134)",
+ "rgb(255, 204, 83)",
+ "rgb(244, 192, 0)",
+ "rgb(255, 166, 17)",
+ "rgb(255, 123, 38)",
+ "rgb(255, 77, 7)"
+ ],
+ "featureimp_bar": {
+ "1": "rgba(244, 192, 0, 1.0)",
+ "2": "rgba(52, 55, 54, 0.7)"
+ },
+ "featureimp_groups": {
+ "0": "rgb(245, 133, 24)",
+ "1": "rgb(49, 99, 149)"
+ },
+ "featureimp_line": "rgba(52, 55, 54, 0.8)",
+ "interaction_discrete": [
+ "rgb(133, 92, 117)",
+ "rgb(217, 175, 107)",
+ "rgb(175, 100, 88)",
+ "rgb(115, 111, 76)",
+ "rgb(82, 106, 131)",
+ "rgb(98, 83, 119)",
+ "rgb(104, 133, 92)",
+ "rgb(156, 156, 94)",
+ "rgb(160, 97, 119)",
+ "rgb(140, 120, 93)",
+ "rgb(124, 124, 124)"
+ ],
+ "interaction_scale": [
+ "rgb(175, 169, 157)",
+ "rgb(255, 255, 255)",
+ "rgb(255, 77, 7)"
+ ],
+ "localplot_bar": {
+ "-1": "rgba(74, 99, 138, 0.7)",
+ "-2": "rgba(52, 55, 54, 0.7)",
+ "0": "rgba(113, 101, 59, 1.0)",
+ "1": "rgba(244, 192, 0, 1.0)"
+ },
+ "localplot_line": {
+ "-1": "rgba(27, 28, 28, 1.0)",
+ "-2": "rgba(27, 28, 28, 1.0)",
+ "0": "rgba(52, 55, 54, 0.8)",
+ "1": "rgba(52, 55, 54, 0.8)"
+ },
+ "prediction_plot": {
+ "0": "rgba(255, 77, 7, 0.9)",
+ "1": "rgba(52, 55, 54, 0.9)"
+ },
+ "report_confusion_matrix": [
+ "rgb(255, 255, 255)",
+ "rgb(244, 192, 0)",
+ "rgb(255, 166, 17)"
+ ],
+ "report_feature_distribution": {
+ "pred": "rgba(244, 192, 0, 1)",
+ "test": "rgba(244, 192, 0, 1)",
+ "train": "rgba(74, 99, 128, 0.7)",
+ "true": "rgba(74, 99, 138, 0.7)"
+ },
+ "stability_bar": {
+ "0": "rgba(117, 152, 189, 0.9)",
+ "1": "rgba(255, 166, 17, 0.9)"
+ },
+ "title_color": "rgb(50, 50, 50)",
+ "violin_area_classif": {
+ "0": "rgba(117, 152, 189, 0.9)",
+ "1": "rgba(255, 166, 17, 0.9)"
+ },
+ "violin_default": "rgba(117, 152, 189, 0.9)",
+ "webapp_bkg": "rgb(52,55,54)",
+ "webapp_button": {
+ "0": "rgb(244, 192, 0)",
+ "1": "rgb(113, 101, 59)"
+ },
+ "webapp_title": "rgb(244, 192, 0)"
+ }
+}
diff --git a/shapash/style/style_utils.py b/shapash/style/style_utils.py
index 4c722fec..36dad34c 100644
--- a/shapash/style/style_utils.py
+++ b/shapash/style/style_utils.py
@@ -9,23 +9,23 @@
def colors_loading():
"""
- colors_loading allows shapash to load a json file which contains different
+ colors_loading allows shapash to load a json file which contains different
palettes of colors that can be used in the plot
Returns
-------
- dict:
+ dict:
contains all available pallets
"""
current_path = os.path.dirname(os.path.abspath(__file__))
jsonfile = os.path.join(current_path, "colors.json")
- with open(jsonfile, 'r') as openfile:
+ with open(jsonfile) as openfile:
colors_dic = json.load(openfile)
return colors_dic
def select_palette(colors_dic, palette_name):
"""
- colors_loading allows shapash to load a json file which contains different
+ colors_loading allows shapash to load a json file which contains different
palettes of colors that can be used in the plot
Parameters
----------
@@ -35,7 +35,7 @@ def select_palette(colors_dic, palette_name):
name of the palette
Returns
-------
- dict:
+ dict:
contains colors of one palette
"""
if palette_name not in colors_dic.keys():
@@ -58,18 +58,18 @@ def convert_str_color_to_plt_format(txt):
>>> convert_str_color_to_plt_format(txt="rgba(244, 192, 0, 1)")
(0.96, 0.75, 0.0, 1.0)
"""
- txt = txt.replace('rgba', '').replace('rgb', '').replace('(', '').replace(')', '')
- list_txt = txt.split(',')
+ txt = txt.replace("rgba", "").replace("rgb", "").replace("(", "").replace(")", "")
+ list_txt = txt.split(",")
if len(list_txt) > 3:
- return [float(list_txt[i])/255 for i in range(3)] + [float(list_txt[3])]
+ return [float(list_txt[i]) / 255 for i in range(3)] + [float(list_txt[3])]
else:
- return [float(x)/255 for x in list_txt]
+ return [float(x) / 255 for x in list_txt]
def define_style(palette):
"""
- the define_style function is a function that uses a palette
- to define the different styles used in the different outputs
+ the define_style function is a function that uses a palette
+ to define the different styles used in the different outputs
of Shapash
Parameters
----------
@@ -77,111 +77,53 @@ def define_style(palette):
contains colors of one palette
Returns
-------
- dict :
+ dict :
contains different style elements
"""
style_dict = dict()
- style_dict['dict_title'] = {
- 'xanchor': "center",
- 'yanchor': "middle",
- 'x': 0.5,
- 'y': 0.9,
- 'font': {
- 'size': 24,
- 'family': "Arial",
- 'color': palette["title_color"]
- }
+ style_dict["dict_title"] = {
+ "xanchor": "center",
+ "yanchor": "middle",
+ "x": 0.5,
+ "y": 0.9,
+ "font": {"size": 24, "family": "Arial", "color": palette["title_color"]},
}
- style_dict['dict_title_stability'] = {
- 'xanchor': "center",
- 'x': 0.5,
+ style_dict["dict_title_stability"] = {
+ "xanchor": "center",
+ "x": 0.5,
"yanchor": "bottom",
"pad": dict(b=50),
- 'font': {
- 'size': 24,
- 'family': "Arial",
- 'color': palette["title_color"]
- }
+ "font": {"size": 24, "family": "Arial", "color": palette["title_color"]},
}
featureimp_bar = convert_string_to_int_keys(palette["featureimp_bar"])
- style_dict['dict_featimp_colors'] = {
- 1: {
- 'color': featureimp_bar[1],
- 'line': {
- 'color': palette["featureimp_line"],
- 'width': 0.5
- }
- },
- 2: {
- 'color': featureimp_bar[2]
- }
- }
- style_dict['featureimp_groups'] = list(palette["featureimp_groups"].values())
- style_dict['init_contrib_colorscale'] = palette["contrib_colorscale"]
- style_dict['violin_area_classif'] = list(palette["violin_area_classif"].values())
- style_dict['prediction_plot'] = list(palette["prediction_plot"].values())
- style_dict['violin_default'] = palette["violin_default"]
- style_dict['dict_title_compacity'] = {
- 'font': {
- 'size': 14,
- 'family': "Arial",
- 'color': palette["title_color"]
- }
- }
- style_dict['dict_xaxis'] = {
- 'font': {
- 'size': 16,
- 'family': "Arial Black",
- 'color': palette["axis_color"]
- }
- }
- style_dict['dict_yaxis'] = {
- 'font': {
- 'size': 16,
- 'family': "Arial Black",
- 'color': palette["axis_color"]
- }
+ style_dict["dict_featimp_colors"] = {
+ 1: {"color": featureimp_bar[1], "line": {"color": palette["featureimp_line"], "width": 0.5}},
+ 2: {"color": featureimp_bar[2]},
}
+ style_dict["featureimp_groups"] = list(palette["featureimp_groups"].values())
+ style_dict["init_contrib_colorscale"] = palette["contrib_colorscale"]
+ style_dict["violin_area_classif"] = list(palette["violin_area_classif"].values())
+ style_dict["prediction_plot"] = list(palette["prediction_plot"].values())
+ style_dict["violin_default"] = palette["violin_default"]
+ style_dict["dict_title_compacity"] = {"font": {"size": 14, "family": "Arial", "color": palette["title_color"]}}
+ style_dict["dict_xaxis"] = {"font": {"size": 16, "family": "Arial Black", "color": palette["axis_color"]}}
+ style_dict["dict_yaxis"] = {"font": {"size": 16, "family": "Arial Black", "color": palette["axis_color"]}}
localplot_bar = convert_string_to_int_keys(palette["localplot_bar"])
localplot_line = convert_string_to_int_keys(palette["localplot_line"])
- style_dict['dict_local_plot_colors'] = {
- 1: {
- 'color': localplot_bar[1],
- 'line': {
- 'color': localplot_line[1],
- 'width': 0.5
- }
- },
- -1: {
- 'color':localplot_bar[-1],
- 'line': {
- 'color': localplot_line[-1],
- 'width': 0.5
- }
- },
- 0: {
- 'color': localplot_bar[0],
- 'line': {
- 'color': localplot_line[0],
- 'width': 0.5
- }
- },
- -2: {
- 'color': localplot_bar[-2],
- 'line': {
- 'color': localplot_line[-2],
- 'width': 0.5
- }
- }
+ style_dict["dict_local_plot_colors"] = {
+ 1: {"color": localplot_bar[1], "line": {"color": localplot_line[1], "width": 0.5}},
+ -1: {"color": localplot_bar[-1], "line": {"color": localplot_line[-1], "width": 0.5}},
+ 0: {"color": localplot_bar[0], "line": {"color": localplot_line[0], "width": 0.5}},
+ -2: {"color": localplot_bar[-2], "line": {"color": localplot_line[-2], "width": 0.5}},
}
- style_dict['dict_compare_colors'] = palette["compare_plot"]
- style_dict['interactions_col_scale'] = palette["interaction_scale"]
- style_dict['interactions_discrete_colors'] = palette["interaction_discrete"]
- style_dict['dict_stability_bar_colors'] = convert_string_to_int_keys(palette["stability_bar"])
- style_dict['dict_compacity_bar_colors'] = convert_string_to_int_keys(palette["compacity_bar"])
- style_dict['webapp_button'] = convert_string_to_int_keys(palette["webapp_button"])
- style_dict['webapp_bkg'] = palette["webapp_bkg"]
- style_dict['webapp_title'] = palette["webapp_title"]
+ style_dict["dict_compare_colors"] = palette["compare_plot"]
+ style_dict["interactions_col_scale"] = palette["interaction_scale"]
+ style_dict["interactions_discrete_colors"] = palette["interaction_discrete"]
+ style_dict["dict_stability_bar_colors"] = convert_string_to_int_keys(palette["stability_bar"])
+ style_dict["dict_compacity_bar_colors"] = convert_string_to_int_keys(palette["compacity_bar"])
+ style_dict["webapp_button"] = convert_string_to_int_keys(palette["webapp_button"])
+ style_dict["webapp_bkg"] = palette["webapp_bkg"]
+ style_dict["webapp_title"] = palette["webapp_title"]
return style_dict
@@ -218,10 +160,7 @@ def get_pyplot_color(colors):
if isinstance(colors, str):
return convert_str_color_to_plt_format(colors)
elif isinstance(colors, dict):
- dict_color_palette = {
- k: convert_str_color_to_plt_format(v)
- for k, v in colors.items()
- }
+ dict_color_palette = {k: convert_str_color_to_plt_format(v) for k, v in colors.items()}
return dict_color_palette
elif isinstance(colors, list):
return [convert_str_color_to_plt_format(v) for v in colors]
diff --git a/shapash/utils/category_encoder_backend.py b/shapash/utils/category_encoder_backend.py
index aea24aae..1ce95c15 100644
--- a/shapash/utils/category_encoder_backend.py
+++ b/shapash/utils/category_encoder_backend.py
@@ -2,9 +2,9 @@
Category_encoder
"""
-import pandas as pd
-import numpy as np
import category_encoders as ce
+import numpy as np
+import pandas as pd
category_encoder_onehot = ""
category_encoder_ordinal = ""
@@ -12,18 +12,17 @@
category_encoder_binary = ""
category_encoder_targetencoder = ""
-dummies_category_encoder = (category_encoder_onehot,
- category_encoder_binary,
- category_encoder_basen)
+dummies_category_encoder = (category_encoder_onehot, category_encoder_binary, category_encoder_basen)
-no_dummies_category_encoder = (category_encoder_ordinal,
- category_encoder_targetencoder)
+no_dummies_category_encoder = (category_encoder_ordinal, category_encoder_targetencoder)
-supported_category_encoder = (category_encoder_onehot,
- category_encoder_binary,
- category_encoder_basen,
- category_encoder_ordinal,
- category_encoder_targetencoder)
+supported_category_encoder = (
+ category_encoder_onehot,
+ category_encoder_binary,
+ category_encoder_basen,
+ category_encoder_ordinal,
+ category_encoder_targetencoder,
+)
def inv_transform_ce(x_in, encoding):
@@ -55,7 +54,7 @@ def inv_transform_ce(x_in, encoding):
rst = inv_transform_ordinal(x, encoding.ordinal_encoder.mapping)
elif str(type(encoding)) == category_encoder_binary:
- if ce.__version__ <= '2.2.2':
+ if ce.__version__ <= "2.2.2":
x = reverse_basen(x_in, encoding.base_n_encoder)
rst = inv_transform_ordinal(x, encoding.base_n_encoder.ordinal_encoder.mapping)
else:
@@ -104,20 +103,22 @@ def inv_transform_target(x_in, enc_target):
The reversed dataframe.
"""
for tgt_enc in enc_target.ordinal_encoder.mapping:
- name_target = tgt_enc.get('col')
+ name_target = tgt_enc.get("col")
mapping_ordinal = enc_target.mapping[name_target]
- mapping_target = tgt_enc.get('mapping')
+ mapping_target = tgt_enc.get("mapping")
reverse_target = pd.Series(mapping_target.index.values, index=mapping_target)
- rst_target = pd.concat([reverse_target, mapping_ordinal], axis=1, join='inner').fillna(value='NaN')
- aggregate = rst_target.groupby(1)[0].apply(lambda x: ' / '.join(map(str, x)))
+ rst_target = pd.concat([reverse_target, mapping_ordinal], axis=1, join="inner").fillna(value="NaN")
+ aggregate = rst_target.groupby(1)[0].apply(lambda x: " / ".join(map(str, x)))
if aggregate.shape[0] != rst_target.shape[0]:
- raise Exception('Multiple label found for the same value in TargetEncoder on col '+str(name_target) + '.')
+ raise Exception("Multiple label found for the same value in TargetEncoder on col " + str(name_target) + ".")
# print("Warning in inverse TargetEncoder - col " + str(name_target) + ": Multiple label for the same value, "
# "each label will be separate using : / ")
- transco = {'col': name_target,
- 'mapping': pd.Series(data=aggregate.index, index=aggregate.values),
- 'data_type': 'object'}
+ transco = {
+ "col": name_target,
+ "mapping": pd.Series(data=aggregate.index, index=aggregate.values),
+ "data_type": "object",
+ }
x_in = inv_transform_ordinal(x_in, [transco])
return x_in
@@ -139,15 +140,15 @@ def inv_transform_ordinal(x_in, encoding):
The reversed dataframe.
"""
for switch in encoding:
- col_name = switch.get('col')
- if not col_name in x_in.columns:
- raise Exception(f'Columns {col_name} not in dataframe.')
- column_mapping = switch.get('mapping')
+ col_name = switch.get("col")
+ if col_name not in x_in.columns:
+ raise Exception(f"Columns {col_name} not in dataframe.")
+ column_mapping = switch.get("mapping")
if isinstance(column_mapping, dict):
inverse = pd.Series(data=column_mapping.keys(), index=column_mapping.values())
else:
inverse = pd.Series(data=column_mapping.index, index=column_mapping.values)
- x_in[col_name] = x_in[col_name].map(inverse).astype(switch.get('data_type'))
+ x_in[col_name] = x_in[col_name].map(inverse).astype(switch.get("data_type"))
return x_in
@@ -170,11 +171,11 @@ def reverse_basen(x_in, encoding):
x = x_in.copy(deep=True)
out_cols = x.columns.values.tolist()
for ind_enc in range(len(encoding.mapping)):
- col_list = encoding.mapping[ind_enc].get('mapping').columns.tolist()
+ col_list = encoding.mapping[ind_enc].get("mapping").columns.tolist()
insert_at = out_cols.index(col_list[0])
if encoding.base == 1:
- value_array = np.array([int(col0.split('_')[-1]) for col0 in col_list])
+ value_array = np.array([int(col0.split("_")[-1]) for col0 in col_list])
else:
len0 = len(col_list)
value_array = np.array([encoding.base ** (len0 - 1 - i) for i in range(len0)])
@@ -205,14 +206,14 @@ def calc_inv_contrib_ce(x_contrib, encoding, agg_columns):
The aggregate contributions depending on which processing is apply.
"""
if str(type(encoding)) in dummies_category_encoder:
- if str(type(encoding)) in category_encoder_binary and ce.__version__ <= '2.2.2':
+ if str(type(encoding)) in category_encoder_binary and ce.__version__ <= "2.2.2":
encoding = encoding.base_n_encoder
drop_col = []
for switch in encoding.mapping:
- col_in = switch.get('col')
- mod = switch.get('mapping').columns.tolist()
+ col_in = switch.get("col")
+ mod = switch.get("mapping").columns.tolist()
insert_at = x_contrib.columns.tolist().index(mod[0])
- if agg_columns == 'first':
+ if agg_columns == "first":
x_contrib.insert(insert_at, col_in, x_contrib[mod[0]])
else:
x_contrib.insert(insert_at, col_in, x_contrib[mod].sum(axis=1))
@@ -240,9 +241,13 @@ def transform_ce(x_in, encoding):
pandas.Dataframe
The dataset preprocessed with the given encoding.
"""
- encoder = [category_encoder_ordinal, category_encoder_onehot,
- category_encoder_basen, category_encoder_binary,
- category_encoder_targetencoder]
+ encoder = [
+ category_encoder_ordinal,
+ category_encoder_onehot,
+ category_encoder_basen,
+ category_encoder_binary,
+ category_encoder_targetencoder,
+ ]
if str(type(encoding)) in encoder:
rst = encoding.transform(x_in)
@@ -273,15 +278,15 @@ def transform_ordinal(x_in, encoding):
The dataframe preprocessed.
"""
for switch in encoding:
- col_name = switch.get('col')
- if not col_name in x_in.columns:
- raise Exception(f'Columns {col_name} not in dataframe.')
- column_mapping = switch.get('mapping')
+ col_name = switch.get("col")
+ if col_name not in x_in.columns:
+ raise Exception(f"Columns {col_name} not in dataframe.")
+ column_mapping = switch.get("mapping")
if isinstance(column_mapping, dict):
transform = pd.Series(data=column_mapping.values(), index=column_mapping.keys())
else:
transform = pd.Series(data=column_mapping.values, index=column_mapping.index)
- x_in[col_name] = x_in[col_name].map(transform).astype(switch.get('mapping').values.dtype)
+ x_in[col_name] = x_in[col_name].map(transform).astype(switch.get("mapping").values.dtype)
return x_in
@@ -299,11 +304,15 @@ def get_col_mapping_ce(encoder):
dict_col_mapping : dict
Dict of mapping between dataframe columns before and after encoding.
"""
- if str(type(encoder)) in [category_encoder_ordinal, category_encoder_onehot, category_encoder_basen,
- category_encoder_targetencoder]:
+ if str(type(encoder)) in [
+ category_encoder_ordinal,
+ category_encoder_onehot,
+ category_encoder_basen,
+ category_encoder_targetencoder,
+ ]:
encoder_mapping = encoder.mapping
elif str(type(encoder)) == category_encoder_binary:
- if ce.__version__ <= '2.2.2':
+ if ce.__version__ <= "2.2.2":
encoder_mapping = encoder.base_n_encoder.mapping
else:
encoder_mapping = encoder.mapping
@@ -316,10 +325,10 @@ def get_col_mapping_ce(encoder):
dict_col_mapping[col] = [col]
elif isinstance(encoder_mapping, list):
for col_enc in encoder_mapping:
- if isinstance(col_enc.get('mapping'), pd.DataFrame):
- dict_col_mapping[col_enc.get('col')] = col_enc.get('mapping').columns.to_list()
+ if isinstance(col_enc.get("mapping"), pd.DataFrame):
+ dict_col_mapping[col_enc.get("col")] = col_enc.get("mapping").columns.to_list()
else:
- dict_col_mapping[col_enc.get('col')] = [col_enc.get('col')]
+ dict_col_mapping[col_enc.get("col")] = [col_enc.get("col")]
else:
raise NotImplementedError
return dict_col_mapping
diff --git a/shapash/utils/check.py b/shapash/utils/check.py
index 2dcdb9ed..69892339 100644
--- a/shapash/utils/check.py
+++ b/shapash/utils/check.py
@@ -2,15 +2,25 @@
Check Module
"""
import copy
+
import numpy as np
import pandas as pd
-from shapash.utils.category_encoder_backend import no_dummies_category_encoder, supported_category_encoder,\
- dummies_category_encoder
-from shapash.utils.columntransformer_backend import no_dummies_sklearn, supported_sklearn
+
+from shapash.utils.category_encoder_backend import (
+ dummies_category_encoder,
+ no_dummies_category_encoder,
+ supported_category_encoder,
+)
+from shapash.utils.columntransformer_backend import (
+ columntransformer,
+ get_feature_names,
+ get_list_features_names,
+ no_dummies_sklearn,
+ supported_sklearn,
+)
from shapash.utils.model import extract_features_model
from shapash.utils.model_synoptic import dict_model_feature
-from shapash.utils.transform import preprocessing_tolist, check_transformers
-from shapash.utils.columntransformer_backend import columntransformer, get_feature_names, get_list_features_names
+from shapash.utils.transform import check_transformers, preprocessing_tolist
def check_preprocessing(preprocessing=None):
@@ -27,6 +37,7 @@ def check_preprocessing(preprocessing=None):
use_ct, use_ce = check_transformers(list_preprocessing)
return use_ct, use_ce
+
def check_model(model):
"""
Check if model has a predict_proba method is a one column dataframe of integer or float
@@ -43,25 +54,25 @@ def check_model(model):
'regression' or 'classification' according to the attributes of the model
"""
_classes = None
- if hasattr(model, 'predict'):
- if hasattr(model, 'predict_proba') or \
- any(hasattr(model, attrib) for attrib in ['classes_', '_classes']):
- if hasattr(model, '_classes'): _classes = model._classes
- if hasattr(model, 'classes_'): _classes = model.classes_
- if isinstance(_classes, np.ndarray): _classes = _classes.tolist()
- if hasattr(model, 'predict_proba') and _classes == []: _classes = [0, 1] # catboost binary
- if hasattr(model, 'predict_proba') and _classes is None:
- raise ValueError(
- "No attribute _classes, classification model not supported"
- )
+ if hasattr(model, "predict"):
+ if hasattr(model, "predict_proba") or any(hasattr(model, attrib) for attrib in ["classes_", "_classes"]):
+ if hasattr(model, "_classes"):
+ _classes = model._classes
+ if hasattr(model, "classes_"):
+ _classes = model.classes_
+ if isinstance(_classes, np.ndarray):
+ _classes = _classes.tolist()
+ if hasattr(model, "predict_proba") and _classes == []:
+ _classes = [0, 1] # catboost binary
+ if hasattr(model, "predict_proba") and _classes is None:
+ raise ValueError("No attribute _classes, classification model not supported")
if _classes not in (None, []):
- return 'classification', _classes
+ return "classification", _classes
else:
- return 'regression', None
+ return "regression", None
else:
- raise ValueError(
- "No method predict in the specified model. Please, check model parameter"
- )
+ raise ValueError("No method predict in the specified model. Please, check model parameter")
+
def check_label_dict(label_dict, case, classes=None):
"""
@@ -76,14 +87,15 @@ def check_label_dict(label_dict, case, classes=None):
classes: list, None
List of labels if the model used is for classification problem, None otherwise.
"""
- if label_dict is not None and case == 'classification':
+ if label_dict is not None and case == "classification":
if set(classes) != set(list(label_dict.keys())):
raise ValueError(
- "label_dict and don't match: \n" +
- f"label_dict keys: {str(list(label_dict.keys()))}\n" +
- f"Classes model values {str(classes)}"
+ "label_dict and don't match: \n"
+ + f"label_dict keys: {str(list(label_dict.keys()))}\n"
+ + f"Classes model values {str(classes)}"
)
+
def check_mask_params(mask_params):
"""
Check if mask_params given respect the expected format.
@@ -96,24 +108,24 @@ def check_mask_params(mask_params):
if not isinstance(mask_params, dict):
raise ValueError(
"""
- mask_params must be a dict
+ mask_params must be a dict
"""
)
else:
conform_arguments = ["features_to_hide", "threshold", "positive", "max_contrib"]
- mask_arguments_not_conform = [argument for argument in mask_params.keys()
- if argument not in conform_arguments]
+ mask_arguments_not_conform = [argument for argument in mask_params.keys() if argument not in conform_arguments]
if len(mask_arguments_not_conform) != 0:
raise ValueError(
- """
+ """
mask_params must only have the following key arguments:
-feature_to_hide
-threshold
-positive
- -max_contrib
+ -max_contrib
"""
)
+
def check_y(x=None, y=None, y_name="y_target"):
"""
Check that ypred given has the right shape and expected value.
@@ -135,16 +147,17 @@ def check_y(x=None, y=None, y_name="y_target"):
if isinstance(y, pd.DataFrame):
if y.shape[1] > 1:
raise ValueError(f"{y_name} must be a one column pd.Dataframe or pd.Series.")
- if (y.dtypes.iloc[0] not in [float, int, np.int32, np.float32, np.int64, np.float64]):
+ if y.dtypes.iloc[0] not in [float, int, np.int32, np.float32, np.int64, np.float64]:
raise ValueError(f"{y_name} must contain int or float only")
if isinstance(y, pd.Series):
- if (y.dtype not in [float, int, np.int32, np.float32, np.int64, np.float64]):
+ if y.dtype not in [float, int, np.int32, np.float32, np.int64, np.float64]:
raise ValueError(f"{y_name} must contain int or float only")
y = y.to_frame()
if isinstance(y.columns[0], (int, float)):
y.columns = [y_name]
return y
+
def check_contribution_object(case, classes, contributions):
"""
Check len of list if _case is "classification"
@@ -162,9 +175,9 @@ def check_contribution_object(case, classes, contributions):
if case == "regression" and isinstance(contributions, (np.ndarray, pd.DataFrame)) == False:
raise ValueError(
"""
- Type of contributions parameter specified is not compatible with
+ Type of contributions parameter specified is not compatible with
regression model.
- Please check model and contributions parameters.
+ Please check model and contributions parameters.
"""
)
elif case == "classification":
@@ -180,15 +193,24 @@ def check_contribution_object(case, classes, contributions):
else:
raise ValueError(
"""
- Type of contributions parameter specified is not compatible with
+ Type of contributions parameter specified is not compatible with
classification model.
Please check model and contributions parameters.
"""
)
-def check_consistency_model_features(features_dict, model, columns_dict, features_types,
- mask_params=None, preprocessing=None, postprocessing=None,
- list_preprocessing=None, features_groups=None):
+
+def check_consistency_model_features(
+ features_dict,
+ model,
+ columns_dict,
+ features_types,
+ mask_params=None,
+ preprocessing=None,
+ postprocessing=None,
+ list_preprocessing=None,
+ features_groups=None,
+):
"""
Check the matching between attributes, features names are same, or include
@@ -229,8 +251,8 @@ def check_consistency_model_features(features_dict, model, columns_dict, feature
raise ValueError("features of features_types and columns_dict must be the same")
if mask_params is not None:
- if mask_params['features_to_hide'] is not None:
- if not all(feature in set(features_types) for feature in mask_params['features_to_hide']):
+ if mask_params["features_to_hide"] is not None:
+ if not all(feature in set(features_types) for feature in mask_params["features_to_hide"]):
raise ValueError("All features of mask_params must be in model")
if preprocessing is not None and str(type(preprocessing)) in (supported_category_encoder):
@@ -252,22 +274,26 @@ def check_consistency_model_features(features_dict, model, columns_dict, feature
if set(columns_dict_feature) != set(feature_expected_model):
raise ValueError("Features of columns_dict and model must be the same.")
else:
- if len(set(columns_dict.values())) != model_expected :
+ if len(set(columns_dict.values())) != model_expected:
raise ValueError("Features of columns_dict and model must have the same length")
if str(type(preprocessing)) in supported_category_encoder and isinstance(feature_expected_model, list):
if set(preprocessing.feature_names_out_) != set(feature_expected_model):
- raise ValueError("""
+ raise ValueError(
+ """
One of features returned by the Category_Encoders preprocessing doesn't
match the model's expected features.
- """)
+ """
+ )
elif preprocessing is not None:
feature_encoded = get_list_features_names(list_preprocessing, columns_dict)
if model_expected != len(feature_encoded):
- raise ValueError("""
+ raise ValueError(
+ """
Number of features returned by the preprocessing step doesn't
match the model's expected features.
- """)
+ """
+ )
if postprocessing:
if not isinstance(postprocessing, dict):
@@ -279,6 +305,7 @@ def check_consistency_model_features(features_dict, model, columns_dict, feature
raise ValueError("Postprocessing and columns_dict must have the same features names.")
check_postprocessing(features_types, postprocessing)
+
def check_preprocessing_options(columns_dict, features_dict, preprocessing=None, list_preprocessing=None):
"""
Check if preprocessing for ColumnTransformer doesn't have "drop" option otherwise compute several
@@ -309,8 +336,7 @@ def check_preprocessing_options(columns_dict, features_dict, preprocessing=None,
if len(feature_to_drop) != 0:
feature_to_drop = [columns_dict[index] for index in feature_to_drop]
- features_dict_op = {key: value for key, value in features_dict.items()
- if key not in feature_to_drop}
+ features_dict_op = {key: value for key, value in features_dict.items() if key not in feature_to_drop}
i = 0
columns_dict_op = dict()
@@ -319,13 +345,16 @@ def check_preprocessing_options(columns_dict, features_dict, preprocessing=None,
columns_dict_op[i] = value
i += 1
- return {"features_to_drop": feature_to_drop,
- "features_dict_op": features_dict_op,
- "columns_dict_op": columns_dict_op}
+ return {
+ "features_to_drop": feature_to_drop,
+ "features_dict_op": features_dict_op,
+ "columns_dict_op": columns_dict_op,
+ }
else:
return None
+
def check_consistency_model_label(columns_dict, label_dict=None):
"""
Check the matching between attributes, features names are same, or include
@@ -342,6 +371,7 @@ def check_consistency_model_label(columns_dict, label_dict=None):
if not all(feat in columns_dict for feat in label_dict):
raise ValueError("All features of label_dict must be in model")
+
def check_postprocessing(x, postprocessing=None):
"""
Check that postprocessing parameter has good attributes matching with x dataset or with dict of types of
@@ -359,21 +389,22 @@ def check_postprocessing(x, postprocessing=None):
raise ValueError("Postprocessing parameter must be a dictionnary")
for key in postprocessing.keys():
-
dict_post = postprocessing[key]
if not isinstance(dict_post, dict):
raise ValueError(f"{key} values must be a dict")
- if not list(dict_post.keys()) == ['type', 'rule']:
+ if list(dict_post.keys()) != ["type", "rule"]:
raise ValueError("Wrong postprocessing keys, you need 'type' and 'rule' keys")
- if not dict_post['type'] in ['prefix', 'suffix', 'transcoding', 'regex', 'case']:
- raise ValueError("Wrong postprocessing method. \n"
- "The available methods are: 'prefix', 'suffix', 'transcoding', 'regex', or 'case'")
+ if dict_post["type"] not in ["prefix", "suffix", "transcoding", "regex", "case"]:
+ raise ValueError(
+ "Wrong postprocessing method. \n"
+ "The available methods are: 'prefix', 'suffix', 'transcoding', 'regex', or 'case'"
+ )
- if dict_post['type'] == 'case':
- if dict_post['rule'] not in ['lower', 'upper']:
+ if dict_post["type"] == "case":
+ if dict_post["rule"] not in ["lower", "upper"]:
raise ValueError("Case modification unknown. Available ones are 'lower', 'upper'.")
if isinstance(x, dict):
@@ -383,17 +414,20 @@ def check_postprocessing(x, postprocessing=None):
if not pd.api.types.is_string_dtype(x[key]):
raise ValueError(f"Expected string object to modify with upper/lower method in {key} dict")
- if dict_post['type'] == 'regex':
- if not set(dict_post['rule'].keys()) == {'in', 'out'}:
- raise ValueError(f"Regex modifications for {key} are not possible, the keys in 'rule' dict"
- f" must be 'in' and 'out'.")
- if isinstance(x,dict):
+ if dict_post["type"] == "regex":
+ if set(dict_post["rule"].keys()) != {"in", "out"}:
+ raise ValueError(
+ f"Regex modifications for {key} are not possible, the keys in 'rule' dict"
+ f" must be 'in' and 'out'."
+ )
+ if isinstance(x, dict):
if x[key] != "object":
raise ValueError(f"Expected string object to modify with regex methods in {key} dict")
else:
if not pd.api.types.is_string_dtype(x[key]):
raise ValueError(f"Expected string object to modify with upper/lower method in {key} dict")
+
def check_features_name(columns_dict, features_dict, features):
"""
Convert a list of feature names (string) or features ids into features ids.
@@ -426,9 +460,7 @@ def check_features_name(columns_dict, features_dict, features):
elif inv_columns_dict and all(f in columns_dict.values() for f in features):
features_ids = [inv_columns_dict[f] for f in features]
else:
- raise ValueError(
- 'All features must came from the same dict of features (technical names or domain names).'
- )
+ raise ValueError("All features must came from the same dict of features (technical names or domain names).")
else:
raise ValueError(
@@ -439,6 +471,7 @@ def check_features_name(columns_dict, features_dict, features):
)
return features_ids
+
def check_additional_data(x, additional_data):
if not isinstance(additional_data, pd.DataFrame):
raise ValueError(f"additional_data must be a pd.Dataframe.")
diff --git a/shapash/utils/columntransformer_backend.py b/shapash/utils/columntransformer_backend.py
index cdf83671..e788597b 100644
--- a/shapash/utils/columntransformer_backend.py
+++ b/shapash/utils/columntransformer_backend.py
@@ -2,17 +2,28 @@
sklearn columntransformer
"""
-import pandas as pd
import numpy as np
-from shapash.utils.category_encoder_backend import inv_transform_ordinal
-from shapash.utils.category_encoder_backend import inv_transform_ce
-from shapash.utils.category_encoder_backend import supported_category_encoder
-from shapash.utils.category_encoder_backend import dummies_category_encoder
-from shapash.utils.category_encoder_backend import category_encoder_binary
-from shapash.utils.category_encoder_backend import transform_ordinal, get_col_mapping_ce
-from shapash.utils.model_synoptic import simple_tree_model_sklearn, catboost_model,\
- linear_model, svm_model, xgboost_model, lightgbm_model, dict_model_feature
+import pandas as pd
+
+from shapash.utils.category_encoder_backend import (
+ category_encoder_binary,
+ dummies_category_encoder,
+ get_col_mapping_ce,
+ inv_transform_ce,
+ inv_transform_ordinal,
+ supported_category_encoder,
+ transform_ordinal,
+)
from shapash.utils.model import extract_features_model
+from shapash.utils.model_synoptic import (
+ catboost_model,
+ dict_model_feature,
+ lightgbm_model,
+ linear_model,
+ simple_tree_model_sklearn,
+ svm_model,
+ xgboost_model,
+)
columntransformer = ""
@@ -26,18 +37,17 @@
other_model = xgboost_model + catboost_model + lightgbm_model
-dummies_sklearn = (sklearn_onehot)
+dummies_sklearn = sklearn_onehot
-no_dummies_sklearn = (sklearn_ordinal,
- sklearn_standardscaler,
- sklearn_quantiletransformer,
- sklearn_powertransformer)
+no_dummies_sklearn = (sklearn_ordinal, sklearn_standardscaler, sklearn_quantiletransformer, sklearn_powertransformer)
-supported_sklearn = (sklearn_onehot,
- sklearn_ordinal,
- sklearn_standardscaler,
- sklearn_quantiletransformer,
- sklearn_powertransformer)
+supported_sklearn = (
+ sklearn_onehot,
+ sklearn_ordinal,
+ sklearn_standardscaler,
+ sklearn_quantiletransformer,
+ sklearn_powertransformer,
+)
def inv_transform_ct(x_in, encoding):
@@ -73,30 +83,22 @@ def inv_transform_ct(x_in, encoding):
col_encoding = enc[2]
# For Scikit encoding we use the associated inverse transform method
if str(type(ct_encoding)) in supported_sklearn:
- frame, init = inv_transform_sklearn_in_ct(x_in,
- init,
- name_encoding,
- col_encoding,
- ct_encoding)
+ frame, init = inv_transform_sklearn_in_ct(x_in, init, name_encoding, col_encoding, ct_encoding)
# For category encoding we use the mapping
elif str(type(ct_encoding)) in supported_category_encoder:
- frame, init = inv_transform_ce_in_ct(x_in,
- init,
- name_encoding,
- col_encoding,
- ct_encoding)
+ frame, init = inv_transform_ce_in_ct(x_in, init, name_encoding, col_encoding, ct_encoding)
# columns not encode
- elif name_encoding == 'remainder':
- if ct_encoding == 'passthrough':
+ elif name_encoding == "remainder":
+ if ct_encoding == "passthrough":
nb_col = len(col_encoding)
- frame = x_in.iloc[:, init:init + nb_col]
+ frame = x_in.iloc[:, init : init + nb_col]
else:
frame = pd.DataFrame()
else:
- raise Exception(f'{ct_encoding} is not supported yet.')
+ raise Exception(f"{ct_encoding} is not supported yet.")
rst = pd.concat([rst, frame], axis=1)
@@ -133,10 +135,10 @@ def inv_transform_ce_in_ct(x_in, init, name_encoding, col_encoding, ct_encoding)
init : np.int
Index of the last column use to make the transformation.
"""
- colname_output = [name_encoding + '_' + val for val in col_encoding]
+ colname_output = [name_encoding + "_" + val for val in col_encoding]
colname_input = ct_encoding.get_feature_names_out()
nb_col = len(colname_input)
- x_to_inverse = x_in.iloc[:, init:init + nb_col].copy()
+ x_to_inverse = x_in.iloc[:, init : init + nb_col].copy()
x_to_inverse.columns = colname_input
frame = inv_transform_ce(x_to_inverse, ct_encoding)
frame.columns = colname_output
@@ -168,13 +170,13 @@ def inv_transform_sklearn_in_ct(x_in, init, name_encoding, col_encoding, ct_enco
init : np.int
Index of the last column use to make the transformation.
"""
- colname_output = [name_encoding + '_' + val for val in col_encoding]
+ colname_output = [name_encoding + "_" + val for val in col_encoding]
if str(type(ct_encoding)) in dummies_sklearn:
colname_input = ct_encoding.get_feature_names_out(col_encoding)
nb_col = len(colname_input)
else:
nb_col = len(colname_output)
- x_inverse = ct_encoding.inverse_transform(x_in.iloc[:, init:init + nb_col])
+ x_inverse = ct_encoding.inverse_transform(x_in.iloc[:, init : init + nb_col])
frame = pd.DataFrame(x_inverse, columns=colname_output, index=x_in.index)
init += nb_col
return frame, init
@@ -212,9 +214,9 @@ def calc_inv_contrib_ct(x_contrib, encoding, agg_columns):
ct_encoding = enc[1]
col_encoding = enc[2]
- if str(type(ct_encoding)) in supported_category_encoder+supported_sklearn:
+ if str(type(ct_encoding)) in supported_category_encoder + supported_sklearn:
# We create new columns names depending on the name of the transformers and the name of the column.
- colname_output = [name_encoding + '_' + val for val in col_encoding]
+ colname_output = [name_encoding + "_" + val for val in col_encoding]
# If the processing create multiple columns we find the number of original categories and aggregate
# the contribution.
@@ -224,32 +226,32 @@ def calc_inv_contrib_ct(x_contrib, encoding, agg_columns):
col_origin = ct_encoding.categories_[i_enc]
elif str(type(ct_encoding)) == category_encoder_binary:
try:
- col_origin = ct_encoding.base_n_encoder.mapping[i_enc].get('mapping').columns.tolist()
+ col_origin = ct_encoding.base_n_encoder.mapping[i_enc].get("mapping").columns.tolist()
except:
- col_origin = ct_encoding.mapping[i_enc].get('mapping').columns.tolist()
+ col_origin = ct_encoding.mapping[i_enc].get("mapping").columns.tolist()
else:
- col_origin = ct_encoding.mapping[i_enc].get('mapping').columns.tolist()
+ col_origin = ct_encoding.mapping[i_enc].get("mapping").columns.tolist()
nb_col = len(col_origin)
- if agg_columns == 'first':
+ if agg_columns == "first":
contrib_inverse = x_contrib.iloc[:, init]
else:
- contrib_inverse = x_contrib.iloc[:, init:init + nb_col].sum(axis=1)
- frame = pd.DataFrame(contrib_inverse,
- columns=[colname_output[i_enc]],
- index=contrib_inverse.index)
+ contrib_inverse = x_contrib.iloc[:, init : init + nb_col].sum(axis=1)
+ frame = pd.DataFrame(
+ contrib_inverse, columns=[colname_output[i_enc]], index=contrib_inverse.index
+ )
rst = pd.concat([rst, frame], axis=1)
init += nb_col
else:
nb_col = len(colname_output)
- frame = x_contrib.iloc[:, init:init + nb_col]
+ frame = x_contrib.iloc[:, init : init + nb_col]
frame.columns = colname_output
rst = pd.concat([rst, frame], axis=1)
init += nb_col
- elif name_encoding == 'remainder':
- if ct_encoding == 'passthrough':
+ elif name_encoding == "remainder":
+ if ct_encoding == "passthrough":
nb_col = len(col_encoding)
- frame = x_contrib.iloc[:, init:init + nb_col]
+ frame = x_contrib.iloc[:, init : init + nb_col]
rst = pd.concat([rst, frame], axis=1)
init += nb_col
else:
@@ -286,14 +288,15 @@ def transform_ct(x_in, model, encoding):
if str(type(encoding)) == columntransformer:
# We use inverse tranform from the encoding method base on columns position
if str(type(model)) in sklearn_model:
- rst = pd.DataFrame(encoding.transform(x_in),
- index=x_in.index)
+ rst = pd.DataFrame(encoding.transform(x_in), index=x_in.index)
rst.columns = ["col_" + str(feature) for feature in rst.columns]
elif str(type(model)) in other_model:
- rst = pd.DataFrame(encoding.transform(x_in),
- columns=extract_features_model(model, dict_model_feature[str(type(model))]),
- index=x_in.index)
+ rst = pd.DataFrame(
+ encoding.transform(x_in),
+ columns=extract_features_model(model, dict_model_feature[str(type(model))]),
+ index=x_in.index,
+ )
else:
raise ValueError("Model specified isn't supported by Shapash.")
@@ -327,20 +330,18 @@ def get_names(name, trans, column, column_transformer):
list:
List of returned features when specific transformer is applied.
"""
- if trans == 'drop' or (
- hasattr(column, '__len__') and not len(column)):
+ if trans == "drop" or (hasattr(column, "__len__") and not len(column)):
return []
- if trans == 'passthrough':
- if hasattr(column_transformer, '_df_columns'):
- if ((not isinstance(column, slice))
- and all(isinstance(col, str) for col in column)):
+ if trans == "passthrough":
+ if hasattr(column_transformer, "_df_columns"):
+ if (not isinstance(column, slice)) and all(isinstance(col, str) for col in column):
return column
else:
return column_transformer._df_columns[column]
else:
indices = np.arange(column_transformer._n_features)
- return ['x%d' % i for i in indices[column]]
- if not hasattr(trans, 'get_feature_names_out'):
+ return ["x%d" % i for i in indices[column]]
+ if not hasattr(trans, "get_feature_names_out"):
if column is None:
return []
else:
@@ -408,9 +409,9 @@ def get_feature_out(estimator, feature_in):
"""
Returns estimator features out if it has get_feature_names_out method, else features_in
"""
- if hasattr(estimator, 'get_feature_names_out') and hasattr(estimator, 'categories_'):
+ if hasattr(estimator, "get_feature_names_out") and hasattr(estimator, "categories_"):
return estimator.get_feature_names_out(), estimator.categories_
- elif hasattr(estimator, 'get_feature_names_out'):
+ elif hasattr(estimator, "get_feature_names_out"):
return estimator.get_feature_names_out(), []
else:
return feature_in, []
@@ -435,38 +436,38 @@ def get_col_mapping_ct(encoder, x_encoded):
dict_col_mapping = dict()
idx_encoded = 0
for name, estimator, features in encoder.transformers_:
- if name != 'remainder':
+ if name != "remainder":
if str(type(estimator)) in dummies_sklearn:
features_out, categories_out = get_feature_out(estimator, features)
for i, f_name in enumerate(features):
- dict_col_mapping[name + '_' + f_name] = list()
+ dict_col_mapping[name + "_" + f_name] = list()
for _ in categories_out[i]:
- dict_col_mapping[name + '_' + f_name].append(x_encoded.columns.to_list()[idx_encoded])
+ dict_col_mapping[name + "_" + f_name].append(x_encoded.columns.to_list()[idx_encoded])
idx_encoded += 1
elif str(type(estimator)) in no_dummies_sklearn:
features_out, categories_out = get_feature_out(estimator, features)
for f_name in features_out:
- dict_col_mapping[name + '_' + f_name] = [x_encoded.columns.to_list()[idx_encoded]]
+ dict_col_mapping[name + "_" + f_name] = [x_encoded.columns.to_list()[idx_encoded]]
idx_encoded += 1
elif str(type(estimator)) in supported_category_encoder:
dict_mapping_ce = get_col_mapping_ce(estimator)
for f_name in dict_mapping_ce.keys():
- dict_col_mapping[name + '_' + f_name] = list()
+ dict_col_mapping[name + "_" + f_name] = list()
for _ in dict_mapping_ce[f_name]:
- dict_col_mapping[name + '_' + f_name].append(x_encoded.columns.to_list()[idx_encoded])
+ dict_col_mapping[name + "_" + f_name].append(x_encoded.columns.to_list()[idx_encoded])
idx_encoded += 1
else:
- raise NotImplementedError(f'Estimator not supported : {estimator}')
+ raise NotImplementedError(f"Estimator not supported : {estimator}")
- elif estimator == 'passthrough':
+ elif estimator == "passthrough":
try:
features_out = encoder.feature_names_in_[features]
except:
- features_out = encoder._feature_names_in[features] #for oldest sklearn version
+ features_out = encoder._feature_names_in[features] # for oldest sklearn version
for f_name in features_out:
dict_col_mapping[f_name] = [x_encoded.columns.to_list()[idx_encoded]]
idx_encoded += 1
diff --git a/shapash/utils/explanation_metrics.py b/shapash/utils/explanation_metrics.py
index 3e8d6bfe..04c18880 100644
--- a/shapash/utils/explanation_metrics.py
+++ b/shapash/utils/explanation_metrics.py
@@ -41,7 +41,7 @@ def _compute_distance(x1, x2, mean_vector, epsilon=0.0000001):
Returns
-------
diff : float
- Returns :math:`\\sum(\\frac{|x1-x2|}{mean\_vector+epsilon})`
+ Returns :math:`\\sum(\\frac{|x1-x2|}{mean\\_vector+epsilon})`
"""
diff = np.sum(np.abs(x1 - x2) / (mean_vector + epsilon))
return diff
@@ -109,7 +109,7 @@ def _get_radius(dataset, n_neighbors, sample_size=500, percentile=95):
similarity_distance[i, j] = dist
similarity_distance[j, i] = dist
# Select top n_neighbors
- ordered_X = np.sort(similarity_distance)[:, 1: n_neighbors + 1]
+ ordered_X = np.sort(similarity_distance)[:, 1 : n_neighbors + 1]
# Select the value of the distance that captures XX% of all distances (percentile)
return np.percentile(ordered_X.flatten(), percentile)
@@ -186,6 +186,7 @@ def find_neighbors(selection, dataset, model, mode, n_neighbors=10):
all_neighbors[i] = neighbors[neighbors[:, -2] < radius]
return all_neighbors
+
def shap_neighbors(instance, x_encoded, contributions, mode):
"""
For an instance and corresponding neighbors, calculate various
@@ -211,8 +212,11 @@ def shap_neighbors(instance, x_encoded, contributions, mode):
"""
# Extract SHAP values for instance and neighbors
# :-2 indicates that two columns are disregarded : distance to instance and model output
- ind = pd.merge(x_encoded.reset_index(), pd.DataFrame(instance[:, :-2], columns=x_encoded.columns), how='inner')\
- .set_index(x_encoded.index.name if x_encoded.index.name is not None else 'index').index
+ ind = (
+ pd.merge(x_encoded.reset_index(), pd.DataFrame(instance[:, :-2], columns=x_encoded.columns), how="inner")
+ .set_index(x_encoded.index.name if x_encoded.index.name is not None else "index")
+ .index
+ )
# If classification, select contrbutions of one class only
if mode == "classification" and len(contributions) == 2:
contributions = contributions[1]
@@ -223,12 +227,16 @@ def shap_neighbors(instance, x_encoded, contributions, mode):
norm_abs_shap_values = normalize(np.abs(shap_values), axis=1, norm="l1")
# Compute the average difference between the instance and its neighbors
# And replace NaN with 0
- average_diff = np.divide(norm_shap_values.std(axis=0), norm_abs_shap_values.mean(axis=0),
- out=np.zeros(norm_abs_shap_values.shape[1]),
- where=norm_abs_shap_values.mean(axis=0) != 0)
+ average_diff = np.divide(
+ norm_shap_values.std(axis=0),
+ norm_abs_shap_values.mean(axis=0),
+ out=np.zeros(norm_abs_shap_values.shape[1]),
+ where=norm_abs_shap_values.mean(axis=0) != 0,
+ )
return norm_shap_values, average_diff, norm_abs_shap_values[0, :]
+
def get_min_nb_features(selection, contributions, mode, distance):
"""
Determine the minimum number of features needed for the prediction \
@@ -237,13 +245,13 @@ def get_min_nb_features(selection, contributions, mode, distance):
The closeness is defined via the following distances:
- * For regression:
+ * For regression:
.. math::
-
+
distance = \\frac{|output_{allFeatures} - output_{currentFeatures}|}{|output_{allFeatures}|}
- * For classification:
+ * For classification:
.. math::
@@ -311,9 +319,9 @@ def get_distance(selection, contributions, mode, nb_features):
-------
distance : array
List of distances for each instance by using top selected features (ex: np.array([0.12, 0.16...])).
-
+
* For regression:
-
+
* normalized distance between the output of current model and output of full model
* For classifciation:
diff --git a/shapash/utils/io.py b/shapash/utils/io.py
index 51168c75..3f359290 100644
--- a/shapash/utils/io.py
+++ b/shapash/utils/io.py
@@ -2,8 +2,10 @@
IO module
"""
import pickle
+
try:
import yaml
+
_is_yaml_available = True
except (ImportError, ModuleNotFoundError):
_is_yaml_available = False
@@ -87,7 +89,7 @@ def load_yml(path):
"""
)
- with open(path, "r") as f:
+ with open(path) as f:
d = yaml.full_load(f)
return d
diff --git a/shapash/utils/load_smartpredictor.py b/shapash/utils/load_smartpredictor.py
index f0d5f99d..13946079 100644
--- a/shapash/utils/load_smartpredictor.py
+++ b/shapash/utils/load_smartpredictor.py
@@ -22,6 +22,4 @@ def load_smartpredictor(path):
if isinstance(predictor, SmartPredictor):
return predictor
else:
- raise ValueError(
- f"{predictor} is not an instance of type SmartPredictor"
- )
+ raise ValueError(f"{predictor} is not an instance of type SmartPredictor")
diff --git a/shapash/utils/model.py b/shapash/utils/model.py
index b3699caa..173837f8 100644
--- a/shapash/utils/model.py
+++ b/shapash/utils/model.py
@@ -2,8 +2,10 @@
Model Module
"""
from inspect import ismethod
+
import pandas as pd
+
def extract_features_model(model, model_attribute):
"""
Extract features of models if it's possible,
@@ -14,7 +16,7 @@ def extract_features_model(model, model_attribute):
model_attribute: String or List
if model can give features, attributes to access features, if not 'length'
"""
- if model_attribute[0] == 'length':
+ if model_attribute[0] == "length":
return model.n_features_in_
else:
if ismethod(getattr(model, model_attribute[0])):
@@ -45,11 +47,10 @@ def predict_proba(model, x_encoded, classes):
pandas.DataFrame
dataset of predicted proba for each label.
"""
- if hasattr(model, 'predict_proba'):
+ if hasattr(model, "predict_proba"):
proba_values = pd.DataFrame(
- model.predict_proba(x_encoded),
- columns=['class_' + str(x) for x in classes],
- index=x_encoded.index)
+ model.predict_proba(x_encoded), columns=["class_" + str(x) for x in classes], index=x_encoded.index
+ )
else:
raise ValueError("model has no predict_proba method")
@@ -72,8 +73,8 @@ def predict(model, x_encoded):
pandas.DataFrame
1-column dataframe containing the predictions.
"""
- if hasattr(model, 'predict'):
- y_pred = pd.DataFrame(model.predict(x_encoded), columns=['pred'], index=x_encoded.index)
+ if hasattr(model, "predict"):
+ y_pred = pd.DataFrame(model.predict(x_encoded), columns=["pred"], index=x_encoded.index)
else:
raise ValueError("model has no predict method")
@@ -100,10 +101,10 @@ def predict_error(y_target, y_pred, case):
1-column dataframe containing the prediction errors.
"""
prediction_error = None
- if y_target is not None and y_pred is not None and case=="regression":
+ if y_target is not None and y_pred is not None and case == "regression":
if (y_target == 0).any()[0]:
- prediction_error = abs(y_target.values-y_pred.values)
+ prediction_error = abs(y_target.values - y_pred.values)
else:
- prediction_error = abs((y_target.values-y_pred.values)/y_target.values)
+ prediction_error = abs((y_target.values - y_pred.values) / y_target.values)
prediction_error = pd.DataFrame(prediction_error, index=y_target.index, columns=["_error_"])
return prediction_error
diff --git a/shapash/utils/model_synoptic.py b/shapash/utils/model_synoptic.py
index ff8a24e1..a3f44d2f 100644
--- a/shapash/utils/model_synoptic.py
+++ b/shapash/utils/model_synoptic.py
@@ -3,55 +3,53 @@
"""
simple_tree_model_sklearn = (
- "",
- "",
- "",
- "",
- "",
- ""
- )
+ "",
+ "",
+ "",
+ "",
+ "",
+ "",
+)
xgboost_model = (
"",
"",
- "")
+ "",
+)
lightgbm_model = (
"",
"",
- ""
+ "",
)
-catboost_model = (
- "",
- "")
+catboost_model = ("", "")
linear_model = (
"",
- "")
+ "",
+)
-svm_model = (
- "",
- "")
+svm_model = ("", "")
simple_tree_model = simple_tree_model_sklearn + xgboost_model + lightgbm_model
-dict_model_feature = {"": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ["booster_","feature_name"],
- "": ["booster_","feature_name"],
- "": ["feature_names"],
- "": ["get_booster","feature_names"],
- "": ["get_booster","feature_names"],
- "": ["feature_names"],
- "": ["feature_names_"],
- "": ["feature_names_"],
- }
-
+dict_model_feature = {
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["booster_", "feature_name"],
+ "": ["booster_", "feature_name"],
+ "": ["feature_names"],
+ "": ["get_booster", "feature_names"],
+ "": ["get_booster", "feature_names"],
+ "": ["feature_names"],
+ "": ["feature_names_"],
+ "": ["feature_names_"],
+}
diff --git a/shapash/utils/threading.py b/shapash/utils/threading.py
index d1c836d1..33065587 100644
--- a/shapash/utils/threading.py
+++ b/shapash/utils/threading.py
@@ -4,6 +4,7 @@
import sys
import threading
+
class CustomThread(threading.Thread):
"""
Python ovveride threading class
@@ -13,6 +14,7 @@ class CustomThread(threading.Thread):
threading : threading.Thread
Thread which you want to instanciate
"""
+
def __init__(self, *args, **keywords):
threading.Thread.__init__(self, *args, **keywords)
self.killed = False
@@ -23,7 +25,6 @@ def start(self):
self.run = self.__run
threading.Thread.start(self)
-
def __run(self):
sys.settrace(self.globaltrace)
self.__run_backup()
@@ -33,7 +34,7 @@ def globaltrace(self, frame, event, arg):
"""
Track the global trace
"""
- if event == 'call':
+ if event == "call":
return self.localtrace
else:
return None
@@ -43,7 +44,7 @@ def localtrace(self, frame, event, arg):
Track the local trace
"""
if self.killed:
- if event == 'line':
+ if event == "line":
raise SystemExit()
return self.localtrace
diff --git a/shapash/utils/transform.py b/shapash/utils/transform.py
index f55dfc22..877a044f 100644
--- a/shapash/utils/transform.py
+++ b/shapash/utils/transform.py
@@ -1,27 +1,30 @@
"""
Transform Module
"""
+import re
+
+import numpy as np
+import pandas as pd
+
+from shapash.utils.category_encoder_backend import (
+ get_col_mapping_ce,
+ inv_transform_ce,
+ supported_category_encoder,
+ transform_ce,
+)
from shapash.utils.columntransformer_backend import (
columntransformer,
+ get_col_mapping_ct,
inv_transform_ct,
supported_sklearn,
transform_ct,
- get_col_mapping_ct
)
-from shapash.utils.category_encoder_backend import (
- transform_ce,
- inv_transform_ce,
- supported_category_encoder,
- get_col_mapping_ce
-)
-import re
-import numpy as np
-import pandas as pd
# TODO
# encode targeted variable ? from sklearn.preprocessing import LabelEncoder
# make an easy version for dict, not writing all mapping
+
def inverse_transform(x_init, preprocessing=None):
"""
Reverse transformation giving a preprocessing.
@@ -72,6 +75,7 @@ def inverse_transform(x_init, preprocessing=None):
x_inverse = inv_transform_ce(x_inverse, encoding)
return x_inverse
+
def apply_preprocessing(x_init, model, preprocessing=None):
"""
Apply preprocessing on a raw dataset giving a preprocessing.
@@ -120,6 +124,7 @@ def apply_preprocessing(x_init, model, preprocessing=None):
x_init = transform_ce(x_init, encoding)
return x_init
+
def preprocessing_tolist(preprocess):
"""
Transform preprocess into a list, if preprocess contains a dict, transform the dict into a list of dict.
@@ -138,6 +143,7 @@ def preprocessing_tolist(preprocess):
list_encoding = [[x] if isinstance(x, dict) else x for x in list_encoding]
return list_encoding
+
def check_transformers(list_encoding):
"""
Check that all transformation are supported.
@@ -176,9 +182,10 @@ def check_transformers(list_encoding):
use_ct = True
for encoding in enc.transformers_:
ct_encoding = encoding[1]
- if (str(type(ct_encoding)) not in supported_sklearn) \
- and (str(type(ct_encoding)) not in supported_category_encoder):
- if str(type(ct_encoding)) != "" :
+ if (str(type(ct_encoding)) not in supported_sklearn) and (
+ str(type(ct_encoding)) not in supported_category_encoder
+ ):
+ if str(type(ct_encoding)) != "":
raise ValueError("One of the encoders used in ColumnTransformers isn't supported.")
elif str(type(enc)) in supported_category_encoder:
@@ -189,32 +196,32 @@ def check_transformers(list_encoding):
for enc_dict in enc:
if isinstance(enc_dict, dict):
# Check dict structure : col - mapping - data_type
- if not all(struct in enc_dict for struct in ('col', 'mapping', 'data_type')):
- raise Exception(f'{enc_dict} should have col, mapping and data_type as keys.')
+ if not all(struct in enc_dict for struct in ("col", "mapping", "data_type")):
+ raise Exception(f"{enc_dict} should have col, mapping and data_type as keys.")
else:
- raise Exception(f'{enc} is not a list of dict.')
+ raise Exception(f"{enc} is not a list of dict.")
else:
- raise Exception(f'{enc} is not supported yet.')
+ raise Exception(f"{enc} is not supported yet.")
# check that encoding don't use ColumnTransformer and Category encoding at the same time
if use_ct and use_ce:
raise Exception(
f"Can't support ColumnTransformer and Category encoding at the same time. "
- f"Use Category encoding in ColumnTransformer")
+ f"Use Category encoding in ColumnTransformer"
+ )
# check that Category encoding is apply on different columns
col = []
for enc in list_encoding:
- if not str(type(enc)) in ("",
- "",
- columntransformer):
+ if not str(type(enc)) in ("", "", columntransformer):
col += enc.cols
- duplicate = set([x for x in col if col.count(x) > 1])
+ duplicate = {x for x in col if col.count(x) > 1}
if duplicate:
- raise Exception('Columns ' + str(duplicate) + ' is used in multiple category encoding')
+ raise Exception("Columns " + str(duplicate) + " is used in multiple category encoding")
return use_ct, use_ce
+
def apply_postprocessing(x_init, postprocessing):
"""
Transforms x_init depending on postprocessing parameters.
@@ -237,36 +244,38 @@ def apply_postprocessing(x_init, postprocessing):
data_modif = new_preds[feature_name]
new_datai = list()
- if dict_postprocessing['type'] == 'prefix':
+ if dict_postprocessing["type"] == "prefix":
for value in data_modif.values:
- new_datai.append(dict_postprocessing['rule'] + str(value))
+ new_datai.append(dict_postprocessing["rule"] + str(value))
new_preds[feature_name] = new_datai
- elif dict_postprocessing['type'] == 'suffix':
+ elif dict_postprocessing["type"] == "suffix":
for value in data_modif.values:
- new_datai.append(str(value) + dict_postprocessing['rule'])
+ new_datai.append(str(value) + dict_postprocessing["rule"])
new_preds[feature_name] = new_datai
- elif dict_postprocessing['type'] == 'transcoding':
+ elif dict_postprocessing["type"] == "transcoding":
unique_values = x_init[feature_name].unique().tolist()
- unique_values = [value for value in unique_values if value not in dict_postprocessing['rule'].keys()]
+ unique_values = [value for value in unique_values if value not in dict_postprocessing["rule"].keys()]
for value in unique_values:
- dict_postprocessing['rule'][value] = value
- new_preds[feature_name] = new_preds[feature_name].map(dict_postprocessing['rule'])
+ dict_postprocessing["rule"][value] = value
+ new_preds[feature_name] = new_preds[feature_name].map(dict_postprocessing["rule"])
- elif dict_postprocessing['type'] == 'regex':
+ elif dict_postprocessing["type"] == "regex":
new_preds[feature_name] = new_preds[feature_name].apply(
- lambda x: re.sub(dict_postprocessing["rule"]['in'], dict_postprocessing["rule"]['out'], x))
+ lambda x: re.sub(dict_postprocessing["rule"]["in"], dict_postprocessing["rule"]["out"], x)
+ )
- elif dict_postprocessing['type'] == 'case':
- if dict_postprocessing['rule'] == 'lower':
+ elif dict_postprocessing["type"] == "case":
+ if dict_postprocessing["rule"] == "lower":
new_preds[feature_name] = new_preds[feature_name].apply(lambda x: x.lower())
- elif dict_postprocessing['rule'] == 'upper':
+ elif dict_postprocessing["rule"] == "upper":
new_preds[feature_name] = new_preds[feature_name].apply(lambda x: x.upper())
return new_preds
-def adapt_contributions(case,contributions):
+
+def adapt_contributions(case, contributions):
"""
If _case is "classification" and contributions a np.array or pd.DataFrame
this function transform contributions matrix in a list of 2 contributions
@@ -288,12 +297,8 @@ def adapt_contributions(case,contributions):
# So that we have the following format : [contributions_class_0, contributions_class_1, ...]
if isinstance(contributions, np.ndarray) and contributions.ndim == 3:
contributions = [contributions[:, :, i] for i in range(contributions.shape[-1])]
- if (
- (isinstance(contributions, pd.DataFrame) and case == 'classification')
- or (
- isinstance(contributions, (np.ndarray, list))
- and case == 'classification'
- and np.array(contributions).ndim == 2)
+ if (isinstance(contributions, pd.DataFrame) and case == "classification") or (
+ isinstance(contributions, (np.ndarray, list)) and case == "classification" and np.array(contributions).ndim == 2
):
return [contributions * -1, contributions]
else:
@@ -374,7 +379,8 @@ def get_features_transform_mapping(x_init, x_encoded, preprocessing=None):
dict_all_cols_mapping[col_name] = [col_name]
return dict_all_cols_mapping
-def handle_categorical_missing(df : pd.DataFrame)-> pd.DataFrame:
+
+def handle_categorical_missing(df: pd.DataFrame) -> pd.DataFrame:
"""
Replace missing values for categorical columns
@@ -383,7 +389,7 @@ def handle_categorical_missing(df : pd.DataFrame)-> pd.DataFrame:
df : pd.DataFrame
Pandas dataframe on which we will replace the missing values
"""
- categorical_cols = df.select_dtypes(include=['object']).columns
+ categorical_cols = df.select_dtypes(include=["object"]).columns
df_handle_missing = df.copy()
df_handle_missing[categorical_cols] = df_handle_missing[categorical_cols].fillna("missing")
return df_handle_missing
diff --git a/shapash/utils/translate.py b/shapash/utils/translate.py
index 776eac57..f5afeaf3 100644
--- a/shapash/utils/translate.py
+++ b/shapash/utils/translate.py
@@ -1,6 +1,8 @@
"""
Translate Module
"""
+
+
def translate(elements, mapping):
"""
Map a dictionary to a list of elements.
diff --git a/shapash/utils/utils.py b/shapash/utils/utils.py
index 6df58a4b..0fc4b673 100644
--- a/shapash/utils/utils.py
+++ b/shapash/utils/utils.py
@@ -1,12 +1,15 @@
"""
Utils is a group of function for the library
"""
+import math
+import socket
+
import numpy as np
import pandas as pd
-import socket
-import math
-from shapash.explainer.smart_state import SmartState
+
from shapash.explainer.multi_decorator import MultiDecorator
+from shapash.explainer.smart_state import SmartState
+
def get_host_name():
"""
@@ -68,6 +71,7 @@ def is_nested_list(object_param):
"""
return any(isinstance(elem, list) for elem in object_param)
+
def add_line_break(text, nbchar, maxlen=150):
"""
adding line break in string if necessary
@@ -86,7 +90,7 @@ def add_line_break(text, nbchar, maxlen=150):
string
original text + line break
"""
- if isinstance(text,str):
+ if isinstance(text, str):
length = 0
tot_length = 0
input_word = text.split()
@@ -97,20 +101,21 @@ def add_line_break(text, nbchar, maxlen=150):
if tot_length <= maxlen:
if length >= nbchar:
length = 0
- final_sep.append('
' % s
@@ -136,4 +140,3 @@ def html_str_df_and_image(df: pd.DataFrame, fig: plt.Figure) -> str:
def print_figure(fig):
print_html(convert_fig_to_html(fig))
-
diff --git a/shapash/style/colors.json b/shapash/style/colors.json
index 513d9922..32999b6e 100644
--- a/shapash/style/colors.json
+++ b/shapash/style/colors.json
@@ -1,200 +1,200 @@
{
- "default": {
- "title_color": "rgb(50, 50, 50)",
- "axis_color": "rgb(50, 50, 50)",
- "featureimp_bar": {
- "1": "rgba(244, 192, 0, 1.0)",
- "2": "rgba(52, 55, 54, 0.7)"
- },
- "featureimp_line": "rgba(52, 55, 54, 0.8)",
- "featureimp_groups": {
- "0": "rgb(245, 133, 24)",
- "1": "rgb(49, 99, 149)"
- },
- "contrib_colorscale": [
- "rgb(52, 55, 54)",
- "rgb(74, 99, 138)",
- "rgb(116, 153, 214)",
- "rgb(162, 188, 213)",
- "rgb(212, 234, 242)",
- "rgb(235, 216, 134)",
- "rgb(255, 204, 83)",
- "rgb(244, 192, 0)",
- "rgb(255, 166, 17)",
- "rgb(255, 123, 38)",
- "rgb(255, 77, 7)"
- ],
- "violin_area_classif": {
- "0": "rgba(117, 152, 189, 0.9)",
- "1": "rgba(255, 166, 17, 0.9)"
- },
- "violin_default": "rgba(117, 152, 189, 0.9)",
- "localplot_bar": {
- "1": "rgba(244, 192, 0, 1.0)",
- "-1": "rgba(74, 99, 138, 0.7)",
- "0": "rgba(113, 101, 59, 1.0)",
- "-2": "rgba(52, 55, 54, 0.7)"
- },
- "localplot_line": {
- "1": "rgba(52, 55, 54, 0.8)",
- "-1": "rgba(27, 28, 28, 1.0)",
- "0": "rgba(52, 55, 54, 0.8)",
- "-2": "rgba(27, 28, 28, 1.0)"
- },
- "compare_plot": [
- "rgba(244, 192, 0, 1.0)",
- "rgba(74, 99, 138, 0.7)",
- "rgba(113, 101, 59, 1.0)",
- "rgba(183, 58, 56, 0.9)",
- "rgba(255, 123, 38, 1.0)",
- "rgba(0, 21, 179, 0.97)",
- "rgba(116, 1, 179, 0.9)"
- ],
- "interaction_scale": [
- "rgb(175, 169, 157)",
- "rgb(255, 255, 255)",
- "rgb(255, 77, 7)"
- ],
- "interaction_discrete": [
- "rgb(133, 92, 117)",
- "rgb(217, 175, 107)",
- "rgb(175, 100, 88)",
- "rgb(115, 111, 76)",
- "rgb(82, 106, 131)",
- "rgb(98, 83, 119)",
- "rgb(104, 133, 92)",
- "rgb(156, 156, 94)",
- "rgb(160, 97, 119)",
- "rgb(140, 120, 93)",
- "rgb(124, 124, 124)"
- ],
- "stability_bar": {
- "1": "rgba(255, 166, 17, 0.9)",
- "0": "rgba(117, 152, 189, 0.9)"
- },
- "compacity_bar": {
- "1": "rgba(255, 166, 17, 0.9)",
- "0": "rgba(117, 152, 189, 0.9)"
- },
- "webapp_button": {
- "0": "rgb(244, 192, 0)",
- "1": "rgb(113, 101, 59)"
- },
- "webapp_bkg": "rgb(52,55,54)",
- "webapp_title": "rgb(244, 192, 0)",
- "report_feature_distribution": {
- "train": "rgba(74, 99, 128, 0.7)",
- "test": "rgba(244, 192, 0, 1)",
- "true": "rgba(74, 99, 138, 0.7)",
- "pred": "rgba(244, 192, 0, 1)"
- },
- "report_confusion_matrix": [
- "rgb(255, 255, 255)",
- "rgb(244, 192, 0)",
- "rgb(255, 166, 17)"
- ],
- "prediction_plot": {
- "1": "rgba(52, 55, 54, 0.9)",
- "0": "rgba(255, 77, 7, 0.9)"
- }
- },
- "blues": {
- "title_color": "rgb(50, 50, 50)",
- "axis_color": "rgb(50, 50, 50)",
- "featureimp_bar": {
- "1": "rgba(0, 154, 203, 1)",
- "2": "rgba(223, 103, 0, 0.8)"
- },
- "featureimp_line": "rgba(52, 55, 54, 0.8)",
- "featureimp_groups": {
- "0": "rgb(10, 204, 143)",
- "1": "rgb(176, 140, 104)"
- },
- "contrib_colorscale": [
- "rgb(168, 84, 0)",
- "rgb(204, 102, 0)",
- "rgb(245, 122, 0)",
- "rgb(240, 150, 67)",
- "rgb(240, 195, 162)",
- "rgb(237, 235, 232)",
- "rgb(161, 221, 254)",
- "rgb(103, 208, 255)",
- "rgb(0, 154, 203)",
- "rgb(0, 98, 128)",
- "rgb(0, 70, 92)"
- ],
- "violin_area_classif": {
- "0": "rgba(223, 103, 0, 0.9)",
- "1": "rgba(0, 154, 203, 0.9)"
- },
- "violin_default": "rgba(0, 98, 128, 0.7)",
- "localplot_bar": {
- "1": "rgba(0, 154, 203, 1)",
- "-1": "rgba(235, 149, 72, 0.9)",
- "0": "rgba(106, 178, 204, 0.8)",
- "-2": "rgba(217, 116, 0, 0.5)"
- },
- "localplot_line": {
- "1": "rgba(52, 55, 54, 0.8)",
- "-1": "rgba(27, 28, 28, 1.0)",
- "0": "rgba(52, 55, 54, 0.8)",
- "-2": "rgba(27, 28, 28, 1.0)"
- },
- "compare_plot": [
- "rgba(0, 154, 203, 1.0)",
- "rgba(223, 103, 0, 0.8)",
- "rgba(11, 60, 217, 1.0)",
- "rgba(204, 50, 10, 0.9)",
- "rgba(11, 217, 152, 1.0)",
- "rgba(129, 64, 0, 0.9)",
- "rgba(0, 98, 128, 0.9)"
- ],
- "interaction_scale": [
- "rgb(175, 169, 157)",
- "rgb(255, 255, 255)",
- "rgb(0, 98, 128)"
- ],
- "interaction_discrete": [
- "rgb(95, 70, 144)",
- "rgb(29, 105, 150)",
- "rgb(56, 166, 165)",
- "rgb(15, 133, 84)",
- "rgb(115, 175, 72)",
- "rgb(237, 173, 8)",
- "rgb(225, 124, 5)",
- "rgb(204, 80, 62)",
- "rgb(148, 52, 110)",
- "rgb(111, 64, 112)",
- "rgb(102, 102, 102)"
- ],
- "stability_bar": {
- "1": "rgba(0, 154, 203, 0.9)",
- "0": "rgba(230, 111, 0, 0.9)"
- },
- "compacity_bar": {
- "1": "rgba(0, 154, 203, 0.9)",
- "0": "rgba(230, 111, 0, 0.9)"
- },
- "webapp_button": {
- "0": "rgb(0, 154, 203)",
- "1": "rgb(50, 90, 117)"
- },
- "webapp_bkg": "rgb(52,55,54)",
- "webapp_title": "rgb(0, 154, 203)",
- "report_feature_distribution": {
- "train": "rgba(0, 154, 203, 0.9)",
- "test": "rgba(230, 111, 0, 0.9)",
- "true": "rgba(0, 154, 203, 0.9)",
- "pred": "rgba(230, 111, 0, 0.9)"
- },
- "report_confusion_matrix": [
- "rgb(255, 255, 255)",
- "rgb(244, 192, 0)",
- "rgb(255, 166, 17)"
- ],
- "prediction_plot": {
- "1": "rgba(168, 84, 0, 0.9)",
- "0": "rgba(0, 70, 92, 0.9)"
- }
- }
-}
\ No newline at end of file
+ "blues": {
+ "axis_color": "rgb(50, 50, 50)",
+ "compacity_bar": {
+ "0": "rgba(230, 111, 0, 0.9)",
+ "1": "rgba(0, 154, 203, 0.9)"
+ },
+ "compare_plot": [
+ "rgba(0, 154, 203, 1.0)",
+ "rgba(223, 103, 0, 0.8)",
+ "rgba(11, 60, 217, 1.0)",
+ "rgba(204, 50, 10, 0.9)",
+ "rgba(11, 217, 152, 1.0)",
+ "rgba(129, 64, 0, 0.9)",
+ "rgba(0, 98, 128, 0.9)"
+ ],
+ "contrib_colorscale": [
+ "rgb(168, 84, 0)",
+ "rgb(204, 102, 0)",
+ "rgb(245, 122, 0)",
+ "rgb(240, 150, 67)",
+ "rgb(240, 195, 162)",
+ "rgb(237, 235, 232)",
+ "rgb(161, 221, 254)",
+ "rgb(103, 208, 255)",
+ "rgb(0, 154, 203)",
+ "rgb(0, 98, 128)",
+ "rgb(0, 70, 92)"
+ ],
+ "featureimp_bar": {
+ "1": "rgba(0, 154, 203, 1)",
+ "2": "rgba(223, 103, 0, 0.8)"
+ },
+ "featureimp_groups": {
+ "0": "rgb(10, 204, 143)",
+ "1": "rgb(176, 140, 104)"
+ },
+ "featureimp_line": "rgba(52, 55, 54, 0.8)",
+ "interaction_discrete": [
+ "rgb(95, 70, 144)",
+ "rgb(29, 105, 150)",
+ "rgb(56, 166, 165)",
+ "rgb(15, 133, 84)",
+ "rgb(115, 175, 72)",
+ "rgb(237, 173, 8)",
+ "rgb(225, 124, 5)",
+ "rgb(204, 80, 62)",
+ "rgb(148, 52, 110)",
+ "rgb(111, 64, 112)",
+ "rgb(102, 102, 102)"
+ ],
+ "interaction_scale": [
+ "rgb(175, 169, 157)",
+ "rgb(255, 255, 255)",
+ "rgb(0, 98, 128)"
+ ],
+ "localplot_bar": {
+ "-1": "rgba(235, 149, 72, 0.9)",
+ "-2": "rgba(217, 116, 0, 0.5)",
+ "0": "rgba(106, 178, 204, 0.8)",
+ "1": "rgba(0, 154, 203, 1)"
+ },
+ "localplot_line": {
+ "-1": "rgba(27, 28, 28, 1.0)",
+ "-2": "rgba(27, 28, 28, 1.0)",
+ "0": "rgba(52, 55, 54, 0.8)",
+ "1": "rgba(52, 55, 54, 0.8)"
+ },
+ "prediction_plot": {
+ "0": "rgba(0, 70, 92, 0.9)",
+ "1": "rgba(168, 84, 0, 0.9)"
+ },
+ "report_confusion_matrix": [
+ "rgb(255, 255, 255)",
+ "rgb(244, 192, 0)",
+ "rgb(255, 166, 17)"
+ ],
+ "report_feature_distribution": {
+ "pred": "rgba(230, 111, 0, 0.9)",
+ "test": "rgba(230, 111, 0, 0.9)",
+ "train": "rgba(0, 154, 203, 0.9)",
+ "true": "rgba(0, 154, 203, 0.9)"
+ },
+ "stability_bar": {
+ "0": "rgba(230, 111, 0, 0.9)",
+ "1": "rgba(0, 154, 203, 0.9)"
+ },
+ "title_color": "rgb(50, 50, 50)",
+ "violin_area_classif": {
+ "0": "rgba(223, 103, 0, 0.9)",
+ "1": "rgba(0, 154, 203, 0.9)"
+ },
+ "violin_default": "rgba(0, 98, 128, 0.7)",
+ "webapp_bkg": "rgb(52,55,54)",
+ "webapp_button": {
+ "0": "rgb(0, 154, 203)",
+ "1": "rgb(50, 90, 117)"
+ },
+ "webapp_title": "rgb(0, 154, 203)"
+ },
+ "default": {
+ "axis_color": "rgb(50, 50, 50)",
+ "compacity_bar": {
+ "0": "rgba(117, 152, 189, 0.9)",
+ "1": "rgba(255, 166, 17, 0.9)"
+ },
+ "compare_plot": [
+ "rgba(244, 192, 0, 1.0)",
+ "rgba(74, 99, 138, 0.7)",
+ "rgba(113, 101, 59, 1.0)",
+ "rgba(183, 58, 56, 0.9)",
+ "rgba(255, 123, 38, 1.0)",
+ "rgba(0, 21, 179, 0.97)",
+ "rgba(116, 1, 179, 0.9)"
+ ],
+ "contrib_colorscale": [
+ "rgb(52, 55, 54)",
+ "rgb(74, 99, 138)",
+ "rgb(116, 153, 214)",
+ "rgb(162, 188, 213)",
+ "rgb(212, 234, 242)",
+ "rgb(235, 216, 134)",
+ "rgb(255, 204, 83)",
+ "rgb(244, 192, 0)",
+ "rgb(255, 166, 17)",
+ "rgb(255, 123, 38)",
+ "rgb(255, 77, 7)"
+ ],
+ "featureimp_bar": {
+ "1": "rgba(244, 192, 0, 1.0)",
+ "2": "rgba(52, 55, 54, 0.7)"
+ },
+ "featureimp_groups": {
+ "0": "rgb(245, 133, 24)",
+ "1": "rgb(49, 99, 149)"
+ },
+ "featureimp_line": "rgba(52, 55, 54, 0.8)",
+ "interaction_discrete": [
+ "rgb(133, 92, 117)",
+ "rgb(217, 175, 107)",
+ "rgb(175, 100, 88)",
+ "rgb(115, 111, 76)",
+ "rgb(82, 106, 131)",
+ "rgb(98, 83, 119)",
+ "rgb(104, 133, 92)",
+ "rgb(156, 156, 94)",
+ "rgb(160, 97, 119)",
+ "rgb(140, 120, 93)",
+ "rgb(124, 124, 124)"
+ ],
+ "interaction_scale": [
+ "rgb(175, 169, 157)",
+ "rgb(255, 255, 255)",
+ "rgb(255, 77, 7)"
+ ],
+ "localplot_bar": {
+ "-1": "rgba(74, 99, 138, 0.7)",
+ "-2": "rgba(52, 55, 54, 0.7)",
+ "0": "rgba(113, 101, 59, 1.0)",
+ "1": "rgba(244, 192, 0, 1.0)"
+ },
+ "localplot_line": {
+ "-1": "rgba(27, 28, 28, 1.0)",
+ "-2": "rgba(27, 28, 28, 1.0)",
+ "0": "rgba(52, 55, 54, 0.8)",
+ "1": "rgba(52, 55, 54, 0.8)"
+ },
+ "prediction_plot": {
+ "0": "rgba(255, 77, 7, 0.9)",
+ "1": "rgba(52, 55, 54, 0.9)"
+ },
+ "report_confusion_matrix": [
+ "rgb(255, 255, 255)",
+ "rgb(244, 192, 0)",
+ "rgb(255, 166, 17)"
+ ],
+ "report_feature_distribution": {
+ "pred": "rgba(244, 192, 0, 1)",
+ "test": "rgba(244, 192, 0, 1)",
+ "train": "rgba(74, 99, 128, 0.7)",
+ "true": "rgba(74, 99, 138, 0.7)"
+ },
+ "stability_bar": {
+ "0": "rgba(117, 152, 189, 0.9)",
+ "1": "rgba(255, 166, 17, 0.9)"
+ },
+ "title_color": "rgb(50, 50, 50)",
+ "violin_area_classif": {
+ "0": "rgba(117, 152, 189, 0.9)",
+ "1": "rgba(255, 166, 17, 0.9)"
+ },
+ "violin_default": "rgba(117, 152, 189, 0.9)",
+ "webapp_bkg": "rgb(52,55,54)",
+ "webapp_button": {
+ "0": "rgb(244, 192, 0)",
+ "1": "rgb(113, 101, 59)"
+ },
+ "webapp_title": "rgb(244, 192, 0)"
+ }
+}
diff --git a/shapash/style/style_utils.py b/shapash/style/style_utils.py
index 4c722fec..36dad34c 100644
--- a/shapash/style/style_utils.py
+++ b/shapash/style/style_utils.py
@@ -9,23 +9,23 @@
def colors_loading():
"""
- colors_loading allows shapash to load a json file which contains different
+ colors_loading allows shapash to load a json file which contains different
palettes of colors that can be used in the plot
Returns
-------
- dict:
+ dict:
contains all available pallets
"""
current_path = os.path.dirname(os.path.abspath(__file__))
jsonfile = os.path.join(current_path, "colors.json")
- with open(jsonfile, 'r') as openfile:
+ with open(jsonfile) as openfile:
colors_dic = json.load(openfile)
return colors_dic
def select_palette(colors_dic, palette_name):
"""
- colors_loading allows shapash to load a json file which contains different
+ colors_loading allows shapash to load a json file which contains different
palettes of colors that can be used in the plot
Parameters
----------
@@ -35,7 +35,7 @@ def select_palette(colors_dic, palette_name):
name of the palette
Returns
-------
- dict:
+ dict:
contains colors of one palette
"""
if palette_name not in colors_dic.keys():
@@ -58,18 +58,18 @@ def convert_str_color_to_plt_format(txt):
>>> convert_str_color_to_plt_format(txt="rgba(244, 192, 0, 1)")
(0.96, 0.75, 0.0, 1.0)
"""
- txt = txt.replace('rgba', '').replace('rgb', '').replace('(', '').replace(')', '')
- list_txt = txt.split(',')
+ txt = txt.replace("rgba", "").replace("rgb", "").replace("(", "").replace(")", "")
+ list_txt = txt.split(",")
if len(list_txt) > 3:
- return [float(list_txt[i])/255 for i in range(3)] + [float(list_txt[3])]
+ return [float(list_txt[i]) / 255 for i in range(3)] + [float(list_txt[3])]
else:
- return [float(x)/255 for x in list_txt]
+ return [float(x) / 255 for x in list_txt]
def define_style(palette):
"""
- the define_style function is a function that uses a palette
- to define the different styles used in the different outputs
+ the define_style function is a function that uses a palette
+ to define the different styles used in the different outputs
of Shapash
Parameters
----------
@@ -77,111 +77,53 @@ def define_style(palette):
contains colors of one palette
Returns
-------
- dict :
+ dict :
contains different style elements
"""
style_dict = dict()
- style_dict['dict_title'] = {
- 'xanchor': "center",
- 'yanchor': "middle",
- 'x': 0.5,
- 'y': 0.9,
- 'font': {
- 'size': 24,
- 'family': "Arial",
- 'color': palette["title_color"]
- }
+ style_dict["dict_title"] = {
+ "xanchor": "center",
+ "yanchor": "middle",
+ "x": 0.5,
+ "y": 0.9,
+ "font": {"size": 24, "family": "Arial", "color": palette["title_color"]},
}
- style_dict['dict_title_stability'] = {
- 'xanchor': "center",
- 'x': 0.5,
+ style_dict["dict_title_stability"] = {
+ "xanchor": "center",
+ "x": 0.5,
"yanchor": "bottom",
"pad": dict(b=50),
- 'font': {
- 'size': 24,
- 'family': "Arial",
- 'color': palette["title_color"]
- }
+ "font": {"size": 24, "family": "Arial", "color": palette["title_color"]},
}
featureimp_bar = convert_string_to_int_keys(palette["featureimp_bar"])
- style_dict['dict_featimp_colors'] = {
- 1: {
- 'color': featureimp_bar[1],
- 'line': {
- 'color': palette["featureimp_line"],
- 'width': 0.5
- }
- },
- 2: {
- 'color': featureimp_bar[2]
- }
- }
- style_dict['featureimp_groups'] = list(palette["featureimp_groups"].values())
- style_dict['init_contrib_colorscale'] = palette["contrib_colorscale"]
- style_dict['violin_area_classif'] = list(palette["violin_area_classif"].values())
- style_dict['prediction_plot'] = list(palette["prediction_plot"].values())
- style_dict['violin_default'] = palette["violin_default"]
- style_dict['dict_title_compacity'] = {
- 'font': {
- 'size': 14,
- 'family': "Arial",
- 'color': palette["title_color"]
- }
- }
- style_dict['dict_xaxis'] = {
- 'font': {
- 'size': 16,
- 'family': "Arial Black",
- 'color': palette["axis_color"]
- }
- }
- style_dict['dict_yaxis'] = {
- 'font': {
- 'size': 16,
- 'family': "Arial Black",
- 'color': palette["axis_color"]
- }
+ style_dict["dict_featimp_colors"] = {
+ 1: {"color": featureimp_bar[1], "line": {"color": palette["featureimp_line"], "width": 0.5}},
+ 2: {"color": featureimp_bar[2]},
}
+ style_dict["featureimp_groups"] = list(palette["featureimp_groups"].values())
+ style_dict["init_contrib_colorscale"] = palette["contrib_colorscale"]
+ style_dict["violin_area_classif"] = list(palette["violin_area_classif"].values())
+ style_dict["prediction_plot"] = list(palette["prediction_plot"].values())
+ style_dict["violin_default"] = palette["violin_default"]
+ style_dict["dict_title_compacity"] = {"font": {"size": 14, "family": "Arial", "color": palette["title_color"]}}
+ style_dict["dict_xaxis"] = {"font": {"size": 16, "family": "Arial Black", "color": palette["axis_color"]}}
+ style_dict["dict_yaxis"] = {"font": {"size": 16, "family": "Arial Black", "color": palette["axis_color"]}}
localplot_bar = convert_string_to_int_keys(palette["localplot_bar"])
localplot_line = convert_string_to_int_keys(palette["localplot_line"])
- style_dict['dict_local_plot_colors'] = {
- 1: {
- 'color': localplot_bar[1],
- 'line': {
- 'color': localplot_line[1],
- 'width': 0.5
- }
- },
- -1: {
- 'color':localplot_bar[-1],
- 'line': {
- 'color': localplot_line[-1],
- 'width': 0.5
- }
- },
- 0: {
- 'color': localplot_bar[0],
- 'line': {
- 'color': localplot_line[0],
- 'width': 0.5
- }
- },
- -2: {
- 'color': localplot_bar[-2],
- 'line': {
- 'color': localplot_line[-2],
- 'width': 0.5
- }
- }
+ style_dict["dict_local_plot_colors"] = {
+ 1: {"color": localplot_bar[1], "line": {"color": localplot_line[1], "width": 0.5}},
+ -1: {"color": localplot_bar[-1], "line": {"color": localplot_line[-1], "width": 0.5}},
+ 0: {"color": localplot_bar[0], "line": {"color": localplot_line[0], "width": 0.5}},
+ -2: {"color": localplot_bar[-2], "line": {"color": localplot_line[-2], "width": 0.5}},
}
- style_dict['dict_compare_colors'] = palette["compare_plot"]
- style_dict['interactions_col_scale'] = palette["interaction_scale"]
- style_dict['interactions_discrete_colors'] = palette["interaction_discrete"]
- style_dict['dict_stability_bar_colors'] = convert_string_to_int_keys(palette["stability_bar"])
- style_dict['dict_compacity_bar_colors'] = convert_string_to_int_keys(palette["compacity_bar"])
- style_dict['webapp_button'] = convert_string_to_int_keys(palette["webapp_button"])
- style_dict['webapp_bkg'] = palette["webapp_bkg"]
- style_dict['webapp_title'] = palette["webapp_title"]
+ style_dict["dict_compare_colors"] = palette["compare_plot"]
+ style_dict["interactions_col_scale"] = palette["interaction_scale"]
+ style_dict["interactions_discrete_colors"] = palette["interaction_discrete"]
+ style_dict["dict_stability_bar_colors"] = convert_string_to_int_keys(palette["stability_bar"])
+ style_dict["dict_compacity_bar_colors"] = convert_string_to_int_keys(palette["compacity_bar"])
+ style_dict["webapp_button"] = convert_string_to_int_keys(palette["webapp_button"])
+ style_dict["webapp_bkg"] = palette["webapp_bkg"]
+ style_dict["webapp_title"] = palette["webapp_title"]
return style_dict
@@ -218,10 +160,7 @@ def get_pyplot_color(colors):
if isinstance(colors, str):
return convert_str_color_to_plt_format(colors)
elif isinstance(colors, dict):
- dict_color_palette = {
- k: convert_str_color_to_plt_format(v)
- for k, v in colors.items()
- }
+ dict_color_palette = {k: convert_str_color_to_plt_format(v) for k, v in colors.items()}
return dict_color_palette
elif isinstance(colors, list):
return [convert_str_color_to_plt_format(v) for v in colors]
diff --git a/shapash/utils/category_encoder_backend.py b/shapash/utils/category_encoder_backend.py
index aea24aae..1ce95c15 100644
--- a/shapash/utils/category_encoder_backend.py
+++ b/shapash/utils/category_encoder_backend.py
@@ -2,9 +2,9 @@
Category_encoder
"""
-import pandas as pd
-import numpy as np
import category_encoders as ce
+import numpy as np
+import pandas as pd
category_encoder_onehot = ""
category_encoder_ordinal = ""
@@ -12,18 +12,17 @@
category_encoder_binary = ""
category_encoder_targetencoder = ""
-dummies_category_encoder = (category_encoder_onehot,
- category_encoder_binary,
- category_encoder_basen)
+dummies_category_encoder = (category_encoder_onehot, category_encoder_binary, category_encoder_basen)
-no_dummies_category_encoder = (category_encoder_ordinal,
- category_encoder_targetencoder)
+no_dummies_category_encoder = (category_encoder_ordinal, category_encoder_targetencoder)
-supported_category_encoder = (category_encoder_onehot,
- category_encoder_binary,
- category_encoder_basen,
- category_encoder_ordinal,
- category_encoder_targetencoder)
+supported_category_encoder = (
+ category_encoder_onehot,
+ category_encoder_binary,
+ category_encoder_basen,
+ category_encoder_ordinal,
+ category_encoder_targetencoder,
+)
def inv_transform_ce(x_in, encoding):
@@ -55,7 +54,7 @@ def inv_transform_ce(x_in, encoding):
rst = inv_transform_ordinal(x, encoding.ordinal_encoder.mapping)
elif str(type(encoding)) == category_encoder_binary:
- if ce.__version__ <= '2.2.2':
+ if ce.__version__ <= "2.2.2":
x = reverse_basen(x_in, encoding.base_n_encoder)
rst = inv_transform_ordinal(x, encoding.base_n_encoder.ordinal_encoder.mapping)
else:
@@ -104,20 +103,22 @@ def inv_transform_target(x_in, enc_target):
The reversed dataframe.
"""
for tgt_enc in enc_target.ordinal_encoder.mapping:
- name_target = tgt_enc.get('col')
+ name_target = tgt_enc.get("col")
mapping_ordinal = enc_target.mapping[name_target]
- mapping_target = tgt_enc.get('mapping')
+ mapping_target = tgt_enc.get("mapping")
reverse_target = pd.Series(mapping_target.index.values, index=mapping_target)
- rst_target = pd.concat([reverse_target, mapping_ordinal], axis=1, join='inner').fillna(value='NaN')
- aggregate = rst_target.groupby(1)[0].apply(lambda x: ' / '.join(map(str, x)))
+ rst_target = pd.concat([reverse_target, mapping_ordinal], axis=1, join="inner").fillna(value="NaN")
+ aggregate = rst_target.groupby(1)[0].apply(lambda x: " / ".join(map(str, x)))
if aggregate.shape[0] != rst_target.shape[0]:
- raise Exception('Multiple label found for the same value in TargetEncoder on col '+str(name_target) + '.')
+ raise Exception("Multiple label found for the same value in TargetEncoder on col " + str(name_target) + ".")
# print("Warning in inverse TargetEncoder - col " + str(name_target) + ": Multiple label for the same value, "
# "each label will be separate using : / ")
- transco = {'col': name_target,
- 'mapping': pd.Series(data=aggregate.index, index=aggregate.values),
- 'data_type': 'object'}
+ transco = {
+ "col": name_target,
+ "mapping": pd.Series(data=aggregate.index, index=aggregate.values),
+ "data_type": "object",
+ }
x_in = inv_transform_ordinal(x_in, [transco])
return x_in
@@ -139,15 +140,15 @@ def inv_transform_ordinal(x_in, encoding):
The reversed dataframe.
"""
for switch in encoding:
- col_name = switch.get('col')
- if not col_name in x_in.columns:
- raise Exception(f'Columns {col_name} not in dataframe.')
- column_mapping = switch.get('mapping')
+ col_name = switch.get("col")
+ if col_name not in x_in.columns:
+ raise Exception(f"Columns {col_name} not in dataframe.")
+ column_mapping = switch.get("mapping")
if isinstance(column_mapping, dict):
inverse = pd.Series(data=column_mapping.keys(), index=column_mapping.values())
else:
inverse = pd.Series(data=column_mapping.index, index=column_mapping.values)
- x_in[col_name] = x_in[col_name].map(inverse).astype(switch.get('data_type'))
+ x_in[col_name] = x_in[col_name].map(inverse).astype(switch.get("data_type"))
return x_in
@@ -170,11 +171,11 @@ def reverse_basen(x_in, encoding):
x = x_in.copy(deep=True)
out_cols = x.columns.values.tolist()
for ind_enc in range(len(encoding.mapping)):
- col_list = encoding.mapping[ind_enc].get('mapping').columns.tolist()
+ col_list = encoding.mapping[ind_enc].get("mapping").columns.tolist()
insert_at = out_cols.index(col_list[0])
if encoding.base == 1:
- value_array = np.array([int(col0.split('_')[-1]) for col0 in col_list])
+ value_array = np.array([int(col0.split("_")[-1]) for col0 in col_list])
else:
len0 = len(col_list)
value_array = np.array([encoding.base ** (len0 - 1 - i) for i in range(len0)])
@@ -205,14 +206,14 @@ def calc_inv_contrib_ce(x_contrib, encoding, agg_columns):
The aggregate contributions depending on which processing is apply.
"""
if str(type(encoding)) in dummies_category_encoder:
- if str(type(encoding)) in category_encoder_binary and ce.__version__ <= '2.2.2':
+ if str(type(encoding)) in category_encoder_binary and ce.__version__ <= "2.2.2":
encoding = encoding.base_n_encoder
drop_col = []
for switch in encoding.mapping:
- col_in = switch.get('col')
- mod = switch.get('mapping').columns.tolist()
+ col_in = switch.get("col")
+ mod = switch.get("mapping").columns.tolist()
insert_at = x_contrib.columns.tolist().index(mod[0])
- if agg_columns == 'first':
+ if agg_columns == "first":
x_contrib.insert(insert_at, col_in, x_contrib[mod[0]])
else:
x_contrib.insert(insert_at, col_in, x_contrib[mod].sum(axis=1))
@@ -240,9 +241,13 @@ def transform_ce(x_in, encoding):
pandas.Dataframe
The dataset preprocessed with the given encoding.
"""
- encoder = [category_encoder_ordinal, category_encoder_onehot,
- category_encoder_basen, category_encoder_binary,
- category_encoder_targetencoder]
+ encoder = [
+ category_encoder_ordinal,
+ category_encoder_onehot,
+ category_encoder_basen,
+ category_encoder_binary,
+ category_encoder_targetencoder,
+ ]
if str(type(encoding)) in encoder:
rst = encoding.transform(x_in)
@@ -273,15 +278,15 @@ def transform_ordinal(x_in, encoding):
The dataframe preprocessed.
"""
for switch in encoding:
- col_name = switch.get('col')
- if not col_name in x_in.columns:
- raise Exception(f'Columns {col_name} not in dataframe.')
- column_mapping = switch.get('mapping')
+ col_name = switch.get("col")
+ if col_name not in x_in.columns:
+ raise Exception(f"Columns {col_name} not in dataframe.")
+ column_mapping = switch.get("mapping")
if isinstance(column_mapping, dict):
transform = pd.Series(data=column_mapping.values(), index=column_mapping.keys())
else:
transform = pd.Series(data=column_mapping.values, index=column_mapping.index)
- x_in[col_name] = x_in[col_name].map(transform).astype(switch.get('mapping').values.dtype)
+ x_in[col_name] = x_in[col_name].map(transform).astype(switch.get("mapping").values.dtype)
return x_in
@@ -299,11 +304,15 @@ def get_col_mapping_ce(encoder):
dict_col_mapping : dict
Dict of mapping between dataframe columns before and after encoding.
"""
- if str(type(encoder)) in [category_encoder_ordinal, category_encoder_onehot, category_encoder_basen,
- category_encoder_targetencoder]:
+ if str(type(encoder)) in [
+ category_encoder_ordinal,
+ category_encoder_onehot,
+ category_encoder_basen,
+ category_encoder_targetencoder,
+ ]:
encoder_mapping = encoder.mapping
elif str(type(encoder)) == category_encoder_binary:
- if ce.__version__ <= '2.2.2':
+ if ce.__version__ <= "2.2.2":
encoder_mapping = encoder.base_n_encoder.mapping
else:
encoder_mapping = encoder.mapping
@@ -316,10 +325,10 @@ def get_col_mapping_ce(encoder):
dict_col_mapping[col] = [col]
elif isinstance(encoder_mapping, list):
for col_enc in encoder_mapping:
- if isinstance(col_enc.get('mapping'), pd.DataFrame):
- dict_col_mapping[col_enc.get('col')] = col_enc.get('mapping').columns.to_list()
+ if isinstance(col_enc.get("mapping"), pd.DataFrame):
+ dict_col_mapping[col_enc.get("col")] = col_enc.get("mapping").columns.to_list()
else:
- dict_col_mapping[col_enc.get('col')] = [col_enc.get('col')]
+ dict_col_mapping[col_enc.get("col")] = [col_enc.get("col")]
else:
raise NotImplementedError
return dict_col_mapping
diff --git a/shapash/utils/check.py b/shapash/utils/check.py
index 2dcdb9ed..69892339 100644
--- a/shapash/utils/check.py
+++ b/shapash/utils/check.py
@@ -2,15 +2,25 @@
Check Module
"""
import copy
+
import numpy as np
import pandas as pd
-from shapash.utils.category_encoder_backend import no_dummies_category_encoder, supported_category_encoder,\
- dummies_category_encoder
-from shapash.utils.columntransformer_backend import no_dummies_sklearn, supported_sklearn
+
+from shapash.utils.category_encoder_backend import (
+ dummies_category_encoder,
+ no_dummies_category_encoder,
+ supported_category_encoder,
+)
+from shapash.utils.columntransformer_backend import (
+ columntransformer,
+ get_feature_names,
+ get_list_features_names,
+ no_dummies_sklearn,
+ supported_sklearn,
+)
from shapash.utils.model import extract_features_model
from shapash.utils.model_synoptic import dict_model_feature
-from shapash.utils.transform import preprocessing_tolist, check_transformers
-from shapash.utils.columntransformer_backend import columntransformer, get_feature_names, get_list_features_names
+from shapash.utils.transform import check_transformers, preprocessing_tolist
def check_preprocessing(preprocessing=None):
@@ -27,6 +37,7 @@ def check_preprocessing(preprocessing=None):
use_ct, use_ce = check_transformers(list_preprocessing)
return use_ct, use_ce
+
def check_model(model):
"""
Check if model has a predict_proba method is a one column dataframe of integer or float
@@ -43,25 +54,25 @@ def check_model(model):
'regression' or 'classification' according to the attributes of the model
"""
_classes = None
- if hasattr(model, 'predict'):
- if hasattr(model, 'predict_proba') or \
- any(hasattr(model, attrib) for attrib in ['classes_', '_classes']):
- if hasattr(model, '_classes'): _classes = model._classes
- if hasattr(model, 'classes_'): _classes = model.classes_
- if isinstance(_classes, np.ndarray): _classes = _classes.tolist()
- if hasattr(model, 'predict_proba') and _classes == []: _classes = [0, 1] # catboost binary
- if hasattr(model, 'predict_proba') and _classes is None:
- raise ValueError(
- "No attribute _classes, classification model not supported"
- )
+ if hasattr(model, "predict"):
+ if hasattr(model, "predict_proba") or any(hasattr(model, attrib) for attrib in ["classes_", "_classes"]):
+ if hasattr(model, "_classes"):
+ _classes = model._classes
+ if hasattr(model, "classes_"):
+ _classes = model.classes_
+ if isinstance(_classes, np.ndarray):
+ _classes = _classes.tolist()
+ if hasattr(model, "predict_proba") and _classes == []:
+ _classes = [0, 1] # catboost binary
+ if hasattr(model, "predict_proba") and _classes is None:
+ raise ValueError("No attribute _classes, classification model not supported")
if _classes not in (None, []):
- return 'classification', _classes
+ return "classification", _classes
else:
- return 'regression', None
+ return "regression", None
else:
- raise ValueError(
- "No method predict in the specified model. Please, check model parameter"
- )
+ raise ValueError("No method predict in the specified model. Please, check model parameter")
+
def check_label_dict(label_dict, case, classes=None):
"""
@@ -76,14 +87,15 @@ def check_label_dict(label_dict, case, classes=None):
classes: list, None
List of labels if the model used is for classification problem, None otherwise.
"""
- if label_dict is not None and case == 'classification':
+ if label_dict is not None and case == "classification":
if set(classes) != set(list(label_dict.keys())):
raise ValueError(
- "label_dict and don't match: \n" +
- f"label_dict keys: {str(list(label_dict.keys()))}\n" +
- f"Classes model values {str(classes)}"
+ "label_dict and don't match: \n"
+ + f"label_dict keys: {str(list(label_dict.keys()))}\n"
+ + f"Classes model values {str(classes)}"
)
+
def check_mask_params(mask_params):
"""
Check if mask_params given respect the expected format.
@@ -96,24 +108,24 @@ def check_mask_params(mask_params):
if not isinstance(mask_params, dict):
raise ValueError(
"""
- mask_params must be a dict
+ mask_params must be a dict
"""
)
else:
conform_arguments = ["features_to_hide", "threshold", "positive", "max_contrib"]
- mask_arguments_not_conform = [argument for argument in mask_params.keys()
- if argument not in conform_arguments]
+ mask_arguments_not_conform = [argument for argument in mask_params.keys() if argument not in conform_arguments]
if len(mask_arguments_not_conform) != 0:
raise ValueError(
- """
+ """
mask_params must only have the following key arguments:
-feature_to_hide
-threshold
-positive
- -max_contrib
+ -max_contrib
"""
)
+
def check_y(x=None, y=None, y_name="y_target"):
"""
Check that ypred given has the right shape and expected value.
@@ -135,16 +147,17 @@ def check_y(x=None, y=None, y_name="y_target"):
if isinstance(y, pd.DataFrame):
if y.shape[1] > 1:
raise ValueError(f"{y_name} must be a one column pd.Dataframe or pd.Series.")
- if (y.dtypes.iloc[0] not in [float, int, np.int32, np.float32, np.int64, np.float64]):
+ if y.dtypes.iloc[0] not in [float, int, np.int32, np.float32, np.int64, np.float64]:
raise ValueError(f"{y_name} must contain int or float only")
if isinstance(y, pd.Series):
- if (y.dtype not in [float, int, np.int32, np.float32, np.int64, np.float64]):
+ if y.dtype not in [float, int, np.int32, np.float32, np.int64, np.float64]:
raise ValueError(f"{y_name} must contain int or float only")
y = y.to_frame()
if isinstance(y.columns[0], (int, float)):
y.columns = [y_name]
return y
+
def check_contribution_object(case, classes, contributions):
"""
Check len of list if _case is "classification"
@@ -162,9 +175,9 @@ def check_contribution_object(case, classes, contributions):
if case == "regression" and isinstance(contributions, (np.ndarray, pd.DataFrame)) == False:
raise ValueError(
"""
- Type of contributions parameter specified is not compatible with
+ Type of contributions parameter specified is not compatible with
regression model.
- Please check model and contributions parameters.
+ Please check model and contributions parameters.
"""
)
elif case == "classification":
@@ -180,15 +193,24 @@ def check_contribution_object(case, classes, contributions):
else:
raise ValueError(
"""
- Type of contributions parameter specified is not compatible with
+ Type of contributions parameter specified is not compatible with
classification model.
Please check model and contributions parameters.
"""
)
-def check_consistency_model_features(features_dict, model, columns_dict, features_types,
- mask_params=None, preprocessing=None, postprocessing=None,
- list_preprocessing=None, features_groups=None):
+
+def check_consistency_model_features(
+ features_dict,
+ model,
+ columns_dict,
+ features_types,
+ mask_params=None,
+ preprocessing=None,
+ postprocessing=None,
+ list_preprocessing=None,
+ features_groups=None,
+):
"""
Check the matching between attributes, features names are same, or include
@@ -229,8 +251,8 @@ def check_consistency_model_features(features_dict, model, columns_dict, feature
raise ValueError("features of features_types and columns_dict must be the same")
if mask_params is not None:
- if mask_params['features_to_hide'] is not None:
- if not all(feature in set(features_types) for feature in mask_params['features_to_hide']):
+ if mask_params["features_to_hide"] is not None:
+ if not all(feature in set(features_types) for feature in mask_params["features_to_hide"]):
raise ValueError("All features of mask_params must be in model")
if preprocessing is not None and str(type(preprocessing)) in (supported_category_encoder):
@@ -252,22 +274,26 @@ def check_consistency_model_features(features_dict, model, columns_dict, feature
if set(columns_dict_feature) != set(feature_expected_model):
raise ValueError("Features of columns_dict and model must be the same.")
else:
- if len(set(columns_dict.values())) != model_expected :
+ if len(set(columns_dict.values())) != model_expected:
raise ValueError("Features of columns_dict and model must have the same length")
if str(type(preprocessing)) in supported_category_encoder and isinstance(feature_expected_model, list):
if set(preprocessing.feature_names_out_) != set(feature_expected_model):
- raise ValueError("""
+ raise ValueError(
+ """
One of features returned by the Category_Encoders preprocessing doesn't
match the model's expected features.
- """)
+ """
+ )
elif preprocessing is not None:
feature_encoded = get_list_features_names(list_preprocessing, columns_dict)
if model_expected != len(feature_encoded):
- raise ValueError("""
+ raise ValueError(
+ """
Number of features returned by the preprocessing step doesn't
match the model's expected features.
- """)
+ """
+ )
if postprocessing:
if not isinstance(postprocessing, dict):
@@ -279,6 +305,7 @@ def check_consistency_model_features(features_dict, model, columns_dict, feature
raise ValueError("Postprocessing and columns_dict must have the same features names.")
check_postprocessing(features_types, postprocessing)
+
def check_preprocessing_options(columns_dict, features_dict, preprocessing=None, list_preprocessing=None):
"""
Check if preprocessing for ColumnTransformer doesn't have "drop" option otherwise compute several
@@ -309,8 +336,7 @@ def check_preprocessing_options(columns_dict, features_dict, preprocessing=None,
if len(feature_to_drop) != 0:
feature_to_drop = [columns_dict[index] for index in feature_to_drop]
- features_dict_op = {key: value for key, value in features_dict.items()
- if key not in feature_to_drop}
+ features_dict_op = {key: value for key, value in features_dict.items() if key not in feature_to_drop}
i = 0
columns_dict_op = dict()
@@ -319,13 +345,16 @@ def check_preprocessing_options(columns_dict, features_dict, preprocessing=None,
columns_dict_op[i] = value
i += 1
- return {"features_to_drop": feature_to_drop,
- "features_dict_op": features_dict_op,
- "columns_dict_op": columns_dict_op}
+ return {
+ "features_to_drop": feature_to_drop,
+ "features_dict_op": features_dict_op,
+ "columns_dict_op": columns_dict_op,
+ }
else:
return None
+
def check_consistency_model_label(columns_dict, label_dict=None):
"""
Check the matching between attributes, features names are same, or include
@@ -342,6 +371,7 @@ def check_consistency_model_label(columns_dict, label_dict=None):
if not all(feat in columns_dict for feat in label_dict):
raise ValueError("All features of label_dict must be in model")
+
def check_postprocessing(x, postprocessing=None):
"""
Check that postprocessing parameter has good attributes matching with x dataset or with dict of types of
@@ -359,21 +389,22 @@ def check_postprocessing(x, postprocessing=None):
raise ValueError("Postprocessing parameter must be a dictionnary")
for key in postprocessing.keys():
-
dict_post = postprocessing[key]
if not isinstance(dict_post, dict):
raise ValueError(f"{key} values must be a dict")
- if not list(dict_post.keys()) == ['type', 'rule']:
+ if list(dict_post.keys()) != ["type", "rule"]:
raise ValueError("Wrong postprocessing keys, you need 'type' and 'rule' keys")
- if not dict_post['type'] in ['prefix', 'suffix', 'transcoding', 'regex', 'case']:
- raise ValueError("Wrong postprocessing method. \n"
- "The available methods are: 'prefix', 'suffix', 'transcoding', 'regex', or 'case'")
+ if dict_post["type"] not in ["prefix", "suffix", "transcoding", "regex", "case"]:
+ raise ValueError(
+ "Wrong postprocessing method. \n"
+ "The available methods are: 'prefix', 'suffix', 'transcoding', 'regex', or 'case'"
+ )
- if dict_post['type'] == 'case':
- if dict_post['rule'] not in ['lower', 'upper']:
+ if dict_post["type"] == "case":
+ if dict_post["rule"] not in ["lower", "upper"]:
raise ValueError("Case modification unknown. Available ones are 'lower', 'upper'.")
if isinstance(x, dict):
@@ -383,17 +414,20 @@ def check_postprocessing(x, postprocessing=None):
if not pd.api.types.is_string_dtype(x[key]):
raise ValueError(f"Expected string object to modify with upper/lower method in {key} dict")
- if dict_post['type'] == 'regex':
- if not set(dict_post['rule'].keys()) == {'in', 'out'}:
- raise ValueError(f"Regex modifications for {key} are not possible, the keys in 'rule' dict"
- f" must be 'in' and 'out'.")
- if isinstance(x,dict):
+ if dict_post["type"] == "regex":
+ if set(dict_post["rule"].keys()) != {"in", "out"}:
+ raise ValueError(
+ f"Regex modifications for {key} are not possible, the keys in 'rule' dict"
+ f" must be 'in' and 'out'."
+ )
+ if isinstance(x, dict):
if x[key] != "object":
raise ValueError(f"Expected string object to modify with regex methods in {key} dict")
else:
if not pd.api.types.is_string_dtype(x[key]):
raise ValueError(f"Expected string object to modify with upper/lower method in {key} dict")
+
def check_features_name(columns_dict, features_dict, features):
"""
Convert a list of feature names (string) or features ids into features ids.
@@ -426,9 +460,7 @@ def check_features_name(columns_dict, features_dict, features):
elif inv_columns_dict and all(f in columns_dict.values() for f in features):
features_ids = [inv_columns_dict[f] for f in features]
else:
- raise ValueError(
- 'All features must came from the same dict of features (technical names or domain names).'
- )
+ raise ValueError("All features must came from the same dict of features (technical names or domain names).")
else:
raise ValueError(
@@ -439,6 +471,7 @@ def check_features_name(columns_dict, features_dict, features):
)
return features_ids
+
def check_additional_data(x, additional_data):
if not isinstance(additional_data, pd.DataFrame):
raise ValueError(f"additional_data must be a pd.Dataframe.")
diff --git a/shapash/utils/columntransformer_backend.py b/shapash/utils/columntransformer_backend.py
index cdf83671..e788597b 100644
--- a/shapash/utils/columntransformer_backend.py
+++ b/shapash/utils/columntransformer_backend.py
@@ -2,17 +2,28 @@
sklearn columntransformer
"""
-import pandas as pd
import numpy as np
-from shapash.utils.category_encoder_backend import inv_transform_ordinal
-from shapash.utils.category_encoder_backend import inv_transform_ce
-from shapash.utils.category_encoder_backend import supported_category_encoder
-from shapash.utils.category_encoder_backend import dummies_category_encoder
-from shapash.utils.category_encoder_backend import category_encoder_binary
-from shapash.utils.category_encoder_backend import transform_ordinal, get_col_mapping_ce
-from shapash.utils.model_synoptic import simple_tree_model_sklearn, catboost_model,\
- linear_model, svm_model, xgboost_model, lightgbm_model, dict_model_feature
+import pandas as pd
+
+from shapash.utils.category_encoder_backend import (
+ category_encoder_binary,
+ dummies_category_encoder,
+ get_col_mapping_ce,
+ inv_transform_ce,
+ inv_transform_ordinal,
+ supported_category_encoder,
+ transform_ordinal,
+)
from shapash.utils.model import extract_features_model
+from shapash.utils.model_synoptic import (
+ catboost_model,
+ dict_model_feature,
+ lightgbm_model,
+ linear_model,
+ simple_tree_model_sklearn,
+ svm_model,
+ xgboost_model,
+)
columntransformer = ""
@@ -26,18 +37,17 @@
other_model = xgboost_model + catboost_model + lightgbm_model
-dummies_sklearn = (sklearn_onehot)
+dummies_sklearn = sklearn_onehot
-no_dummies_sklearn = (sklearn_ordinal,
- sklearn_standardscaler,
- sklearn_quantiletransformer,
- sklearn_powertransformer)
+no_dummies_sklearn = (sklearn_ordinal, sklearn_standardscaler, sklearn_quantiletransformer, sklearn_powertransformer)
-supported_sklearn = (sklearn_onehot,
- sklearn_ordinal,
- sklearn_standardscaler,
- sklearn_quantiletransformer,
- sklearn_powertransformer)
+supported_sklearn = (
+ sklearn_onehot,
+ sklearn_ordinal,
+ sklearn_standardscaler,
+ sklearn_quantiletransformer,
+ sklearn_powertransformer,
+)
def inv_transform_ct(x_in, encoding):
@@ -73,30 +83,22 @@ def inv_transform_ct(x_in, encoding):
col_encoding = enc[2]
# For Scikit encoding we use the associated inverse transform method
if str(type(ct_encoding)) in supported_sklearn:
- frame, init = inv_transform_sklearn_in_ct(x_in,
- init,
- name_encoding,
- col_encoding,
- ct_encoding)
+ frame, init = inv_transform_sklearn_in_ct(x_in, init, name_encoding, col_encoding, ct_encoding)
# For category encoding we use the mapping
elif str(type(ct_encoding)) in supported_category_encoder:
- frame, init = inv_transform_ce_in_ct(x_in,
- init,
- name_encoding,
- col_encoding,
- ct_encoding)
+ frame, init = inv_transform_ce_in_ct(x_in, init, name_encoding, col_encoding, ct_encoding)
# columns not encode
- elif name_encoding == 'remainder':
- if ct_encoding == 'passthrough':
+ elif name_encoding == "remainder":
+ if ct_encoding == "passthrough":
nb_col = len(col_encoding)
- frame = x_in.iloc[:, init:init + nb_col]
+ frame = x_in.iloc[:, init : init + nb_col]
else:
frame = pd.DataFrame()
else:
- raise Exception(f'{ct_encoding} is not supported yet.')
+ raise Exception(f"{ct_encoding} is not supported yet.")
rst = pd.concat([rst, frame], axis=1)
@@ -133,10 +135,10 @@ def inv_transform_ce_in_ct(x_in, init, name_encoding, col_encoding, ct_encoding)
init : np.int
Index of the last column use to make the transformation.
"""
- colname_output = [name_encoding + '_' + val for val in col_encoding]
+ colname_output = [name_encoding + "_" + val for val in col_encoding]
colname_input = ct_encoding.get_feature_names_out()
nb_col = len(colname_input)
- x_to_inverse = x_in.iloc[:, init:init + nb_col].copy()
+ x_to_inverse = x_in.iloc[:, init : init + nb_col].copy()
x_to_inverse.columns = colname_input
frame = inv_transform_ce(x_to_inverse, ct_encoding)
frame.columns = colname_output
@@ -168,13 +170,13 @@ def inv_transform_sklearn_in_ct(x_in, init, name_encoding, col_encoding, ct_enco
init : np.int
Index of the last column use to make the transformation.
"""
- colname_output = [name_encoding + '_' + val for val in col_encoding]
+ colname_output = [name_encoding + "_" + val for val in col_encoding]
if str(type(ct_encoding)) in dummies_sklearn:
colname_input = ct_encoding.get_feature_names_out(col_encoding)
nb_col = len(colname_input)
else:
nb_col = len(colname_output)
- x_inverse = ct_encoding.inverse_transform(x_in.iloc[:, init:init + nb_col])
+ x_inverse = ct_encoding.inverse_transform(x_in.iloc[:, init : init + nb_col])
frame = pd.DataFrame(x_inverse, columns=colname_output, index=x_in.index)
init += nb_col
return frame, init
@@ -212,9 +214,9 @@ def calc_inv_contrib_ct(x_contrib, encoding, agg_columns):
ct_encoding = enc[1]
col_encoding = enc[2]
- if str(type(ct_encoding)) in supported_category_encoder+supported_sklearn:
+ if str(type(ct_encoding)) in supported_category_encoder + supported_sklearn:
# We create new columns names depending on the name of the transformers and the name of the column.
- colname_output = [name_encoding + '_' + val for val in col_encoding]
+ colname_output = [name_encoding + "_" + val for val in col_encoding]
# If the processing create multiple columns we find the number of original categories and aggregate
# the contribution.
@@ -224,32 +226,32 @@ def calc_inv_contrib_ct(x_contrib, encoding, agg_columns):
col_origin = ct_encoding.categories_[i_enc]
elif str(type(ct_encoding)) == category_encoder_binary:
try:
- col_origin = ct_encoding.base_n_encoder.mapping[i_enc].get('mapping').columns.tolist()
+ col_origin = ct_encoding.base_n_encoder.mapping[i_enc].get("mapping").columns.tolist()
except:
- col_origin = ct_encoding.mapping[i_enc].get('mapping').columns.tolist()
+ col_origin = ct_encoding.mapping[i_enc].get("mapping").columns.tolist()
else:
- col_origin = ct_encoding.mapping[i_enc].get('mapping').columns.tolist()
+ col_origin = ct_encoding.mapping[i_enc].get("mapping").columns.tolist()
nb_col = len(col_origin)
- if agg_columns == 'first':
+ if agg_columns == "first":
contrib_inverse = x_contrib.iloc[:, init]
else:
- contrib_inverse = x_contrib.iloc[:, init:init + nb_col].sum(axis=1)
- frame = pd.DataFrame(contrib_inverse,
- columns=[colname_output[i_enc]],
- index=contrib_inverse.index)
+ contrib_inverse = x_contrib.iloc[:, init : init + nb_col].sum(axis=1)
+ frame = pd.DataFrame(
+ contrib_inverse, columns=[colname_output[i_enc]], index=contrib_inverse.index
+ )
rst = pd.concat([rst, frame], axis=1)
init += nb_col
else:
nb_col = len(colname_output)
- frame = x_contrib.iloc[:, init:init + nb_col]
+ frame = x_contrib.iloc[:, init : init + nb_col]
frame.columns = colname_output
rst = pd.concat([rst, frame], axis=1)
init += nb_col
- elif name_encoding == 'remainder':
- if ct_encoding == 'passthrough':
+ elif name_encoding == "remainder":
+ if ct_encoding == "passthrough":
nb_col = len(col_encoding)
- frame = x_contrib.iloc[:, init:init + nb_col]
+ frame = x_contrib.iloc[:, init : init + nb_col]
rst = pd.concat([rst, frame], axis=1)
init += nb_col
else:
@@ -286,14 +288,15 @@ def transform_ct(x_in, model, encoding):
if str(type(encoding)) == columntransformer:
# We use inverse tranform from the encoding method base on columns position
if str(type(model)) in sklearn_model:
- rst = pd.DataFrame(encoding.transform(x_in),
- index=x_in.index)
+ rst = pd.DataFrame(encoding.transform(x_in), index=x_in.index)
rst.columns = ["col_" + str(feature) for feature in rst.columns]
elif str(type(model)) in other_model:
- rst = pd.DataFrame(encoding.transform(x_in),
- columns=extract_features_model(model, dict_model_feature[str(type(model))]),
- index=x_in.index)
+ rst = pd.DataFrame(
+ encoding.transform(x_in),
+ columns=extract_features_model(model, dict_model_feature[str(type(model))]),
+ index=x_in.index,
+ )
else:
raise ValueError("Model specified isn't supported by Shapash.")
@@ -327,20 +330,18 @@ def get_names(name, trans, column, column_transformer):
list:
List of returned features when specific transformer is applied.
"""
- if trans == 'drop' or (
- hasattr(column, '__len__') and not len(column)):
+ if trans == "drop" or (hasattr(column, "__len__") and not len(column)):
return []
- if trans == 'passthrough':
- if hasattr(column_transformer, '_df_columns'):
- if ((not isinstance(column, slice))
- and all(isinstance(col, str) for col in column)):
+ if trans == "passthrough":
+ if hasattr(column_transformer, "_df_columns"):
+ if (not isinstance(column, slice)) and all(isinstance(col, str) for col in column):
return column
else:
return column_transformer._df_columns[column]
else:
indices = np.arange(column_transformer._n_features)
- return ['x%d' % i for i in indices[column]]
- if not hasattr(trans, 'get_feature_names_out'):
+ return ["x%d" % i for i in indices[column]]
+ if not hasattr(trans, "get_feature_names_out"):
if column is None:
return []
else:
@@ -408,9 +409,9 @@ def get_feature_out(estimator, feature_in):
"""
Returns estimator features out if it has get_feature_names_out method, else features_in
"""
- if hasattr(estimator, 'get_feature_names_out') and hasattr(estimator, 'categories_'):
+ if hasattr(estimator, "get_feature_names_out") and hasattr(estimator, "categories_"):
return estimator.get_feature_names_out(), estimator.categories_
- elif hasattr(estimator, 'get_feature_names_out'):
+ elif hasattr(estimator, "get_feature_names_out"):
return estimator.get_feature_names_out(), []
else:
return feature_in, []
@@ -435,38 +436,38 @@ def get_col_mapping_ct(encoder, x_encoded):
dict_col_mapping = dict()
idx_encoded = 0
for name, estimator, features in encoder.transformers_:
- if name != 'remainder':
+ if name != "remainder":
if str(type(estimator)) in dummies_sklearn:
features_out, categories_out = get_feature_out(estimator, features)
for i, f_name in enumerate(features):
- dict_col_mapping[name + '_' + f_name] = list()
+ dict_col_mapping[name + "_" + f_name] = list()
for _ in categories_out[i]:
- dict_col_mapping[name + '_' + f_name].append(x_encoded.columns.to_list()[idx_encoded])
+ dict_col_mapping[name + "_" + f_name].append(x_encoded.columns.to_list()[idx_encoded])
idx_encoded += 1
elif str(type(estimator)) in no_dummies_sklearn:
features_out, categories_out = get_feature_out(estimator, features)
for f_name in features_out:
- dict_col_mapping[name + '_' + f_name] = [x_encoded.columns.to_list()[idx_encoded]]
+ dict_col_mapping[name + "_" + f_name] = [x_encoded.columns.to_list()[idx_encoded]]
idx_encoded += 1
elif str(type(estimator)) in supported_category_encoder:
dict_mapping_ce = get_col_mapping_ce(estimator)
for f_name in dict_mapping_ce.keys():
- dict_col_mapping[name + '_' + f_name] = list()
+ dict_col_mapping[name + "_" + f_name] = list()
for _ in dict_mapping_ce[f_name]:
- dict_col_mapping[name + '_' + f_name].append(x_encoded.columns.to_list()[idx_encoded])
+ dict_col_mapping[name + "_" + f_name].append(x_encoded.columns.to_list()[idx_encoded])
idx_encoded += 1
else:
- raise NotImplementedError(f'Estimator not supported : {estimator}')
+ raise NotImplementedError(f"Estimator not supported : {estimator}")
- elif estimator == 'passthrough':
+ elif estimator == "passthrough":
try:
features_out = encoder.feature_names_in_[features]
except:
- features_out = encoder._feature_names_in[features] #for oldest sklearn version
+ features_out = encoder._feature_names_in[features] # for oldest sklearn version
for f_name in features_out:
dict_col_mapping[f_name] = [x_encoded.columns.to_list()[idx_encoded]]
idx_encoded += 1
diff --git a/shapash/utils/explanation_metrics.py b/shapash/utils/explanation_metrics.py
index 3e8d6bfe..04c18880 100644
--- a/shapash/utils/explanation_metrics.py
+++ b/shapash/utils/explanation_metrics.py
@@ -41,7 +41,7 @@ def _compute_distance(x1, x2, mean_vector, epsilon=0.0000001):
Returns
-------
diff : float
- Returns :math:`\\sum(\\frac{|x1-x2|}{mean\_vector+epsilon})`
+ Returns :math:`\\sum(\\frac{|x1-x2|}{mean\\_vector+epsilon})`
"""
diff = np.sum(np.abs(x1 - x2) / (mean_vector + epsilon))
return diff
@@ -109,7 +109,7 @@ def _get_radius(dataset, n_neighbors, sample_size=500, percentile=95):
similarity_distance[i, j] = dist
similarity_distance[j, i] = dist
# Select top n_neighbors
- ordered_X = np.sort(similarity_distance)[:, 1: n_neighbors + 1]
+ ordered_X = np.sort(similarity_distance)[:, 1 : n_neighbors + 1]
# Select the value of the distance that captures XX% of all distances (percentile)
return np.percentile(ordered_X.flatten(), percentile)
@@ -186,6 +186,7 @@ def find_neighbors(selection, dataset, model, mode, n_neighbors=10):
all_neighbors[i] = neighbors[neighbors[:, -2] < radius]
return all_neighbors
+
def shap_neighbors(instance, x_encoded, contributions, mode):
"""
For an instance and corresponding neighbors, calculate various
@@ -211,8 +212,11 @@ def shap_neighbors(instance, x_encoded, contributions, mode):
"""
# Extract SHAP values for instance and neighbors
# :-2 indicates that two columns are disregarded : distance to instance and model output
- ind = pd.merge(x_encoded.reset_index(), pd.DataFrame(instance[:, :-2], columns=x_encoded.columns), how='inner')\
- .set_index(x_encoded.index.name if x_encoded.index.name is not None else 'index').index
+ ind = (
+ pd.merge(x_encoded.reset_index(), pd.DataFrame(instance[:, :-2], columns=x_encoded.columns), how="inner")
+ .set_index(x_encoded.index.name if x_encoded.index.name is not None else "index")
+ .index
+ )
# If classification, select contrbutions of one class only
if mode == "classification" and len(contributions) == 2:
contributions = contributions[1]
@@ -223,12 +227,16 @@ def shap_neighbors(instance, x_encoded, contributions, mode):
norm_abs_shap_values = normalize(np.abs(shap_values), axis=1, norm="l1")
# Compute the average difference between the instance and its neighbors
# And replace NaN with 0
- average_diff = np.divide(norm_shap_values.std(axis=0), norm_abs_shap_values.mean(axis=0),
- out=np.zeros(norm_abs_shap_values.shape[1]),
- where=norm_abs_shap_values.mean(axis=0) != 0)
+ average_diff = np.divide(
+ norm_shap_values.std(axis=0),
+ norm_abs_shap_values.mean(axis=0),
+ out=np.zeros(norm_abs_shap_values.shape[1]),
+ where=norm_abs_shap_values.mean(axis=0) != 0,
+ )
return norm_shap_values, average_diff, norm_abs_shap_values[0, :]
+
def get_min_nb_features(selection, contributions, mode, distance):
"""
Determine the minimum number of features needed for the prediction \
@@ -237,13 +245,13 @@ def get_min_nb_features(selection, contributions, mode, distance):
The closeness is defined via the following distances:
- * For regression:
+ * For regression:
.. math::
-
+
distance = \\frac{|output_{allFeatures} - output_{currentFeatures}|}{|output_{allFeatures}|}
- * For classification:
+ * For classification:
.. math::
@@ -311,9 +319,9 @@ def get_distance(selection, contributions, mode, nb_features):
-------
distance : array
List of distances for each instance by using top selected features (ex: np.array([0.12, 0.16...])).
-
+
* For regression:
-
+
* normalized distance between the output of current model and output of full model
* For classifciation:
diff --git a/shapash/utils/io.py b/shapash/utils/io.py
index 51168c75..3f359290 100644
--- a/shapash/utils/io.py
+++ b/shapash/utils/io.py
@@ -2,8 +2,10 @@
IO module
"""
import pickle
+
try:
import yaml
+
_is_yaml_available = True
except (ImportError, ModuleNotFoundError):
_is_yaml_available = False
@@ -87,7 +89,7 @@ def load_yml(path):
"""
)
- with open(path, "r") as f:
+ with open(path) as f:
d = yaml.full_load(f)
return d
diff --git a/shapash/utils/load_smartpredictor.py b/shapash/utils/load_smartpredictor.py
index f0d5f99d..13946079 100644
--- a/shapash/utils/load_smartpredictor.py
+++ b/shapash/utils/load_smartpredictor.py
@@ -22,6 +22,4 @@ def load_smartpredictor(path):
if isinstance(predictor, SmartPredictor):
return predictor
else:
- raise ValueError(
- f"{predictor} is not an instance of type SmartPredictor"
- )
+ raise ValueError(f"{predictor} is not an instance of type SmartPredictor")
diff --git a/shapash/utils/model.py b/shapash/utils/model.py
index b3699caa..173837f8 100644
--- a/shapash/utils/model.py
+++ b/shapash/utils/model.py
@@ -2,8 +2,10 @@
Model Module
"""
from inspect import ismethod
+
import pandas as pd
+
def extract_features_model(model, model_attribute):
"""
Extract features of models if it's possible,
@@ -14,7 +16,7 @@ def extract_features_model(model, model_attribute):
model_attribute: String or List
if model can give features, attributes to access features, if not 'length'
"""
- if model_attribute[0] == 'length':
+ if model_attribute[0] == "length":
return model.n_features_in_
else:
if ismethod(getattr(model, model_attribute[0])):
@@ -45,11 +47,10 @@ def predict_proba(model, x_encoded, classes):
pandas.DataFrame
dataset of predicted proba for each label.
"""
- if hasattr(model, 'predict_proba'):
+ if hasattr(model, "predict_proba"):
proba_values = pd.DataFrame(
- model.predict_proba(x_encoded),
- columns=['class_' + str(x) for x in classes],
- index=x_encoded.index)
+ model.predict_proba(x_encoded), columns=["class_" + str(x) for x in classes], index=x_encoded.index
+ )
else:
raise ValueError("model has no predict_proba method")
@@ -72,8 +73,8 @@ def predict(model, x_encoded):
pandas.DataFrame
1-column dataframe containing the predictions.
"""
- if hasattr(model, 'predict'):
- y_pred = pd.DataFrame(model.predict(x_encoded), columns=['pred'], index=x_encoded.index)
+ if hasattr(model, "predict"):
+ y_pred = pd.DataFrame(model.predict(x_encoded), columns=["pred"], index=x_encoded.index)
else:
raise ValueError("model has no predict method")
@@ -100,10 +101,10 @@ def predict_error(y_target, y_pred, case):
1-column dataframe containing the prediction errors.
"""
prediction_error = None
- if y_target is not None and y_pred is not None and case=="regression":
+ if y_target is not None and y_pred is not None and case == "regression":
if (y_target == 0).any()[0]:
- prediction_error = abs(y_target.values-y_pred.values)
+ prediction_error = abs(y_target.values - y_pred.values)
else:
- prediction_error = abs((y_target.values-y_pred.values)/y_target.values)
+ prediction_error = abs((y_target.values - y_pred.values) / y_target.values)
prediction_error = pd.DataFrame(prediction_error, index=y_target.index, columns=["_error_"])
return prediction_error
diff --git a/shapash/utils/model_synoptic.py b/shapash/utils/model_synoptic.py
index ff8a24e1..a3f44d2f 100644
--- a/shapash/utils/model_synoptic.py
+++ b/shapash/utils/model_synoptic.py
@@ -3,55 +3,53 @@
"""
simple_tree_model_sklearn = (
- "",
- "",
- "",
- "",
- "",
- ""
- )
+ "",
+ "",
+ "",
+ "",
+ "",
+ "",
+)
xgboost_model = (
"",
"",
- "")
+ "",
+)
lightgbm_model = (
"",
"",
- ""
+ "",
)
-catboost_model = (
- "",
- "")
+catboost_model = ("", "")
linear_model = (
"",
- "")
+ "",
+)
-svm_model = (
- "",
- "")
+svm_model = ("", "")
simple_tree_model = simple_tree_model_sklearn + xgboost_model + lightgbm_model
-dict_model_feature = {"": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ['length'],
- "": ["booster_","feature_name"],
- "": ["booster_","feature_name"],
- "": ["feature_names"],
- "": ["get_booster","feature_names"],
- "": ["get_booster","feature_names"],
- "": ["feature_names"],
- "": ["feature_names_"],
- "": ["feature_names_"],
- }
-
+dict_model_feature = {
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["length"],
+ "": ["booster_", "feature_name"],
+ "": ["booster_", "feature_name"],
+ "": ["feature_names"],
+ "": ["get_booster", "feature_names"],
+ "": ["get_booster", "feature_names"],
+ "": ["feature_names"],
+ "": ["feature_names_"],
+ "": ["feature_names_"],
+}
diff --git a/shapash/utils/threading.py b/shapash/utils/threading.py
index d1c836d1..33065587 100644
--- a/shapash/utils/threading.py
+++ b/shapash/utils/threading.py
@@ -4,6 +4,7 @@
import sys
import threading
+
class CustomThread(threading.Thread):
"""
Python ovveride threading class
@@ -13,6 +14,7 @@ class CustomThread(threading.Thread):
threading : threading.Thread
Thread which you want to instanciate
"""
+
def __init__(self, *args, **keywords):
threading.Thread.__init__(self, *args, **keywords)
self.killed = False
@@ -23,7 +25,6 @@ def start(self):
self.run = self.__run
threading.Thread.start(self)
-
def __run(self):
sys.settrace(self.globaltrace)
self.__run_backup()
@@ -33,7 +34,7 @@ def globaltrace(self, frame, event, arg):
"""
Track the global trace
"""
- if event == 'call':
+ if event == "call":
return self.localtrace
else:
return None
@@ -43,7 +44,7 @@ def localtrace(self, frame, event, arg):
Track the local trace
"""
if self.killed:
- if event == 'line':
+ if event == "line":
raise SystemExit()
return self.localtrace
diff --git a/shapash/utils/transform.py b/shapash/utils/transform.py
index f55dfc22..877a044f 100644
--- a/shapash/utils/transform.py
+++ b/shapash/utils/transform.py
@@ -1,27 +1,30 @@
"""
Transform Module
"""
+import re
+
+import numpy as np
+import pandas as pd
+
+from shapash.utils.category_encoder_backend import (
+ get_col_mapping_ce,
+ inv_transform_ce,
+ supported_category_encoder,
+ transform_ce,
+)
from shapash.utils.columntransformer_backend import (
columntransformer,
+ get_col_mapping_ct,
inv_transform_ct,
supported_sklearn,
transform_ct,
- get_col_mapping_ct
)
-from shapash.utils.category_encoder_backend import (
- transform_ce,
- inv_transform_ce,
- supported_category_encoder,
- get_col_mapping_ce
-)
-import re
-import numpy as np
-import pandas as pd
# TODO
# encode targeted variable ? from sklearn.preprocessing import LabelEncoder
# make an easy version for dict, not writing all mapping
+
def inverse_transform(x_init, preprocessing=None):
"""
Reverse transformation giving a preprocessing.
@@ -72,6 +75,7 @@ def inverse_transform(x_init, preprocessing=None):
x_inverse = inv_transform_ce(x_inverse, encoding)
return x_inverse
+
def apply_preprocessing(x_init, model, preprocessing=None):
"""
Apply preprocessing on a raw dataset giving a preprocessing.
@@ -120,6 +124,7 @@ def apply_preprocessing(x_init, model, preprocessing=None):
x_init = transform_ce(x_init, encoding)
return x_init
+
def preprocessing_tolist(preprocess):
"""
Transform preprocess into a list, if preprocess contains a dict, transform the dict into a list of dict.
@@ -138,6 +143,7 @@ def preprocessing_tolist(preprocess):
list_encoding = [[x] if isinstance(x, dict) else x for x in list_encoding]
return list_encoding
+
def check_transformers(list_encoding):
"""
Check that all transformation are supported.
@@ -176,9 +182,10 @@ def check_transformers(list_encoding):
use_ct = True
for encoding in enc.transformers_:
ct_encoding = encoding[1]
- if (str(type(ct_encoding)) not in supported_sklearn) \
- and (str(type(ct_encoding)) not in supported_category_encoder):
- if str(type(ct_encoding)) != "" :
+ if (str(type(ct_encoding)) not in supported_sklearn) and (
+ str(type(ct_encoding)) not in supported_category_encoder
+ ):
+ if str(type(ct_encoding)) != "":
raise ValueError("One of the encoders used in ColumnTransformers isn't supported.")
elif str(type(enc)) in supported_category_encoder:
@@ -189,32 +196,32 @@ def check_transformers(list_encoding):
for enc_dict in enc:
if isinstance(enc_dict, dict):
# Check dict structure : col - mapping - data_type
- if not all(struct in enc_dict for struct in ('col', 'mapping', 'data_type')):
- raise Exception(f'{enc_dict} should have col, mapping and data_type as keys.')
+ if not all(struct in enc_dict for struct in ("col", "mapping", "data_type")):
+ raise Exception(f"{enc_dict} should have col, mapping and data_type as keys.")
else:
- raise Exception(f'{enc} is not a list of dict.')
+ raise Exception(f"{enc} is not a list of dict.")
else:
- raise Exception(f'{enc} is not supported yet.')
+ raise Exception(f"{enc} is not supported yet.")
# check that encoding don't use ColumnTransformer and Category encoding at the same time
if use_ct and use_ce:
raise Exception(
f"Can't support ColumnTransformer and Category encoding at the same time. "
- f"Use Category encoding in ColumnTransformer")
+ f"Use Category encoding in ColumnTransformer"
+ )
# check that Category encoding is apply on different columns
col = []
for enc in list_encoding:
- if not str(type(enc)) in ("",
- "",
- columntransformer):
+ if not str(type(enc)) in ("", "", columntransformer):
col += enc.cols
- duplicate = set([x for x in col if col.count(x) > 1])
+ duplicate = {x for x in col if col.count(x) > 1}
if duplicate:
- raise Exception('Columns ' + str(duplicate) + ' is used in multiple category encoding')
+ raise Exception("Columns " + str(duplicate) + " is used in multiple category encoding")
return use_ct, use_ce
+
def apply_postprocessing(x_init, postprocessing):
"""
Transforms x_init depending on postprocessing parameters.
@@ -237,36 +244,38 @@ def apply_postprocessing(x_init, postprocessing):
data_modif = new_preds[feature_name]
new_datai = list()
- if dict_postprocessing['type'] == 'prefix':
+ if dict_postprocessing["type"] == "prefix":
for value in data_modif.values:
- new_datai.append(dict_postprocessing['rule'] + str(value))
+ new_datai.append(dict_postprocessing["rule"] + str(value))
new_preds[feature_name] = new_datai
- elif dict_postprocessing['type'] == 'suffix':
+ elif dict_postprocessing["type"] == "suffix":
for value in data_modif.values:
- new_datai.append(str(value) + dict_postprocessing['rule'])
+ new_datai.append(str(value) + dict_postprocessing["rule"])
new_preds[feature_name] = new_datai
- elif dict_postprocessing['type'] == 'transcoding':
+ elif dict_postprocessing["type"] == "transcoding":
unique_values = x_init[feature_name].unique().tolist()
- unique_values = [value for value in unique_values if value not in dict_postprocessing['rule'].keys()]
+ unique_values = [value for value in unique_values if value not in dict_postprocessing["rule"].keys()]
for value in unique_values:
- dict_postprocessing['rule'][value] = value
- new_preds[feature_name] = new_preds[feature_name].map(dict_postprocessing['rule'])
+ dict_postprocessing["rule"][value] = value
+ new_preds[feature_name] = new_preds[feature_name].map(dict_postprocessing["rule"])
- elif dict_postprocessing['type'] == 'regex':
+ elif dict_postprocessing["type"] == "regex":
new_preds[feature_name] = new_preds[feature_name].apply(
- lambda x: re.sub(dict_postprocessing["rule"]['in'], dict_postprocessing["rule"]['out'], x))
+ lambda x: re.sub(dict_postprocessing["rule"]["in"], dict_postprocessing["rule"]["out"], x)
+ )
- elif dict_postprocessing['type'] == 'case':
- if dict_postprocessing['rule'] == 'lower':
+ elif dict_postprocessing["type"] == "case":
+ if dict_postprocessing["rule"] == "lower":
new_preds[feature_name] = new_preds[feature_name].apply(lambda x: x.lower())
- elif dict_postprocessing['rule'] == 'upper':
+ elif dict_postprocessing["rule"] == "upper":
new_preds[feature_name] = new_preds[feature_name].apply(lambda x: x.upper())
return new_preds
-def adapt_contributions(case,contributions):
+
+def adapt_contributions(case, contributions):
"""
If _case is "classification" and contributions a np.array or pd.DataFrame
this function transform contributions matrix in a list of 2 contributions
@@ -288,12 +297,8 @@ def adapt_contributions(case,contributions):
# So that we have the following format : [contributions_class_0, contributions_class_1, ...]
if isinstance(contributions, np.ndarray) and contributions.ndim == 3:
contributions = [contributions[:, :, i] for i in range(contributions.shape[-1])]
- if (
- (isinstance(contributions, pd.DataFrame) and case == 'classification')
- or (
- isinstance(contributions, (np.ndarray, list))
- and case == 'classification'
- and np.array(contributions).ndim == 2)
+ if (isinstance(contributions, pd.DataFrame) and case == "classification") or (
+ isinstance(contributions, (np.ndarray, list)) and case == "classification" and np.array(contributions).ndim == 2
):
return [contributions * -1, contributions]
else:
@@ -374,7 +379,8 @@ def get_features_transform_mapping(x_init, x_encoded, preprocessing=None):
dict_all_cols_mapping[col_name] = [col_name]
return dict_all_cols_mapping
-def handle_categorical_missing(df : pd.DataFrame)-> pd.DataFrame:
+
+def handle_categorical_missing(df: pd.DataFrame) -> pd.DataFrame:
"""
Replace missing values for categorical columns
@@ -383,7 +389,7 @@ def handle_categorical_missing(df : pd.DataFrame)-> pd.DataFrame:
df : pd.DataFrame
Pandas dataframe on which we will replace the missing values
"""
- categorical_cols = df.select_dtypes(include=['object']).columns
+ categorical_cols = df.select_dtypes(include=["object"]).columns
df_handle_missing = df.copy()
df_handle_missing[categorical_cols] = df_handle_missing[categorical_cols].fillna("missing")
return df_handle_missing
diff --git a/shapash/utils/translate.py b/shapash/utils/translate.py
index 776eac57..f5afeaf3 100644
--- a/shapash/utils/translate.py
+++ b/shapash/utils/translate.py
@@ -1,6 +1,8 @@
"""
Translate Module
"""
+
+
def translate(elements, mapping):
"""
Map a dictionary to a list of elements.
diff --git a/shapash/utils/utils.py b/shapash/utils/utils.py
index 6df58a4b..0fc4b673 100644
--- a/shapash/utils/utils.py
+++ b/shapash/utils/utils.py
@@ -1,12 +1,15 @@
"""
Utils is a group of function for the library
"""
+import math
+import socket
+
import numpy as np
import pandas as pd
-import socket
-import math
-from shapash.explainer.smart_state import SmartState
+
from shapash.explainer.multi_decorator import MultiDecorator
+from shapash.explainer.smart_state import SmartState
+
def get_host_name():
"""
@@ -68,6 +71,7 @@ def is_nested_list(object_param):
"""
return any(isinstance(elem, list) for elem in object_param)
+
def add_line_break(text, nbchar, maxlen=150):
"""
adding line break in string if necessary
@@ -86,7 +90,7 @@ def add_line_break(text, nbchar, maxlen=150):
string
original text + line break
"""
- if isinstance(text,str):
+ if isinstance(text, str):
length = 0
tot_length = 0
input_word = text.split()
@@ -97,20 +101,21 @@ def add_line_break(text, nbchar, maxlen=150):
if tot_length <= maxlen:
if length >= nbchar:
length = 0
- final_sep.append('
')
+ final_sep.append("
")
else:
- final_sep.append(' ')
+ final_sep.append(" ")
if len(final_sep) == len(input_word) - 1:
- last_char=''
- else :
- last_char=('...')
+ last_char = ""
+ else:
+ last_char = "..."
- new_string = "".join(sum(zip(input_word, final_sep+['']), ())[:-1]) + last_char
+ new_string = "".join(sum(zip(input_word, final_sep + [""]), ())[:-1]) + last_char
return new_string
else:
return text
-def truncate_str(text, maxlen= 40):
+
+def truncate_str(text, maxlen=40):
"""
truncate a string
@@ -137,9 +142,10 @@ def truncate_str(text, maxlen= 40):
text = " ".join(output_words)
if len(input_words) > len(output_words):
- text = text + '...'
+ text = text + "..."
return text
+
def compute_digit_number(value):
"""
return int, number of digits to display
@@ -155,14 +161,15 @@ def compute_digit_number(value):
number of digits
"""
# fix for 0 value
- if(value == 0):
+ if value == 0:
first_nz = 1
else:
first_nz = int(math.log10(abs(value)))
digit = abs(min(3, first_nz) - 3)
return digit
-def add_text(text_list,sep):
+
+def add_text(text_list, sep):
"""
return int, number of digits to display
@@ -178,9 +185,10 @@ def add_text(text_list,sep):
int
number of digits
"""
- clean_list = [x for x in text_list if x not in ['', None]]
+ clean_list = [x for x in text_list if x not in ["", None]]
return sep.join(clean_list)
+
def maximum_difference_sort_value(contributions):
"""
Auxiliary function to sort the contributions for the compare_plot.
@@ -232,7 +240,9 @@ def compute_sorted_variables_interactions_list_indices(interaction_values):
for i in range(tmp.shape[0]):
tmp[i, i:] = 0
- interaction_contrib_sorted_indices = np.dstack(np.unravel_index(np.argsort(tmp.ravel(), kind="stable"), tmp.shape))[0][::-1]
+ interaction_contrib_sorted_indices = np.dstack(np.unravel_index(np.argsort(tmp.ravel(), kind="stable"), tmp.shape))[
+ 0
+ ][::-1]
return interaction_contrib_sorted_indices
@@ -295,7 +305,7 @@ def choose_state(contributions):
return SmartState()
-def convert_string_to_int_keys(input_dict: dict) -> dict:
+def convert_string_to_int_keys(input_dict: dict) -> dict:
"""
Returns the dict with integer keys instead of string keys
@@ -307,4 +317,4 @@ def convert_string_to_int_keys(input_dict: dict) -> dict:
-------
dict
"""
- return {int(k): v for k,v in input_dict.items()}
+ return {int(k): v for k, v in input_dict.items()}
diff --git a/shapash/webapp/assets/jquery.js b/shapash/webapp/assets/jquery.js
index 07c00cd2..a1c07fd8 100644
--- a/shapash/webapp/assets/jquery.js
+++ b/shapash/webapp/assets/jquery.js
@@ -1,2 +1,2 @@
/*! jQuery v3.4.1 | (c) JS Foundation and other contributors | jquery.org/license */
-!function(e,t){"use strict";"object"==typeof module&&"object"==typeof module.exports?module.exports=e.document?t(e,!0):function(e){if(!e.document)throw new Error("jQuery requires a window with a document");return t(e)}:t(e)}("undefined"!=typeof window?window:this,function(C,e){"use strict";var t=[],E=C.document,r=Object.getPrototypeOf,s=t.slice,g=t.concat,u=t.push,i=t.indexOf,n={},o=n.toString,v=n.hasOwnProperty,a=v.toString,l=a.call(Object),y={},m=function(e){return"function"==typeof e&&"number"!=typeof e.nodeType},x=function(e){return null!=e&&e===e.window},c={type:!0,src:!0,nonce:!0,noModule:!0};function b(e,t,n){var r,i,o=(n=n||E).createElement("script");if(o.text=e,t)for(r in c)(i=t[r]||t.getAttribute&&t.getAttribute(r))&&o.setAttribute(r,i);n.head.appendChild(o).parentNode.removeChild(o)}function w(e){return null==e?e+"":"object"==typeof e||"function"==typeof e?n[o.call(e)]||"object":typeof e}var f="3.4.1",k=function(e,t){return new k.fn.init(e,t)},p=/^[\s\uFEFF\xA0]+|[\s\uFEFF\xA0]+$/g;function d(e){var t=!!e&&"length"in e&&e.length,n=w(e);return!m(e)&&!x(e)&&("array"===n||0===t||"number"==typeof t&&0+~]|"+M+")"+M+"*"),U=new RegExp(M+"|>"),X=new RegExp($),V=new RegExp("^"+I+"$"),G={ID:new RegExp("^#("+I+")"),CLASS:new RegExp("^\\.("+I+")"),TAG:new RegExp("^("+I+"|[*])"),ATTR:new RegExp("^"+W),PSEUDO:new RegExp("^"+$),CHILD:new RegExp("^:(only|first|last|nth|nth-last)-(child|of-type)(?:\\("+M+"*(even|odd|(([+-]|)(\\d*)n|)"+M+"*(?:([+-]|)"+M+"*(\\d+)|))"+M+"*\\)|)","i"),bool:new RegExp("^(?:"+R+")$","i"),needsContext:new RegExp("^"+M+"*[>+~]|:(even|odd|eq|gt|lt|nth|first|last)(?:\\("+M+"*((?:-\\d)?\\d*)"+M+"*\\)|)(?=[^-]|$)","i")},Y=/HTML$/i,Q=/^(?:input|select|textarea|button)$/i,J=/^h\d$/i,K=/^[^{]+\{\s*\[native \w/,Z=/^(?:#([\w-]+)|(\w+)|\.([\w-]+))$/,ee=/[+~]/,te=new RegExp("\\\\([\\da-f]{1,6}"+M+"?|("+M+")|.)","ig"),ne=function(e,t,n){var r="0x"+t-65536;return r!=r||n?t:r<0?String.fromCharCode(r+65536):String.fromCharCode(r>>10|55296,1023&r|56320)},re=/([\0-\x1f\x7f]|^-?\d)|^-$|[^\0-\x1f\x7f-\uFFFF\w-]/g,ie=function(e,t){return t?"\0"===e?"\ufffd":e.slice(0,-1)+"\\"+e.charCodeAt(e.length-1).toString(16)+" ":"\\"+e},oe=function(){T()},ae=be(function(e){return!0===e.disabled&&"fieldset"===e.nodeName.toLowerCase()},{dir:"parentNode",next:"legend"});try{H.apply(t=O.call(m.childNodes),m.childNodes),t[m.childNodes.length].nodeType}catch(e){H={apply:t.length?function(e,t){L.apply(e,O.call(t))}:function(e,t){var n=e.length,r=0;while(e[n++]=t[r++]);e.length=n-1}}}function se(t,e,n,r){var i,o,a,s,u,l,c,f=e&&e.ownerDocument,p=e?e.nodeType:9;if(n=n||[],"string"!=typeof t||!t||1!==p&&9!==p&&11!==p)return n;if(!r&&((e?e.ownerDocument||e:m)!==C&&T(e),e=e||C,E)){if(11!==p&&(u=Z.exec(t)))if(i=u[1]){if(9===p){if(!(a=e.getElementById(i)))return n;if(a.id===i)return n.push(a),n}else if(f&&(a=f.getElementById(i))&&y(e,a)&&a.id===i)return n.push(a),n}else{if(u[2])return H.apply(n,e.getElementsByTagName(t)),n;if((i=u[3])&&d.getElementsByClassName&&e.getElementsByClassName)return H.apply(n,e.getElementsByClassName(i)),n}if(d.qsa&&!A[t+" "]&&(!v||!v.test(t))&&(1!==p||"object"!==e.nodeName.toLowerCase())){if(c=t,f=e,1===p&&U.test(t)){(s=e.getAttribute("id"))?s=s.replace(re,ie):e.setAttribute("id",s=k),o=(l=h(t)).length;while(o--)l[o]="#"+s+" "+xe(l[o]);c=l.join(","),f=ee.test(t)&&ye(e.parentNode)||e}try{return H.apply(n,f.querySelectorAll(c)),n}catch(e){A(t,!0)}finally{s===k&&e.removeAttribute("id")}}}return g(t.replace(B,"$1"),e,n,r)}function ue(){var r=[];return function e(t,n){return r.push(t+" ")>b.cacheLength&&delete e[r.shift()],e[t+" "]=n}}function le(e){return e[k]=!0,e}function ce(e){var t=C.createElement("fieldset");try{return!!e(t)}catch(e){return!1}finally{t.parentNode&&t.parentNode.removeChild(t),t=null}}function fe(e,t){var n=e.split("|"),r=n.length;while(r--)b.attrHandle[n[r]]=t}function pe(e,t){var n=t&&e,r=n&&1===e.nodeType&&1===t.nodeType&&e.sourceIndex-t.sourceIndex;if(r)return r;if(n)while(n=n.nextSibling)if(n===t)return-1;return e?1:-1}function de(t){return function(e){return"input"===e.nodeName.toLowerCase()&&e.type===t}}function he(n){return function(e){var t=e.nodeName.toLowerCase();return("input"===t||"button"===t)&&e.type===n}}function ge(t){return function(e){return"form"in e?e.parentNode&&!1===e.disabled?"label"in e?"label"in e.parentNode?e.parentNode.disabled===t:e.disabled===t:e.isDisabled===t||e.isDisabled!==!t&&ae(e)===t:e.disabled===t:"label"in e&&e.disabled===t}}function ve(a){return le(function(o){return o=+o,le(function(e,t){var n,r=a([],e.length,o),i=r.length;while(i--)e[n=r[i]]&&(e[n]=!(t[n]=e[n]))})})}function ye(e){return e&&"undefined"!=typeof e.getElementsByTagName&&e}for(e in d=se.support={},i=se.isXML=function(e){var t=e.namespaceURI,n=(e.ownerDocument||e).documentElement;return!Y.test(t||n&&n.nodeName||"HTML")},T=se.setDocument=function(e){var t,n,r=e?e.ownerDocument||e:m;return r!==C&&9===r.nodeType&&r.documentElement&&(a=(C=r).documentElement,E=!i(C),m!==C&&(n=C.defaultView)&&n.top!==n&&(n.addEventListener?n.addEventListener("unload",oe,!1):n.attachEvent&&n.attachEvent("onunload",oe)),d.attributes=ce(function(e){return e.className="i",!e.getAttribute("className")}),d.getElementsByTagName=ce(function(e){return e.appendChild(C.createComment("")),!e.getElementsByTagName("*").length}),d.getElementsByClassName=K.test(C.getElementsByClassName),d.getById=ce(function(e){return a.appendChild(e).id=k,!C.getElementsByName||!C.getElementsByName(k).length}),d.getById?(b.filter.ID=function(e){var t=e.replace(te,ne);return function(e){return e.getAttribute("id")===t}},b.find.ID=function(e,t){if("undefined"!=typeof t.getElementById&&E){var n=t.getElementById(e);return n?[n]:[]}}):(b.filter.ID=function(e){var n=e.replace(te,ne);return function(e){var t="undefined"!=typeof e.getAttributeNode&&e.getAttributeNode("id");return t&&t.value===n}},b.find.ID=function(e,t){if("undefined"!=typeof t.getElementById&&E){var n,r,i,o=t.getElementById(e);if(o){if((n=o.getAttributeNode("id"))&&n.value===e)return[o];i=t.getElementsByName(e),r=0;while(o=i[r++])if((n=o.getAttributeNode("id"))&&n.value===e)return[o]}return[]}}),b.find.TAG=d.getElementsByTagName?function(e,t){return"undefined"!=typeof t.getElementsByTagName?t.getElementsByTagName(e):d.qsa?t.querySelectorAll(e):void 0}:function(e,t){var n,r=[],i=0,o=t.getElementsByTagName(e);if("*"===e){while(n=o[i++])1===n.nodeType&&r.push(n);return r}return o},b.find.CLASS=d.getElementsByClassName&&function(e,t){if("undefined"!=typeof t.getElementsByClassName&&E)return t.getElementsByClassName(e)},s=[],v=[],(d.qsa=K.test(C.querySelectorAll))&&(ce(function(e){a.appendChild(e).innerHTML="",e.querySelectorAll("[msallowcapture^='']").length&&v.push("[*^$]="+M+"*(?:''|\"\")"),e.querySelectorAll("[selected]").length||v.push("\\["+M+"*(?:value|"+R+")"),e.querySelectorAll("[id~="+k+"-]").length||v.push("~="),e.querySelectorAll(":checked").length||v.push(":checked"),e.querySelectorAll("a#"+k+"+*").length||v.push(".#.+[+~]")}),ce(function(e){e.innerHTML="";var t=C.createElement("input");t.setAttribute("type","hidden"),e.appendChild(t).setAttribute("name","D"),e.querySelectorAll("[name=d]").length&&v.push("name"+M+"*[*^$|!~]?="),2!==e.querySelectorAll(":enabled").length&&v.push(":enabled",":disabled"),a.appendChild(e).disabled=!0,2!==e.querySelectorAll(":disabled").length&&v.push(":enabled",":disabled"),e.querySelectorAll("*,:x"),v.push(",.*:")})),(d.matchesSelector=K.test(c=a.matches||a.webkitMatchesSelector||a.mozMatchesSelector||a.oMatchesSelector||a.msMatchesSelector))&&ce(function(e){d.disconnectedMatch=c.call(e,"*"),c.call(e,"[s!='']:x"),s.push("!=",$)}),v=v.length&&new RegExp(v.join("|")),s=s.length&&new RegExp(s.join("|")),t=K.test(a.compareDocumentPosition),y=t||K.test(a.contains)?function(e,t){var n=9===e.nodeType?e.documentElement:e,r=t&&t.parentNode;return e===r||!(!r||1!==r.nodeType||!(n.contains?n.contains(r):e.compareDocumentPosition&&16&e.compareDocumentPosition(r)))}:function(e,t){if(t)while(t=t.parentNode)if(t===e)return!0;return!1},D=t?function(e,t){if(e===t)return l=!0,0;var n=!e.compareDocumentPosition-!t.compareDocumentPosition;return n||(1&(n=(e.ownerDocument||e)===(t.ownerDocument||t)?e.compareDocumentPosition(t):1)||!d.sortDetached&&t.compareDocumentPosition(e)===n?e===C||e.ownerDocument===m&&y(m,e)?-1:t===C||t.ownerDocument===m&&y(m,t)?1:u?P(u,e)-P(u,t):0:4&n?-1:1)}:function(e,t){if(e===t)return l=!0,0;var n,r=0,i=e.parentNode,o=t.parentNode,a=[e],s=[t];if(!i||!o)return e===C?-1:t===C?1:i?-1:o?1:u?P(u,e)-P(u,t):0;if(i===o)return pe(e,t);n=e;while(n=n.parentNode)a.unshift(n);n=t;while(n=n.parentNode)s.unshift(n);while(a[r]===s[r])r++;return r?pe(a[r],s[r]):a[r]===m?-1:s[r]===m?1:0}),C},se.matches=function(e,t){return se(e,null,null,t)},se.matchesSelector=function(e,t){if((e.ownerDocument||e)!==C&&T(e),d.matchesSelector&&E&&!A[t+" "]&&(!s||!s.test(t))&&(!v||!v.test(t)))try{var n=c.call(e,t);if(n||d.disconnectedMatch||e.document&&11!==e.document.nodeType)return n}catch(e){A(t,!0)}return 0":{dir:"parentNode",first:!0}," ":{dir:"parentNode"},"+":{dir:"previousSibling",first:!0},"~":{dir:"previousSibling"}},preFilter:{ATTR:function(e){return e[1]=e[1].replace(te,ne),e[3]=(e[3]||e[4]||e[5]||"").replace(te,ne),"~="===e[2]&&(e[3]=" "+e[3]+" "),e.slice(0,4)},CHILD:function(e){return e[1]=e[1].toLowerCase(),"nth"===e[1].slice(0,3)?(e[3]||se.error(e[0]),e[4]=+(e[4]?e[5]+(e[6]||1):2*("even"===e[3]||"odd"===e[3])),e[5]=+(e[7]+e[8]||"odd"===e[3])):e[3]&&se.error(e[0]),e},PSEUDO:function(e){var t,n=!e[6]&&e[2];return G.CHILD.test(e[0])?null:(e[3]?e[2]=e[4]||e[5]||"":n&&X.test(n)&&(t=h(n,!0))&&(t=n.indexOf(")",n.length-t)-n.length)&&(e[0]=e[0].slice(0,t),e[2]=n.slice(0,t)),e.slice(0,3))}},filter:{TAG:function(e){var t=e.replace(te,ne).toLowerCase();return"*"===e?function(){return!0}:function(e){return e.nodeName&&e.nodeName.toLowerCase()===t}},CLASS:function(e){var t=p[e+" "];return t||(t=new RegExp("(^|"+M+")"+e+"("+M+"|$)"))&&p(e,function(e){return t.test("string"==typeof e.className&&e.className||"undefined"!=typeof e.getAttribute&&e.getAttribute("class")||"")})},ATTR:function(n,r,i){return function(e){var t=se.attr(e,n);return null==t?"!="===r:!r||(t+="","="===r?t===i:"!="===r?t!==i:"^="===r?i&&0===t.indexOf(i):"*="===r?i&&-1:\x20\t\r\n\f]*)[\x20\t\r\n\f]*\/?>(?:<\/\1>|)$/i;function j(e,n,r){return m(n)?k.grep(e,function(e,t){return!!n.call(e,t,e)!==r}):n.nodeType?k.grep(e,function(e){return e===n!==r}):"string"!=typeof n?k.grep(e,function(e){return-1)[^>]*|#([\w-]+))$/;(k.fn.init=function(e,t,n){var r,i;if(!e)return this;if(n=n||q,"string"==typeof e){if(!(r="<"===e[0]&&">"===e[e.length-1]&&3<=e.length?[null,e,null]:L.exec(e))||!r[1]&&t)return!t||t.jquery?(t||n).find(e):this.constructor(t).find(e);if(r[1]){if(t=t instanceof k?t[0]:t,k.merge(this,k.parseHTML(r[1],t&&t.nodeType?t.ownerDocument||t:E,!0)),D.test(r[1])&&k.isPlainObject(t))for(r in t)m(this[r])?this[r](t[r]):this.attr(r,t[r]);return this}return(i=E.getElementById(r[2]))&&(this[0]=i,this.length=1),this}return e.nodeType?(this[0]=e,this.length=1,this):m(e)?void 0!==n.ready?n.ready(e):e(k):k.makeArray(e,this)}).prototype=k.fn,q=k(E);var H=/^(?:parents|prev(?:Until|All))/,O={children:!0,contents:!0,next:!0,prev:!0};function P(e,t){while((e=e[t])&&1!==e.nodeType);return e}k.fn.extend({has:function(e){var t=k(e,this),n=t.length;return this.filter(function(){for(var e=0;e\x20\t\r\n\f]*)/i,he=/^$|^module$|\/(?:java|ecma)script/i,ge={option:[1,""],thead:[1,""],col:[2,""],tr:[2,""],td:[3,""],_default:[0,"",""]};function ve(e,t){var n;return n="undefined"!=typeof e.getElementsByTagName?e.getElementsByTagName(t||"*"):"undefined"!=typeof e.querySelectorAll?e.querySelectorAll(t||"*"):[],void 0===t||t&&A(e,t)?k.merge([e],n):n}function ye(e,t){for(var n=0,r=e.length;nx",y.noCloneChecked=!!me.cloneNode(!0).lastChild.defaultValue;var Te=/^key/,Ce=/^(?:mouse|pointer|contextmenu|drag|drop)|click/,Ee=/^([^.]*)(?:\.(.+)|)/;function ke(){return!0}function Se(){return!1}function Ne(e,t){return e===function(){try{return E.activeElement}catch(e){}}()==("focus"===t)}function Ae(e,t,n,r,i,o){var a,s;if("object"==typeof t){for(s in"string"!=typeof n&&(r=r||n,n=void 0),t)Ae(e,s,n,r,t[s],o);return e}if(null==r&&null==i?(i=n,r=n=void 0):null==i&&("string"==typeof n?(i=r,r=void 0):(i=r,r=n,n=void 0)),!1===i)i=Se;else if(!i)return e;return 1===o&&(a=i,(i=function(e){return k().off(e),a.apply(this,arguments)}).guid=a.guid||(a.guid=k.guid++)),e.each(function(){k.event.add(this,t,i,r,n)})}function De(e,i,o){o?(Q.set(e,i,!1),k.event.add(e,i,{namespace:!1,handler:function(e){var t,n,r=Q.get(this,i);if(1&e.isTrigger&&this[i]){if(r.length)(k.event.special[i]||{}).delegateType&&e.stopPropagation();else if(r=s.call(arguments),Q.set(this,i,r),t=o(this,i),this[i](),r!==(n=Q.get(this,i))||t?Q.set(this,i,!1):n={},r!==n)return e.stopImmediatePropagation(),e.preventDefault(),n.value}else r.length&&(Q.set(this,i,{value:k.event.trigger(k.extend(r[0],k.Event.prototype),r.slice(1),this)}),e.stopImmediatePropagation())}})):void 0===Q.get(e,i)&&k.event.add(e,i,ke)}k.event={global:{},add:function(t,e,n,r,i){var o,a,s,u,l,c,f,p,d,h,g,v=Q.get(t);if(v){n.handler&&(n=(o=n).handler,i=o.selector),i&&k.find.matchesSelector(ie,i),n.guid||(n.guid=k.guid++),(u=v.events)||(u=v.events={}),(a=v.handle)||(a=v.handle=function(e){return"undefined"!=typeof k&&k.event.triggered!==e.type?k.event.dispatch.apply(t,arguments):void 0}),l=(e=(e||"").match(R)||[""]).length;while(l--)d=g=(s=Ee.exec(e[l])||[])[1],h=(s[2]||"").split(".").sort(),d&&(f=k.event.special[d]||{},d=(i?f.delegateType:f.bindType)||d,f=k.event.special[d]||{},c=k.extend({type:d,origType:g,data:r,handler:n,guid:n.guid,selector:i,needsContext:i&&k.expr.match.needsContext.test(i),namespace:h.join(".")},o),(p=u[d])||((p=u[d]=[]).delegateCount=0,f.setup&&!1!==f.setup.call(t,r,h,a)||t.addEventListener&&t.addEventListener(d,a)),f.add&&(f.add.call(t,c),c.handler.guid||(c.handler.guid=n.guid)),i?p.splice(p.delegateCount++,0,c):p.push(c),k.event.global[d]=!0)}},remove:function(e,t,n,r,i){var o,a,s,u,l,c,f,p,d,h,g,v=Q.hasData(e)&&Q.get(e);if(v&&(u=v.events)){l=(t=(t||"").match(R)||[""]).length;while(l--)if(d=g=(s=Ee.exec(t[l])||[])[1],h=(s[2]||"").split(".").sort(),d){f=k.event.special[d]||{},p=u[d=(r?f.delegateType:f.bindType)||d]||[],s=s[2]&&new RegExp("(^|\\.)"+h.join("\\.(?:.*\\.|)")+"(\\.|$)"),a=o=p.length;while(o--)c=p[o],!i&&g!==c.origType||n&&n.guid!==c.guid||s&&!s.test(c.namespace)||r&&r!==c.selector&&("**"!==r||!c.selector)||(p.splice(o,1),c.selector&&p.delegateCount--,f.remove&&f.remove.call(e,c));a&&!p.length&&(f.teardown&&!1!==f.teardown.call(e,h,v.handle)||k.removeEvent(e,d,v.handle),delete u[d])}else for(d in u)k.event.remove(e,d+t[l],n,r,!0);k.isEmptyObject(u)&&Q.remove(e,"handle events")}},dispatch:function(e){var t,n,r,i,o,a,s=k.event.fix(e),u=new Array(arguments.length),l=(Q.get(this,"events")||{})[s.type]||[],c=k.event.special[s.type]||{};for(u[0]=s,t=1;t\x20\t\r\n\f]*)[^>]*)\/>/gi,qe=/\s*$/g;function Oe(e,t){return A(e,"table")&&A(11!==t.nodeType?t:t.firstChild,"tr")&&k(e).children("tbody")[0]||e}function Pe(e){return e.type=(null!==e.getAttribute("type"))+"/"+e.type,e}function Re(e){return"true/"===(e.type||"").slice(0,5)?e.type=e.type.slice(5):e.removeAttribute("type"),e}function Me(e,t){var n,r,i,o,a,s,u,l;if(1===t.nodeType){if(Q.hasData(e)&&(o=Q.access(e),a=Q.set(t,o),l=o.events))for(i in delete a.handle,a.events={},l)for(n=0,r=l[i].length;n")},clone:function(e,t,n){var r,i,o,a,s,u,l,c=e.cloneNode(!0),f=oe(e);if(!(y.noCloneChecked||1!==e.nodeType&&11!==e.nodeType||k.isXMLDoc(e)))for(a=ve(c),r=0,i=(o=ve(e)).length;r").attr(n.scriptAttrs||{}).prop({charset:n.scriptCharset,src:n.url}).on("load error",i=function(e){r.remove(),i=null,e&&t("error"===e.type?404:200,e.type)}),E.head.appendChild(r[0])},abort:function(){i&&i()}}});var Vt,Gt=[],Yt=/(=)\?(?=&|$)|\?\?/;k.ajaxSetup({jsonp:"callback",jsonpCallback:function(){var e=Gt.pop()||k.expando+"_"+kt++;return this[e]=!0,e}}),k.ajaxPrefilter("json jsonp",function(e,t,n){var r,i,o,a=!1!==e.jsonp&&(Yt.test(e.url)?"url":"string"==typeof e.data&&0===(e.contentType||"").indexOf("application/x-www-form-urlencoded")&&Yt.test(e.data)&&"data");if(a||"jsonp"===e.dataTypes[0])return r=e.jsonpCallback=m(e.jsonpCallback)?e.jsonpCallback():e.jsonpCallback,a?e[a]=e[a].replace(Yt,"$1"+r):!1!==e.jsonp&&(e.url+=(St.test(e.url)?"&":"?")+e.jsonp+"="+r),e.converters["script json"]=function(){return o||k.error(r+" was not called"),o[0]},e.dataTypes[0]="json",i=C[r],C[r]=function(){o=arguments},n.always(function(){void 0===i?k(C).removeProp(r):C[r]=i,e[r]&&(e.jsonpCallback=t.jsonpCallback,Gt.push(r)),o&&m(i)&&i(o[0]),o=i=void 0}),"script"}),y.createHTMLDocument=((Vt=E.implementation.createHTMLDocument("").body).innerHTML="",2===Vt.childNodes.length),k.parseHTML=function(e,t,n){return"string"!=typeof e?[]:("boolean"==typeof t&&(n=t,t=!1),t||(y.createHTMLDocument?((r=(t=E.implementation.createHTMLDocument("")).createElement("base")).href=E.location.href,t.head.appendChild(r)):t=E),o=!n&&[],(i=D.exec(e))?[t.createElement(i[1])]:(i=we([e],t,o),o&&o.length&&k(o).remove(),k.merge([],i.childNodes)));var r,i,o},k.fn.load=function(e,t,n){var r,i,o,a=this,s=e.indexOf(" ");return-1").append(k.parseHTML(e)).find(r):e)}).always(n&&function(e,t){a.each(function(){n.apply(this,o||[e.responseText,t,e])})}),this},k.each(["ajaxStart","ajaxStop","ajaxComplete","ajaxError","ajaxSuccess","ajaxSend"],function(e,t){k.fn[t]=function(e){return this.on(t,e)}}),k.expr.pseudos.animated=function(t){return k.grep(k.timers,function(e){return t===e.elem}).length},k.offset={setOffset:function(e,t,n){var r,i,o,a,s,u,l=k.css(e,"position"),c=k(e),f={};"static"===l&&(e.style.position="relative"),s=c.offset(),o=k.css(e,"top"),u=k.css(e,"left"),("absolute"===l||"fixed"===l)&&-1<(o+u).indexOf("auto")?(a=(r=c.position()).top,i=r.left):(a=parseFloat(o)||0,i=parseFloat(u)||0),m(t)&&(t=t.call(e,n,k.extend({},s))),null!=t.top&&(f.top=t.top-s.top+a),null!=t.left&&(f.left=t.left-s.left+i),"using"in t?t.using.call(e,f):c.css(f)}},k.fn.extend({offset:function(t){if(arguments.length)return void 0===t?this:this.each(function(e){k.offset.setOffset(this,t,e)});var e,n,r=this[0];return r?r.getClientRects().length?(e=r.getBoundingClientRect(),n=r.ownerDocument.defaultView,{top:e.top+n.pageYOffset,left:e.left+n.pageXOffset}):{top:0,left:0}:void 0},position:function(){if(this[0]){var e,t,n,r=this[0],i={top:0,left:0};if("fixed"===k.css(r,"position"))t=r.getBoundingClientRect();else{t=this.offset(),n=r.ownerDocument,e=r.offsetParent||n.documentElement;while(e&&(e===n.body||e===n.documentElement)&&"static"===k.css(e,"position"))e=e.parentNode;e&&e!==r&&1===e.nodeType&&((i=k(e).offset()).top+=k.css(e,"borderTopWidth",!0),i.left+=k.css(e,"borderLeftWidth",!0))}return{top:t.top-i.top-k.css(r,"marginTop",!0),left:t.left-i.left-k.css(r,"marginLeft",!0)}}},offsetParent:function(){return this.map(function(){var e=this.offsetParent;while(e&&"static"===k.css(e,"position"))e=e.offsetParent;return e||ie})}}),k.each({scrollLeft:"pageXOffset",scrollTop:"pageYOffset"},function(t,i){var o="pageYOffset"===i;k.fn[t]=function(e){return _(this,function(e,t,n){var r;if(x(e)?r=e:9===e.nodeType&&(r=e.defaultView),void 0===n)return r?r[i]:e[t];r?r.scrollTo(o?r.pageXOffset:n,o?n:r.pageYOffset):e[t]=n},t,e,arguments.length)}}),k.each(["top","left"],function(e,n){k.cssHooks[n]=ze(y.pixelPosition,function(e,t){if(t)return t=_e(e,n),$e.test(t)?k(e).position()[n]+"px":t})}),k.each({Height:"height",Width:"width"},function(a,s){k.each({padding:"inner"+a,content:s,"":"outer"+a},function(r,o){k.fn[o]=function(e,t){var n=arguments.length&&(r||"boolean"!=typeof e),i=r||(!0===e||!0===t?"margin":"border");return _(this,function(e,t,n){var r;return x(e)?0===o.indexOf("outer")?e["inner"+a]:e.document.documentElement["client"+a]:9===e.nodeType?(r=e.documentElement,Math.max(e.body["scroll"+a],r["scroll"+a],e.body["offset"+a],r["offset"+a],r["client"+a])):void 0===n?k.css(e,t,i):k.style(e,t,n,i)},s,n?e:void 0,n)}})}),k.each("blur focus focusin focusout resize scroll click dblclick mousedown mouseup mousemove mouseover mouseout mouseenter mouseleave change select submit keydown keypress keyup contextmenu".split(" "),function(e,n){k.fn[n]=function(e,t){return 0+~]|"+M+")"+M+"*"),U=new RegExp(M+"|>"),X=new RegExp($),V=new RegExp("^"+I+"$"),G={ID:new RegExp("^#("+I+")"),CLASS:new RegExp("^\\.("+I+")"),TAG:new RegExp("^("+I+"|[*])"),ATTR:new RegExp("^"+W),PSEUDO:new RegExp("^"+$),CHILD:new RegExp("^:(only|first|last|nth|nth-last)-(child|of-type)(?:\\("+M+"*(even|odd|(([+-]|)(\\d*)n|)"+M+"*(?:([+-]|)"+M+"*(\\d+)|))"+M+"*\\)|)","i"),bool:new RegExp("^(?:"+R+")$","i"),needsContext:new RegExp("^"+M+"*[>+~]|:(even|odd|eq|gt|lt|nth|first|last)(?:\\("+M+"*((?:-\\d)?\\d*)"+M+"*\\)|)(?=[^-]|$)","i")},Y=/HTML$/i,Q=/^(?:input|select|textarea|button)$/i,J=/^h\d$/i,K=/^[^{]+\{\s*\[native \w/,Z=/^(?:#([\w-]+)|(\w+)|\.([\w-]+))$/,ee=/[+~]/,te=new RegExp("\\\\([\\da-f]{1,6}"+M+"?|("+M+")|.)","ig"),ne=function(e,t,n){var r="0x"+t-65536;return r!=r||n?t:r<0?String.fromCharCode(r+65536):String.fromCharCode(r>>10|55296,1023&r|56320)},re=/([\0-\x1f\x7f]|^-?\d)|^-$|[^\0-\x1f\x7f-\uFFFF\w-]/g,ie=function(e,t){return t?"\0"===e?"\ufffd":e.slice(0,-1)+"\\"+e.charCodeAt(e.length-1).toString(16)+" ":"\\"+e},oe=function(){T()},ae=be(function(e){return!0===e.disabled&&"fieldset"===e.nodeName.toLowerCase()},{dir:"parentNode",next:"legend"});try{H.apply(t=O.call(m.childNodes),m.childNodes),t[m.childNodes.length].nodeType}catch(e){H={apply:t.length?function(e,t){L.apply(e,O.call(t))}:function(e,t){var n=e.length,r=0;while(e[n++]=t[r++]);e.length=n-1}}}function se(t,e,n,r){var i,o,a,s,u,l,c,f=e&&e.ownerDocument,p=e?e.nodeType:9;if(n=n||[],"string"!=typeof t||!t||1!==p&&9!==p&&11!==p)return n;if(!r&&((e?e.ownerDocument||e:m)!==C&&T(e),e=e||C,E)){if(11!==p&&(u=Z.exec(t)))if(i=u[1]){if(9===p){if(!(a=e.getElementById(i)))return n;if(a.id===i)return n.push(a),n}else if(f&&(a=f.getElementById(i))&&y(e,a)&&a.id===i)return n.push(a),n}else{if(u[2])return H.apply(n,e.getElementsByTagName(t)),n;if((i=u[3])&&d.getElementsByClassName&&e.getElementsByClassName)return H.apply(n,e.getElementsByClassName(i)),n}if(d.qsa&&!A[t+" "]&&(!v||!v.test(t))&&(1!==p||"object"!==e.nodeName.toLowerCase())){if(c=t,f=e,1===p&&U.test(t)){(s=e.getAttribute("id"))?s=s.replace(re,ie):e.setAttribute("id",s=k),o=(l=h(t)).length;while(o--)l[o]="#"+s+" "+xe(l[o]);c=l.join(","),f=ee.test(t)&&ye(e.parentNode)||e}try{return H.apply(n,f.querySelectorAll(c)),n}catch(e){A(t,!0)}finally{s===k&&e.removeAttribute("id")}}}return g(t.replace(B,"$1"),e,n,r)}function ue(){var r=[];return function e(t,n){return r.push(t+" ")>b.cacheLength&&delete e[r.shift()],e[t+" "]=n}}function le(e){return e[k]=!0,e}function ce(e){var t=C.createElement("fieldset");try{return!!e(t)}catch(e){return!1}finally{t.parentNode&&t.parentNode.removeChild(t),t=null}}function fe(e,t){var n=e.split("|"),r=n.length;while(r--)b.attrHandle[n[r]]=t}function pe(e,t){var n=t&&e,r=n&&1===e.nodeType&&1===t.nodeType&&e.sourceIndex-t.sourceIndex;if(r)return r;if(n)while(n=n.nextSibling)if(n===t)return-1;return e?1:-1}function de(t){return function(e){return"input"===e.nodeName.toLowerCase()&&e.type===t}}function he(n){return function(e){var t=e.nodeName.toLowerCase();return("input"===t||"button"===t)&&e.type===n}}function ge(t){return function(e){return"form"in e?e.parentNode&&!1===e.disabled?"label"in e?"label"in e.parentNode?e.parentNode.disabled===t:e.disabled===t:e.isDisabled===t||e.isDisabled!==!t&&ae(e)===t:e.disabled===t:"label"in e&&e.disabled===t}}function ve(a){return le(function(o){return o=+o,le(function(e,t){var n,r=a([],e.length,o),i=r.length;while(i--)e[n=r[i]]&&(e[n]=!(t[n]=e[n]))})})}function ye(e){return e&&"undefined"!=typeof e.getElementsByTagName&&e}for(e in d=se.support={},i=se.isXML=function(e){var t=e.namespaceURI,n=(e.ownerDocument||e).documentElement;return!Y.test(t||n&&n.nodeName||"HTML")},T=se.setDocument=function(e){var t,n,r=e?e.ownerDocument||e:m;return r!==C&&9===r.nodeType&&r.documentElement&&(a=(C=r).documentElement,E=!i(C),m!==C&&(n=C.defaultView)&&n.top!==n&&(n.addEventListener?n.addEventListener("unload",oe,!1):n.attachEvent&&n.attachEvent("onunload",oe)),d.attributes=ce(function(e){return e.className="i",!e.getAttribute("className")}),d.getElementsByTagName=ce(function(e){return e.appendChild(C.createComment("")),!e.getElementsByTagName("*").length}),d.getElementsByClassName=K.test(C.getElementsByClassName),d.getById=ce(function(e){return a.appendChild(e).id=k,!C.getElementsByName||!C.getElementsByName(k).length}),d.getById?(b.filter.ID=function(e){var t=e.replace(te,ne);return function(e){return e.getAttribute("id")===t}},b.find.ID=function(e,t){if("undefined"!=typeof t.getElementById&&E){var n=t.getElementById(e);return n?[n]:[]}}):(b.filter.ID=function(e){var n=e.replace(te,ne);return function(e){var t="undefined"!=typeof e.getAttributeNode&&e.getAttributeNode("id");return t&&t.value===n}},b.find.ID=function(e,t){if("undefined"!=typeof t.getElementById&&E){var n,r,i,o=t.getElementById(e);if(o){if((n=o.getAttributeNode("id"))&&n.value===e)return[o];i=t.getElementsByName(e),r=0;while(o=i[r++])if((n=o.getAttributeNode("id"))&&n.value===e)return[o]}return[]}}),b.find.TAG=d.getElementsByTagName?function(e,t){return"undefined"!=typeof t.getElementsByTagName?t.getElementsByTagName(e):d.qsa?t.querySelectorAll(e):void 0}:function(e,t){var n,r=[],i=0,o=t.getElementsByTagName(e);if("*"===e){while(n=o[i++])1===n.nodeType&&r.push(n);return r}return o},b.find.CLASS=d.getElementsByClassName&&function(e,t){if("undefined"!=typeof t.getElementsByClassName&&E)return t.getElementsByClassName(e)},s=[],v=[],(d.qsa=K.test(C.querySelectorAll))&&(ce(function(e){a.appendChild(e).innerHTML="",e.querySelectorAll("[msallowcapture^='']").length&&v.push("[*^$]="+M+"*(?:''|\"\")"),e.querySelectorAll("[selected]").length||v.push("\\["+M+"*(?:value|"+R+")"),e.querySelectorAll("[id~="+k+"-]").length||v.push("~="),e.querySelectorAll(":checked").length||v.push(":checked"),e.querySelectorAll("a#"+k+"+*").length||v.push(".#.+[+~]")}),ce(function(e){e.innerHTML="";var t=C.createElement("input");t.setAttribute("type","hidden"),e.appendChild(t).setAttribute("name","D"),e.querySelectorAll("[name=d]").length&&v.push("name"+M+"*[*^$|!~]?="),2!==e.querySelectorAll(":enabled").length&&v.push(":enabled",":disabled"),a.appendChild(e).disabled=!0,2!==e.querySelectorAll(":disabled").length&&v.push(":enabled",":disabled"),e.querySelectorAll("*,:x"),v.push(",.*:")})),(d.matchesSelector=K.test(c=a.matches||a.webkitMatchesSelector||a.mozMatchesSelector||a.oMatchesSelector||a.msMatchesSelector))&&ce(function(e){d.disconnectedMatch=c.call(e,"*"),c.call(e,"[s!='']:x"),s.push("!=",$)}),v=v.length&&new RegExp(v.join("|")),s=s.length&&new RegExp(s.join("|")),t=K.test(a.compareDocumentPosition),y=t||K.test(a.contains)?function(e,t){var n=9===e.nodeType?e.documentElement:e,r=t&&t.parentNode;return e===r||!(!r||1!==r.nodeType||!(n.contains?n.contains(r):e.compareDocumentPosition&&16&e.compareDocumentPosition(r)))}:function(e,t){if(t)while(t=t.parentNode)if(t===e)return!0;return!1},D=t?function(e,t){if(e===t)return l=!0,0;var n=!e.compareDocumentPosition-!t.compareDocumentPosition;return n||(1&(n=(e.ownerDocument||e)===(t.ownerDocument||t)?e.compareDocumentPosition(t):1)||!d.sortDetached&&t.compareDocumentPosition(e)===n?e===C||e.ownerDocument===m&&y(m,e)?-1:t===C||t.ownerDocument===m&&y(m,t)?1:u?P(u,e)-P(u,t):0:4&n?-1:1)}:function(e,t){if(e===t)return l=!0,0;var n,r=0,i=e.parentNode,o=t.parentNode,a=[e],s=[t];if(!i||!o)return e===C?-1:t===C?1:i?-1:o?1:u?P(u,e)-P(u,t):0;if(i===o)return pe(e,t);n=e;while(n=n.parentNode)a.unshift(n);n=t;while(n=n.parentNode)s.unshift(n);while(a[r]===s[r])r++;return r?pe(a[r],s[r]):a[r]===m?-1:s[r]===m?1:0}),C},se.matches=function(e,t){return se(e,null,null,t)},se.matchesSelector=function(e,t){if((e.ownerDocument||e)!==C&&T(e),d.matchesSelector&&E&&!A[t+" "]&&(!s||!s.test(t))&&(!v||!v.test(t)))try{var n=c.call(e,t);if(n||d.disconnectedMatch||e.document&&11!==e.document.nodeType)return n}catch(e){A(t,!0)}return 0":{dir:"parentNode",first:!0}," ":{dir:"parentNode"},"+":{dir:"previousSibling",first:!0},"~":{dir:"previousSibling"}},preFilter:{ATTR:function(e){return e[1]=e[1].replace(te,ne),e[3]=(e[3]||e[4]||e[5]||"").replace(te,ne),"~="===e[2]&&(e[3]=" "+e[3]+" "),e.slice(0,4)},CHILD:function(e){return e[1]=e[1].toLowerCase(),"nth"===e[1].slice(0,3)?(e[3]||se.error(e[0]),e[4]=+(e[4]?e[5]+(e[6]||1):2*("even"===e[3]||"odd"===e[3])),e[5]=+(e[7]+e[8]||"odd"===e[3])):e[3]&&se.error(e[0]),e},PSEUDO:function(e){var t,n=!e[6]&&e[2];return G.CHILD.test(e[0])?null:(e[3]?e[2]=e[4]||e[5]||"":n&&X.test(n)&&(t=h(n,!0))&&(t=n.indexOf(")",n.length-t)-n.length)&&(e[0]=e[0].slice(0,t),e[2]=n.slice(0,t)),e.slice(0,3))}},filter:{TAG:function(e){var t=e.replace(te,ne).toLowerCase();return"*"===e?function(){return!0}:function(e){return e.nodeName&&e.nodeName.toLowerCase()===t}},CLASS:function(e){var t=p[e+" "];return t||(t=new RegExp("(^|"+M+")"+e+"("+M+"|$)"))&&p(e,function(e){return t.test("string"==typeof e.className&&e.className||"undefined"!=typeof e.getAttribute&&e.getAttribute("class")||"")})},ATTR:function(n,r,i){return function(e){var t=se.attr(e,n);return null==t?"!="===r:!r||(t+="","="===r?t===i:"!="===r?t!==i:"^="===r?i&&0===t.indexOf(i):"*="===r?i&&-1:\x20\t\r\n\f]*)[\x20\t\r\n\f]*\/?>(?:<\/\1>|)$/i;function j(e,n,r){return m(n)?k.grep(e,function(e,t){return!!n.call(e,t,e)!==r}):n.nodeType?k.grep(e,function(e){return e===n!==r}):"string"!=typeof n?k.grep(e,function(e){return-1)[^>]*|#([\w-]+))$/;(k.fn.init=function(e,t,n){var r,i;if(!e)return this;if(n=n||q,"string"==typeof e){if(!(r="<"===e[0]&&">"===e[e.length-1]&&3<=e.length?[null,e,null]:L.exec(e))||!r[1]&&t)return!t||t.jquery?(t||n).find(e):this.constructor(t).find(e);if(r[1]){if(t=t instanceof k?t[0]:t,k.merge(this,k.parseHTML(r[1],t&&t.nodeType?t.ownerDocument||t:E,!0)),D.test(r[1])&&k.isPlainObject(t))for(r in t)m(this[r])?this[r](t[r]):this.attr(r,t[r]);return this}return(i=E.getElementById(r[2]))&&(this[0]=i,this.length=1),this}return e.nodeType?(this[0]=e,this.length=1,this):m(e)?void 0!==n.ready?n.ready(e):e(k):k.makeArray(e,this)}).prototype=k.fn,q=k(E);var H=/^(?:parents|prev(?:Until|All))/,O={children:!0,contents:!0,next:!0,prev:!0};function P(e,t){while((e=e[t])&&1!==e.nodeType);return e}k.fn.extend({has:function(e){var t=k(e,this),n=t.length;return this.filter(function(){for(var e=0;e\x20\t\r\n\f]*)/i,he=/^$|^module$|\/(?:java|ecma)script/i,ge={option:[1,""],thead:[1,""],col:[2,""],tr:[2,""],td:[3,""],_default:[0,"",""]};function ve(e,t){var n;return n="undefined"!=typeof e.getElementsByTagName?e.getElementsByTagName(t||"*"):"undefined"!=typeof e.querySelectorAll?e.querySelectorAll(t||"*"):[],void 0===t||t&&A(e,t)?k.merge([e],n):n}function ye(e,t){for(var n=0,r=e.length;nx",y.noCloneChecked=!!me.cloneNode(!0).lastChild.defaultValue;var Te=/^key/,Ce=/^(?:mouse|pointer|contextmenu|drag|drop)|click/,Ee=/^([^.]*)(?:\.(.+)|)/;function ke(){return!0}function Se(){return!1}function Ne(e,t){return e===function(){try{return E.activeElement}catch(e){}}()==("focus"===t)}function Ae(e,t,n,r,i,o){var a,s;if("object"==typeof t){for(s in"string"!=typeof n&&(r=r||n,n=void 0),t)Ae(e,s,n,r,t[s],o);return e}if(null==r&&null==i?(i=n,r=n=void 0):null==i&&("string"==typeof n?(i=r,r=void 0):(i=r,r=n,n=void 0)),!1===i)i=Se;else if(!i)return e;return 1===o&&(a=i,(i=function(e){return k().off(e),a.apply(this,arguments)}).guid=a.guid||(a.guid=k.guid++)),e.each(function(){k.event.add(this,t,i,r,n)})}function De(e,i,o){o?(Q.set(e,i,!1),k.event.add(e,i,{namespace:!1,handler:function(e){var t,n,r=Q.get(this,i);if(1&e.isTrigger&&this[i]){if(r.length)(k.event.special[i]||{}).delegateType&&e.stopPropagation();else if(r=s.call(arguments),Q.set(this,i,r),t=o(this,i),this[i](),r!==(n=Q.get(this,i))||t?Q.set(this,i,!1):n={},r!==n)return e.stopImmediatePropagation(),e.preventDefault(),n.value}else r.length&&(Q.set(this,i,{value:k.event.trigger(k.extend(r[0],k.Event.prototype),r.slice(1),this)}),e.stopImmediatePropagation())}})):void 0===Q.get(e,i)&&k.event.add(e,i,ke)}k.event={global:{},add:function(t,e,n,r,i){var o,a,s,u,l,c,f,p,d,h,g,v=Q.get(t);if(v){n.handler&&(n=(o=n).handler,i=o.selector),i&&k.find.matchesSelector(ie,i),n.guid||(n.guid=k.guid++),(u=v.events)||(u=v.events={}),(a=v.handle)||(a=v.handle=function(e){return"undefined"!=typeof k&&k.event.triggered!==e.type?k.event.dispatch.apply(t,arguments):void 0}),l=(e=(e||"").match(R)||[""]).length;while(l--)d=g=(s=Ee.exec(e[l])||[])[1],h=(s[2]||"").split(".").sort(),d&&(f=k.event.special[d]||{},d=(i?f.delegateType:f.bindType)||d,f=k.event.special[d]||{},c=k.extend({type:d,origType:g,data:r,handler:n,guid:n.guid,selector:i,needsContext:i&&k.expr.match.needsContext.test(i),namespace:h.join(".")},o),(p=u[d])||((p=u[d]=[]).delegateCount=0,f.setup&&!1!==f.setup.call(t,r,h,a)||t.addEventListener&&t.addEventListener(d,a)),f.add&&(f.add.call(t,c),c.handler.guid||(c.handler.guid=n.guid)),i?p.splice(p.delegateCount++,0,c):p.push(c),k.event.global[d]=!0)}},remove:function(e,t,n,r,i){var o,a,s,u,l,c,f,p,d,h,g,v=Q.hasData(e)&&Q.get(e);if(v&&(u=v.events)){l=(t=(t||"").match(R)||[""]).length;while(l--)if(d=g=(s=Ee.exec(t[l])||[])[1],h=(s[2]||"").split(".").sort(),d){f=k.event.special[d]||{},p=u[d=(r?f.delegateType:f.bindType)||d]||[],s=s[2]&&new RegExp("(^|\\.)"+h.join("\\.(?:.*\\.|)")+"(\\.|$)"),a=o=p.length;while(o--)c=p[o],!i&&g!==c.origType||n&&n.guid!==c.guid||s&&!s.test(c.namespace)||r&&r!==c.selector&&("**"!==r||!c.selector)||(p.splice(o,1),c.selector&&p.delegateCount--,f.remove&&f.remove.call(e,c));a&&!p.length&&(f.teardown&&!1!==f.teardown.call(e,h,v.handle)||k.removeEvent(e,d,v.handle),delete u[d])}else for(d in u)k.event.remove(e,d+t[l],n,r,!0);k.isEmptyObject(u)&&Q.remove(e,"handle events")}},dispatch:function(e){var t,n,r,i,o,a,s=k.event.fix(e),u=new Array(arguments.length),l=(Q.get(this,"events")||{})[s.type]||[],c=k.event.special[s.type]||{};for(u[0]=s,t=1;t\x20\t\r\n\f]*)[^>]*)\/>/gi,qe=/\s*$/g;function Oe(e,t){return A(e,"table")&&A(11!==t.nodeType?t:t.firstChild,"tr")&&k(e).children("tbody")[0]||e}function Pe(e){return e.type=(null!==e.getAttribute("type"))+"/"+e.type,e}function Re(e){return"true/"===(e.type||"").slice(0,5)?e.type=e.type.slice(5):e.removeAttribute("type"),e}function Me(e,t){var n,r,i,o,a,s,u,l;if(1===t.nodeType){if(Q.hasData(e)&&(o=Q.access(e),a=Q.set(t,o),l=o.events))for(i in delete a.handle,a.events={},l)for(n=0,r=l[i].length;n")},clone:function(e,t,n){var r,i,o,a,s,u,l,c=e.cloneNode(!0),f=oe(e);if(!(y.noCloneChecked||1!==e.nodeType&&11!==e.nodeType||k.isXMLDoc(e)))for(a=ve(c),r=0,i=(o=ve(e)).length;r").attr(n.scriptAttrs||{}).prop({charset:n.scriptCharset,src:n.url}).on("load error",i=function(e){r.remove(),i=null,e&&t("error"===e.type?404:200,e.type)}),E.head.appendChild(r[0])},abort:function(){i&&i()}}});var Vt,Gt=[],Yt=/(=)\?(?=&|$)|\?\?/;k.ajaxSetup({jsonp:"callback",jsonpCallback:function(){var e=Gt.pop()||k.expando+"_"+kt++;return this[e]=!0,e}}),k.ajaxPrefilter("json jsonp",function(e,t,n){var r,i,o,a=!1!==e.jsonp&&(Yt.test(e.url)?"url":"string"==typeof e.data&&0===(e.contentType||"").indexOf("application/x-www-form-urlencoded")&&Yt.test(e.data)&&"data");if(a||"jsonp"===e.dataTypes[0])return r=e.jsonpCallback=m(e.jsonpCallback)?e.jsonpCallback():e.jsonpCallback,a?e[a]=e[a].replace(Yt,"$1"+r):!1!==e.jsonp&&(e.url+=(St.test(e.url)?"&":"?")+e.jsonp+"="+r),e.converters["script json"]=function(){return o||k.error(r+" was not called"),o[0]},e.dataTypes[0]="json",i=C[r],C[r]=function(){o=arguments},n.always(function(){void 0===i?k(C).removeProp(r):C[r]=i,e[r]&&(e.jsonpCallback=t.jsonpCallback,Gt.push(r)),o&&m(i)&&i(o[0]),o=i=void 0}),"script"}),y.createHTMLDocument=((Vt=E.implementation.createHTMLDocument("").body).innerHTML="",2===Vt.childNodes.length),k.parseHTML=function(e,t,n){return"string"!=typeof e?[]:("boolean"==typeof t&&(n=t,t=!1),t||(y.createHTMLDocument?((r=(t=E.implementation.createHTMLDocument("")).createElement("base")).href=E.location.href,t.head.appendChild(r)):t=E),o=!n&&[],(i=D.exec(e))?[t.createElement(i[1])]:(i=we([e],t,o),o&&o.length&&k(o).remove(),k.merge([],i.childNodes)));var r,i,o},k.fn.load=function(e,t,n){var r,i,o,a=this,s=e.indexOf(" ");return-1").append(k.parseHTML(e)).find(r):e)}).always(n&&function(e,t){a.each(function(){n.apply(this,o||[e.responseText,t,e])})}),this},k.each(["ajaxStart","ajaxStop","ajaxComplete","ajaxError","ajaxSuccess","ajaxSend"],function(e,t){k.fn[t]=function(e){return this.on(t,e)}}),k.expr.pseudos.animated=function(t){return k.grep(k.timers,function(e){return t===e.elem}).length},k.offset={setOffset:function(e,t,n){var r,i,o,a,s,u,l=k.css(e,"position"),c=k(e),f={};"static"===l&&(e.style.position="relative"),s=c.offset(),o=k.css(e,"top"),u=k.css(e,"left"),("absolute"===l||"fixed"===l)&&-1<(o+u).indexOf("auto")?(a=(r=c.position()).top,i=r.left):(a=parseFloat(o)||0,i=parseFloat(u)||0),m(t)&&(t=t.call(e,n,k.extend({},s))),null!=t.top&&(f.top=t.top-s.top+a),null!=t.left&&(f.left=t.left-s.left+i),"using"in t?t.using.call(e,f):c.css(f)}},k.fn.extend({offset:function(t){if(arguments.length)return void 0===t?this:this.each(function(e){k.offset.setOffset(this,t,e)});var e,n,r=this[0];return r?r.getClientRects().length?(e=r.getBoundingClientRect(),n=r.ownerDocument.defaultView,{top:e.top+n.pageYOffset,left:e.left+n.pageXOffset}):{top:0,left:0}:void 0},position:function(){if(this[0]){var e,t,n,r=this[0],i={top:0,left:0};if("fixed"===k.css(r,"position"))t=r.getBoundingClientRect();else{t=this.offset(),n=r.ownerDocument,e=r.offsetParent||n.documentElement;while(e&&(e===n.body||e===n.documentElement)&&"static"===k.css(e,"position"))e=e.parentNode;e&&e!==r&&1===e.nodeType&&((i=k(e).offset()).top+=k.css(e,"borderTopWidth",!0),i.left+=k.css(e,"borderLeftWidth",!0))}return{top:t.top-i.top-k.css(r,"marginTop",!0),left:t.left-i.left-k.css(r,"marginLeft",!0)}}},offsetParent:function(){return this.map(function(){var e=this.offsetParent;while(e&&"static"===k.css(e,"position"))e=e.offsetParent;return e||ie})}}),k.each({scrollLeft:"pageXOffset",scrollTop:"pageYOffset"},function(t,i){var o="pageYOffset"===i;k.fn[t]=function(e){return _(this,function(e,t,n){var r;if(x(e)?r=e:9===e.nodeType&&(r=e.defaultView),void 0===n)return r?r[i]:e[t];r?r.scrollTo(o?r.pageXOffset:n,o?n:r.pageYOffset):e[t]=n},t,e,arguments.length)}}),k.each(["top","left"],function(e,n){k.cssHooks[n]=ze(y.pixelPosition,function(e,t){if(t)return t=_e(e,n),$e.test(t)?k(e).position()[n]+"px":t})}),k.each({Height:"height",Width:"width"},function(a,s){k.each({padding:"inner"+a,content:s,"":"outer"+a},function(r,o){k.fn[o]=function(e,t){var n=arguments.length&&(r||"boolean"!=typeof e),i=r||(!0===e||!0===t?"margin":"border");return _(this,function(e,t,n){var r;return x(e)?0===o.indexOf("outer")?e["inner"+a]:e.document.documentElement["client"+a]:9===e.nodeType?(r=e.documentElement,Math.max(e.body["scroll"+a],r["scroll"+a],e.body["offset"+a],r["offset"+a],r["client"+a])):void 0===n?k.css(e,t,i):k.style(e,t,n,i)},s,n?e:void 0,n)}})}),k.each("blur focus focusin focusout resize scroll click dblclick mousedown mouseup mousemove mouseover mouseout mouseenter mouseleave change select submit keydown keypress keyup contextmenu".split(" "),function(e,n){k.fn[n]=function(e,t){return 0 1)):
+ elif (
+ (ctx.triggered[0]["prop_id"] == "prediction_picking.selectedData")
+ and (selected_data is not None)
+ and (len(selected_data) > 1)
+ ):
df = select_data_from_prediction_picking(self.round_dataframe, selected_data)
# If click on reset button
- elif ctx.triggered[0]['prop_id'] == 'reset_dropdown_button.n_clicks':
+ elif ctx.triggered[0]["prop_id"] == "reset_dropdown_button.n_clicks":
df = self.round_dataframe
# If click on Apply filter
- elif ((ctx.triggered[0]['prop_id'] == 'apply_filter.n_clicks') | (
- (ctx.triggered[0]['prop_id'] == 'prediction_picking.selectedData') and
- ((selected_data is None))) | (
- (ctx.triggered[0]['prop_id'] == 'prediction_picking.selectedData') and
- (selected_data is not None and len(selected_data) == 1 and
- len(selected_data['points'])!=0 and selected_data['points'][0]['curveNumber'] > 0)
- )):
+ elif (
+ (ctx.triggered[0]["prop_id"] == "apply_filter.n_clicks")
+ | ((ctx.triggered[0]["prop_id"] == "prediction_picking.selectedData") and (selected_data is None))
+ | (
+ (ctx.triggered[0]["prop_id"] == "prediction_picking.selectedData")
+ and (
+ selected_data is not None
+ and len(selected_data) == 1
+ and len(selected_data["points"]) != 0
+ and selected_data["points"][0]["curveNumber"] > 0
+ )
+ )
+ ):
df = self.round_dataframe.copy()
- feature_id = [id_feature[i]['index'] for i in range(len(id_feature))]
+ feature_id = [id_feature[i]["index"] for i in range(len(id_feature))]
df = select_data_from_str_filters(
df,
- feature_id,
- id_str_modality,
- val_feature,
+ feature_id,
+ id_str_modality,
+ val_feature,
val_str_modality,
)
df = select_data_from_bool_filters(
@@ -1500,25 +1588,25 @@ def update_datatable(selected_data,
df = select_data_from_date_filters(
df,
feature_id,
- id_date,
+ id_date,
val_feature,
start_date,
end_date,
)
- filtered_subset_info = f"Subset length: {len(df)} ({int(round(100*len(df)/self.explainer.x_init.shape[0]))}%)"
- if len(df)==0:
- filtered_subset_color="danger"
-
+ filtered_subset_info = (
+ f"Subset length: {len(df)} ({int(round(100*len(df)/self.explainer.x_init.shape[0]))}%)"
+ )
+ if len(df) == 0:
+ filtered_subset_color = "danger"
+
elif None not in nclicks_del:
df = self.round_dataframe
else:
raise dash.exceptions.PreventUpdate
- data = df.to_dict('records')
+ data = df.to_dict("records")
tooltip_data = [
- {
- column: {'value': str(value), 'type': 'text'}
- for column, value in row.items()
- } for row in df.to_dict('index').values()
+ {column: {"value": str(value), "type": "text"} for column, value in row.items()}
+ for row in df.to_dict("index").values()
]
return (
data,
@@ -1530,41 +1618,43 @@ def update_datatable(selected_data,
@app.callback(
[
- Output('global_feature_importance', 'figure'),
- Output('global_feature_importance', 'clickData'),
- Output('clickdata-store', 'data')
+ Output("global_feature_importance", "figure"),
+ Output("global_feature_importance", "clickData"),
+ Output("clickdata-store", "data"),
],
[
- Input('select_label', 'value'),
- Input('dataset', 'data'),
- Input('prediction_picking', 'selectedData'),
- Input('apply_filter', 'n_clicks'),
- Input('reset_dropdown_button', 'n_clicks'),
- Input({'type': 'del_dropdown_button', 'index': ALL}, 'n_clicks'),
- Input('card_global_feature_importance', 'n_clicks'),
- Input('bool_groups', 'on'),
- Input('ember_global_feature_importance', 'n_clicks')
+ Input("select_label", "value"),
+ Input("dataset", "data"),
+ Input("prediction_picking", "selectedData"),
+ Input("apply_filter", "n_clicks"),
+ Input("reset_dropdown_button", "n_clicks"),
+ Input({"type": "del_dropdown_button", "index": ALL}, "n_clicks"),
+ Input("card_global_feature_importance", "n_clicks"),
+ Input("bool_groups", "on"),
+ Input("ember_global_feature_importance", "n_clicks"),
],
[
- State('global_feature_importance', 'clickData'),
- State('global_feature_importance', 'figure'),
- State('features', 'value'),
- State('clickdata-store', 'data')
- ]
+ State("global_feature_importance", "clickData"),
+ State("global_feature_importance", "figure"),
+ State("features", "value"),
+ State("clickdata-store", "data"),
+ ],
)
- def update_feature_importance(label,
- data,
- selected_data,
- apply_filters,
- reset_filter,
- nclicks_del,
- n_clicks,
- bool_group,
- click_zoom,
- clickData,
- figure,
- features,
- clickData_store):
+ def update_feature_importance(
+ label,
+ data,
+ selected_data,
+ apply_filters,
+ reset_filter,
+ nclicks_del,
+ n_clicks,
+ bool_group,
+ click_zoom,
+ clickData,
+ figure,
+ features,
+ clickData_store,
+ ):
"""
update feature importance plot according label, click on graph,
filters applied and subset selected in prediction picking graph.
@@ -1593,29 +1683,32 @@ def update_feature_importance(label,
zoom_active = get_figure_zoom(click_zoom)
selection = None
list_index = self.list_index
- if clickData is not None and (ctx.triggered[0]['prop_id'] in [
- 'apply_filter.n_clicks',
- 'reset_dropdown_button.n_clicks',
- 'dataset.data'
- ] or ('del_dropdown_button' in ctx.triggered[0]['prop_id'] and
- None not in nclicks_del)):
+ if clickData is not None and (
+ ctx.triggered[0]["prop_id"]
+ in ["apply_filter.n_clicks", "reset_dropdown_button.n_clicks", "dataset.data"]
+ or ("del_dropdown_button" in ctx.triggered[0]["prop_id"] and None not in nclicks_del)
+ ):
clickData = update_click_data_on_subset_changes(clickData)
- selected_feature = self.explainer.inv_features_dict.get(
- get_feature_from_clicked_data(clickData)
- ) if clickData else None
+ selected_feature = (
+ self.explainer.inv_features_dict.get(get_feature_from_clicked_data(clickData)) if clickData else None
+ )
if self.explainer.features_groups and bool_group:
- if ctx.triggered[0]['prop_id'] == 'card_global_feature_importance.n_clicks':
+ if ctx.triggered[0]["prop_id"] == "card_global_feature_importance.n_clicks":
# When we click twice on the same bar this will reset the graph
if clickData_store == clickData:
selected_feature = None
- selected_feature = get_feature_from_features_groups(selected_feature, self.explainer.features_groups)
- elif ctx.triggered[0]['prop_id'] == 'ember_global_feature_importance.n_clicks':
- selected_feature = get_feature_from_features_groups(selected_feature, self.explainer.features_groups)
+ selected_feature = get_feature_from_features_groups(
+ selected_feature, self.explainer.features_groups
+ )
+ elif ctx.triggered[0]["prop_id"] == "ember_global_feature_importance.n_clicks":
+ selected_feature = get_feature_from_features_groups(
+ selected_feature, self.explainer.features_groups
+ )
else:
selected_feature = None
-
+
selection = get_indexes_from_datatable(data, list_index)
group_name = get_group_name(selected_feature, self.explainer.features_groups)
@@ -1626,44 +1719,35 @@ def update_feature_importance(label,
label=label,
group_name=group_name,
display_groups=bool_group,
- zoom=zoom_active
+ zoom=zoom_active,
)
# Adjust graph with adding x axis title
- MyGraph.adjust_graph_static(figure, x_ax='Mean absolute Contribution')
- figure.layout.clickmode = 'event+select'
+ MyGraph.adjust_graph_static(figure, x_ax="Mean absolute Contribution")
+ figure.layout.clickmode = "event+select"
if selected_feature:
self.select_point(figure, clickData)
# font size can be adapted to screen size
- nb_car = max([len(figure.data[0].y[i]) for i in
- range(len(figure.data[0].y))])
- figure.update_layout(
- yaxis=dict(tickfont={'size': min(round(500 / nb_car), 12)})
- )
+ nb_car = max([len(figure.data[0].y[i]) for i in range(len(figure.data[0].y))])
+ figure.update_layout(yaxis=dict(tickfont={"size": min(round(500 / nb_car), 12)}))
clickData_store = clickData.copy() if clickData is not None else None
return figure, clickData, clickData_store
@app.callback(
- Output(component_id='feature_selector', component_property='figure'),
+ Output(component_id="feature_selector", component_property="figure"),
[
- Input('global_feature_importance', 'clickData'),
- Input('dataset', 'data'),
- Input('select_label', 'value'),
- Input('ember_feature_selector', 'n_clicks')
+ Input("global_feature_importance", "clickData"),
+ Input("dataset", "data"),
+ Input("select_label", "value"),
+ Input("ember_feature_selector", "n_clicks"),
],
[
- State('points', 'value'),
- State('violin', 'value'),
- State('global_feature_importance', 'figure'),
- ]
+ State("points", "value"),
+ State("violin", "value"),
+ State("global_feature_importance", "figure"),
+ ],
)
- def update_feature_selector(feature,
- data,
- label,
- click_zoom,
- points,
- violin,
- gfi_figure):
+ def update_feature_selector(feature, data, label, click_zoom, points, violin, gfi_figure):
"""
Update feature plot according to label, data,
selected feature on features importance graph,
@@ -1689,55 +1773,45 @@ def update_feature_selector(feature,
else:
selected_feature = self.selected_feature
- if feature is not None and feature['points'][0]['curveNumber'] == 0 and \
- len(gfi_figure['data']) == 2:
+ if feature is not None and feature["points"][0]["curveNumber"] == 0 and len(gfi_figure["data"]) == 2:
subset = get_indexes_from_datatable(data, list_index)
else:
subset = None
fs_figure = self.explainer.plot.contribution_plot(
- col=selected_feature,
- selection=subset,
- label=label,
- violin_maxf=violin,
- max_points=points,
- zoom=zoom_active
- )
+ col=selected_feature,
+ selection=subset,
+ label=label,
+ violin_maxf=violin,
+ max_points=points,
+ zoom=zoom_active,
+ )
- fs_figure['layout'].clickmode = 'event+select'
+ fs_figure["layout"].clickmode = "event+select"
# Adjust graph with adding x and y axis titles
- MyGraph.adjust_graph_static(fs_figure,
- #x_ax=truncate_str(self.layout.selected_feature, 110),
+ MyGraph.adjust_graph_static(
+ fs_figure,
+ # x_ax=truncate_str(self.layout.selected_feature, 110),
x_ax=truncate_str(selected_feature, 110),
- y_ax='Contribution')
+ y_ax="Contribution",
+ )
return fs_figure
@app.callback(
+ [Output("index_id", "value"), Output("index_id", "n_submit")],
[
- Output('index_id', 'value'),
- Output("index_id", "n_submit")
+ Input("feature_selector", "clickData"),
+ Input("prediction_picking", "clickData"),
+ Input("dataset", "active_cell"),
+ Input("apply_filter", "n_clicks"),
+ Input("reset_dropdown_button", "n_clicks"),
+ Input({"type": "del_dropdown_button", "index": ALL}, "n_clicks"),
],
- [
- Input('feature_selector', 'clickData'),
- Input('prediction_picking', 'clickData'),
- Input('dataset', 'active_cell'),
- Input('apply_filter', 'n_clicks'),
- Input('reset_dropdown_button', 'n_clicks'),
- Input({'type': 'del_dropdown_button', 'index': ALL}, 'n_clicks')
- ],
- [
- State('dataset', 'data'),
- State('index_id', 'value') # Get the current value of the index
- ]
+ [State("dataset", "data"), State("index_id", "value")], # Get the current value of the index
)
- def update_index_id(click_data,
- prediction_picking,
- cell,
- apply_filters,
- reset_filter,
- nclicks_del,
- data,
- current_index_id):
+ def update_index_id(
+ click_data, prediction_picking, cell, apply_filters, reset_filter, nclicks_del, data, current_index_id
+ ):
"""
This function is used to update index value according to
active cell, filters and click data on feature plot or on
@@ -1758,91 +1832,75 @@ def update_index_id(click_data,
"""
ctx = dash.callback_context
selected = None
- if ctx.triggered[0]['prop_id'] == 'feature_selector.clickData':
- selected = click_data['points'][0]['customdata'][1]
- elif ctx.triggered[0]['prop_id'] == 'prediction_picking.clickData':
- selected = prediction_picking['points'][0]['customdata']
- elif ctx.triggered[0]['prop_id'] == 'dataset.active_cell':
- selected = data[cell['row']]['_index_']
- elif (('del_dropdown_button' in ctx.triggered[0]['prop_id']) &
- (None in nclicks_del)):
+ if ctx.triggered[0]["prop_id"] == "feature_selector.clickData":
+ selected = click_data["points"][0]["customdata"][1]
+ elif ctx.triggered[0]["prop_id"] == "prediction_picking.clickData":
+ selected = prediction_picking["points"][0]["customdata"]
+ elif ctx.triggered[0]["prop_id"] == "dataset.active_cell":
+ selected = data[cell["row"]]["_index_"]
+ elif ("del_dropdown_button" in ctx.triggered[0]["prop_id"]) & (None in nclicks_del):
selected = current_index_id
return selected, True
- @app.callback(
- Output('threshold_label', 'children'),
- [Input('threshold_id', 'value')])
+ @app.callback(Output("threshold_label", "children"), [Input("threshold_id", "value")])
def update_threshold_label(value):
"""
update threshold label
"""
- return f'Threshold: {value}'
+ return f"Threshold: {value}"
- @app.callback(
- Output('max_contrib_label', 'children'),
- [Input('max_contrib_id', 'value')])
+ @app.callback(Output("max_contrib_label", "children"), [Input("max_contrib_id", "value")])
def update_max_contrib_label(value):
"""
update max_contrib label
"""
- return f'Features to display: {value}'
+ return f"Features to display: {value}"
@app.callback(
- [Output('max_contrib_id', 'value'),
- Output('max_contrib_id', 'max'),
- Output('max_contrib_id', 'marks')
- ],
- [Input('modal', 'is_open')],
- [State('features', 'value'),
- State('max_contrib_id', 'value')]
+ [Output("max_contrib_id", "value"), Output("max_contrib_id", "max"), Output("max_contrib_id", "marks")],
+ [Input("modal", "is_open")],
+ [State("features", "value"), State("max_contrib_id", "value")],
)
- def update_max_contrib_id(is_open,
- features,
- value):
+ def update_max_contrib_id(is_open, features, value):
"""
update max contrib component layout after settings modifications
"""
ctx = dash.callback_context
- if ctx.triggered[0]['prop_id'] == 'modal.is_open':
+ if ctx.triggered[0]["prop_id"] == "modal.is_open":
if is_open:
raise PreventUpdate
else:
- value, max, marks = update_features_to_display(
- features,
- len(self.explainer.x_init.columns),
- value
- )
+ value, max, marks = update_features_to_display(features, len(self.explainer.x_init.columns), value)
return value, max, marks
@app.callback(
- Output(component_id='detail_feature', component_property='figure'),
+ Output(component_id="detail_feature", component_property="figure"),
[
- Input('threshold_id', 'value'),
- Input('max_contrib_id', 'value'),
- Input('check_id_positive', 'value'),
- Input('check_id_negative', 'value'),
- Input('masked_contrib_id', 'value'),
- Input('select_label', 'value'),
+ Input("threshold_id", "value"),
+ Input("max_contrib_id", "value"),
+ Input("check_id_positive", "value"),
+ Input("check_id_negative", "value"),
+ Input("masked_contrib_id", "value"),
+ Input("select_label", "value"),
Input("validation", "n_clicks"),
- Input('bool_groups', 'on'),
- Input('ember_detail_feature', 'n_clicks'),
+ Input("bool_groups", "on"),
+ Input("ember_detail_feature", "n_clicks"),
],
- [
- State('index_id', 'value'),
- State('dataset', 'data')
- ]
+ [State("index_id", "value"), State("dataset", "data")],
)
- def update_detail_feature(threshold,
- max_contrib,
- positive,
- negative,
- masked,
- label,
- validation_click,
- bool_group,
- click_zoom,
- index,
- data):
+ def update_detail_feature(
+ threshold,
+ max_contrib,
+ positive,
+ negative,
+ masked,
+ label,
+ validation_click,
+ bool_group,
+ click_zoom,
+ index,
+ data,
+ ):
"""
update local explanation plot according to app changes.
-------------------------------------------------------
@@ -1871,39 +1929,36 @@ def update_detail_feature(threshold,
selected = None
threshold = threshold if threshold != 0 else None
sign = get_feature_contributions_sign_to_show(positive, negative)
- self.explainer.filter(threshold=threshold,
- features_to_hide=masked,
- positive=sign,
- max_contrib=max_contrib,
- display_groups=bool_group)
+ self.explainer.filter(
+ threshold=threshold,
+ features_to_hide=masked,
+ positive=sign,
+ max_contrib=max_contrib,
+ display_groups=bool_group,
+ )
figure = self.explainer.plot.local_plot(
index=selected,
label=label,
show_masked=True,
yaxis_max_label=8,
display_groups=bool_group,
- zoom=zoom_active
+ zoom=zoom_active,
)
if selected is not None:
# Adjust graph with adding x axis titles
- MyGraph.adjust_graph_static(figure, x_ax='Contribution')
+ MyGraph.adjust_graph_static(figure, x_ax="Contribution")
# font size can be adapted to screen size
- list_yaxis = [figure.data[i].y[0] for i in
- range(len(figure.data))]
+ list_yaxis = [figure.data[i].y[0] for i in range(len(figure.data))]
# exclude new line with labels of y axis
if list_yaxis != []:
- list_yaxis = [x.split('
')[0] for x in list_yaxis]
+ list_yaxis = [x.split("
")[0] for x in list_yaxis]
nb_car = max([len(x) for x in list_yaxis])
- figure.update_layout(
- yaxis=dict(tickfont={'size': min(round(500 / nb_car), 12)})
- )
+ figure.update_layout(yaxis=dict(tickfont={"size": min(round(500 / nb_car), 12)}))
return figure
@app.callback(
Output("validation", "n_clicks"),
- [
- Input("index_id", "n_submit")
- ],
+ [Input("index_id", "n_submit")],
)
def click_validation(n_submit):
"""
@@ -1913,20 +1968,20 @@ def click_validation(n_submit):
return 1
else:
raise PreventUpdate
-
+
@app.callback(
- Output('id_card', 'style'),
- Output('id_card_body', 'children'),
- Output('id_card_title_contrib', 'children'),
+ Output("id_card", "style"),
+ Output("id_card_body", "children"),
+ Output("id_card_title_contrib", "children"),
[
- Input('index_id', 'n_submit'),
- Input('select_label', 'value'),
- Input('select_id_card_sorting', 'value'),
- Input('select_id_card_order', 'value'),
+ Input("index_id", "n_submit"),
+ Input("select_label", "value"),
+ Input("select_id_card_sorting", "value"),
+ Input("select_id_card_order", "value"),
],
[
- State('dataset', 'data'),
- State('index_id', 'value'),
+ State("dataset", "data"),
+ State("index_id", "value"),
],
)
def update_id_card(n_submit, label, sort_by, order, data, index):
@@ -1948,16 +2003,12 @@ def update_id_card(n_submit, label, sort_by, order, data, index):
title_contrib = "Contribution"
if n_submit and selected is not None:
selected_row = get_id_card_features(data, selected, self.special_cols, self.features_dict)
- if self.explainer._case == 'classification':
+ if self.explainer._case == "classification":
if label is None:
label = -1
label_num, _, label_value = self.explainer.check_label_name(label)
selected_contrib = get_id_card_contrib(
- self.explainer.data,
- index,
- self.explainer.features_dict,
- self.explainer.columns_dict,
- label_num
+ self.explainer.data, index, self.explainer.features_dict, self.explainer.columns_dict, label_num
)
proba = self.explainer.plot.local_pred(index, label_num)
title_contrib = f"Contribution: {label_value} ({proba.round(2):.2f})"
@@ -1967,36 +2018,31 @@ def update_id_card(n_submit, label, sort_by, order, data, index):
selected_row.loc["_predict_", "feature_value"] = predicted_label_value
else:
selected_contrib = get_id_card_contrib(
- self.explainer.data,
+ self.explainer.data,
index,
self.explainer.features_dict,
self.explainer.columns_dict,
)
-
+
selected_data = create_id_card_data(
selected_row,
selected_contrib,
sort_by,
order,
self.special_cols,
- self.explainer.additional_features_dict
+ self.explainer.additional_features_dict,
)
children = create_id_card_layout(selected_data, self.explainer.additional_features_dict)
- return {"display":"flex", "margin-left":"auto", "margin-right":0}, children, title_contrib
+ return {"display": "flex", "margin-left": "auto", "margin-right": 0}, children, title_contrib
else:
- return {"display":"none"}, [], title_contrib
-
+ return {"display": "none"}, [], title_contrib
+
@app.callback(
Output("modal_id_card", "is_open"),
- [
- Input("id_card", "n_clicks"),
- Input("close_id_card", "n_clicks")
- ],
- [
- State("modal_id_card", "is_open")
- ],
+ [Input("id_card", "n_clicks"), Input("close_id_card", "n_clicks")],
+ [State("modal_id_card", "is_open")],
)
def toggle_modal_id_card(n1, n2, is_open):
"""
@@ -2016,49 +2062,36 @@ def toggle_modal_id_card(n1, n2, is_open):
@app.callback(
[
- Output('dataset', 'style_data_conditional'),
- Output('dataset', 'style_filter_conditional'),
- Output('dataset', 'style_header_conditional'),
- Output('dataset', 'style_cell_conditional'),
+ Output("dataset", "style_data_conditional"),
+ Output("dataset", "style_filter_conditional"),
+ Output("dataset", "style_header_conditional"),
+ Output("dataset", "style_cell_conditional"),
],
- [
- Input("validation", "n_clicks")
- ],
- [
- State('dataset', 'data'),
- State('index_id', 'value')
- ]
+ [Input("validation", "n_clicks")],
+ [State("dataset", "data"), State("index_id", "value")],
)
- def datatable_layout(validation,
- data,
- index):
+ def datatable_layout(validation, data, index):
ctx = dash.callback_context
- if ctx.triggered[0]['prop_id'] == 'validation.n_clicks' and validation is not None:
+ if ctx.triggered[0]["prop_id"] == "validation.n_clicks" and validation is not None:
pass
else:
raise PreventUpdate
style_data_conditional = [
+ {"if": {"row_index": "odd"}, "backgroundColor": "rgb(248, 248, 248)"},
{
- 'if': {'row_index': 'odd'},
- 'backgroundColor': 'rgb(248, 248, 248)'
- },
- {
- "if": {"state": "selected"},
+ "if": {"state": "selected"},
"border": f"1px solid {self.color[0]}",
- }
+ },
]
style_filter_conditional = []
- style_header_conditional = [
- {'if': {'column_id': c}, 'fontWeight': 'bold'}
- for c in self.special_cols
- ] + [
- {'if': {'column_id': c}, 'font-style': 'italic'}
- for c in self.dataframe if c in self.explainer.additional_features_dict
+ style_header_conditional = [{"if": {"column_id": c}, "fontWeight": "bold"} for c in self.special_cols] + [
+ {"if": {"column_id": c}, "font-style": "italic"}
+ for c in self.dataframe
+ if c in self.explainer.additional_features_dict
]
style_cell_conditional = [
- {'if': {'column_id': c},
- 'width': '70px', 'fontWeight': 'bold'} for c in self.special_cols
+ {"if": {"column_id": c}, "width": "70px", "fontWeight": "bold"} for c in self.special_cols
]
selected = check_row(data, index)
@@ -2068,33 +2101,22 @@ def datatable_layout(validation,
return style_data_conditional, style_filter_conditional, style_header_conditional, style_cell_conditional
@app.callback(
- Output('prediction_picking', 'figure'),
- Output('prediction_picking', 'selectedData'),
+ Output("prediction_picking", "figure"),
+ Output("prediction_picking", "selectedData"),
[
- Input('dataset', 'data'),
- Input('apply_filter', 'n_clicks'),
- Input('reset_dropdown_button', 'n_clicks'),
- Input({'type': 'del_dropdown_button', 'index': ALL}, 'n_clicks'),
- Input('select_label', 'value'),
- Input('modal', 'is_open'),
- Input('ember_prediction_picking', 'n_clicks')
+ Input("dataset", "data"),
+ Input("apply_filter", "n_clicks"),
+ Input("reset_dropdown_button", "n_clicks"),
+ Input({"type": "del_dropdown_button", "index": ALL}, "n_clicks"),
+ Input("select_label", "value"),
+ Input("modal", "is_open"),
+ Input("ember_prediction_picking", "n_clicks"),
],
- [
- State('points', 'value'),
- State('violin', 'value'),
- State('prediction_picking','selectedData')
- ]
+ [State("points", "value"), State("violin", "value"), State("prediction_picking", "selectedData")],
)
- def update_prediction_picking(data,
- apply_filters,
- reset_filter,
- nclicks_del,
- label,
- is_open,
- click_zoom,
- points,
- violin,
- selectedData):
+ def update_prediction_picking(
+ data, apply_filters, reset_filter, nclicks_del, label, is_open, click_zoom, points, violin, selectedData
+ ):
"""
Update feature plot according to label, data,
selected feature and settings modifications
@@ -2115,39 +2137,31 @@ def update_prediction_picking(data,
ctx = dash.callback_context
# Filter subset
subset = None
- if (ctx.triggered[0]['prop_id'] == 'apply_filter.n_clicks' or
- ctx.triggered[0]['prop_id'] == 'reset_dropdown_button.n_clicks' or
- ('del_dropdown_button' in ctx.triggered[0]['prop_id'] and
- None not in nclicks_del)):
+ if (
+ ctx.triggered[0]["prop_id"] == "apply_filter.n_clicks"
+ or ctx.triggered[0]["prop_id"] == "reset_dropdown_button.n_clicks"
+ or ("del_dropdown_button" in ctx.triggered[0]["prop_id"] and None not in nclicks_del)
+ ):
selectedData = None
- if selectedData is not None and len(selectedData['points'])>0:
+ if selectedData is not None and len(selectedData["points"]) > 0:
raise PreventUpdate
else:
subset = get_indexes_from_datatable(data)
- figure = self.explainer.plot.scatter_plot_prediction(
- selection=subset,
- max_points=points,
- label=label
- )
+ figure = self.explainer.plot.scatter_plot_prediction(selection=subset, max_points=points, label=label)
if self.explainer.y_target is not None:
- figure['layout'].clickmode = 'event+select'
+ figure["layout"].clickmode = "event+select"
# Adjust graph with adding x and y axis titles
- MyGraph.adjust_graph_static(figure,
- x_ax="True Values",
- y_ax="Predicted Values")
+ MyGraph.adjust_graph_static(figure, x_ax="True Values", y_ax="Predicted Values")
return figure, selectedData
@app.callback(
Output("modal_feature_importance", "is_open"),
- [Input("open_feature_importance", "n_clicks"),
- Input("close_feature_importance", "n_clicks")],
+ [Input("open_feature_importance", "n_clicks"), Input("close_feature_importance", "n_clicks")],
[State("modal_feature_importance", "is_open")],
)
- def toggle_modal_feature_importancet(n1,
- n2,
- is_open):
+ def toggle_modal_feature_importancet(n1, n2, is_open):
"""
Function used to open and close modal explication when we click
on "?" button on feature_importance graph
@@ -2163,13 +2177,10 @@ def toggle_modal_feature_importancet(n1,
@app.callback(
Output("modal_feature_selector", "is_open"),
- [Input("open_feature_selector", "n_clicks"),
- Input("close_feature_selector", "n_clicks")],
+ [Input("open_feature_selector", "n_clicks"), Input("close_feature_selector", "n_clicks")],
[State("modal_feature_selector", "is_open")],
)
- def toggle_modal_feature_selector(n1,
- n2,
- is_open):
+ def toggle_modal_feature_selector(n1, n2, is_open):
"""
Function used to open and close modal explication when we click
on "?" button on feature_selector graph
@@ -2185,13 +2196,10 @@ def toggle_modal_feature_selector(n1,
@app.callback(
Output("modal_detail_feature", "is_open"),
- [Input("open_detail_feature", "n_clicks"),
- Input("close_detail_feature", "n_clicks")],
+ [Input("open_detail_feature", "n_clicks"), Input("close_detail_feature", "n_clicks")],
[State("modal_detail_feature", "is_open")],
)
- def toggle_modal_detail_feature(n1,
- n2,
- is_open):
+ def toggle_modal_detail_feature(n1, n2, is_open):
"""
Function used to open and close modal explication when we click
on "?" button on detail_feature graph
@@ -2207,13 +2215,10 @@ def toggle_modal_detail_feature(n1,
@app.callback(
Output("modal_prediction_picking", "is_open"),
- [Input("open_prediction_picking", "n_clicks"),
- Input("close_prediction_picking", "n_clicks")],
+ [Input("open_prediction_picking", "n_clicks"), Input("close_prediction_picking", "n_clicks")],
[State("modal_prediction_picking", "is_open")],
)
- def toggle_modal_prediction_picking(n1,
- n2,
- is_open):
+ def toggle_modal_prediction_picking(n1, n2, is_open):
"""
Function used to open and close modal explication when we click
on "?" button on prediction_picking graph
@@ -2229,13 +2234,10 @@ def toggle_modal_prediction_picking(n1,
@app.callback(
Output("modal_filter", "is_open"),
- [Input("open_filter", "n_clicks"),
- Input("close_filter", "n_clicks")],
+ [Input("open_filter", "n_clicks"), Input("close_filter", "n_clicks")],
[State("modal_filter", "is_open")],
)
- def toggle_modal_filters(n1,
- n2,
- is_open):
+ def toggle_modal_filters(n1, n2, is_open):
"""
Function used to open and close modal explication when we click
on "?" button on Dataset Filters Tab
@@ -2251,23 +2253,15 @@ def toggle_modal_filters(n1,
# Add or remove plot blocs in the 'dropdowns_container'
@app.callback(
- Output('dropdowns_container', 'children'),
+ Output("dropdowns_container", "children"),
[
- Input('add_dropdown_button', 'n_clicks'),
- Input('reset_dropdown_button', 'n_clicks'),
- Input({'type': 'del_dropdown_button', 'index': ALL}, 'n_clicks')
+ Input("add_dropdown_button", "n_clicks"),
+ Input("reset_dropdown_button", "n_clicks"),
+ Input({"type": "del_dropdown_button", "index": ALL}, "n_clicks"),
],
- [
- State('dropdowns_container', 'children'),
- State('name', 'value')
- ]
+ [State("dropdowns_container", "children"), State("name", "value")],
)
- def layout_filter(n_clicks_add,
- n_clicks_rm,
- n_clicks_reset,
- currents_filters,
- name
- ):
+ def layout_filter(n_clicks_add, n_clicks_rm, n_clicks_reset, currents_filters, name):
"""
Function used to create filter blocs in the dropdowns_container.
Each bloc will contains:
@@ -2289,35 +2283,31 @@ def layout_filter(n_clicks_add,
ctx = dash.callback_context
if not ctx.triggered:
raise dash.exceptions.PreventUpdate
- button_id = ctx.triggered[0]['prop_id'].split('.')[0]
+ button_id = ctx.triggered[0]["prop_id"].split(".")[0]
options = get_feature_filter_options(self.dataframe, self.features_dict, self.special_cols)
# Creation of a new graph
- if button_id == 'add_dropdown_button':
+ if button_id == "add_dropdown_button":
subset_filter = create_dropdown_feature_filter(n_clicks_add, options)
return currents_filters + [subset_filter]
# Removal of all existing filters
- elif button_id == 'reset_dropdown_button':
- return [html.Div(
- id={'type': 'bloc_div',
- 'index': 0},
- children=[])]
+ elif button_id == "reset_dropdown_button":
+ return [html.Div(id={"type": "bloc_div", "index": 0}, children=[])]
# Removal of an existing filter
else:
- filter_id_to_remove = eval(button_id)['index']
- return [gr for gr in currents_filters
- if gr['props']['id']['index'] != filter_id_to_remove]
+ filter_id_to_remove = eval(button_id)["index"]
+ return [gr for gr in currents_filters if gr["props"]["id"]["index"] != filter_id_to_remove]
@app.callback(
- Output('reset_dropdown_button', 'disabled'),
- [Input('add_dropdown_button', 'n_clicks'),
- Input('reset_dropdown_button', 'n_clicks'),
- Input({'type': 'del_dropdown_button', 'index': ALL}, 'n_clicks')]
+ Output("reset_dropdown_button", "disabled"),
+ [
+ Input("add_dropdown_button", "n_clicks"),
+ Input("reset_dropdown_button", "n_clicks"),
+ Input({"type": "del_dropdown_button", "index": ALL}, "n_clicks"),
+ ],
)
- def update_disabled_reset_button(n_click_add,
- n_click_reset,
- n_click_del):
+ def update_disabled_reset_button(n_click_add, n_click_reset, n_click_del):
"""
Function used to disabled or not the reset filter button.
This button is disabled if there is no filter added.
@@ -2329,9 +2319,9 @@ def update_disabled_reset_button(n_click_add,
return disabled style
"""
ctx = dash.callback_context
- if ctx.triggered[0]['prop_id'] == 'add_dropdown_button.n_clicks':
+ if ctx.triggered[0]["prop_id"] == "add_dropdown_button.n_clicks":
disabled = False
- elif ctx.triggered[0]['prop_id'] == 'reset_dropdown_button.n_clicks':
+ elif ctx.triggered[0]["prop_id"] == "reset_dropdown_button.n_clicks":
disabled = True
elif None not in n_click_del:
disabled = True
@@ -2340,15 +2330,15 @@ def update_disabled_reset_button(n_click_add,
return disabled
@app.callback(
- Output('apply_filter', 'style'),
- Output('filtered_subset_info', 'style'),
- [Input('add_dropdown_button', 'n_clicks'),
- Input('reset_dropdown_button', 'n_clicks'),
- Input({'type': 'del_dropdown_button', 'index': ALL}, 'n_clicks')]
+ Output("apply_filter", "style"),
+ Output("filtered_subset_info", "style"),
+ [
+ Input("add_dropdown_button", "n_clicks"),
+ Input("reset_dropdown_button", "n_clicks"),
+ Input({"type": "del_dropdown_button", "index": ALL}, "n_clicks"),
+ ],
)
- def update_style_apply_filter_button(n_click_add,
- n_click_reset,
- n_click_del):
+ def update_style_apply_filter_button(n_click_add, n_click_reset, n_click_del):
"""
Function used to display or not the apply filter button.
This button is only display if almost one filter was added.
@@ -2360,18 +2350,18 @@ def update_style_apply_filter_button(n_click_add,
return style of apply filter button
"""
ctx = dash.callback_context
- if ctx.triggered[0]['prop_id'] == 'add_dropdown_button.n_clicks':
- return {}, {'margin-left': '20px'}
- elif ctx.triggered[0]['prop_id'] == 'reset_dropdown_button.n_clicks':
- return {'display': 'none'}, {'display': 'none'}
+ if ctx.triggered[0]["prop_id"] == "add_dropdown_button.n_clicks":
+ return {}, {"margin-left": "20px"}
+ elif ctx.triggered[0]["prop_id"] == "reset_dropdown_button.n_clicks":
+ return {"display": "none"}, {"display": "none"}
elif None not in n_click_del:
- return {'display': 'none'}, {'display': 'none'}
+ return {"display": "none"}, {"display": "none"}
else:
- return {}, {'margin-left': '20px'}
+ return {}, {"margin-left": "20px"}
@app.callback(
- Output({'type': 'dynamic-output-label', 'index': MATCH}, 'children'),
- Input({'type': 'var_dropdown', 'index': MATCH}, 'value'),
+ Output({"type": "dynamic-output-label", "index": MATCH}, "children"),
+ Input({"type": "var_dropdown", "index": MATCH}, "value"),
)
def update_label_filter(value):
"""
@@ -2383,19 +2373,19 @@ def update_label_filter(value):
return label
"""
if value is not None:
- return html.Label("Variable {} is filtered".format(value))
+ return html.Label(f"Variable {value} is filtered")
else:
- return html.Label('Select variable to filter')
+ return html.Label("Select variable to filter")
@app.callback(
- Output({'type': 'dynamic-output', 'index': MATCH}, 'children'),
- [Input({'type': 'var_dropdown', 'index': MATCH}, 'value'),
- Input({'type': 'var_dropdown', 'index': MATCH}, 'id'),
- Input('add_dropdown_button', 'n_clicks')],
+ Output({"type": "dynamic-output", "index": MATCH}, "children"),
+ [
+ Input({"type": "var_dropdown", "index": MATCH}, "value"),
+ Input({"type": "var_dropdown", "index": MATCH}, "id"),
+ Input("add_dropdown_button", "n_clicks"),
+ ],
)
- def display_output(value,
- id,
- add_click):
+ def display_output(value, id, add_click):
"""
Function used to create modalities choices. Componenents are different
according to the type of the selected variable.
@@ -2418,7 +2408,7 @@ def display_output(value,
if not ctx.triggered:
raise PreventUpdate
# No update last modalities values if we click on add button
- if ctx.triggered[0]['prop_id'] == 'add_dropdown_button.n_clicks':
+ if ctx.triggered[0]["prop_id"] == "add_dropdown_button.n_clicks":
raise PreventUpdate
# Creation on modalities dropdown button
else:
diff --git a/shapash/webapp/utils/MyGraph.py b/shapash/webapp/utils/MyGraph.py
index 10e43612..e77da014 100644
--- a/shapash/webapp/utils/MyGraph.py
+++ b/shapash/webapp/utils/MyGraph.py
@@ -2,51 +2,53 @@
Class inherited from dcc.Graph. Add one method for updating graph layout.
"""
-from dash import dcc
import re
+from dash import dcc
+
class MyGraph(dcc.Graph):
-
def __init__(self, figure, id, style={}, **kwds):
super().__init__(**kwds)
self.figure = figure
self.style = style
self.id = id
# self.config = {'modeBarButtons': {'pan2d': True}}
- if id == 'prediction_picking':
+ if id == "prediction_picking":
self.config = {
# Graph is responsive
- 'responsive': True,
- 'modeBarButtonsToRemove': ['zoomOut2d',
- 'zoomIn2d',
- 'resetScale2d',
- 'hoverClosestCartesian',
- 'hoverCompareCartesian',
- 'toggleSpikelines'],
- 'displaylogo': False
- }
+ "responsive": True,
+ "modeBarButtonsToRemove": [
+ "zoomOut2d",
+ "zoomIn2d",
+ "resetScale2d",
+ "hoverClosestCartesian",
+ "hoverCompareCartesian",
+ "toggleSpikelines",
+ ],
+ "displaylogo": False,
+ }
# 'modeBarStyle': {'orientation': 'v'}, # Deprecated in Dash 1.17.0
else:
self.config = {
# Graph is responsive
- 'responsive': True,
+ "responsive": True,
# Graph don't have select box button
- 'modeBarButtonsToRemove': ['lasso2d',
- 'zoomOut2d',
- 'zoomIn2d',
- 'resetScale2d',
- 'hoverClosestCartesian',
- 'hoverCompareCartesian',
- 'toggleSpikelines',
- 'select'],
- 'displaylogo': False
- }
+ "modeBarButtonsToRemove": [
+ "lasso2d",
+ "zoomOut2d",
+ "zoomIn2d",
+ "resetScale2d",
+ "hoverClosestCartesian",
+ "hoverCompareCartesian",
+ "toggleSpikelines",
+ "select",
+ ],
+ "displaylogo": False,
+ }
@staticmethod
- def adjust_graph_static(figure,
- x_ax="",
- y_ax=""):
+ def adjust_graph_static(figure, x_ax="", y_ax=""):
"""
Override graph layout for app use
----------------------------------------
@@ -57,32 +59,22 @@ def adjust_graph_static(figure,
new_title = update_title(figure.layout.title.text)
figure.update_layout(
autosize=True,
- margin=dict(
- l=50,
- r=10,
- b=10,
- t=67,
- pad=0
- ),
+ margin=dict(l=50, r=10, b=10, t=67, pad=0),
width=None,
height=None,
title={
- 'y': 0.95,
- 'x': 0.5,
- 'xanchor': 'center',
- 'yanchor': 'top',
+ "y": 0.95,
+ "x": 0.5,
+ "xanchor": "center",
+ "yanchor": "top",
# update title and font-size of the title
- 'text': '' + new_title + ''
- }
+ "text": '' + new_title + "",
+ },
)
# update x title and font-size of the title
- figure.update_xaxes(title='' + x_ax + '',
- automargin=True
- )
+ figure.update_xaxes(title='' + x_ax + "", automargin=True)
# update y title and font-size of the title
- figure.update_yaxes(title='' + y_ax + '',
- automargin=True
- )
+ figure.update_yaxes(title='' + y_ax + "", automargin=True)
def update_title(title):
@@ -96,10 +88,10 @@ def update_title(title):
-------
str
"""
- patt = re.compile('^(.+)$')
+ patt = re.compile("^(.+)$")
try:
- list_non_empty_str_matches = [x for x in patt.findall(title)[0] if x != '']
- updated = ' - '.join(map(str, list_non_empty_str_matches))
+ list_non_empty_str_matches = [x for x in patt.findall(title)[0] if x != ""]
+ updated = " - ".join(map(str, list_non_empty_str_matches))
except:
updated = title
return updated
diff --git a/shapash/webapp/utils/callbacks.py b/shapash/webapp/utils/callbacks.py
index 3275e29c..c5748e4a 100644
--- a/shapash/webapp/utils/callbacks.py
+++ b/shapash/webapp/utils/callbacks.py
@@ -1,10 +1,10 @@
+import datetime
from typing import Optional, Tuple
+import dash_bootstrap_components as dbc
import numpy as np
import pandas as pd
-import datetime
from dash import dcc, html
-import dash_bootstrap_components as dbc
def select_data_from_prediction_picking(round_dataframe: pd.DataFrame, selected_data: dict) -> pd.DataFrame:
@@ -23,18 +23,18 @@ def select_data_from_prediction_picking(round_dataframe: pd.DataFrame, selected_
Subset dataframe
"""
row_ids = []
- for p in selected_data['points']:
- row_ids.append(p['customdata'])
+ for p in selected_data["points"]:
+ row_ids.append(p["customdata"])
df = round_dataframe.loc[row_ids]
-
+
return df
def select_data_from_str_filters(
df: pd.DataFrame,
- feature_id: list,
- id_str_modality: list,
- val_feature: list,
+ feature_id: list,
+ id_str_modality: list,
+ val_feature: list,
val_str_modality: list,
) -> pd.DataFrame:
"""Create a subset dataframe from filters.
@@ -58,23 +58,23 @@ def select_data_from_str_filters(
Subset dataframe
"""
# get list of ID
- str_id = [id_str_modality[i]['index'] for i in range(len(id_str_modality))]
+ str_id = [id_str_modality[i]["index"] for i in range(len(id_str_modality))]
# If there is some filters
if len(str_id) > 0:
for i in range(len(feature_id)):
if feature_id[i] in str_id:
position = np.where(np.array(str_id) == feature_id[i])[0][0]
- if ((position is not None) & (val_str_modality[position] is not None)):
+ if (position is not None) & (val_str_modality[position] is not None):
df = df[df[val_feature[i]].isin(val_str_modality[position])]
-
+
return df
def select_data_from_bool_filters(
df: pd.DataFrame,
- feature_id: list,
- id_bool_modality: list,
- val_feature: list,
+ feature_id: list,
+ id_bool_modality: list,
+ val_feature: list,
val_bool_modality: list,
) -> pd.DataFrame:
"""Create a subset dataframe from filters.
@@ -98,23 +98,23 @@ def select_data_from_bool_filters(
Subset dataframe
"""
# get list of ID
- bool_id = [id_bool_modality[i]['index'] for i in range(len(id_bool_modality))]
+ bool_id = [id_bool_modality[i]["index"] for i in range(len(id_bool_modality))]
# If there is some filters
if len(bool_id) > 0:
for i in range(len(feature_id)):
if feature_id[i] in bool_id:
position = np.where(np.array(bool_id) == feature_id[i])[0][0]
- if ((position is not None) & (val_bool_modality[position] is not None)):
+ if (position is not None) & (val_bool_modality[position] is not None):
df = df[df[val_feature[i]] == val_bool_modality[position]]
-
+
return df
def select_data_from_numeric_filters(
df: pd.DataFrame,
- feature_id: list,
- id_lower_modality: list,
- val_feature: list,
+ feature_id: list,
+ id_lower_modality: list,
+ val_feature: list,
val_lower_modality: list,
val_upper_modality: list,
) -> pd.DataFrame:
@@ -141,26 +141,31 @@ def select_data_from_numeric_filters(
Subset dataframe
"""
# get list of ID
- lower_id = [id_lower_modality[i]['index'] for i in range(len(id_lower_modality))]
+ lower_id = [id_lower_modality[i]["index"] for i in range(len(id_lower_modality))]
# If there is some filters
if len(lower_id) > 0:
for i in range(len(feature_id)):
if feature_id[i] in lower_id:
position = np.where(np.array(lower_id) == feature_id[i])[0][0]
- if((position is not None) & (val_lower_modality[position] is not None) &
- (val_upper_modality[position] is not None)):
- if (val_lower_modality[position] < val_upper_modality[position]):
- df = df[(df[val_feature[i]] >= val_lower_modality[position]) &
- (df[val_feature[i]] <= val_upper_modality[position])]
-
+ if (
+ (position is not None)
+ & (val_lower_modality[position] is not None)
+ & (val_upper_modality[position] is not None)
+ ):
+ if val_lower_modality[position] < val_upper_modality[position]:
+ df = df[
+ (df[val_feature[i]] >= val_lower_modality[position])
+ & (df[val_feature[i]] <= val_upper_modality[position])
+ ]
+
return df
def select_data_from_date_filters(
df: pd.DataFrame,
- feature_id: list,
- id_date: list,
- val_feature: list,
+ feature_id: list,
+ id_date: list,
+ val_feature: list,
start_date: list,
end_date: list,
) -> pd.DataFrame:
@@ -187,17 +192,15 @@ def select_data_from_date_filters(
Subset dataframe
"""
# get list of ID
- date_id = [id_date[i]['index'] for i in range(len(id_date))]
+ date_id = [id_date[i]["index"] for i in range(len(id_date))]
# If there is some filters
if len(date_id) > 0:
for i in range(len(feature_id)):
if feature_id[i] in date_id:
position = np.where(np.array(date_id) == feature_id[i])[0][0]
- if((position is not None) &
- (start_date[position] < end_date[position])):
- df = df[((df[val_feature[i]] >= start_date[position]) &
- (df[val_feature[i]] <= end_date[position]))]
-
+ if (position is not None) & (start_date[position] < end_date[position]):
+ df = df[((df[val_feature[i]] >= start_date[position]) & (df[val_feature[i]] <= end_date[position]))]
+
return df
@@ -214,7 +217,7 @@ def get_feature_from_clicked_data(click_data: dict) -> str:
str
Selected feature
"""
- selected_feature = click_data['points'][0]['label'].replace('', '').replace('', '')
+ selected_feature = click_data["points"][0]["label"].replace("", "").replace("", "")
return selected_feature
@@ -233,8 +236,7 @@ def get_feature_from_features_groups(selected_feature: Optional[str], features_g
Optional[str]
Group feature name or selected feature if not in a group.
"""
- list_sub_features = [f for group_features in features_groups.values()
- for f in group_features]
+ list_sub_features = [f for group_features in features_groups.values() for f in group_features]
if selected_feature in list_sub_features:
for k, v in features_groups.items():
if selected_feature in v:
@@ -257,14 +259,14 @@ def get_group_name(selected_feature: Optional[str], features_groups: Optional[di
Optional[str]
Group feature name
"""
- group_name = selected_feature if (
- features_groups is not None and selected_feature in features_groups.keys()
- ) else None
+ group_name = (
+ selected_feature if (features_groups is not None and selected_feature in features_groups.keys()) else None
+ )
return group_name
def get_indexes_from_datatable(data: list, list_index: Optional[list] = None) -> Optional[list]:
- """Get the indexes of the data. If list_index is given and is the same length than
+ """Get the indexes of the data. If list_index is given and is the same length than
the indexes, there is no subset selected.
Parameters
@@ -279,8 +281,8 @@ def get_indexes_from_datatable(data: list, list_index: Optional[list] = None) ->
Optional[list]
Indexes of the data
"""
- indexes = [d['_index_'] for d in data]
- if list_index is not None and (len(indexes)==len(list_index) or len(indexes)==0):
+ indexes = [d["_index_"] for d in data]
+ if list_index is not None and (len(indexes) == len(list_index) or len(indexes) == 0):
indexes = None
return indexes
@@ -298,13 +300,13 @@ def update_click_data_on_subset_changes(click_data: dict) -> dict:
dict
Updated feature importance click data
"""
- point = click_data['points'][0]
- point['curveNumber'] = 0
- click_data = {'points':[point]}
+ point = click_data["points"][0]
+ point["curveNumber"] = 0
+ click_data = {"points": [point]}
return click_data
-def get_figure_zoom(click_zoom: int) -> bool :
+def get_figure_zoom(click_zoom: int) -> bool:
"""Get figure zoom from n_clicks
Parameters
@@ -341,9 +343,9 @@ def get_feature_contributions_sign_to_show(positive: list, negative: list) -> Op
Sign to show on plot
"""
if positive == [1]:
- sign = (None if negative == [1] else True)
+ sign = None if negative == [1] else True
else:
- sign = (False if negative == [1] else None)
+ sign = False if negative == [1] else None
return sign
@@ -375,9 +377,11 @@ def update_features_to_display(features: int, nb_columns: int, value: int) -> Tu
nb_marks = min(int(max_value // 7), 10)
else:
nb_marks = 2
- marks = {f'{round(max_value * feat / nb_marks)}': f'{round(max_value * feat / nb_marks)}'
- for feat in range(1, nb_marks + 1)}
- marks['1'] = '1'
+ marks = {
+ f"{round(max_value * feat / nb_marks)}": f"{round(max_value * feat / nb_marks)}"
+ for feat in range(1, nb_marks + 1)
+ }
+ marks["1"] = "1"
if max_value < value:
value = max_value
@@ -404,18 +408,12 @@ def get_id_card_features(data: list, selected: int, special_cols: list, features
Dataframe of the features
"""
selected_row = pd.DataFrame([data[selected]], index=["feature_value"]).T
- selected_row["feature_name"] = selected_row.index.map(
- lambda x: x if x in special_cols else features_dict[x]
- )
+ selected_row["feature_name"] = selected_row.index.map(lambda x: x if x in special_cols else features_dict[x])
return selected_row
def get_id_card_contrib(
- data: dict,
- index: int,
- features_dict: dict,
- columns_dict: dict,
- label_num: int = None
+ data: dict, index: int, features_dict: dict, columns_dict: dict, label_num: int = None
) -> pd.DataFrame:
"""Get the contributions of the selected index for the identity card.
@@ -438,11 +436,11 @@ def get_id_card_contrib(
Dataframe of the contributions
"""
if label_num is not None:
- contrib = data['contrib_sorted'][label_num].loc[index, :].values
- var_dict = data['var_dict'][label_num].loc[index, :].values
+ contrib = data["contrib_sorted"][label_num].loc[index, :].values
+ var_dict = data["var_dict"][label_num].loc[index, :].values
else:
- contrib = data['contrib_sorted'].loc[index, :].values
- var_dict = data['var_dict'].loc[index, :].values
+ contrib = data["contrib_sorted"].loc[index, :].values
+ var_dict = data["var_dict"].loc[index, :].values
var_dict = [features_dict[columns_dict[x]] for x in var_dict]
selected_contrib = pd.DataFrame([var_dict, contrib], index=["feature_name", "feature_contrib"]).T
@@ -453,11 +451,11 @@ def get_id_card_contrib(
def create_id_card_data(
selected_row: pd.DataFrame,
- selected_contrib: pd.DataFrame,
- sort_by: str,
+ selected_contrib: pd.DataFrame,
+ sort_by: str,
order: bool,
- special_cols: list,
- additional_features_dict: dict
+ special_cols: list,
+ additional_features_dict: dict,
) -> pd.DataFrame:
"""Merge and sort features and contributions dataframes for the identity card.
@@ -483,11 +481,15 @@ def create_id_card_data(
"""
selected_data = selected_row.merge(selected_contrib, how="left", on="feature_name")
selected_data.index = selected_row.index
- selected_data = pd.concat([
- selected_data.loc[special_cols],
- selected_data.drop(index=special_cols+list(additional_features_dict.keys())).sort_values(sort_by, ascending=order),
- selected_data.loc[list(additional_features_dict.keys())].sort_values(sort_by, ascending=order)
- ])
+ selected_data = pd.concat(
+ [
+ selected_data.loc[special_cols],
+ selected_data.drop(index=special_cols + list(additional_features_dict.keys())).sort_values(
+ sort_by, ascending=order
+ ),
+ selected_data.loc[list(additional_features_dict.keys())].sort_values(sort_by, ascending=order),
+ ]
+ )
return selected_data
@@ -509,26 +511,31 @@ def create_id_card_layout(selected_data: pd.DataFrame, additional_features_dict:
"""
children = []
for _, row in selected_data.iterrows():
- label_style = {
- 'fontWeight': 'bold',
- 'font-style': 'italic'
- } if row["feature_name"] in additional_features_dict.values() else {'fontWeight': 'bold'}
+ label_style = (
+ {"fontWeight": "bold", "font-style": "italic"}
+ if row["feature_name"] in additional_features_dict.values()
+ else {"fontWeight": "bold"}
+ )
children.append(
- dbc.Row([
- dbc.Col(dbc.Label(row["feature_name"]), width=3, style=label_style),
- dbc.Col(dbc.Label(row["feature_value"]), width=5, className="id_card_solid"),
- dbc.Col(width=1),
- dbc.Col(
- dbc.Row(
- dbc.Label(format(row["feature_contrib"], '.4f'), width="auto", style={"padding-top":0}),
- justify="end"
- ),
- width=2,
- className="id_card_solid",
- ) if not np.isnan(row["feature_contrib"]) else None,
- ])
+ dbc.Row(
+ [
+ dbc.Col(dbc.Label(row["feature_name"]), width=3, style=label_style),
+ dbc.Col(dbc.Label(row["feature_value"]), width=5, className="id_card_solid"),
+ dbc.Col(width=1),
+ dbc.Col(
+ dbc.Row(
+ dbc.Label(format(row["feature_contrib"], ".4f"), width="auto", style={"padding-top": 0}),
+ justify="end",
+ ),
+ width=2,
+ className="id_card_solid",
+ )
+ if not np.isnan(row["feature_contrib"])
+ else None,
+ ]
+ )
)
-
+
return children
@@ -550,19 +557,16 @@ def get_feature_filter_options(dataframe: pd.DataFrame, features_dict: dict, spe
Options for the filter
"""
# We use domain name for feature name
- dict_name = [features_dict[i]
- for i in dataframe.drop(special_cols, axis=1).columns]
+ dict_name = [features_dict[i] for i in dataframe.drop(special_cols, axis=1).columns]
dict_id = [i for i in dataframe.drop(special_cols, axis=1).columns]
# Create dataframe to sort it by feature_name
- df_feature_name = pd.DataFrame({'feature_name': dict_name,
- 'feature_id': dict_id})
- df_feature_name = df_feature_name.sort_values(
- by='feature_name').reset_index(drop=True)
+ df_feature_name = pd.DataFrame({"feature_name": dict_name, "feature_id": dict_id})
+ df_feature_name = df_feature_name.sort_values(by="feature_name").reset_index(drop=True)
# Options are sorted by feature_name
- options = [{"label": i, "value": i} for i in special_cols] + \
- [{"label": df_feature_name.loc[i, 'feature_name'],
- "value": df_feature_name.loc[i, 'feature_id']}
- for i in range(len(df_feature_name))]
+ options = [{"label": i, "value": i} for i in special_cols] + [
+ {"label": df_feature_name.loc[i, "feature_name"], "value": df_feature_name.loc[i, "feature_id"]}
+ for i in range(len(df_feature_name))
+ ]
return options
@@ -589,41 +593,36 @@ def create_dropdown_feature_filter(n_clicks_add: Optional[int], options: list) -
index_id = n_clicks_add
# Appending a dropdown block to 'dropdowns_container'children
subset_filter = html.Div(
- id={'type': 'bloc_div',
- 'index': index_id},
+ id={"type": "bloc_div", "index": index_id},
children=[
- html.Div([
- html.Br(),
- # div which will contains label
- html.Div(
- id={'type': 'dynamic-output-label',
- 'index': index_id},
- )
- ]),
- html.Div([
- # div with dopdown button to select feature to filter
- html.Div(dcc.Dropdown(
- id={'type': 'var_dropdown',
- 'index': index_id},
- options=options,
- placeholder="Variable"
- ), style={"width": "30%"}),
- # div which will contains modalities
- html.Div(
- id={'type': 'dynamic-output',
- 'index': index_id},
- style={"width": "50%"}
+ html.Div(
+ [
+ html.Br(),
+ # div which will contains label
+ html.Div(
+ id={"type": "dynamic-output-label", "index": index_id},
),
- # Button to delete bloc
- dbc.Button(
- id={'type': 'del_dropdown_button',
- 'index': index_id},
- children='X',
- color='warning',
- size='sm'
- )
- ], style={'display': 'flex'})
- ]
+ ]
+ ),
+ html.Div(
+ [
+ # div with dopdown button to select feature to filter
+ html.Div(
+ dcc.Dropdown(
+ id={"type": "var_dropdown", "index": index_id}, options=options, placeholder="Variable"
+ ),
+ style={"width": "30%"},
+ ),
+ # div which will contains modalities
+ html.Div(id={"type": "dynamic-output", "index": index_id}, style={"width": "50%"}),
+ # Button to delete bloc
+ dbc.Button(
+ id={"type": "del_dropdown_button", "index": index_id}, children="X", color="warning", size="sm"
+ ),
+ ],
+ style={"display": "flex"},
+ ),
+ ],
)
return subset_filter
@@ -647,64 +646,61 @@ def create_filter_modalities_selection(value: str, id: dict, round_dataframe: pd
Div containing the modalities selection options
"""
if type(round_dataframe[value].iloc[0]) == bool:
- new_element = html.Div(dcc.RadioItems(
- [{'label': val, 'value': val} for
- val in round_dataframe[value].unique()],
- id={'type': 'dynamic-bool',
- 'index': id['index']},
- value=round_dataframe[value].iloc[0],
- inline=False
- ), style={"width": "65%", 'margin-left': '20px'})
- elif (type(round_dataframe[value].iloc[0]) == str) | \
- ((type(round_dataframe[value].iloc[0]) == np.int64) &
- (len(round_dataframe[value].unique()) <= 20)):
- new_element = html.Div(dcc.Dropdown(
- id={
- 'type': 'dynamic-str',
- 'index': id['index']
- },
- options=[{'label': i, 'value': i} for
- i in np.sort(round_dataframe[value].unique())],
- multi=True,
- ), style={"width": "65%", 'margin-left': '20px'})
- elif ((type(round_dataframe[value].iloc[0]) is pd.Timestamp) |
- (type(round_dataframe[value].iloc[0]) is datetime.datetime)):
new_element = html.Div(
- dcc.DatePickerRange(
- id={
- 'type': 'dynamic-date',
- 'index': id['index']
- },
- min_date_allowed=round_dataframe[value].min(),
- max_date_allowed=round_dataframe[value].max(),
- start_date=round_dataframe[value].min(),
- end_date=round_dataframe[value].max()
- ), style={'width': '65%', 'margin-left': '20px'}),
+ dcc.RadioItems(
+ [{"label": val, "value": val} for val in round_dataframe[value].unique()],
+ id={"type": "dynamic-bool", "index": id["index"]},
+ value=round_dataframe[value].iloc[0],
+ inline=False,
+ ),
+ style={"width": "65%", "margin-left": "20px"},
+ )
+ elif (type(round_dataframe[value].iloc[0]) == str) | (
+ (type(round_dataframe[value].iloc[0]) == np.int64) & (len(round_dataframe[value].unique()) <= 20)
+ ):
+ new_element = html.Div(
+ dcc.Dropdown(
+ id={"type": "dynamic-str", "index": id["index"]},
+ options=[{"label": i, "value": i} for i in np.sort(round_dataframe[value].unique())],
+ multi=True,
+ ),
+ style={"width": "65%", "margin-left": "20px"},
+ )
+ elif (type(round_dataframe[value].iloc[0]) is pd.Timestamp) | (
+ type(round_dataframe[value].iloc[0]) is datetime.datetime
+ ):
+ new_element = (
+ html.Div(
+ dcc.DatePickerRange(
+ id={"type": "dynamic-date", "index": id["index"]},
+ min_date_allowed=round_dataframe[value].min(),
+ max_date_allowed=round_dataframe[value].max(),
+ start_date=round_dataframe[value].min(),
+ end_date=round_dataframe[value].max(),
+ ),
+ style={"width": "65%", "margin-left": "20px"},
+ ),
+ )
else:
lower_value = 0
upper_value = 0
- new_element = html.Div([
- dcc.Input(
- id={
- 'type': 'lower',
- 'index': id['index']
- },
- value=lower_value,
- type="number",
- style={'width': '60px'}),
- ' <= {} in [{}, {}]<= '.format(
- value,
- round_dataframe[value].min(),
- round_dataframe[value].max()),
- dcc.Input(
- id={
- 'type': 'upper',
- 'index': id['index']
- },
- value=upper_value,
- type="number",
- style={'width': '60px'}
- )
- ], style={'margin-left': '20px'})
+ new_element = html.Div(
+ [
+ dcc.Input(
+ id={"type": "lower", "index": id["index"]},
+ value=lower_value,
+ type="number",
+ style={"width": "60px"},
+ ),
+ " <= {} in [{}, {}]<= ".format(value, round_dataframe[value].min(), round_dataframe[value].max()),
+ dcc.Input(
+ id={"type": "upper", "index": id["index"]},
+ value=upper_value,
+ type="number",
+ style={"width": "60px"},
+ ),
+ ],
+ style={"margin-left": "20px"},
+ )
return new_element
diff --git a/shapash/webapp/utils/explanations.py b/shapash/webapp/utils/explanations.py
index 1ac70abe..5ab21ee6 100644
--- a/shapash/webapp/utils/explanations.py
+++ b/shapash/webapp/utils/explanations.py
@@ -1,19 +1,18 @@
-# -*- coding: utf-8 -*-
"""
Created on Mon Oct 3 15:12:08 2022
@author: Florine
"""
+
class Explanations:
"""
- Contains the explanations of all "?" buttons in the app.
- ----------
- explanations: object
- SmartApp instance to point to.
+ Contains the explanations of all "?" buttons in the app.
+ ----------
+ explanations: object
+ SmartApp instance to point to.
"""
-
def __init__(self):
self.detail_feature = """
**Local interpretability:** the understanding of the decision
diff --git a/shapash/webapp/utils/utils.py b/shapash/webapp/utils/utils.py
index 5a5726eb..3d84bf48 100644
--- a/shapash/webapp/utils/utils.py
+++ b/shapash/webapp/utils/utils.py
@@ -1,5 +1,5 @@
-import pandas as pd
import numpy as np
+import pandas as pd
def round_to_k(x, k):
@@ -18,7 +18,7 @@ def round_to_k(x, k):
"""
x = float(x)
- new_x = float('%s' % float(f'%.{k}g' % x)) # Rounding to k important figures
+ new_x = float("%s" % float(f"%.{k}g" % x)) # Rounding to k important figures
if new_x % 1 == 0:
return int(new_x) # Avoid the '.0' that can mislead the user that it may be a round number
else:
@@ -60,7 +60,7 @@ def check_row(data, index):
row number corresponding to index
"""
if index is not None:
- df = pd.DataFrame.from_records(data, index='_index_')
+ df = pd.DataFrame.from_records(data, index="_index_")
row = df.index.get_loc(index) if index in list(df.index) else None
else:
row = None
@@ -81,24 +81,26 @@ def split_filter_part(filter_part):
column, operator, value of the filter part
"""
- operators = [['ge ', '>='],
- ['le ', '<='],
- ['lt ', '<'],
- ['gt ', '>'],
- ['ne ', '!='],
- ['eq ', '='],
- ['contains '],
- ['datestartswith ']]
+ operators = [
+ ["ge ", ">="],
+ ["le ", "<="],
+ ["lt ", "<"],
+ ["gt ", ">"],
+ ["ne ", "!="],
+ ["eq ", "="],
+ ["contains "],
+ ["datestartswith "],
+ ]
for operator_type in operators:
for operator in operator_type:
if operator in filter_part:
name_part, value_part = filter_part.split(operator, 1)
- name = name_part[name_part.find('{') + 1: name_part.rfind('}')]
+ name = name_part[name_part.find("{") + 1 : name_part.rfind("}")]
value_part = value_part.strip()
v0 = value_part[0]
- if v0 == value_part[-1] and v0 in ("'", '"', '`'):
- value = value_part[1: -1].replace('\\' + v0, v0)
+ if v0 == value_part[-1] and v0 in ("'", '"', "`"):
+ value = value_part[1:-1].replace("\\" + v0, v0)
else:
try:
value = float(value_part)
@@ -128,17 +130,16 @@ def apply_filter(df, filter_query):
pandas.DataFrame
"""
- filtering_expressions = filter_query.split(' && ')
+ filtering_expressions = filter_query.split(" && ")
for filter_part in filtering_expressions:
col_name, operator, filter_value = split_filter_part(filter_part)
- if operator in ('eq', 'ne', 'lt', 'le', 'gt', 'ge'):
+ if operator in ("eq", "ne", "lt", "le", "gt", "ge"):
# these operators match pandas series operator method names
df = df.loc[getattr(df[col_name], operator)(filter_value)]
- elif operator == 'contains':
+ elif operator == "contains":
df = df.loc[df[col_name].str.contains(filter_value)]
- elif operator == 'datestartswith':
+ elif operator == "datestartswith":
# this is a simplification of the front-end filtering logic,
# only works with complete fields in standard format
df = df.loc[df[col_name].str.startswith(filter_value)]
return df
-
diff --git a/shapash/webapp/webapp_launch.py b/shapash/webapp/webapp_launch.py
index d03cb408..1c768311 100644
--- a/shapash/webapp/webapp_launch.py
+++ b/shapash/webapp/webapp_launch.py
@@ -2,41 +2,42 @@
Webapp launch module
This is an example in python how to launch app from explainer
"""
-from lightgbm import LGBMClassifier, LGBMRegressor
import pandas as pd
+from category_encoders import one_hot
+from lightgbm import LGBMClassifier, LGBMRegressor
from sklearn.model_selection import train_test_split
+
from shapash import SmartExplainer
-from category_encoders import one_hot
cases = {
- 1: 'Titanic regression',
- 2: 'Titanic binary classification',
- 3: 'Titanic multi class classification',
+ 1: "Titanic regression",
+ 2: "Titanic binary classification",
+ 3: "Titanic multi class classification",
}
CASE = 2
-titanic = pd.read_pickle('tests/data/clean_titanic.pkl')
+titanic = pd.read_pickle("tests/data/clean_titanic.pkl")
if CASE == 1:
- features = ['Pclass', 'Survived', 'Embarked', 'Sex']
- encoder = one_hot.OneHotEncoder(titanic, cols=['Embarked', 'Sex'])
+ features = ["Pclass", "Survived", "Embarked", "Sex"]
+ encoder = one_hot.OneHotEncoder(titanic, cols=["Embarked", "Sex"])
X = titanic[features]
- y = titanic['Age'].to_frame()
+ y = titanic["Age"].to_frame()
model = LGBMRegressor()
elif CASE == 2:
- features = ['Pclass', 'Age', 'Embarked', 'Sex']
- encoder = one_hot.OneHotEncoder(titanic, cols=['Embarked', 'Sex'])
+ features = ["Pclass", "Age", "Embarked", "Sex"]
+ encoder = one_hot.OneHotEncoder(titanic, cols=["Embarked", "Sex"])
X = titanic[features]
- y = titanic['Survived'].to_frame()
+ y = titanic["Survived"].to_frame()
model = LGBMClassifier()
else:
- features = ['Survived', 'Age', 'Embarked', 'Sex']
- encoder = one_hot.OneHotEncoder(titanic, cols=['Embarked', 'Sex'])
+ features = ["Survived", "Age", "Embarked", "Sex"]
+ encoder = one_hot.OneHotEncoder(titanic, cols=["Embarked", "Sex"])
X = titanic[features]
- y = titanic['Pclass'].to_frame()
+ y = titanic["Pclass"].to_frame()
model = LGBMClassifier()
titanic_enc = encoder.fit_transform(X)
@@ -55,21 +56,12 @@
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
-y_pred = pd.DataFrame(data=y_pred,
- columns=y.columns.to_list(),
- index=X_test.index)
+y_pred = pd.DataFrame(data=y_pred, columns=y.columns.to_list(), index=X_test.index)
-y_target = pd.DataFrame(data=y_test,
- columns=y.columns.to_list(),
- index=X_test.index)
+y_target = pd.DataFrame(data=y_test, columns=y.columns.to_list(), index=X_test.index)
-response_dict = {0: 'Death', 1: 'Survival'}
-xpl = SmartExplainer(
- model,
- preprocessing=encoder,
- title_story=cases[CASE],
- label_dict=response_dict
-)
+response_dict = {0: "Death", 1: "Survival"}
+xpl = SmartExplainer(model, preprocessing=encoder, title_story=cases[CASE], label_dict=response_dict)
xpl.compile(X_test, y_pred=y_pred, y_target=y_target)
diff --git a/shapash/webapp/webapp_launch_DVF.py b/shapash/webapp/webapp_launch_DVF.py
index 718dfded..c82436d6 100644
--- a/shapash/webapp/webapp_launch_DVF.py
+++ b/shapash/webapp/webapp_launch_DVF.py
@@ -4,39 +4,31 @@
"""
import pandas as pd
+from category_encoders import OrdinalEncoder
from lightgbm import LGBMRegressor
from sklearn.model_selection import train_test_split
-from category_encoders import OrdinalEncoder
-from shapash.data.data_loader import data_loading
from shapash import SmartExplainer
+from shapash.data.data_loader import data_loading
-house_df, house_dict = data_loading('house_prices')
-y_df = house_df['SalePrice'].to_frame()
-X_df = house_df[house_df.columns.difference(['SalePrice'])]
+house_df, house_dict = data_loading("house_prices")
+y_df = house_df["SalePrice"].to_frame()
+X_df = house_df[house_df.columns.difference(["SalePrice"])]
house_df.head()
-categorical_features = [col for col in X_df.columns if X_df[col].dtype == 'object']
-encoder = OrdinalEncoder(
- cols=categorical_features,
- handle_unknown='ignore',
- return_df=True).fit(X_df)
+categorical_features = [col for col in X_df.columns if X_df[col].dtype == "object"]
+encoder = OrdinalEncoder(cols=categorical_features, handle_unknown="ignore", return_df=True).fit(X_df)
X_df = encoder.transform(X_df)
Xtrain, Xtest, ytrain, ytest = train_test_split(X_df, y_df, train_size=0.75, random_state=1)
regressor = LGBMRegressor(n_estimators=200).fit(Xtrain, ytrain)
-y_pred = pd.DataFrame(regressor.predict(Xtest), columns=['pred'], index=Xtest.index)
-y_target = pd.DataFrame(data=ytest,
- columns=y_df.columns.to_list(),
- index=Xtest.index)
+y_pred = pd.DataFrame(regressor.predict(Xtest), columns=["pred"], index=Xtest.index)
+y_target = pd.DataFrame(data=ytest, columns=y_df.columns.to_list(), index=Xtest.index)
xpl = SmartExplainer(
- model=regressor,
- features_dict=house_dict,
- preprocessing=encoder,
- title_story='House Prices - Lightgbm Regressor'
+ model=regressor, features_dict=house_dict, preprocessing=encoder, title_story="House Prices - Lightgbm Regressor"
)
xpl.compile(x=Xtest, y_pred=y_pred, y_target=y_target)
diff --git a/tests/integration_tests/test_contributions_binary.py b/tests/integration_tests/test_contributions_binary.py
index 0c693ddd..f9ce1d52 100644
--- a/tests/integration_tests/test_contributions_binary.py
+++ b/tests/integration_tests/test_contributions_binary.py
@@ -2,10 +2,12 @@
Unit test for contributions binary
"""
import unittest
-from os.path import dirname, abspath, join
-import pandas as pd
-import numpy as np
+from os.path import abspath, dirname, join
+
import category_encoders as ce
+import numpy as np
+import pandas as pd
+
from shapash.decomposition.contributions import inverse_transform_contributions
@@ -18,6 +20,7 @@ class TestContributions(unittest.TestCase):
unittest : [type]
[description]
"""
+
def setUp(self):
"""
Setup
@@ -27,18 +30,18 @@ def setUp(self):
[description]
"""
data_path = dirname(dirname(abspath(__file__)))
- self.ds_titanic_clean = pd.read_pickle(join(data_path, 'data', 'clean_titanic.pkl'))
+ self.ds_titanic_clean = pd.read_pickle(join(data_path, "data", "clean_titanic.pkl"))
def test_inverse_transform_contributions_ce_basen(self):
"""
Unit test inverse transform contributions ce base n
"""
- preprocessing = ce.BaseNEncoder(cols=['Age', 'Sex'], return_df=True, base=3)
+ preprocessing = ce.BaseNEncoder(cols=["Age", "Sex"], return_df=True, base=3)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
contributions = pd.DataFrame(
data=np.random.rand(fitted_dataset.shape[0], fitted_dataset.shape[1]),
columns=fitted_dataset.columns,
- index=self.ds_titanic_clean.index
+ index=self.ds_titanic_clean.index,
)
output = inverse_transform_contributions(contributions, preprocessing)
assert isinstance(output, pd.DataFrame)
@@ -49,12 +52,12 @@ def test_inverse_transform_contributions_ce_binary(self):
"""
Unit test inverse transform contributions ce binary
"""
- preprocessing = ce.BinaryEncoder(cols=['Pclass', 'Age', 'Sex'], return_df=True)
+ preprocessing = ce.BinaryEncoder(cols=["Pclass", "Age", "Sex"], return_df=True)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
contributions = pd.DataFrame(
data=np.random.rand(fitted_dataset.shape[0], fitted_dataset.shape[1]),
columns=fitted_dataset.columns,
- index=self.ds_titanic_clean.index
+ index=self.ds_titanic_clean.index,
)
output = inverse_transform_contributions(contributions, preprocessing)
assert isinstance(output, pd.DataFrame)
@@ -65,12 +68,12 @@ def test_inverse_transform_contributions_ce_onehot(self):
"""
Unit test inverse transform contributions ce onehot
"""
- preprocessing = ce.OneHotEncoder(cols=['Pclass', 'Sex'], return_df=True)
+ preprocessing = ce.OneHotEncoder(cols=["Pclass", "Sex"], return_df=True)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
contributions = pd.DataFrame(
data=np.random.rand(fitted_dataset.shape[0], fitted_dataset.shape[1]),
columns=fitted_dataset.columns,
- index=self.ds_titanic_clean.index
+ index=self.ds_titanic_clean.index,
)
output = inverse_transform_contributions(contributions, preprocessing)
assert isinstance(output, pd.DataFrame)
@@ -81,12 +84,12 @@ def test_inverse_transform_contributions_ce_ordinal(self):
"""
Unit test inverse transform contributions ce ordinal
"""
- preprocessing = ce.OrdinalEncoder(cols=['Pclass', 'Age'], return_df=True)
+ preprocessing = ce.OrdinalEncoder(cols=["Pclass", "Age"], return_df=True)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
contributions = pd.DataFrame(
data=np.random.rand(fitted_dataset.shape[0], fitted_dataset.shape[1]),
columns=fitted_dataset.columns,
- index=self.ds_titanic_clean.index
+ index=self.ds_titanic_clean.index,
)
output = inverse_transform_contributions(contributions, preprocessing)
assert isinstance(output, pd.DataFrame)
diff --git a/tests/integration_tests/test_contributions_multiclass.py b/tests/integration_tests/test_contributions_multiclass.py
index 20bd2e8e..0a2e7bc1 100644
--- a/tests/integration_tests/test_contributions_multiclass.py
+++ b/tests/integration_tests/test_contributions_multiclass.py
@@ -2,12 +2,14 @@
Unit test contributions multiclass
"""
import unittest
+
+import numpy as np
+import pandas as pd
import shap
from sklearn.datasets import load_iris
-from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
-import numpy as np
-import pandas as pd
+from sklearn.model_selection import train_test_split
+
from shapash.decomposition.contributions import rank_contributions
@@ -24,6 +26,7 @@ class TestContributions(unittest.TestCase):
[type]
[description]
"""
+
def setUp(self):
"""
Setup
@@ -31,11 +34,7 @@ def setUp(self):
iris = load_iris()
x_df = pd.DataFrame(data=iris.data, columns=iris.feature_names)
y_df = pd.DataFrame(data=iris.target, columns=["target"])
- self.x_train, self.x_test, self.y_train, self.y_test = train_test_split(
- x_df,
- y_df,
- random_state=1
- )
+ self.x_train, self.x_test, self.y_train, self.y_test = train_test_split(x_df, y_df, random_state=1)
def check_compute_contributions(self, slist, blist, x_test):
"""
@@ -103,7 +102,4 @@ def test_rank_contributions_1(self):
for i in range(3):
s_ord, x_ord, s_dict = rank_contributions(slist[i], pd.DataFrame(data=self.x_test))
assert np.all(np.diff(np.abs(s_ord), axis=1) <= 0) == 1
- assert np.array_equal(
- x_ord.values,
- np.take_along_axis(self.x_test.values, s_dict.values, axis=1)
- )
+ assert np.array_equal(x_ord.values, np.take_along_axis(self.x_test.values, s_dict.values, axis=1))
diff --git a/tests/integration_tests/test_integration_inverse_tranform.py b/tests/integration_tests/test_integration_inverse_tranform.py
index 75eca8cb..f6a545e5 100644
--- a/tests/integration_tests/test_integration_inverse_tranform.py
+++ b/tests/integration_tests/test_integration_inverse_tranform.py
@@ -2,11 +2,14 @@
Unit test inverse transform
"""
import unittest
-from os.path import dirname, abspath, join
-import pandas as pd
+from os.path import abspath, dirname, join
+
import category_encoders as ce
+import pandas as pd
+
from shapash.utils.transform import inverse_transform
+
class TestInverseTranform(unittest.TestCase):
"""
Unit test invers transform class
@@ -16,6 +19,7 @@ class TestInverseTranform(unittest.TestCase):
unittest : [type]
[description]
"""
+
def setUp(self):
"""
Setup
@@ -25,13 +29,13 @@ def setUp(self):
[description]
"""
data_path = dirname(dirname(abspath(__file__)))
- self.ds_titanic_clean = pd.read_pickle(join(data_path, 'data', 'clean_titanic.pkl'))
+ self.ds_titanic_clean = pd.read_pickle(join(data_path, "data", "clean_titanic.pkl"))
def test_inverse_transform_ce_basen(self):
"""
Unit test inverse transform base n
"""
- preprocessing = ce.BaseNEncoder(cols=['Age', 'Sex'], return_df=True, base=3)
+ preprocessing = ce.BaseNEncoder(cols=["Age", "Sex"], return_df=True, base=3)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
output = inverse_transform(fitted_dataset, preprocessing)
pd.testing.assert_frame_equal(output, self.ds_titanic_clean)
@@ -40,7 +44,7 @@ def test_inverse_transform_ce_onehot(self):
"""
Unit test inverse transform ce onehot
"""
- preprocessing = ce.OneHotEncoder(cols=['Age', 'Sex'], return_df=True)
+ preprocessing = ce.OneHotEncoder(cols=["Age", "Sex"], return_df=True)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
output = inverse_transform(fitted_dataset, preprocessing)
pd.testing.assert_frame_equal(output, self.ds_titanic_clean)
@@ -49,7 +53,7 @@ def test_inverse_transform_ce_binary(self):
"""
Unit test inverse transform ce binary
"""
- preprocessing = ce.BinaryEncoder(cols=['Age', 'Sex'], return_df=True)
+ preprocessing = ce.BinaryEncoder(cols=["Age", "Sex"], return_df=True)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
output = inverse_transform(fitted_dataset, preprocessing)
pd.testing.assert_frame_equal(output, self.ds_titanic_clean)
@@ -58,7 +62,7 @@ def test_inverse_transform_ce_ordinal(self):
"""
Unit test inverse transform ce ordinal
"""
- preprocessing = ce.OrdinalEncoder(cols=['Age', 'Sex'], return_df=True)
+ preprocessing = ce.OrdinalEncoder(cols=["Age", "Sex"], return_df=True)
fitted_dataset = preprocessing.fit_transform(self.ds_titanic_clean)
output = inverse_transform(fitted_dataset, preprocessing)
pd.testing.assert_frame_equal(output, self.ds_titanic_clean)
diff --git a/tests/integration_tests/test_report_generation.py b/tests/integration_tests/test_report_generation.py
index 4bfbedd9..91a987f7 100644
--- a/tests/integration_tests/test_report_generation.py
+++ b/tests/integration_tests/test_report_generation.py
@@ -1,35 +1,35 @@
-import unittest
-import tempfile
-import shutil
-import numpy as np
import os
-import pandas as pd
+import shutil
+import tempfile
+import unittest
+
import catboost as cb
import category_encoders as ce
+import numpy as np
+import pandas as pd
+from category_encoders import OrdinalEncoder
from shapash import SmartExplainer
from shapash.report.generation import execute_report, export_and_save_report
-from category_encoders import OrdinalEncoder
current_path = os.path.dirname(os.path.abspath(__file__))
class TestGeneration(unittest.TestCase):
-
def setUp(self):
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 10 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df['x3'] = np.random.choice(['A', 'B', 'C', 'D'], df.shape[0])
- df['x4'] = np.random.choice(['A', 'B', 'C', np.nan], df.shape[0])
- df = df.set_index('id')
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 10 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df["x3"] = np.random.choice(["A", "B", "C", "D"], df.shape[0])
+ df["x4"] = np.random.choice(["A", "B", "C", np.nan], df.shape[0])
+ df = df.set_index("id")
encoder = ce.OrdinalEncoder(cols=["x3", "x4"], handle_unknown="None")
encoder_fitted = encoder.fit(df)
df_encoded = encoder_fitted.transform(df)
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[['x1', 'x2', 'x3', 'x4']], df_encoded['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[["x1", "x2", "x3", "x4"]], df_encoded["y"])
self.xpl = SmartExplainer(model=clf, preprocessing=encoder)
- self.xpl.compile(x=df_encoded[['x1', 'x2', 'x3', 'x4']])
+ self.xpl.compile(x=df_encoded[["x1", "x2", "x3", "x4"]])
self.df = df_encoded
def test_execute_report_1(self):
@@ -38,12 +38,12 @@ def test_execute_report_1(self):
execute_report(
working_dir=tmp_dir_path,
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../data/metadata.yaml"),
config=None,
- notebook_path=None
+ notebook_path=None,
)
- assert os.path.exists(os.path.join(tmp_dir_path, 'smart_explainer.pickle'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'base_report.ipynb'))
+ assert os.path.exists(os.path.join(tmp_dir_path, "smart_explainer.pickle"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "base_report.ipynb"))
shutil.rmtree(tmp_dir_path)
@@ -53,14 +53,14 @@ def test_execute_report_2(self):
execute_report(
working_dir=tmp_dir_path,
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2', 'x3', 'x4']],
+ project_info_file=os.path.join(current_path, "../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2", "x3", "x4"]],
config=None,
- notebook_path=None
+ notebook_path=None,
)
- assert os.path.exists(os.path.join(tmp_dir_path, 'x_train.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'smart_explainer.pickle'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'base_report.ipynb'))
+ assert os.path.exists(os.path.join(tmp_dir_path, "x_train.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "smart_explainer.pickle"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "base_report.ipynb"))
shutil.rmtree(tmp_dir_path)
@@ -70,16 +70,16 @@ def test_execute_report_3(self):
execute_report(
working_dir=tmp_dir_path,
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2', 'x3', 'x4']],
- y_test=self.df['y'],
+ project_info_file=os.path.join(current_path, "../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2", "x3", "x4"]],
+ y_test=self.df["y"],
config=None,
- notebook_path=None
+ notebook_path=None,
)
- assert os.path.exists(os.path.join(tmp_dir_path, 'x_train.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'y_test.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'smart_explainer.pickle'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'base_report.ipynb'))
+ assert os.path.exists(os.path.join(tmp_dir_path, "x_train.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "y_test.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "smart_explainer.pickle"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "base_report.ipynb"))
shutil.rmtree(tmp_dir_path)
@@ -89,18 +89,18 @@ def test_execute_report_4(self):
execute_report(
working_dir=tmp_dir_path,
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2', 'x3', 'x4']],
- y_train=self.df['y'],
- y_test=self.df['y'],
+ project_info_file=os.path.join(current_path, "../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2", "x3", "x4"]],
+ y_train=self.df["y"],
+ y_test=self.df["y"],
config=None,
- notebook_path=None
+ notebook_path=None,
)
- assert os.path.exists(os.path.join(tmp_dir_path, 'x_train.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'y_test.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'y_train.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'smart_explainer.pickle'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'base_report.ipynb'))
+ assert os.path.exists(os.path.join(tmp_dir_path, "x_train.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "y_test.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "y_train.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "smart_explainer.pickle"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "base_report.ipynb"))
shutil.rmtree(tmp_dir_path)
@@ -111,18 +111,18 @@ def test_execute_report_5(self):
execute_report(
working_dir=tmp_dir_path,
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2', 'x3', 'x4']],
- y_train=self.df['y'],
- y_test=self.df['y'],
- notebook_path=None
+ project_info_file=os.path.join(current_path, "../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2", "x3", "x4"]],
+ y_train=self.df["y"],
+ y_test=self.df["y"],
+ notebook_path=None,
)
self.xpl.palette_name = "default"
- assert os.path.exists(os.path.join(tmp_dir_path, 'x_train.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'y_test.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'y_train.csv'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'smart_explainer.pickle'))
- assert os.path.exists(os.path.join(tmp_dir_path, 'base_report.ipynb'))
+ assert os.path.exists(os.path.join(tmp_dir_path, "x_train.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "y_test.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "y_train.csv"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "smart_explainer.pickle"))
+ assert os.path.exists(os.path.join(tmp_dir_path, "base_report.ipynb"))
shutil.rmtree(tmp_dir_path)
@@ -132,10 +132,10 @@ def test_export_and_save_report_1(self):
execute_report(
working_dir=tmp_dir_path,
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../data/metadata.yaml"),
)
- outfile = os.path.join(tmp_dir_path, 'report.html')
+ outfile = os.path.join(tmp_dir_path, "report.html")
export_and_save_report(working_dir=tmp_dir_path, output_file=outfile)
assert os.path.exists(outfile)
shutil.rmtree(tmp_dir_path)
diff --git a/tests/unit_tests/backend/test_base_backend.py b/tests/unit_tests/backend/test_base_backend.py
index 818858e4..1c22bb74 100644
--- a/tests/unit_tests/backend/test_base_backend.py
+++ b/tests/unit_tests/backend/test_base_backend.py
@@ -1,20 +1,20 @@
+import types
import unittest
from unittest.mock import patch
-import types
+import category_encoders as ce
import numpy as np
import pandas as pd
-import category_encoders as ce
-from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from pandas.testing import assert_frame_equal, assert_series_equal
+from sklearn.ensemble import RandomForestClassifier, RandomForestRegressor
from shapash.backend.base_backend import BaseBackend, _needs_preprocessing
from shapash.explainer.smart_state import SmartState
class TestBackend(BaseBackend):
- column_aggregation = 'sum'
- name = 'test'
+ column_aggregation = "sum"
+ name = "test"
def run_explainer(self, x):
return [[0, 1]]
@@ -22,15 +22,13 @@ def run_explainer(self, x):
class TestBaseBackend(unittest.TestCase):
def predict(self, arg1, arg2):
- matrx = np.array(
- [12, 3, 7]
- )
+ matrx = np.array([12, 3, 7])
return matrx
def setUp(self):
self.model = lambda: None
self.model.predict = types.MethodType(self.predict, self.model)
- self.preprocessing = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2'])
+ self.preprocessing = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"])
self.test_backend = TestBackend(model=self.model, preprocessing=self.preprocessing)
@@ -45,7 +43,7 @@ def test_init(self):
def test_run_explainer(self):
assert self.test_backend.run_explainer(pd.DataFrame([0])) == [[0, 1]]
- @patch('shapash.backend.base_backend.BaseBackend.format_and_aggregate_local_contributions')
+ @patch("shapash.backend.base_backend.BaseBackend.format_and_aggregate_local_contributions")
def test_get_local_contributions(self, mock_format_contrib):
mock_format_contrib.return_value = [0, 4]
res = self.test_backend.get_local_contributions(pd.DataFrame([0]), dict(contributions=[np.array([0])]))
@@ -67,51 +65,47 @@ def test_get_global_features_importance(self):
assert_series_equal(res, pd.Series([0.1, 0.2, 0.3, 0.4]))
def test_format_and_aggregate_local_contributions(self):
- self.test_backend._case = 'classification'
+ self.test_backend._case = "classification"
self.test_backend._classes = [0, 1]
self.test_backend.preprocessing = None
- x = pd.DataFrame([[0, 1, 2], [3, 4, 5]], columns=['a', 'b', 'c'])
+ x = pd.DataFrame([[0, 1, 2], [3, 4, 5]], columns=["a", "b", "c"])
contributions = [np.array([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]]), np.array([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]])]
res_contributions = self.test_backend.format_and_aggregate_local_contributions(x, contributions)
assert len(res_contributions) == 2
assert_frame_equal(
- res_contributions[0],
- pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]], columns=['a', 'b', 'c'])
+ res_contributions[0], pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]], columns=["a", "b", "c"])
)
assert_frame_equal(
- res_contributions[1],
- pd.DataFrame([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]], columns=['a', 'b', 'c'])
+ res_contributions[1], pd.DataFrame([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]], columns=["a", "b", "c"])
)
def test_format_and_aggregate_local_contributions_2(self):
- self.test_backend._case = 'classification'
+ self.test_backend._case = "classification"
self.test_backend._classes = [0, 1]
self.test_backend.preprocessing = None
- x = pd.DataFrame([[0, 1, 2], [3, 4, 5]], columns=['a', 'b', 'c'])
+ x = pd.DataFrame([[0, 1, 2], [3, 4, 5]], columns=["a", "b", "c"])
contributions = [
- pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]], columns=['a', 'b', 'c']),
- pd.DataFrame([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]], columns=['a', 'b', 'c'])
+ pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]], columns=["a", "b", "c"]),
+ pd.DataFrame([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]], columns=["a", "b", "c"]),
]
res_contributions = self.test_backend.format_and_aggregate_local_contributions(x, contributions)
assert len(res_contributions) == 2
assert_frame_equal(
- res_contributions[0],
- pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]], columns=['a', 'b', 'c'])
+ res_contributions[0], pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]], columns=["a", "b", "c"])
)
assert_frame_equal(
- res_contributions[1],
- pd.DataFrame([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]], columns=['a', 'b', 'c'])
+ res_contributions[1], pd.DataFrame([[0.1, 0.2, 0.3], [0.1, 0.2, 0.3]], columns=["a", "b", "c"])
)
def test_needs_preprocessing(self):
- df = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D']})
- encoder = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2']).fit(df)
+ df = pd.DataFrame({"Onehot1": ["A", "B", "A", "B"], "Onehot2": ["C", "D", "C", "D"]})
+ encoder = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"]).fit(df)
df_ohe = encoder.transform(df)
res = _needs_preprocessing(df_ohe.columns, df_ohe, preprocessing=encoder)
@@ -119,8 +113,8 @@ def test_needs_preprocessing(self):
assert res is True
def test_needs_preprocessing_2(self):
- df = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D']})
- encoder = ce.OrdinalEncoder(cols=['Onehot1', 'Onehot2']).fit(df)
+ df = pd.DataFrame({"Onehot1": ["A", "B", "A", "B"], "Onehot2": ["C", "D", "C", "D"]})
+ encoder = ce.OrdinalEncoder(cols=["Onehot1", "Onehot2"]).fit(df)
df_ord = encoder.transform(df)
res = _needs_preprocessing(df_ord.columns, df_ord, preprocessing=encoder)
diff --git a/tests/unit_tests/backend/test_lime_backend.py b/tests/unit_tests/backend/test_lime_backend.py
index 9d4a719b..887eaa28 100644
--- a/tests/unit_tests/backend/test_lime_backend.py
+++ b/tests/unit_tests/backend/test_lime_backend.py
@@ -3,11 +3,13 @@
"""
import unittest
+
+import category_encoders as ce
import numpy as np
import pandas as pd
import sklearn.ensemble as ske
import xgboost as xgb
-import category_encoders as ce
+
from shapash.backend.lime_backend import LimeBackend
@@ -17,29 +19,29 @@ def setUp(self):
xgb.XGBClassifier(n_estimators=1),
ske.RandomForestClassifier(n_estimators=1),
ske.RandomForestRegressor(n_estimators=1),
- ske.GradientBoostingRegressor(n_estimators=1)
+ ske.GradientBoostingRegressor(n_estimators=1),
]
- df = pd.DataFrame(range(0, 4), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 3 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- self.x_df = df[['x1', 'x2']]
- self.y_df = df['y'].to_frame()
+ df = pd.DataFrame(range(0, 4), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 3 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ self.x_df = df[["x1", "x2"]]
+ self.y_df = df["y"].to_frame()
def test_init(self):
for model in self.model_list:
print(type(model))
model.fit(self.x_df, self.y_df)
backend_xpl = LimeBackend(model)
- assert hasattr(backend_xpl, 'explainer')
+ assert hasattr(backend_xpl, "explainer")
backend_xpl = LimeBackend(model, data=self.x_df)
- assert hasattr(backend_xpl, 'data')
+ assert hasattr(backend_xpl, "data")
backend_xpl = LimeBackend(model, preprocessing=ce.OrdinalEncoder())
- assert hasattr(backend_xpl, 'preprocessing')
+ assert hasattr(backend_xpl, "preprocessing")
assert isinstance(backend_xpl.preprocessing, ce.OrdinalEncoder)
def test_get_global_contributions(self):
diff --git a/tests/unit_tests/backend/test_shap_backend.py b/tests/unit_tests/backend/test_shap_backend.py
index fe87e265..49790cd7 100644
--- a/tests/unit_tests/backend/test_shap_backend.py
+++ b/tests/unit_tests/backend/test_shap_backend.py
@@ -1,10 +1,11 @@
import unittest
+
+import catboost as cb
+import lightgbm as lgb
import numpy as np
import pandas as pd
import sklearn.ensemble as ske
import xgboost as xgb
-import lightgbm as lgb
-import catboost as cb
from shapash.backend.shap_backend import ShapBackend
@@ -12,21 +13,27 @@
class TestShapBackend(unittest.TestCase):
def setUp(self):
self.model_list = [
- lgb.LGBMRegressor(n_estimators=1), lgb.LGBMClassifier(n_estimators=1),
- xgb.XGBRegressor(n_estimators=1), xgb.XGBClassifier(n_estimators=1),
- cb.CatBoostRegressor(n_estimators=1), cb.CatBoostClassifier(n_estimators=1),
- ske.GradientBoostingRegressor(n_estimators=1), ske.GradientBoostingClassifier(n_estimators=1),
- ske.ExtraTreesRegressor(n_estimators=1), ske.ExtraTreesClassifier(n_estimators=1),
- ske.RandomForestRegressor(n_estimators=1), ske.RandomForestClassifier(n_estimators=1)
+ lgb.LGBMRegressor(n_estimators=1),
+ lgb.LGBMClassifier(n_estimators=1),
+ xgb.XGBRegressor(n_estimators=1),
+ xgb.XGBClassifier(n_estimators=1),
+ cb.CatBoostRegressor(n_estimators=1),
+ cb.CatBoostClassifier(n_estimators=1),
+ ske.GradientBoostingRegressor(n_estimators=1),
+ ske.GradientBoostingClassifier(n_estimators=1),
+ ske.ExtraTreesRegressor(n_estimators=1),
+ ske.ExtraTreesClassifier(n_estimators=1),
+ ske.RandomForestRegressor(n_estimators=1),
+ ske.RandomForestClassifier(n_estimators=1),
]
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 10 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- self.x_df = df[['x1', 'x2']]
- self.y_df = df['y'].to_frame()
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 10 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ self.x_df = df[["x1", "x2"]]
+ self.y_df = df["y"].to_frame()
def test_shap_backend_init(self):
"""
@@ -36,7 +43,7 @@ def test_shap_backend_init(self):
print(type(model))
model.fit(self.x_df, self.y_df)
backend_xpl = ShapBackend(model)
- assert hasattr(backend_xpl, 'explainer')
+ assert hasattr(backend_xpl, "explainer")
def test_run_explainer(self):
for model in self.model_list:
diff --git a/tests/unit_tests/data/test_data_loader.py b/tests/unit_tests/data/test_data_loader.py
index ceaaf9ec..2a9d19fb 100644
--- a/tests/unit_tests/data/test_data_loader.py
+++ b/tests/unit_tests/data/test_data_loader.py
@@ -2,17 +2,18 @@
Unit test data loader
"""
import unittest
-from shapash.data.data_loader import data_loading
+
import pandas as pd
+from shapash.data.data_loader import data_loading
-class Test_load_data(unittest.TestCase):
+class Test_load_data(unittest.TestCase):
def test_load_house(self):
"""
Unit test house_prices
"""
- house_df, house_dict = data_loading('house_prices')
+ house_df, house_dict = data_loading("house_prices")
assert isinstance(house_df, pd.DataFrame)
assert isinstance(house_dict, dict)
@@ -20,7 +21,7 @@ def test_load_titanic(self):
"""
Unit test load_titanic
"""
- titanic_df, titanic_dict = data_loading('titanic')
+ titanic_df, titanic_dict = data_loading("titanic")
assert isinstance(titanic_df, pd.DataFrame)
assert isinstance(titanic_dict, dict)
@@ -28,12 +29,12 @@ def test_load_telco(self):
"""
Unit test telco_customer
"""
- telco_df = data_loading('telco_customer_churn')
+ telco_df = data_loading("telco_customer_churn")
assert isinstance(telco_df, pd.DataFrame)
def test_load_accidents(self):
"""
Unit test us_car_accident
"""
- accidents_df = data_loading('us_car_accident')
+ accidents_df = data_loading("us_car_accident")
assert isinstance(accidents_df, pd.DataFrame)
diff --git a/tests/unit_tests/decomposition/test_contributions.py b/tests/unit_tests/decomposition/test_contributions.py
index 65aafe79..12ae3957 100644
--- a/tests/unit_tests/decomposition/test_contributions.py
+++ b/tests/unit_tests/decomposition/test_contributions.py
@@ -2,11 +2,13 @@
Unit test of contributions
"""
import unittest
-import pandas as pd
+
import numpy as np
+import pandas as pd
+
from shapash.decomposition.contributions import rank_contributions
-
+
class TestContributions(unittest.TestCase):
"""
Unit test class of contributions
@@ -22,38 +24,29 @@ def test_rank_contributions_1(self):
Unit test rank contributions 1
"""
dataframe_s = pd.DataFrame(
- [[3.4, 1, -9, 4],
- [-45, 3, 43, -9]],
- columns=["Phi_" + str(i) for i in range(4)],
- index=['raw_1', 'raw_2']
+ [[3.4, 1, -9, 4], [-45, 3, 43, -9]], columns=["Phi_" + str(i) for i in range(4)], index=["raw_1", "raw_2"]
)
dataframe_x = pd.DataFrame(
- [['Male', 'House', 'Married', 'PhD'],
- ['Female', 'Flat', 'Married', 'Master']],
+ [["Male", "House", "Married", "PhD"], ["Female", "Flat", "Married", "Master"]],
columns=["X" + str(i) for i in range(4)],
- index=['raw_1', 'raw_2']
+ index=["raw_1", "raw_2"],
)
expected_s_ord = pd.DataFrame(
- data=[[-9, 4, 3.4, 1],
- [-45, 43, -9, 3]],
- columns=['contribution_' + str(i) for i in range(4)],
- index=['raw_1', 'raw_2']
+ data=[[-9, 4, 3.4, 1], [-45, 43, -9, 3]],
+ columns=["contribution_" + str(i) for i in range(4)],
+ index=["raw_1", "raw_2"],
)
expected_x_ord = pd.DataFrame(
- data=[['Married', 'PhD', 'Male', 'House'],
- ['Female', 'Married', 'Master', 'Flat']],
- columns=['feature_' + str(i) for i in range(4)],
- index=['raw_1', 'raw_2']
+ data=[["Married", "PhD", "Male", "House"], ["Female", "Married", "Master", "Flat"]],
+ columns=["feature_" + str(i) for i in range(4)],
+ index=["raw_1", "raw_2"],
)
expected_s_dict = pd.DataFrame(
- data=[[2, 3, 0, 1],
- [0, 2, 3, 1]],
- columns=['feature_' + str(i) for i in range(4)],
- index=['raw_1', 'raw_2']
+ data=[[2, 3, 0, 1], [0, 2, 3, 1]], columns=["feature_" + str(i) for i in range(4)], index=["raw_1", "raw_2"]
)
s_ord, x_ord, s_dict = rank_contributions(dataframe_s, dataframe_x)
@@ -68,4 +61,4 @@ def test_rank_contributions_1(self):
assert pd.Index.equals(s_ord.index, expected_s_ord.index)
assert pd.Index.equals(x_ord.index, expected_x_ord.index)
- assert pd.Index.equals(s_dict.index, expected_s_dict.index)
\ No newline at end of file
+ assert pd.Index.equals(s_dict.index, expected_s_dict.index)
diff --git a/tests/unit_tests/decomposition/test_inverse_contribution.py b/tests/unit_tests/decomposition/test_inverse_contribution.py
index 4ab358a1..dea484d3 100644
--- a/tests/unit_tests/decomposition/test_inverse_contribution.py
+++ b/tests/unit_tests/decomposition/test_inverse_contribution.py
@@ -2,11 +2,13 @@
Unit test of Inverse Transform
"""
import unittest
-import pandas as pd
-import numpy as np
+
import category_encoders as ce
+import numpy as np
+import pandas as pd
import sklearn.preprocessing as skp
from sklearn.compose import ColumnTransformer
+
from shapash.decomposition.contributions import inverse_transform_contributions
@@ -24,51 +26,73 @@ def test_multiple_encoding_category_encoder(self):
"""
Test multiple preprocessing
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B'], 'Onehot2': ['C', 'D'],
- 'Binary1': ['E', 'F'], 'Binary2': ['G', 'H'],
- 'Ordinal1': ['I', 'J'], 'Ordinal2': ['K', 'L'],
- 'BaseN1': ['M', 'N'], 'BaseN2': ['O', 'P'],
- 'Target1': ['Q', 'R'], 'Target2': ['S', 'T'],
- 'other': ['other', np.nan]})
-
- contributions = pd.DataFrame([[1, 0, 1, 1, 3, 0, -3.5, 0, 4, 5, 0, 6, 7, 0, 8, 9, 10],
- [.5, .5, 2, 0, 1.5, 1.5, 5.5, -2, -4, -5, 8.5, -2.5, -7, 14, -8, -9, -10]],
- index=['index1', 'index2'])
-
- expected_contrib = pd.DataFrame({'Onehot1': [1., 1.], 'Onehot2': [2, 2],
- 'Binary1': [3., 3.], 'Binary2': [-3.5, 3.5],
- 'Ordinal1': [4, -4], 'Ordinal2': [5, -5],
- 'BaseN1': [6., 6.], 'BaseN2': [7, 7],
- 'Target1': [8, -8], 'Target2': [9, -9],
- 'other': [10, -10]},
- index=['index1', 'index2'])
-
- y = pd.DataFrame(data=[0, 1], columns=['y'])
-
- enc_onehot = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2']).fit(train)
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B"],
+ "Onehot2": ["C", "D"],
+ "Binary1": ["E", "F"],
+ "Binary2": ["G", "H"],
+ "Ordinal1": ["I", "J"],
+ "Ordinal2": ["K", "L"],
+ "BaseN1": ["M", "N"],
+ "BaseN2": ["O", "P"],
+ "Target1": ["Q", "R"],
+ "Target2": ["S", "T"],
+ "other": ["other", np.nan],
+ }
+ )
+
+ contributions = pd.DataFrame(
+ [
+ [1, 0, 1, 1, 3, 0, -3.5, 0, 4, 5, 0, 6, 7, 0, 8, 9, 10],
+ [0.5, 0.5, 2, 0, 1.5, 1.5, 5.5, -2, -4, -5, 8.5, -2.5, -7, 14, -8, -9, -10],
+ ],
+ index=["index1", "index2"],
+ )
+
+ expected_contrib = pd.DataFrame(
+ {
+ "Onehot1": [1.0, 1.0],
+ "Onehot2": [2, 2],
+ "Binary1": [3.0, 3.0],
+ "Binary2": [-3.5, 3.5],
+ "Ordinal1": [4, -4],
+ "Ordinal2": [5, -5],
+ "BaseN1": [6.0, 6.0],
+ "BaseN2": [7, 7],
+ "Target1": [8, -8],
+ "Target2": [9, -9],
+ "other": [10, -10],
+ },
+ index=["index1", "index2"],
+ )
+
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
+
+ enc_onehot = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"]).fit(train)
train_onehot = enc_onehot.transform(train)
- enc_binary = ce.BinaryEncoder(cols=['Binary1', 'Binary2']).fit(train_onehot)
+ enc_binary = ce.BinaryEncoder(cols=["Binary1", "Binary2"]).fit(train_onehot)
train_binary = enc_binary.transform(train_onehot)
- enc_ordinal = ce.OrdinalEncoder(cols=['Ordinal1', 'Ordinal2']).fit(train_binary)
+ enc_ordinal = ce.OrdinalEncoder(cols=["Ordinal1", "Ordinal2"]).fit(train_binary)
train_ordinal = enc_ordinal.transform(train_binary)
- enc_basen = ce.BaseNEncoder(cols=['BaseN1', 'BaseN2']).fit(train_ordinal)
+ enc_basen = ce.BaseNEncoder(cols=["BaseN1", "BaseN2"]).fit(train_ordinal)
train_basen = enc_basen.transform(train_ordinal)
- enc_target = ce.TargetEncoder(cols=['Target1', 'Target2']).fit(train_basen, y)
+ enc_target = ce.TargetEncoder(cols=["Target1", "Target2"]).fit(train_basen, y)
input_dict1 = dict()
- input_dict1['col'] = 'Onehot2'
- input_dict1['mapping'] = pd.Series(data=['C', 'D', np.nan], index=['C', 'D', 'missing'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "Onehot2"
+ input_dict1["mapping"] = pd.Series(data=["C", "D", np.nan], index=["C", "D", "missing"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'Binary2'
- input_dict2['mapping'] = pd.Series(data=['G', 'H', np.nan], index=['G', 'H', 'missing'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "Binary2"
+ input_dict2["mapping"] = pd.Series(data=["G", "H", np.nan], index=["G", "H", "missing"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'Ordinal2'
- input_dict3['mapping'] = pd.Series(data=['K', 'L', np.nan], index=['K', 'L', 'missing'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "Ordinal2"
+ input_dict3["mapping"] = pd.Series(data=["K", "L", np.nan], index=["K", "L", "missing"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
result1 = enc_onehot.transform(train)
@@ -79,10 +103,9 @@ def test_multiple_encoding_category_encoder(self):
contributions.columns = result5.columns
- original = inverse_transform_contributions(contributions,
- [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target,
- input_dict1,
- list_dict])
+ original = inverse_transform_contributions(
+ contributions, [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1, list_dict]
+ )
pd.testing.assert_frame_equal(expected_contrib, original)
@@ -90,61 +113,83 @@ def test_multiple_encoding_columntransfomers(self):
"""
Test multiple preprocessing columntransformers
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B'], 'Onehot2': ['C', 'D'],
- 'Binary1': ['E', 'F'], 'Binary2': ['G', 'H'],
- 'Ordinal1': ['I', 'J'], 'Ordinal2': ['K', 'L'],
- 'BaseN1': ['M', 'N'], 'BaseN2': ['O', 'P'],
- 'Target1': ['Q', 'R'], 'Target2': ['S', 'T'],
- 'other': ['other', np.nan]})
-
- contributions = pd.DataFrame([[1, 0, 1, 1, 1, 0, 1, 1, 3, 0, -3.5, 0, 4, 4, 5, 5, 0, 6, 7, 0, 8, 9, 10],
- [.5, .5, 2, 0, .5, .5, 2, 0, 1.5, 1.5, 5.5, -2, -4, -4, -5, -5, 8.5, -2.5, -7, 14, -8, -9, -10]],
- index=['index1', 'index2'])
-
- expected_contrib = pd.DataFrame({'onehot_skp_Onehot1': [1., 1.], 'onehot_skp_Onehot2': [2, 2],
- 'onehot_ce_Onehot1': [1., 1.], 'onehot_ce_Onehot2': [2, 2],
- 'binary_ce_Binary1': [3., 3.], 'binary_ce_Binary2': [-3.5, 3.5],
- 'ordinal_ce_Ordinal1': [4, -4], 'ordinal_ce_Ordinal2': [4, -4],
- 'ordinal_skp_Ordinal1': [5, -5], 'ordinal_skp_Ordinal2': [5, -5],
- 'basen_ce_BaseN1': [6., 6.], 'basen_ce_BaseN2': [7, 7],
- 'target_ce_Target1': [8, -8], 'target_ce_Target2': [9, -9],
- 22: [10, -10]},
- index=['index1', 'index2'])
-
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B"],
+ "Onehot2": ["C", "D"],
+ "Binary1": ["E", "F"],
+ "Binary2": ["G", "H"],
+ "Ordinal1": ["I", "J"],
+ "Ordinal2": ["K", "L"],
+ "BaseN1": ["M", "N"],
+ "BaseN2": ["O", "P"],
+ "Target1": ["Q", "R"],
+ "Target2": ["S", "T"],
+ "other": ["other", np.nan],
+ }
+ )
+
+ contributions = pd.DataFrame(
+ [
+ [1, 0, 1, 1, 1, 0, 1, 1, 3, 0, -3.5, 0, 4, 4, 5, 5, 0, 6, 7, 0, 8, 9, 10],
+ [0.5, 0.5, 2, 0, 0.5, 0.5, 2, 0, 1.5, 1.5, 5.5, -2, -4, -4, -5, -5, 8.5, -2.5, -7, 14, -8, -9, -10],
+ ],
+ index=["index1", "index2"],
+ )
+
+ expected_contrib = pd.DataFrame(
+ {
+ "onehot_skp_Onehot1": [1.0, 1.0],
+ "onehot_skp_Onehot2": [2, 2],
+ "onehot_ce_Onehot1": [1.0, 1.0],
+ "onehot_ce_Onehot2": [2, 2],
+ "binary_ce_Binary1": [3.0, 3.0],
+ "binary_ce_Binary2": [-3.5, 3.5],
+ "ordinal_ce_Ordinal1": [4, -4],
+ "ordinal_ce_Ordinal2": [4, -4],
+ "ordinal_skp_Ordinal1": [5, -5],
+ "ordinal_skp_Ordinal2": [5, -5],
+ "basen_ce_BaseN1": [6.0, 6.0],
+ "basen_ce_BaseN2": [7, 7],
+ "target_ce_Target1": [8, -8],
+ "target_ce_Target2": [9, -9],
+ 22: [10, -10],
+ },
+ index=["index1", "index2"],
+ )
+
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
enc = ColumnTransformer(
transformers=[
- ('onehot_skp', skp.OneHotEncoder(), ['Onehot1', 'Onehot2']),
- ('onehot_ce', ce.OneHotEncoder(), ['Onehot1', 'Onehot2']),
- ('binary_ce', ce.BinaryEncoder(), ['Binary1', 'Binary2']),
- ('ordinal_ce', ce.OrdinalEncoder(), ['Ordinal1', 'Ordinal2']),
- ('ordinal_skp', skp.OrdinalEncoder(), ['Ordinal1', 'Ordinal2']),
- ('basen_ce', ce.BaseNEncoder(), ['BaseN1', 'BaseN2']),
- ('target_ce', ce.TargetEncoder(), ['Target1', 'Target2'])
+ ("onehot_skp", skp.OneHotEncoder(), ["Onehot1", "Onehot2"]),
+ ("onehot_ce", ce.OneHotEncoder(), ["Onehot1", "Onehot2"]),
+ ("binary_ce", ce.BinaryEncoder(), ["Binary1", "Binary2"]),
+ ("ordinal_ce", ce.OrdinalEncoder(), ["Ordinal1", "Ordinal2"]),
+ ("ordinal_skp", skp.OrdinalEncoder(), ["Ordinal1", "Ordinal2"]),
+ ("basen_ce", ce.BaseNEncoder(), ["BaseN1", "BaseN2"]),
+ ("target_ce", ce.TargetEncoder(), ["Target1", "Target2"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
enc.fit(train, y)
input_dict1 = dict()
- input_dict1['col'] = 'Onehot2'
- input_dict1['mapping'] = pd.Series(data=['C', 'D', np.nan], index=['C', 'D', 'missing'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "Onehot2"
+ input_dict1["mapping"] = pd.Series(data=["C", "D", np.nan], index=["C", "D", "missing"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'Binary2'
- input_dict2['mapping'] = pd.Series(data=['G', 'H', np.nan], index=['G', 'H', 'missing'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "Binary2"
+ input_dict2["mapping"] = pd.Series(data=["G", "H", np.nan], index=["G", "H", "missing"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'Ordinal2'
- input_dict3['mapping'] = pd.Series(data=['K', 'L', np.nan], index=['K', 'L', 'missing'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "Ordinal2"
+ input_dict3["mapping"] = pd.Series(data=["K", "L", np.nan], index=["K", "L", "missing"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
- original = inverse_transform_contributions(contributions,
- [enc,
- input_dict1,
- list_dict])
+ original = inverse_transform_contributions(contributions, [enc, input_dict1, list_dict])
- pd.testing.assert_frame_equal(expected_contrib, original)
\ No newline at end of file
+ pd.testing.assert_frame_equal(expected_contrib, original)
diff --git a/tests/unit_tests/explainer/test_consistency.py b/tests/unit_tests/explainer/test_consistency.py
index ec6d215d..13cc6b9a 100644
--- a/tests/unit_tests/explainer/test_consistency.py
+++ b/tests/unit_tests/explainer/test_consistency.py
@@ -1,8 +1,10 @@
import itertools
+import unittest
+
import numpy as np
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
-import unittest
+
from shapash.explainer.consistency import Consistency
@@ -10,37 +12,31 @@ class TestConsistency(unittest.TestCase):
"""
Unit test consistency
"""
+
def setUp(self):
self.df = pd.DataFrame(
- data=np.array([[1, 2, 3, 0],
- [2, 4, 6, 1],
- [2, 2, 1, 0],
- [2, 4, 1, 1]]),
- columns=['X1', 'X2', 'X3', 'y'])
+ data=np.array([[1, 2, 3, 0], [2, 4, 6, 1], [2, 2, 1, 0], [2, 4, 1, 1]]), columns=["X1", "X2", "X3", "y"]
+ )
self.X = self.df.iloc[:, :-1]
self.y = self.df.iloc[:, -1]
- self.w1 = pd.DataFrame(np.array([[0.14, 0.04, 0.17],
- [0.02, 0.01, 0.33],
- [0.56, 0.12, 0.29],
- [0.03, 0.01, 0.04]]),
- columns=['X1', 'X2', 'X3'])
- self.w2 = pd.DataFrame(np.array([[0.38, 0.35, 0.01],
- [0.01, 0.30, 0.05],
- [0.45, 0.41, 0.12],
- [0.07, 0.30, 0.21]]),
- columns=['X1', 'X2', 'X3'])
- self.w3 = pd.DataFrame(np.array([[0.49, 0.17, 0.02],
- [0.25, 0.12, 0.25],
- [0.01, 0.06, 0.06],
- [0.19, 0.02, 0.18]]),
- columns=['X1', 'X2', 'X3'])
+ self.w1 = pd.DataFrame(
+ np.array([[0.14, 0.04, 0.17], [0.02, 0.01, 0.33], [0.56, 0.12, 0.29], [0.03, 0.01, 0.04]]),
+ columns=["X1", "X2", "X3"],
+ )
+ self.w2 = pd.DataFrame(
+ np.array([[0.38, 0.35, 0.01], [0.01, 0.30, 0.05], [0.45, 0.41, 0.12], [0.07, 0.30, 0.21]]),
+ columns=["X1", "X2", "X3"],
+ )
+ self.w3 = pd.DataFrame(
+ np.array([[0.49, 0.17, 0.02], [0.25, 0.12, 0.25], [0.01, 0.06, 0.06], [0.19, 0.02, 0.18]]),
+ columns=["X1", "X2", "X3"],
+ )
self.contributions = {"contrib_1": self.w1, "contrib_2": self.w2, "contrib_3": self.w3}
self.cns = Consistency()
- self.cns.compile(contributions = self.contributions, x=self.X)
-
+ self.cns.compile(contributions=self.contributions, x=self.X)
def test_compile(self):
assert isinstance(self.cns.methods, list)
@@ -49,34 +45,33 @@ def test_compile(self):
assert self.cns.weights[0].shape == self.w1.shape
assert all(x.shape == self.cns.weights[0].shape for x in self.cns.weights)
-
def test_check_consistency_contributions(self):
weights = [self.w1, self.w2, self.w3]
if weights[0].ndim == 1:
- raise ValueError('Multiple datapoints are required to compute the metric')
+ raise ValueError("Multiple datapoints are required to compute the metric")
if not all(isinstance(x, pd.DataFrame) for x in weights):
- raise ValueError('Contributions must be pandas DataFrames')
+ raise ValueError("Contributions must be pandas DataFrames")
if not all(x.shape == weights[0].shape for x in weights):
- raise ValueError('Contributions must be of same shape')
+ raise ValueError("Contributions must be of same shape")
if not all(x.columns.tolist() == weights[0].columns.tolist() for x in weights):
- raise ValueError('Columns names are different between contributions')
+ raise ValueError("Columns names are different between contributions")
if not all(x.index.tolist() == weights[0].index.tolist() for x in weights):
- raise ValueError('Index names are different between contributions')
+ raise ValueError("Index names are different between contributions")
def test_calculate_all_distances(self):
all_comparisons, mean_distances = self.cns.calculate_all_distances(self.cns.methods, self.cns.weights)
num_comb = len(list(itertools.combinations(self.cns.methods, 2)))
- assert all_comparisons.shape == (num_comb*self.X.shape[0], 4)
+ assert all_comparisons.shape == (num_comb * self.X.shape[0], 4)
assert isinstance(mean_distances, pd.DataFrame)
assert mean_distances.shape == (len(self.cns.methods), len(self.cns.methods))
def test_calculate_pairwise_distances(self):
l2_dist = self.cns.calculate_pairwise_distances(self.cns.weights, 0, 1)
- assert l2_dist.shape == (self.X.shape[0], )
+ assert l2_dist.shape == (self.X.shape[0],)
def test_calculate_mean_distances(self):
_, mean_distances = self.cns.calculate_all_distances(self.cns.methods, self.cns.weights)
@@ -85,8 +80,10 @@ def test_calculate_mean_distances(self):
self.cns.calculate_mean_distances(self.cns.methods, mean_distances, 0, 1, l2_dist)
assert mean_distances.shape == (len(self.cns.methods), len(self.cns.methods))
- assert mean_distances.loc[self.cns.methods[0], self.cns.methods[1]] == \
- mean_distances.loc[self.cns.methods[1], self.cns.methods[0]]
+ assert (
+ mean_distances.loc[self.cns.methods[0], self.cns.methods[1]]
+ == mean_distances.loc[self.cns.methods[1], self.cns.methods[0]]
+ )
def test_find_examples(self):
weights = [weight.values for weight in self.cns.weights]
@@ -109,9 +106,7 @@ def test_pairwise_consistency_plot(self):
methods = ["contrib_1", "contrib_3"]
max_features = 2
max_points = 100
- output = self.cns.pairwise_consistency_plot(methods=methods,
- max_features=max_features,
- max_points=max_points)
+ output = self.cns.pairwise_consistency_plot(methods=methods, max_features=max_features, max_points=max_points)
assert len(output.data[0].x) == min(max_points, len(self.X))
assert len(output.data) == 2 * max_features + 1
diff --git a/tests/unit_tests/explainer/test_multi_decorator.py b/tests/unit_tests/explainer/test_multi_decorator.py
index a9a37008..f2b03a59 100644
--- a/tests/unit_tests/explainer/test_multi_decorator.py
+++ b/tests/unit_tests/explainer/test_multi_decorator.py
@@ -2,9 +2,11 @@
Unit test for multi decorator
"""
import unittest
-from unittest.mock import patch, Mock
+from unittest.mock import Mock, patch
+
import numpy as np
import pandas as pd
+
from shapash.explainer.multi_decorator import MultiDecorator
@@ -32,7 +34,7 @@ def dummy_function(self, xparam, yparam):
[type]
[description]
"""
- return '+'.join([str(xparam), str(yparam)])
+ return "+".join([str(xparam), str(yparam)])
class TestMultiDecorator(unittest.TestCase):
@@ -40,13 +42,14 @@ class TestMultiDecorator(unittest.TestCase):
Unit test multi decorator Class
TODO: Docstring
"""
+
def test_delegate_1(self):
"""
Unit test delegate 1
"""
state = MultiDecorator(DummyState())
- output = state.delegate('dummy_function', [1, 2, 3], 9)
- expected = ['1+9', '2+9', '3+9']
+ output = state.delegate("dummy_function", [1, 2, 3], 9)
+ expected = ["1+9", "2+9", "3+9"]
self.assertListEqual(output, expected)
def test_delegate_2(self):
@@ -55,7 +58,7 @@ def test_delegate_2(self):
"""
state = MultiDecorator(DummyState())
with self.assertRaises(ValueError):
- state.delegate('dummy_function')
+ state.delegate("dummy_function")
def test_delegate_3(self):
"""
@@ -63,7 +66,7 @@ def test_delegate_3(self):
"""
state = MultiDecorator(DummyState())
with self.assertRaises(AttributeError):
- state.delegate('non_existing_function', [1, 2, 3], 9)
+ state.delegate("non_existing_function", [1, 2, 3], 9)
def test_delegate_4(self):
"""
@@ -71,7 +74,7 @@ def test_delegate_4(self):
"""
state = MultiDecorator(DummyState())
with self.assertRaises(ValueError):
- state.delegate('dummy_member', [1, 2, 3], 9)
+ state.delegate("dummy_member", [1, 2, 3], 9)
def test_delegate_5(self):
"""
@@ -79,7 +82,7 @@ def test_delegate_5(self):
"""
state = MultiDecorator(DummyState())
with self.assertRaises(ValueError):
- state.delegate('dummy_function', 1, 9)
+ state.delegate("dummy_function", 1, 9)
def test_assign_contributions(self):
"""
@@ -88,17 +91,14 @@ def test_assign_contributions(self):
backend = Mock()
backend.assign_contributions = Mock(
side_effect=[
- {'a': 1, 'b': 2},
- {'a': 3, 'b': 4},
- {'a': 5, 'b': 6},
+ {"a": 1, "b": 2},
+ {"a": 3, "b": 4},
+ {"a": 5, "b": 6},
]
)
state = MultiDecorator(backend)
output = state.assign_contributions([1, 2, 3])
- expected = {
- 'a': [1, 3, 5],
- 'b': [2, 4, 6]
- }
+ expected = {"a": [1, 3, 5], "b": [2, 4, 6]}
self.assertDictEqual(output, expected)
def test_check_contributions_1(self):
@@ -121,7 +121,7 @@ def test_check_contributions_2(self):
output = state.check_contributions([1, 2, 3], Mock())
assert not output
- @patch('shapash.explainer.multi_decorator.MultiDecorator.delegate')
+ @patch("shapash.explainer.multi_decorator.MultiDecorator.delegate")
def test_combine_masks(self, mock_delegate):
"""
Unit test combine masks
@@ -143,7 +143,7 @@ def test_combine_masks(self, mock_delegate):
backend.combine_masks = Mock()
state = MultiDecorator(backend)
state.combine_masks(masks)
- mock_delegate.assert_called_with('combine_masks', transposed_masks)
+ mock_delegate.assert_called_with("combine_masks", transposed_masks)
def test_compute_masked_contributions(self):
"""
@@ -174,12 +174,12 @@ def test_summarize_1(self):
var_dict1 = pd.DataFrame([[1, 0, 2, 3], [1, 0, 3, 2]])
var_dict2 = pd.DataFrame([[1, 0, 2, 3], [1, 0, 3, 2]])
var_dict = [var_dict1, var_dict2]
- x_sorted1 = pd.DataFrame([[1., 3., 22., 1.], [2., 1., 2., 38.]])
- x_sorted2 = pd.DataFrame([[1., 3., 22., 1.], [2., 1., 2., 38.]])
+ x_sorted1 = pd.DataFrame([[1.0, 3.0, 22.0, 1.0], [2.0, 1.0, 2.0, 38.0]])
+ x_sorted2 = pd.DataFrame([[1.0, 3.0, 22.0, 1.0], [2.0, 1.0, 2.0, 38.0]])
x_sorted = [x_sorted1, x_sorted2]
mask = pd.DataFrame([[True, True, False, False], [True, True, False, False]])
- columns_dict = {0: 'Pclass', 1: 'Sex', 2: 'Age', 3: 'Embarked'}
- features_dict = {'Pclass': 'Pclass', 'Sex': 'Sex', 'Age': 'Age (years)', 'Embarked': 'emb'}
+ columns_dict = {0: "Pclass", 1: "Sex", 2: "Age", 3: "Embarked"}
+ features_dict = {"Pclass": "Pclass", "Sex": "Sex", "Age": "Age (years)", "Embarked": "emb"}
state = MultiDecorator(backend)
state.summarize(contrib_sorted, var_dict, x_sorted, mask, columns_dict, features_dict)
assert backend.summarize.call_count == 2
diff --git a/tests/unit_tests/explainer/test_smart_explainer.py b/tests/unit_tests/explainer/test_smart_explainer.py
index dff25970..97269c09 100644
--- a/tests/unit_tests/explainer/test_smart_explainer.py
+++ b/tests/unit_tests/explainer/test_smart_explainer.py
@@ -1,27 +1,30 @@
"""
Unit test for smart explainer
"""
-import unittest
-from unittest.mock import patch, Mock
import os
import sys
+import types
+import unittest
from os import path
from pathlib import Path
-import types
-import pandas as pd
-import numpy as np
+from unittest.mock import Mock, patch
+
import catboost as cb
+import category_encoders as ce
+import numpy as np
+import pandas as pd
+import shap
+from catboost import CatBoostClassifier, CatBoostRegressor
from pandas.testing import assert_frame_equal
-from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
-from catboost import CatBoostClassifier, CatBoostRegressor
+from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
+
from shapash import SmartExplainer
-from shapash.explainer.multi_decorator import MultiDecorator
from shapash.backend import ShapBackend
+from shapash.explainer.multi_decorator import MultiDecorator
from shapash.explainer.smart_state import SmartState
-import category_encoders as ce
from shapash.utils.check import check_model
-import shap
+
def init_sme_to_pickle_test():
"""
@@ -33,9 +36,9 @@ def init_sme_to_pickle_test():
[description]
"""
current = Path(path.abspath(__file__)).parent.parent.parent
- pkl_file = path.join(current, 'data/xpl.pkl')
+ pkl_file = path.join(current, "data/xpl.pkl")
contributions = pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]])
- y_pred = pd.DataFrame(data=np.array([1, 2]), columns=['pred'])
+ y_pred = pd.DataFrame(data=np.array([1, 2]), columns=["pred"])
dataframe_x = pd.DataFrame([[1, 2, 3], [1, 2, 3]])
model = DecisionTreeRegressor().fit(dataframe_x, y_pred)
xpl = SmartExplainer(model=model)
@@ -43,6 +46,7 @@ def init_sme_to_pickle_test():
xpl.filter(max_contrib=2)
return pkl_file, xpl
+
class TestSmartExplainer(unittest.TestCase):
"""
Unit test smart explainer
@@ -50,12 +54,7 @@ class TestSmartExplainer(unittest.TestCase):
"""
def setUp(self) -> None:
- x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
+ x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
self.model = CatBoostRegressor().fit(x_init, [0, 1])
def test_init(self):
@@ -63,36 +62,27 @@ def test_init(self):
test init smart explainer
"""
xpl = SmartExplainer(self.model)
- assert hasattr(xpl, 'plot')
+ assert hasattr(xpl, "plot")
def assertRaisesWithMessage(self, msg, func, *args, **kwargs):
try:
func(*args, **kwargs)
self.assertFail()
except Exception as inst:
- self.assertEqual(inst.args[0]['message'], msg)
+ self.assertEqual(inst.args[0]["message"], msg)
def test_modify_postprocessing_1(self):
"""
Unit test modify postprocessing 1
"""
xpl = SmartExplainer(self.model)
- xpl.x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
- xpl.features_dict = {'Col1': 'Column1', 'Col2': 'Column2'}
- xpl.columns_dict = {0: 'Col1', 1:'Col2'}
- xpl.inv_features_dict = {'Column1': 'Col1', 'Column2': 'Col2'}
- postprocessing = {0: {'type' : 'suffix', 'rule':' t'},
- 'Column2': {'type' : 'prefix', 'rule' : 'test'}}
-
- expected_output = {
- 'Col1': {'type' : 'suffix', 'rule':' t'},
- 'Col2': {'type' : 'prefix', 'rule' : 'test'}
- }
+ xpl.x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
+ xpl.features_dict = {"Col1": "Column1", "Col2": "Column2"}
+ xpl.columns_dict = {0: "Col1", 1: "Col2"}
+ xpl.inv_features_dict = {"Column1": "Col1", "Column2": "Col2"}
+ postprocessing = {0: {"type": "suffix", "rule": " t"}, "Column2": {"type": "prefix", "rule": "test"}}
+
+ expected_output = {"Col1": {"type": "suffix", "rule": " t"}, "Col2": {"type": "prefix", "rule": "test"}}
output = xpl.modify_postprocessing(postprocessing)
assert output == expected_output
@@ -101,16 +91,11 @@ def test_modify_postprocessing_2(self):
Unit test modify postprocessing 2
"""
xpl = SmartExplainer(self.model)
- xpl.x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
- xpl.features_dict = {'Col1': 'Column1', 'Col2': 'Column2'}
- xpl.columns_dict = {0: 'Col1', 1: 'Col2'}
- xpl.inv_features_dict = {'Column1': 'Col1', 'Column2': 'Col2'}
- postprocessing = {'Error': {'type': 'suffix', 'rule': ' t'}}
+ xpl.x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
+ xpl.features_dict = {"Col1": "Column1", "Col2": "Column2"}
+ xpl.columns_dict = {0: "Col1", 1: "Col2"}
+ xpl.inv_features_dict = {"Column1": "Col1", "Column2": "Col2"}
+ postprocessing = {"Error": {"type": "suffix", "rule": " t"}}
with self.assertRaises(ValueError):
xpl.modify_postprocessing(postprocessing)
@@ -119,15 +104,10 @@ def test_apply_postprocessing_1(self):
Unit test apply_postprocessing 1
"""
xpl = SmartExplainer(self.model)
- xpl.x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
- xpl.features_dict = {'Col1': 'Column1', 'Col2': 'Column2'}
- xpl.columns_dict = {0: 'Col1', 1: 'Col2'}
- xpl.inv_features_dict = {'Column1': 'Col1', 'Column2': 'Col2'}
+ xpl.x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
+ xpl.features_dict = {"Col1": "Column1", "Col2": "Column2"}
+ xpl.columns_dict = {0: "Col1", 1: "Col2"}
+ xpl.inv_features_dict = {"Column1": "Col1", "Column2": "Col2"}
assert np.array_equal(xpl.x_init, xpl.apply_postprocessing())
def test_apply_postprocessing_2(self):
@@ -135,22 +115,13 @@ def test_apply_postprocessing_2(self):
Unit test apply_postprocessing 2
"""
xpl = SmartExplainer(self.model)
- xpl.x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
- xpl.features_dict = {'Col1': 'Column1', 'Col2': 'Column2'}
- xpl.columns_dict = {0: 'Col1', 1: 'Col2'}
- xpl.inv_features_dict = {'Column1': 'Col1', 'Column2': 'Col2'}
- postprocessing = {'Col1': {'type': 'suffix', 'rule': ' t'},
- 'Col2': {'type': 'prefix', 'rule': 'test'}}
+ xpl.x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
+ xpl.features_dict = {"Col1": "Column1", "Col2": "Column2"}
+ xpl.columns_dict = {0: "Col1", 1: "Col2"}
+ xpl.inv_features_dict = {"Column1": "Col1", "Column2": "Col2"}
+ postprocessing = {"Col1": {"type": "suffix", "rule": " t"}, "Col2": {"type": "prefix", "rule": "test"}}
expected_output = pd.DataFrame(
- data=[['1 t', 'test2'],
- ['3 t', 'test4']],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
+ data=[["1 t", "test2"], ["3 t", "test4"]], columns=["Col1", "Col2"], index=["Id1", "Id2"]
)
output = xpl.apply_postprocessing(postprocessing)
assert np.array_equal(output, expected_output)
@@ -181,9 +152,9 @@ def test_check_label_dict_1(self):
"""
Unit test check label dict 1
"""
- xpl = SmartExplainer(self.model, label_dict={1: 'Yes', 0: 'No'})
+ xpl = SmartExplainer(self.model, label_dict={1: "Yes", 0: "No"})
xpl._classes = [0, 1]
- xpl._case = 'classification'
+ xpl._case = "classification"
xpl.check_label_dict()
def test_check_label_dict_2(self):
@@ -191,32 +162,32 @@ def test_check_label_dict_2(self):
Unit test check label dict 2
"""
xpl = SmartExplainer(self.model)
- xpl._case = 'regression'
+ xpl._case = "regression"
xpl.check_label_dict()
def test_check_features_dict_1(self):
"""
Unit test check features dict 1
"""
- xpl = SmartExplainer(self.model, features_dict={'Age': 'Age (Years Old)'})
- xpl.columns_dict = {0: 'Age', 1: 'Education', 2: 'Sex'}
+ xpl = SmartExplainer(self.model, features_dict={"Age": "Age (Years Old)"})
+ xpl.columns_dict = {0: "Age", 1: "Education", 2: "Sex"}
xpl.check_features_dict()
- assert xpl.features_dict['Age'] == 'Age (Years Old)'
- assert xpl.features_dict['Education'] == 'Education'
+ assert xpl.features_dict["Age"] == "Age (Years Old)"
+ assert xpl.features_dict["Education"] == "Education"
def test_compile_1(self):
"""
Unit test compile 1
checking compile method without model
"""
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 10 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 10 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
xpl = SmartExplainer(clf)
- xpl.compile(x=df[['x1', 'x2']])
+ xpl.compile(x=df[["x1", "x2"]])
assert xpl._case == "classification"
self.assertListEqual(xpl._classes, [0, 1])
@@ -225,39 +196,35 @@ def test_compile_2(self):
Unit test compile 2
checking new attributes added to the compile method
"""
- df = pd.DataFrame(range(0, 5), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 2 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = ["S", "M", "S", "D", "M"]
- df = df.set_index('id')
+ df = pd.DataFrame(range(0, 5), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 2 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = ["S", "M", "S", "D", "M"]
+ df = df.set_index("id")
encoder = ce.OrdinalEncoder(cols=["x2"], handle_unknown="None")
encoder_fitted = encoder.fit(df)
df_encoded = encoder_fitted.transform(df)
output = df[["x1", "x2"]].copy()
output["x2"] = ["single", "married", "single", "divorced", "married"]
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[['x1', 'x2']], df_encoded['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[["x1", "x2"]], df_encoded["y"])
- postprocessing_1 = {"x2": {
- "type": "transcoding",
- "rule": {"S": "single", "M": "married", "D": "divorced"}}}
+ postprocessing_1 = {"x2": {"type": "transcoding", "rule": {"S": "single", "M": "married", "D": "divorced"}}}
postprocessing_2 = {
- "family_situation": {
- "type": "transcoding",
- "rule": {"S": "single", "M": "married", "D": "divorced"}}}
-
- xpl_postprocessing1 = SmartExplainer(clf, preprocessing=encoder_fitted,
- postprocessing=postprocessing_1)
- xpl_postprocessing2 = SmartExplainer(clf, features_dict={"x1": "age",
- "x2": "family_situation"},
- preprocessing=encoder_fitted,
- postprocessing=postprocessing_2
- )
- xpl_postprocessing3 = SmartExplainer(clf, preprocessing=None,
- postprocessing=None)
-
- xpl_postprocessing1.compile(x=df_encoded[['x1', 'x2']])
- xpl_postprocessing2.compile(x=df_encoded[['x1', 'x2']])
- xpl_postprocessing3.compile(x=df_encoded[['x1', 'x2']])
+ "family_situation": {"type": "transcoding", "rule": {"S": "single", "M": "married", "D": "divorced"}}
+ }
+
+ xpl_postprocessing1 = SmartExplainer(clf, preprocessing=encoder_fitted, postprocessing=postprocessing_1)
+ xpl_postprocessing2 = SmartExplainer(
+ clf,
+ features_dict={"x1": "age", "x2": "family_situation"},
+ preprocessing=encoder_fitted,
+ postprocessing=postprocessing_2,
+ )
+ xpl_postprocessing3 = SmartExplainer(clf, preprocessing=None, postprocessing=None)
+
+ xpl_postprocessing1.compile(x=df_encoded[["x1", "x2"]])
+ xpl_postprocessing2.compile(x=df_encoded[["x1", "x2"]])
+ xpl_postprocessing3.compile(x=df_encoded[["x1", "x2"]])
assert hasattr(xpl_postprocessing1, "preprocessing")
assert hasattr(xpl_postprocessing1, "postprocessing")
@@ -277,24 +244,22 @@ def test_compile_3(self):
Unit test compile 3
checking compile method without model
"""
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 10 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 10 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
contrib = pd.DataFrame(
- [[1, 2, 3, 4],
- [5, 6, 7, 8],
- [9, 10, 11, 12]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ [[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
xpl = SmartExplainer(clf)
with self.assertRaises(ValueError):
- xpl.compile(x=df[['x1', 'x2']], contributions=contrib)
+ xpl.compile(x=df[["x1", "x2"]], contributions=contrib)
def test_compile_4(self):
"""
@@ -302,31 +267,31 @@ def test_compile_4(self):
checking compile method with lime backend
"""
np.random.seed(1)
- df = pd.DataFrame(range(0, 5), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 2 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = RandomForestClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 5), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 2 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = RandomForestClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
- xpl = SmartExplainer(clf, data=df[['x1', 'x2']], backend="lime")
- xpl.compile(x=df[['x1', 'x2']])
+ xpl = SmartExplainer(clf, data=df[["x1", "x2"]], backend="lime")
+ xpl.compile(x=df[["x1", "x2"]])
def test_compile_5(self):
"""
Unit test compile 5
checking compile method with y_target
"""
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 10 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 10 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
xpl = SmartExplainer(clf)
- xpl.compile(x=df[['x1', 'x2']], y_target=df['y'])
+ xpl.compile(x=df[["x1", "x2"]], y_target=df["y"])
assert xpl._case == "classification"
- assert_frame_equal(xpl.y_target, df[['y']])
+ assert_frame_equal(xpl.y_target, df[["y"]])
self.assertListEqual(xpl._classes, [0, 1])
def test_compile_6(self):
@@ -335,24 +300,24 @@ def test_compile_6(self):
checking compile method with additional_data
"""
np.random.seed(1)
- df = pd.DataFrame(range(0, 5), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 2 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df['x3'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = RandomForestClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 5), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 2 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df["x3"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = RandomForestClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
xpl = SmartExplainer(clf)
- xpl.compile(x=df[['x1', 'x2']], additional_data=df[['x3']])
- assert len(xpl.additional_features_dict)==1
+ xpl.compile(x=df[["x1", "x2"]], additional_data=df[["x3"]])
+ assert len(xpl.additional_features_dict) == 1
def test_filter_0(self):
"""
Unit test filter 0
"""
xpl = SmartExplainer(self.model)
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
@@ -363,11 +328,11 @@ def test_filter_0(self):
mockstate.sign_contributions.assert_not_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_not_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
- @patch('shapash.explainer.smart_explainer.SmartExplainer.check_features_name')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.check_features_name")
def test_filter_1(self, mock_check_features_name):
"""
Unit test filter 1
@@ -378,27 +343,27 @@ def test_filter_1(self, mock_check_features_name):
"""
xpl = SmartExplainer(self.model)
mock_check_features_name.return_value = [1, 2]
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
- xpl.filter(features_to_hide=['X1', 'X2'])
+ xpl.filter(features_to_hide=["X1", "X2"])
mockstate.init_mask.assert_called()
mockstate.hide_contributions.assert_called()
mockstate.cap_contributions.assert_not_called()
mockstate.sign_contributions.assert_not_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_not_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
def test_filter_2(self):
"""
Unit test filter 2
"""
xpl = SmartExplainer(self.model)
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
@@ -409,16 +374,16 @@ def test_filter_2(self):
mockstate.sign_contributions.assert_not_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_not_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
def test_filter_3(self):
"""
Unit test filter 3
"""
xpl = SmartExplainer(self.model)
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
@@ -429,16 +394,16 @@ def test_filter_3(self):
mockstate.sign_contributions.assert_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_not_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
def test_filter_4(self):
"""
Unit test filter 4
"""
xpl = SmartExplainer(self.model)
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
@@ -449,16 +414,16 @@ def test_filter_4(self):
mockstate.sign_contributions.assert_not_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
def test_filter_5(self):
"""
Unit test filter 5
"""
xpl = SmartExplainer(self.model)
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
@@ -469,16 +434,16 @@ def test_filter_5(self):
mockstate.sign_contributions.assert_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
def test_filter_6(self):
"""
Unit test filter 6
"""
xpl = SmartExplainer(self.model)
- mock_data = {'var_dict': 1, 'contrib_sorted': 2, 'x_sorted': 3}
+ mock_data = {"var_dict": 1, "contrib_sorted": 2, "x_sorted": 3}
xpl.data = mock_data
mockstate = Mock()
xpl.state = mockstate
@@ -489,9 +454,9 @@ def test_filter_6(self):
mockstate.sign_contributions.assert_not_called()
mockstate.combine_masks.assert_called()
mockstate.cutoff_contributions.assert_not_called()
- assert hasattr(xpl, 'mask')
+ assert hasattr(xpl, "mask")
mockstate.compute_masked_contributions.assert_called()
- assert hasattr(xpl, 'masked_contributions')
+ assert hasattr(xpl, "masked_contributions")
def test_filter_7(self):
"""
@@ -499,66 +464,48 @@ def test_filter_7(self):
"""
xpl = SmartExplainer(self.model)
contributions = [
- pd.DataFrame(
- data=[[0.5, 0.4, 0.3], [0.9, 0.8, 0.7]],
- columns=['Col1', 'Col2', 'Col3']
- ),
- pd.DataFrame(
- data=[[0.3, 0.2, 0.1], [0.6, 0.5, 0.4]],
- columns=['Col1', 'Col2', 'Col3']
- )
+ pd.DataFrame(data=[[0.5, 0.4, 0.3], [0.9, 0.8, 0.7]], columns=["Col1", "Col2", "Col3"]),
+ pd.DataFrame(data=[[0.3, 0.2, 0.1], [0.6, 0.5, 0.4]], columns=["Col1", "Col2", "Col3"]),
]
- xpl.data = {'var_dict': 1, 'contrib_sorted': contributions, 'x_sorted': 3}
+ xpl.data = {"var_dict": 1, "contrib_sorted": contributions, "x_sorted": 3}
xpl.state = MultiDecorator(SmartState())
xpl.filter(threshold=0.5, max_contrib=2)
expected_mask = [
pd.DataFrame(
- data=[[True, False, False], [True, True, False]],
- columns=['contrib_1', 'contrib_2', 'contrib_3']
+ data=[[True, False, False], [True, True, False]], columns=["contrib_1", "contrib_2", "contrib_3"]
),
pd.DataFrame(
- data=[[False, False, False], [True, True, False]],
- columns=['contrib_1', 'contrib_2', 'contrib_3']
- )
+ data=[[False, False, False], [True, True, False]], columns=["contrib_1", "contrib_2", "contrib_3"]
+ ),
]
assert len(expected_mask) == len(xpl.mask)
test_list = [pd.testing.assert_frame_equal(e, m) for e, m in zip(expected_mask, xpl.mask)]
assert all(x is None for x in test_list)
expected_masked_contributions = [
- pd.DataFrame(
- data=[[0.0, 0.7], [0.0, 0.7]],
- columns=['masked_neg', 'masked_pos']
- ),
- pd.DataFrame(
- data=[[0.0, 0.6], [0.0, 0.4]],
- columns=['masked_neg', 'masked_pos']
- )
+ pd.DataFrame(data=[[0.0, 0.7], [0.0, 0.7]], columns=["masked_neg", "masked_pos"]),
+ pd.DataFrame(data=[[0.0, 0.6], [0.0, 0.4]], columns=["masked_neg", "masked_pos"]),
]
assert len(expected_masked_contributions) == len(xpl.masked_contributions)
- test_list = [pd.testing.assert_frame_equal(e, m) for e, m in
- zip(expected_masked_contributions, xpl.masked_contributions)]
+ test_list = [
+ pd.testing.assert_frame_equal(e, m) for e, m in zip(expected_masked_contributions, xpl.masked_contributions)
+ ]
assert all(x is None for x in test_list)
- expected_param_dict = {
- 'features_to_hide': None,
- 'threshold': 0.5,
- 'positive': None,
- 'max_contrib': 2
- }
+ expected_param_dict = {"features_to_hide": None, "threshold": 0.5, "positive": None, "max_contrib": 2}
self.assertDictEqual(expected_param_dict, xpl.mask_params)
def test_check_label_name_1(self):
"""
Unit test check label name 1
"""
- label_dict = {1: 'Age', 2: 'Education'}
+ label_dict = {1: "Age", 2: "Education"}
xpl = SmartExplainer(self.model, label_dict=label_dict)
xpl.inv_label_dict = {v: k for k, v in xpl.label_dict.items()}
xpl._classes = [1, 2]
- entry = 'Age'
+ entry = "Age"
expected_num = 0
expected_code = 1
- expected_value = 'Age'
- label_num, label_code, label_value = xpl.check_label_name(entry, 'value')
+ expected_value = "Age"
+ label_num, label_code, label_value = xpl.check_label_name(entry, "value")
assert expected_num == label_num
assert expected_code == label_code
assert expected_value == label_value
@@ -567,13 +514,13 @@ def test_check_label_name_2(self):
"""
Unit test check label name 2
"""
- xpl = SmartExplainer(self.model, label_dict = None)
+ xpl = SmartExplainer(self.model, label_dict=None)
xpl._classes = [1, 2]
entry = 1
expected_num = 0
expected_code = 1
expected_value = 1
- label_num, label_code, label_value = xpl.check_label_name(entry, 'code')
+ label_num, label_code, label_value = xpl.check_label_name(entry, "code")
assert expected_num == label_num
assert expected_code == label_code
assert expected_value == label_value
@@ -582,15 +529,15 @@ def test_check_label_name_3(self):
"""
Unit test check label name 3
"""
- label_dict = {1: 'Age', 2: 'Education'}
+ label_dict = {1: "Age", 2: "Education"}
xpl = SmartExplainer(self.model, label_dict=label_dict)
xpl.inv_label_dict = {v: k for k, v in xpl.label_dict.items()}
xpl._classes = [1, 2]
entry = 0
expected_num = 0
expected_code = 1
- expected_value = 'Age'
- label_num, label_code, label_value = xpl.check_label_name(entry, 'num')
+ expected_value = "Age"
+ label_num, label_code, label_value = xpl.check_label_name(entry, "num")
assert expected_num == label_num
assert expected_code == label_code
assert expected_value == label_value
@@ -601,34 +548,34 @@ def test_check_label_name_4(self):
"""
xpl = SmartExplainer(self.model)
label = 0
- origin = 'error'
+ origin = "error"
expected_msg = "Origin must be 'num', 'code' or 'value'."
- self.assertRaisesWithMessage(expected_msg, xpl.check_label_name, **{'label': label, 'origin': origin})
+ self.assertRaisesWithMessage(expected_msg, xpl.check_label_name, **{"label": label, "origin": origin})
def test_check_label_name_5(self):
"""
Unit test check label name 5
"""
- label_dict = {1: 'Age', 2: 'Education'}
+ label_dict = {1: "Age", 2: "Education"}
xpl = SmartExplainer(self.model, label_dict=label_dict)
xpl.inv_label_dict = {v: k for k, v in xpl.label_dict.items()}
xpl._classes = [1, 2]
- label = 'Absent'
+ label = "Absent"
expected_msg = f"Label (Absent) not found for origin (value)"
- origin = 'value'
- self.assertRaisesWithMessage(expected_msg, xpl.check_label_name, **{'label': label, 'origin': origin})
+ origin = "value"
+ self.assertRaisesWithMessage(expected_msg, xpl.check_label_name, **{"label": label, "origin": origin})
def test_check_features_name_1(self):
"""
Unit test check features name 1
"""
xpl = SmartExplainer(self.model)
- xpl.features_dict = {'tech_0': 'domain_0', 'tech_1': 'domain_1', 'tech_2': 'domain_2'}
+ xpl.features_dict = {"tech_0": "domain_0", "tech_1": "domain_1", "tech_2": "domain_2"}
xpl.inv_features_dict = {v: k for k, v in xpl.features_dict.items()}
- xpl.columns_dict = {0: 'tech_0', 1: 'tech_1', 2: 'tech_2'}
+ xpl.columns_dict = {0: "tech_0", 1: "tech_1", 2: "tech_2"}
xpl.inv_columns_dict = {v: k for k, v in xpl.columns_dict.items()}
- feature_list_1 = ['domain_0', 'tech_1']
- feature_list_2 = ['domain_0', 0]
+ feature_list_1 = ["domain_0", "tech_1"]
+ feature_list_2 = ["domain_0", 0]
self.assertRaises(ValueError, xpl.check_features_name, feature_list_1)
self.assertRaises(ValueError, xpl.check_features_name, feature_list_2)
@@ -637,11 +584,11 @@ def test_check_features_name_2(self):
Unit test check features name 2
"""
xpl = SmartExplainer(self.model)
- xpl.features_dict = {'tech_0': 'domain_0', 'tech_1': 'domain_1', 'tech_2': 'domain_2'}
+ xpl.features_dict = {"tech_0": "domain_0", "tech_1": "domain_1", "tech_2": "domain_2"}
xpl.inv_features_dict = {v: k for k, v in xpl.features_dict.items()}
- xpl.columns_dict = {0: 'tech_0', 1: 'tech_1', 2: 'tech_2'}
+ xpl.columns_dict = {0: "tech_0", 1: "tech_1", 2: "tech_2"}
xpl.inv_columns_dict = {v: k for k, v in xpl.columns_dict.items()}
- feature_list = ['domain_0', 'domain_2']
+ feature_list = ["domain_0", "domain_2"]
output = xpl.check_features_name(feature_list)
expected_output = [0, 2]
np.testing.assert_array_equal(output, expected_output)
@@ -651,9 +598,9 @@ def test_check_features_name_3(self):
Unit test check features name 3
"""
xpl = SmartExplainer(self.model)
- xpl.columns_dict = {0: 'tech_0', 1: 'tech_1', 2: 'tech_2'}
+ xpl.columns_dict = {0: "tech_0", 1: "tech_1", 2: "tech_2"}
xpl.inv_columns_dict = {v: k for k, v in xpl.columns_dict.items()}
- feature_list = ['tech_2']
+ feature_list = ["tech_2"]
output = xpl.check_features_name(feature_list)
expected_output = [2]
np.testing.assert_array_equal(output, expected_output)
@@ -686,14 +633,14 @@ def test_load_1(self):
temp, xpl = init_sme_to_pickle_test()
current = Path(path.abspath(__file__)).parent.parent.parent
- if str(sys.version)[0:3] == '3.8':
- pkl_file = path.join(current, 'data/xpl_to_load_38.pkl')
- elif str(sys.version)[0:3] == '3.9':
- pkl_file = path.join(current, 'data/xpl_to_load_39.pkl')
- elif str(sys.version)[0:4] == '3.10':
- pkl_file = path.join(current, 'data/xpl_to_load_310.pkl')
- elif str(sys.version)[0:4] == '3.11':
- pkl_file = path.join(current, 'data/xpl_to_load_311.pkl')
+ if str(sys.version)[0:3] == "3.8":
+ pkl_file = path.join(current, "data/xpl_to_load_38.pkl")
+ elif str(sys.version)[0:3] == "3.9":
+ pkl_file = path.join(current, "data/xpl_to_load_39.pkl")
+ elif str(sys.version)[0:4] == "3.10":
+ pkl_file = path.join(current, "data/xpl_to_load_310.pkl")
+ elif str(sys.version)[0:4] == "3.11":
+ pkl_file = path.join(current, "data/xpl_to_load_311.pkl")
else:
raise NotImplementedError
@@ -727,8 +674,8 @@ def test_predict_1(self):
Test predict method 1
"""
X = pd.DataFrame([[1, 1, 1], [2, 2, 2], [3, 3, 3]])
- y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=['pred'])
- y_false = pd.DataFrame(data=np.array([1, 2, 4]), columns=['pred'])
+ y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=["pred"])
+ y_false = pd.DataFrame(data=np.array([1, 2, 4]), columns=["pred"])
model = DecisionTreeRegressor().fit(X, y_true)
xpl = SmartExplainer(model)
xpl.compile(x=X, y_pred=y_false)
@@ -741,7 +688,7 @@ def test_predict_2(self):
Test predict method 2
"""
X = pd.DataFrame([[1, 1, 1], [2, 2, 2], [3, 3, 3]])
- y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=['pred'])
+ y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=["pred"])
model = DecisionTreeRegressor().fit(X, y_true)
xpl = SmartExplainer(model)
xpl.compile(x=X)
@@ -754,17 +701,17 @@ def test_predict_3(self):
Test predict method 3
"""
X = pd.DataFrame([[1, 1, 1], [2, 2, 2], [3, 3, 3]])
- y_target = pd.DataFrame(data=np.array([1, 2, 3]), columns=['pred'])
+ y_target = pd.DataFrame(data=np.array([1, 2, 3]), columns=["pred"])
model = DecisionTreeRegressor().fit(X, y_target)
xpl = SmartExplainer(model)
xpl.compile(x=X, y_target=y_target)
- xpl.predict() # prediction errors should be computed
+ xpl.predict() # prediction errors should be computed
assert xpl.prediction_error is not None
def test_add_1(self):
xpl = SmartExplainer(self.model)
- dataframe_yp = pd.Series([1, 3, 1], name='pred', index=[0, 1, 2])
+ dataframe_yp = pd.Series([1, 3, 1], name="pred", index=[0, 1, 2])
xpl.x_init = dataframe_yp
xpl.add(y_pred=dataframe_yp)
expected = SmartExplainer(self.model)
@@ -778,19 +725,19 @@ def test_add_2(self):
xpl = SmartExplainer(self.model)
xpl._classes = [0, 1]
xpl._case = "classification"
- xpl.add(label_dict={0: 'Zero', 1: 'One'})
- assert xpl.label_dict[0] == 'Zero'
- assert xpl.label_dict[1] == 'One'
+ xpl.add(label_dict={0: "Zero", 1: "One"})
+ assert xpl.label_dict[0] == "Zero"
+ assert xpl.label_dict[1] == "One"
def test_add_3(self):
"""
Unit test add 3
"""
xpl = SmartExplainer(self.model)
- xpl.columns_dict = {0: 'Age', 1: 'Education', 2: 'Sex'}
- xpl.add(features_dict={'Age': 'Age (Years Old)'})
- assert xpl.features_dict['Age'] == 'Age (Years Old)'
- assert xpl.features_dict['Education'] == 'Education'
+ xpl.columns_dict = {0: "Age", 1: "Education", 2: "Sex"}
+ xpl.add(features_dict={"Age": "Age (Years Old)"})
+ assert xpl.features_dict["Age"] == "Age (Years Old)"
+ assert xpl.features_dict["Education"] == "Education"
def test_to_pandas_1(self):
"""
@@ -799,81 +746,67 @@ def test_to_pandas_1(self):
xpl = SmartExplainer(self.model)
xpl.state = SmartState()
data = {}
- data['contrib_sorted'] = pd.DataFrame(
- [[0.32230754, 0.1550689, 0.10183475, 0.05471339],
- [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
- [-0.48666675, 0.25507156, -0.16968889, 0.0757443]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ data["contrib_sorted"] = pd.DataFrame(
+ [
+ [0.32230754, 0.1550689, 0.10183475, 0.05471339],
+ [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
+ [-0.48666675, 0.25507156, -0.16968889, 0.0757443],
+ ],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
- data['var_dict'] = pd.DataFrame(
- [[1, 0, 2, 3],
- [1, 0, 3, 2],
- [1, 0, 2, 3]],
- columns=['feature_0', 'feature_1', 'feature_2', 'feature_3'],
- index=[0, 1, 2]
+ data["var_dict"] = pd.DataFrame(
+ [[1, 0, 2, 3], [1, 0, 3, 2], [1, 0, 2, 3]],
+ columns=["feature_0", "feature_1", "feature_2", "feature_3"],
+ index=[0, 1, 2],
)
- data['x_sorted'] = pd.DataFrame(
- [[1., 3., 22., 1.],
- [2., 1., 2., 38.],
- [2., 3., 26., 1.]],
- columns=['feature_0', 'feature_1', 'feature_2', 'feature_3'],
- index=[0, 1, 2]
+ data["x_sorted"] = pd.DataFrame(
+ [[1.0, 3.0, 22.0, 1.0], [2.0, 1.0, 2.0, 38.0], [2.0, 3.0, 26.0, 1.0]],
+ columns=["feature_0", "feature_1", "feature_2", "feature_3"],
+ index=[0, 1, 2],
)
xpl.data = data
- xpl.columns_dict = {0: 'Pclass', 1: 'Sex', 2: 'Age', 3: 'Embarked'}
- xpl.features_dict = {'Pclass': 'Pclass', 'Sex': 'Sex', 'Age': 'Age', 'Embarked': 'Embarked'}
+ xpl.columns_dict = {0: "Pclass", 1: "Sex", 2: "Age", 3: "Embarked"}
+ xpl.features_dict = {"Pclass": "Pclass", "Sex": "Sex", "Age": "Age", "Embarked": "Embarked"}
xpl.x = pd.DataFrame(
- [[3., 1., 22., 1.],
- [1., 2., 38., 2.],
- [3., 2., 26., 1.]],
- columns=['Pclass', 'Sex', 'Age', 'Embarked'],
- index=[0, 1, 2]
+ [[3.0, 1.0, 22.0, 1.0], [1.0, 2.0, 38.0, 2.0], [3.0, 2.0, 26.0, 1.0]],
+ columns=["Pclass", "Sex", "Age", "Embarked"],
+ index=[0, 1, 2],
)
xpl.x_init = xpl.x
- xpl.contributions = data['contrib_sorted']
- xpl.y_pred = pd.DataFrame([1, 2, 3], columns=['pred'], index=[0, 1, 2])
- model = lambda : None
+ xpl.contributions = data["contrib_sorted"]
+ xpl.y_pred = pd.DataFrame([1, 2, 3], columns=["pred"], index=[0, 1, 2])
+ model = lambda: None
model.predict = types.MethodType(self.predict, model)
xpl.model = model
xpl._case, xpl._classes = check_model(model)
xpl.state = SmartState()
output = xpl.to_pandas(max_contrib=2)
expected = pd.DataFrame(
- [[1, 'Sex', 1.0, 0.32230754, 'Pclass', 3.0, 0.1550689],
- [2, 'Sex', 2.0, -0.58547512, 'Pclass', 1.0, -0.37050409],
- [3, 'Sex', 2.0, -0.48666675, 'Pclass', 3.0, 0.25507156]],
- columns=['pred',
- 'feature_1',
- 'value_1',
- 'contribution_1',
- 'feature_2',
- 'value_2',
- 'contribution_2'],
+ [
+ [1, "Sex", 1.0, 0.32230754, "Pclass", 3.0, 0.1550689],
+ [2, "Sex", 2.0, -0.58547512, "Pclass", 1.0, -0.37050409],
+ [3, "Sex", 2.0, -0.48666675, "Pclass", 3.0, 0.25507156],
+ ],
+ columns=["pred", "feature_1", "value_1", "contribution_1", "feature_2", "value_2", "contribution_2"],
index=[0, 1, 2],
- dtype=object
+ dtype=object,
)
- expected['pred'] = expected['pred'].astype(int)
+ expected["pred"] = expected["pred"].astype(int)
assert not pd.testing.assert_frame_equal(expected, output)
def predict_proba(self, arg1, arg2):
"""
predict_proba method
"""
- matrx = np.array(
- [[0.2, 0.8],
- [0.3, 0.7],
- [0.4, 0.6]]
- )
+ matrx = np.array([[0.2, 0.8], [0.3, 0.7], [0.4, 0.6]])
return matrx
def predict(self, arg1, arg2):
"""
predict method
"""
- matrx = np.array(
- [12, 3, 7]
- )
+ matrx = np.array([12, 3, 7])
return matrx
def test_to_pandas_2(self):
@@ -883,35 +816,45 @@ def test_to_pandas_2(self):
predict_proba output and column_dict attribute
"""
contrib = pd.DataFrame(
- [[0.32230754, 0.1550689, 0.10183475, 0.05471339],
- [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
- [-0.48666675, 0.25507156, -0.16968889, 0.0757443]],
- index=[0, 1, 2]
- )
- x = pd.DataFrame(
- [[3., 1., 22., 1.],
- [1., 2., 38., 2.],
- [3., 2., 26., 1.]],
- index=[0, 1, 2]
+ [
+ [0.32230754, 0.1550689, 0.10183475, 0.05471339],
+ [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
+ [-0.48666675, 0.25507156, -0.16968889, 0.0757443],
+ ],
+ index=[0, 1, 2],
)
- pred = pd.DataFrame([3, 1, 1], columns=['pred'], index=[0, 1, 2])
+ x = pd.DataFrame([[3.0, 1.0, 22.0, 1.0], [1.0, 2.0, 38.0, 2.0], [3.0, 2.0, 26.0, 1.0]], index=[0, 1, 2])
+ pred = pd.DataFrame([3, 1, 1], columns=["pred"], index=[0, 1, 2])
model = CatBoostClassifier().fit(x, pred)
xpl = SmartExplainer(model)
xpl.compile(contributions=contrib, x=x, y_pred=pred)
- xpl.columns_dict = {0: 'Pclass', 1: 'Sex', 2: 'Age', 3: 'Embarked'}
- xpl.features_dict = {'Pclass': 'Pclass', 'Sex': 'Sex', 'Age': 'Age', 'Embarked': 'Embarked'}
+ xpl.columns_dict = {0: "Pclass", 1: "Sex", 2: "Age", 3: "Embarked"}
+ xpl.features_dict = {"Pclass": "Pclass", "Sex": "Sex", "Age": "Age", "Embarked": "Embarked"}
output = xpl.to_pandas(max_contrib=3, positive=True, proba=True)
expected = pd.DataFrame(
- [[3, 0.8, 'Pclass', 3.0, 0.32230754, 'Sex', 1.0, 0.1550689, 'Age', 22.0, 0.10183475],
- [1, 0.3, 'Pclass', 1.0, 0.58547512, 'Sex', 2.0, 0.37050409, 'Age', 38.0, 0.07249285],
- [1, 0.4, 'Pclass', 3.0, 0.48666675, 'Age', 26.0, 0.16968889, np.nan, np.nan, np.nan]],
- columns=['pred', 'proba', 'feature_1', 'value_1', 'contribution_1', 'feature_2',
- 'value_2', 'contribution_2', 'feature_3', 'value_3', 'contribution_3'],
+ [
+ [3, 0.8, "Pclass", 3.0, 0.32230754, "Sex", 1.0, 0.1550689, "Age", 22.0, 0.10183475],
+ [1, 0.3, "Pclass", 1.0, 0.58547512, "Sex", 2.0, 0.37050409, "Age", 38.0, 0.07249285],
+ [1, 0.4, "Pclass", 3.0, 0.48666675, "Age", 26.0, 0.16968889, np.nan, np.nan, np.nan],
+ ],
+ columns=[
+ "pred",
+ "proba",
+ "feature_1",
+ "value_1",
+ "contribution_1",
+ "feature_2",
+ "value_2",
+ "contribution_2",
+ "feature_3",
+ "value_3",
+ "contribution_3",
+ ],
index=[0, 1, 2],
- dtype=object
+ dtype=object,
)
- expected['pred'] = expected['pred'].astype(int)
- expected['proba'] = expected['proba'].astype(float)
+ expected["pred"] = expected["pred"].astype(int)
+ expected["proba"] = expected["proba"].astype(float)
pd.testing.assert_series_equal(expected.dtypes, output.dtypes)
def test_to_pandas_3(self):
@@ -921,73 +864,63 @@ def test_to_pandas_3(self):
xpl = SmartExplainer(self.model)
xpl.state = SmartState()
data = {}
- data['contrib_sorted'] = pd.DataFrame(
- [[0.32230754, 0.1550689, 0.10183475, 0.05471339],
- [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
- [-0.48666675, 0.25507156, -0.16968889, 0.0757443]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ data["contrib_sorted"] = pd.DataFrame(
+ [
+ [0.32230754, 0.1550689, 0.10183475, 0.05471339],
+ [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
+ [-0.48666675, 0.25507156, -0.16968889, 0.0757443],
+ ],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
- data['var_dict'] = pd.DataFrame(
- [[1, 0, 2, 3],
- [1, 0, 3, 2],
- [1, 0, 2, 3]],
- columns=['feature_0', 'feature_1', 'feature_2', 'feature_3'],
- index=[0, 1, 2]
+ data["var_dict"] = pd.DataFrame(
+ [[1, 0, 2, 3], [1, 0, 3, 2], [1, 0, 2, 3]],
+ columns=["feature_0", "feature_1", "feature_2", "feature_3"],
+ index=[0, 1, 2],
)
- data['x_sorted'] = pd.DataFrame(
- [[1., 3., 22., 1.],
- [2., 1., 2., 38.],
- [2., 3., 26., 1.]],
- columns=['feature_0', 'feature_1', 'feature_2', 'feature_3'],
- index=[0, 1, 2]
+ data["x_sorted"] = pd.DataFrame(
+ [[1.0, 3.0, 22.0, 1.0], [2.0, 1.0, 2.0, 38.0], [2.0, 3.0, 26.0, 1.0]],
+ columns=["feature_0", "feature_1", "feature_2", "feature_3"],
+ index=[0, 1, 2],
)
data_groups = dict()
- data_groups['contrib_sorted'] = pd.DataFrame(
- [[0.32230754, 0.10183475, 0.05471339],
- [-0.58547512, -0.07249285, 0.00171975],
- [-0.48666675, -0.16968889, 0.0757443]],
- columns=['contribution_0', 'contribution_1', 'contribution_2'],
- index=[0, 1, 2]
+ data_groups["contrib_sorted"] = pd.DataFrame(
+ [
+ [0.32230754, 0.10183475, 0.05471339],
+ [-0.58547512, -0.07249285, 0.00171975],
+ [-0.48666675, -0.16968889, 0.0757443],
+ ],
+ columns=["contribution_0", "contribution_1", "contribution_2"],
+ index=[0, 1, 2],
)
- data_groups['var_dict'] = pd.DataFrame(
- [[0, 1, 2],
- [0, 2, 1],
- [0, 1, 2]],
- columns=['feature_0', 'feature_1', 'feature_2'],
- index=[0, 1, 2]
+ data_groups["var_dict"] = pd.DataFrame(
+ [[0, 1, 2], [0, 2, 1], [0, 1, 2]], columns=["feature_0", "feature_1", "feature_2"], index=[0, 1, 2]
)
- data_groups['x_sorted'] = pd.DataFrame(
- [[1., 22., 1.],
- [2., 2., 38.],
- [2., 26., 1.]],
- columns=['feature_0', 'feature_1', 'feature_2'],
- index=[0, 1, 2]
+ data_groups["x_sorted"] = pd.DataFrame(
+ [[1.0, 22.0, 1.0], [2.0, 2.0, 38.0], [2.0, 26.0, 1.0]],
+ columns=["feature_0", "feature_1", "feature_2"],
+ index=[0, 1, 2],
)
xpl.data = data
xpl.data_groups = data_groups
- xpl.columns_dict = {0: 'Pclass', 1: 'Sex', 2: 'Age', 3: 'Embarked'}
- xpl.features_dict = {'Pclass': 'Pclass', 'Sex': 'Sex', 'Age': 'Age', 'Embarked': 'Embarked', 'group1': 'group1'}
- xpl.features_groups = {'group1': ['Pclass', 'Sex']}
+ xpl.columns_dict = {0: "Pclass", 1: "Sex", 2: "Age", 3: "Embarked"}
+ xpl.features_dict = {"Pclass": "Pclass", "Sex": "Sex", "Age": "Age", "Embarked": "Embarked", "group1": "group1"}
+ xpl.features_groups = {"group1": ["Pclass", "Sex"]}
xpl.x = pd.DataFrame(
- [[3., 1., 22., 1.],
- [1., 2., 38., 2.],
- [3., 2., 26., 1.]],
- columns=['Pclass', 'Sex', 'Age', 'Embarked'],
- index=[0, 1, 2]
+ [[3.0, 1.0, 22.0, 1.0], [1.0, 2.0, 38.0, 2.0], [3.0, 2.0, 26.0, 1.0]],
+ columns=["Pclass", "Sex", "Age", "Embarked"],
+ index=[0, 1, 2],
)
xpl.x_init_groups = pd.DataFrame(
- [[3., 22., 1.],
- [1., 38., 2.],
- [3., 26., 1.]],
- columns=['group1', 'Age', 'Embarked'],
- index=[0, 1, 2]
+ [[3.0, 22.0, 1.0], [1.0, 38.0, 2.0], [3.0, 26.0, 1.0]],
+ columns=["group1", "Age", "Embarked"],
+ index=[0, 1, 2],
)
xpl.x_init = xpl.x
- xpl.contributions = data['contrib_sorted']
- xpl.y_pred = pd.DataFrame([1, 2, 3], columns=['pred'], index=[0, 1, 2])
+ xpl.contributions = data["contrib_sorted"]
+ xpl.y_pred = pd.DataFrame([1, 2, 3], columns=["pred"], index=[0, 1, 2])
model = lambda: None
model.predict = types.MethodType(self.predict, model)
xpl.model = model
@@ -996,20 +929,16 @@ def test_to_pandas_3(self):
output = xpl.to_pandas(max_contrib=2, use_groups=True)
expected = pd.DataFrame(
- [[1, 'group1', 1.0, 0.322308, 'Age', 22.0, 0.101835],
- [2, 'group1', 2.0, -0.585475, 'Embarked', 2.0, -0.072493],
- [3, 'group1', 2.0, -0.486667, 'Age', 26.0, -0.169689]],
- columns=['pred',
- 'feature_1',
- 'value_1',
- 'contribution_1',
- 'feature_2',
- 'value_2',
- 'contribution_2'],
+ [
+ [1, "group1", 1.0, 0.322308, "Age", 22.0, 0.101835],
+ [2, "group1", 2.0, -0.585475, "Embarked", 2.0, -0.072493],
+ [3, "group1", 2.0, -0.486667, "Age", 26.0, -0.169689],
+ ],
+ columns=["pred", "feature_1", "value_1", "contribution_1", "feature_2", "value_2", "contribution_2"],
index=[0, 1, 2],
- dtype=object
+ dtype=object,
)
- expected['pred'] = expected['pred'].astype(int)
+ expected["pred"] = expected["pred"].astype(int)
pd.testing.assert_frame_equal(expected, output)
def test_compute_features_import_1(self):
@@ -1019,18 +948,16 @@ def test_compute_features_import_1(self):
"""
xpl = SmartExplainer(self.model)
contributions = pd.DataFrame(
- [[1, 2, 3, 4],
- [5, 6, 7, 8],
- [9, 10, 11, 12]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ [[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
xpl.features_imp = None
xpl.contributions = contributions
xpl.backend = ShapBackend(model=DecisionTreeClassifier().fit([[0]], [[0]]))
xpl.backend.state = SmartState()
xpl.explain_data = None
- xpl._case = 'regression'
+ xpl._case = "regression"
xpl.compute_features_import()
expected = contributions.abs().sum().sort_values(ascending=True)
expected = expected / expected.sum()
@@ -1043,18 +970,14 @@ def test_compute_features_import_2(self):
"""
xpl = SmartExplainer(self.model)
contrib1 = pd.DataFrame(
- [[1, 2, 3, 4],
- [5, 6, 7, 8],
- [9, 10, 11, 12]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ [[1, 2, 3, 4], [5, 6, 7, 8], [9, 10, 11, 12]],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
contrib2 = pd.DataFrame(
- [[13, 14, 15, 16],
- [17, 18, 19, 20],
- [21, 22, 23, 24]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ [[13, 14, 15, 16], [17, 18, 19, 20], [21, 22, 23, 24]],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
contributions = [contrib1, contrib2]
xpl.features_imp = None
@@ -1075,47 +998,43 @@ def test_to_smartpredictor_1(self):
"""
Unit test 1 to_smartpredictor
"""
- df = pd.DataFrame(range(0, 5), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 2 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = ["S", "M", "S", "D", "M"]
- df = df.set_index('id')
+ df = pd.DataFrame(range(0, 5), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 2 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = ["S", "M", "S", "D", "M"]
+ df = df.set_index("id")
encoder = ce.OrdinalEncoder(cols=["x2"], handle_unknown="None")
encoder_fitted = encoder.fit(df[["x1", "x2"]])
df_encoded = encoder_fitted.transform(df[["x1", "x2"]])
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[['x1', 'x2']], df['y'])
-
- postprocessing = {"x2": {
- "type": "transcoding",
- "rule": {"S": "single", "M": "married", "D": "divorced"}}}
- xpl = SmartExplainer(clf, features_dict={"x1": "age", "x2": "family_situation"},
- preprocessing=encoder_fitted,
- postprocessing=postprocessing)
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[["x1", "x2"]], df["y"])
+
+ postprocessing = {"x2": {"type": "transcoding", "rule": {"S": "single", "M": "married", "D": "divorced"}}}
+ xpl = SmartExplainer(
+ clf,
+ features_dict={"x1": "age", "x2": "family_situation"},
+ preprocessing=encoder_fitted,
+ postprocessing=postprocessing,
+ )
- xpl.compile(x=df_encoded[['x1', 'x2']])
+ xpl.compile(x=df_encoded[["x1", "x2"]])
predictor_1 = xpl.to_smartpredictor()
- xpl.mask_params = {
- 'features_to_hide': None,
- 'threshold': None,
- 'positive': True,
- 'max_contrib': 1
- }
+ xpl.mask_params = {"features_to_hide": None, "threshold": None, "positive": True, "max_contrib": 1}
predictor_2 = xpl.to_smartpredictor()
- assert hasattr(predictor_1, 'model')
- assert hasattr(predictor_1, 'backend')
- assert hasattr(predictor_1, 'features_dict')
- assert hasattr(predictor_1, 'label_dict')
- assert hasattr(predictor_1, '_case')
- assert hasattr(predictor_1, '_classes')
- assert hasattr(predictor_1, 'columns_dict')
- assert hasattr(predictor_1, 'features_types')
- assert hasattr(predictor_1, 'preprocessing')
- assert hasattr(predictor_1, 'postprocessing')
- assert hasattr(predictor_1, 'mask_params')
- assert hasattr(predictor_2, 'mask_params')
+ assert hasattr(predictor_1, "model")
+ assert hasattr(predictor_1, "backend")
+ assert hasattr(predictor_1, "features_dict")
+ assert hasattr(predictor_1, "label_dict")
+ assert hasattr(predictor_1, "_case")
+ assert hasattr(predictor_1, "_classes")
+ assert hasattr(predictor_1, "columns_dict")
+ assert hasattr(predictor_1, "features_types")
+ assert hasattr(predictor_1, "preprocessing")
+ assert hasattr(predictor_1, "postprocessing")
+ assert hasattr(predictor_1, "mask_params")
+ assert hasattr(predictor_2, "mask_params")
assert predictor_1.model == xpl.model
assert predictor_1.backend == xpl.backend
@@ -1126,25 +1045,27 @@ def test_to_smartpredictor_1(self):
assert predictor_1.columns_dict == xpl.columns_dict
assert predictor_1.preprocessing == xpl.preprocessing
assert predictor_1.postprocessing == xpl.postprocessing
- assert all(predictor_1.features_types[feature] == str(xpl.x_init[feature].dtypes)
- for feature in xpl.x_init.columns)
+ assert all(
+ predictor_1.features_types[feature] == str(xpl.x_init[feature].dtypes) for feature in xpl.x_init.columns
+ )
assert predictor_2.mask_params == xpl.mask_params
def test_get_interaction_values_1(self):
- df = pd.DataFrame({
- "y": np.random.randint(2, size=10),
- "a": np.random.rand(10)*10,
- "b": np.random.rand(10),
- })
+ df = pd.DataFrame(
+ {
+ "y": np.random.randint(2, size=10),
+ "a": np.random.rand(10) * 10,
+ "b": np.random.rand(10),
+ }
+ )
- clf = RandomForestClassifier(n_estimators=5).fit(df[['a', 'b']], df['y'])
+ clf = RandomForestClassifier(n_estimators=5).fit(df[["a", "b"]], df["y"])
- xpl = SmartExplainer(clf,
- backend='shap',
- explainer_args={'explainer': shap.explainers.TreeExplainer,
- 'model': clf})
- xpl.compile(x=df.drop('y', axis=1))
+ xpl = SmartExplainer(
+ clf, backend="shap", explainer_args={"explainer": shap.explainers.TreeExplainer, "model": clf}
+ )
+ xpl.compile(x=df.drop("y", axis=1))
shap_interaction_values = xpl.get_interaction_values(n_samples_max=10)
assert shap_interaction_values.shape[0] == 10
@@ -1152,17 +1073,16 @@ def test_get_interaction_values_1(self):
shap_interaction_values = xpl.get_interaction_values()
assert shap_interaction_values.shape[0] == df.shape[0]
-
- @patch('shapash.explainer.smart_explainer.SmartApp')
- @patch('shapash.explainer.smart_explainer.CustomThread')
- @patch('shapash.explainer.smart_explainer.get_host_name')
+ @patch("shapash.explainer.smart_explainer.SmartApp")
+ @patch("shapash.explainer.smart_explainer.CustomThread")
+ @patch("shapash.explainer.smart_explainer.get_host_name")
def test_run_app_1(self, mock_get_host_name, mock_custom_thread, mock_smartapp):
"""
Test that when y_pred is not given, y_pred is automatically computed.
"""
X = pd.DataFrame([[1, 1, 1], [2, 2, 2], [3, 3, 3]])
contributions = pd.DataFrame([[0.1, -0.2, 0.3], [0.1, -0.2, 0.3], [0.1, -0.2, 0.3]])
- y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=['pred'])
+ y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=["pred"])
model = DecisionTreeRegressor().fit(X, y_true)
xpl = SmartExplainer(model)
xpl.compile(contributions=contributions, x=X)
@@ -1170,42 +1090,42 @@ def test_run_app_1(self, mock_get_host_name, mock_custom_thread, mock_smartapp):
assert xpl.y_pred is not None
- @patch('shapash.explainer.smart_explainer.SmartApp')
- @patch('shapash.explainer.smart_explainer.CustomThread')
- @patch('shapash.explainer.smart_explainer.get_host_name')
+ @patch("shapash.explainer.smart_explainer.SmartApp")
+ @patch("shapash.explainer.smart_explainer.CustomThread")
+ @patch("shapash.explainer.smart_explainer.get_host_name")
def test_run_app_2(self, mock_get_host_name, mock_custom_thread, mock_smartapp):
"""
Test that when y_target is given
"""
X = pd.DataFrame([[1, 1, 1], [2, 2, 2], [3, 3, 3]])
contributions = pd.DataFrame([[0.1, -0.2, 0.3], [0.1, -0.2, 0.3], [0.1, -0.2, 0.3]])
- y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=['pred'])
+ y_true = pd.DataFrame(data=np.array([1, 2, 3]), columns=["pred"])
model = DecisionTreeRegressor().fit(X, y_true)
xpl = SmartExplainer(model)
xpl.compile(contributions=contributions, x=X, y_target=y_true)
xpl.run_app()
assert xpl.y_target is not None
- @patch('shapash.report.generation.export_and_save_report')
- @patch('shapash.report.generation.execute_report')
+ @patch("shapash.report.generation.export_and_save_report")
+ @patch("shapash.report.generation.execute_report")
def test_generate_report(self, mock_execute_report, mock_export_and_save_report):
"""
Test generate report method
"""
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 10 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 10 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
xpl = SmartExplainer(clf)
- xpl.compile(x=df[['x1', 'x2']])
- xpl.generate_report(output_file='test', project_info_file='test')
+ xpl.compile(x=df[["x1", "x2"]])
+ xpl.generate_report(output_file="test", project_info_file="test")
mock_execute_report.assert_called_once()
mock_export_and_save_report.assert_called_once()
def test_compute_features_stability_1(self):
- df = pd.DataFrame(np.random.randint(1, 100, size=(15, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(1, 100, size=(15, 4)), columns=list("ABCD"))
selection = [1, 3]
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
@@ -1221,7 +1141,7 @@ def test_compute_features_stability_1(self):
assert xpl.features_stability["amplitude"].shape == expected
def test_compute_features_stability_2(self):
- df = pd.DataFrame(np.random.randint(1, 100, size=(15, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(1, 100, size=(15, 4)), columns=list("ABCD"))
selection = [1]
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
@@ -1236,7 +1156,7 @@ def test_compute_features_stability_2(self):
assert xpl.local_neighbors["norm_shap"].shape[1] == expected
def test_compute_features_compacity(self):
- df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list("ABCD"))
selection = [1, 3]
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
diff --git a/tests/unit_tests/explainer/test_smart_plotter.py b/tests/unit_tests/explainer/test_smart_plotter.py
index 4b0652f1..57f279a0 100644
--- a/tests/unit_tests/explainer/test_smart_plotter.py
+++ b/tests/unit_tests/explainer/test_smart_plotter.py
@@ -5,19 +5,21 @@
import types
import unittest
from unittest.mock import patch
-import pandas as pd
+
+import category_encoders as ce
import numpy as np
-import plotly.graph_objects as go
+import pandas as pd
import plotly.express as px
-from sklearn.tree import DecisionTreeRegressor, DecisionTreeClassifier
+import plotly.graph_objects as go
from catboost import CatBoostClassifier
-import category_encoders as ce
+from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
+
from shapash import SmartExplainer
from shapash.backend import ShapBackend
-from shapash.utils.check import check_model
from shapash.explainer.multi_decorator import MultiDecorator
from shapash.explainer.smart_state import SmartState
from shapash.style.style_utils import get_palette
+from shapash.utils.check import check_model
class TestSmartPlotter(unittest.TestCase):
@@ -29,6 +31,7 @@ class TestSmartPlotter(unittest.TestCase):
unittest : [type]
[description]
"""
+
def predict_proba(self, arg1, X):
"""
predict_proba method
@@ -44,9 +47,7 @@ def predict(self, arg1, arg2):
"""
predict method
"""
- matrx = np.array(
- [1, 3]
- )
+ matrx = np.array([1, 3])
return matrx
def setUp(self):
@@ -54,75 +55,50 @@ def setUp(self):
SetUp
"""
self.x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
self.contrib0 = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78],
- [1.2, 3.6]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[-3.4, 0.78], [1.2, 3.6]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
self.contrib1 = pd.DataFrame(
- data=np.array(
- [[-0.3, 0.89],
- [4.7, 0.6]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[-0.3, 0.89], [4.7, 0.6]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
self.contrib_sorted = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78],
- [3.6, 1.2]]),
- columns=['contrib_0', 'contrib_1'],
- index=['person_A', 'person_B']
+ data=np.array([[-3.4, 0.78], [3.6, 1.2]]),
+ columns=["contrib_0", "contrib_1"],
+ index=["person_A", "person_B"],
)
self.x_sorted = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- [27, 'Master']]),
- columns=['feature_0', 'feature_1'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34], [27, "Master"]]),
+ columns=["feature_0", "feature_1"],
+ index=["person_A", "person_B"],
)
self.var_dict = pd.DataFrame(
- data=np.array(
- [[0, 1],
- [1, 0]]),
- columns=['feature_0', 'feature_1'],
- index=['person_A', 'person_B']
+ data=np.array([[0, 1], [1, 0]]), columns=["feature_0", "feature_1"], index=["person_A", "person_B"]
)
self.mask = pd.DataFrame(
- data=np.array(
- [[True, False],
- [True, True]]),
- columns=['feature_0', 'feature_1'],
- index=['person_A', 'person_B']
- )
- self.features_compacity = {
- "features_needed": [1, 1],
- "distance_reached": np.array([0.12, 0.16])
- }
+ data=np.array([[True, False], [True, True]]),
+ columns=["feature_0", "feature_1"],
+ index=["person_A", "person_B"],
+ )
+ self.features_compacity = {"features_needed": [1, 1], "distance_reached": np.array([0.12, 0.16])}
encoder = ce.OrdinalEncoder(cols=["X1"], handle_unknown="None").fit(self.x_init)
model = CatBoostClassifier().fit(encoder.transform(self.x_init), [0, 1])
self.model = model
# Declare explainer object
- self.feature_dictionary = {'X1': 'Education', 'X2': 'Age'}
+ self.feature_dictionary = {"X1": "Education", "X2": "Age"}
self.smart_explainer = SmartExplainer(model, features_dict=self.feature_dictionary, preprocessing=encoder)
self.smart_explainer.data = dict()
- self.smart_explainer.data['contrib_sorted'] = self.contrib_sorted
- self.smart_explainer.data['x_sorted'] = self.x_sorted
- self.smart_explainer.data['var_dict'] = self.var_dict
+ self.smart_explainer.data["contrib_sorted"] = self.contrib_sorted
+ self.smart_explainer.data["x_sorted"] = self.x_sorted
+ self.smart_explainer.data["var_dict"] = self.var_dict
self.smart_explainer.x_encoded = encoder.transform(self.x_init)
self.smart_explainer.x_init = self.x_init
self.smart_explainer.postprocessing_modifications = False
self.smart_explainer.backend = ShapBackend(model=model)
self.smart_explainer.backend.state = MultiDecorator(SmartState())
self.smart_explainer.explain_data = None
- #self.smart_explainer.x_contrib_plot = self.x_contrib_plot
+ # self.smart_explainer.x_contrib_plot = self.x_contrib_plot
self.smart_explainer.columns_dict = {i: col for i, col in enumerate(self.smart_explainer.x_init.columns)}
self.smart_explainer.inv_columns_dict = {v: k for k, v in self.smart_explainer.columns_dict.items()}
self.smart_explainer.mask = self.mask
@@ -139,14 +115,14 @@ def test_define_style_attributes(self):
# clear style attributes
del self.smart_explainer.plot._style_dict
- colors_dict = get_palette('default')
+ colors_dict = get_palette("default")
self.smart_explainer.plot.define_style_attributes(colors_dict=colors_dict)
- assert hasattr(self.smart_explainer.plot, '_style_dict')
- assert len(list(self.smart_explainer.plot._style_dict.keys())) > 0
+ assert hasattr(self.smart_explainer.plot, "_style_dict")
+ assert len(list(self.smart_explainer.plot._style_dict.keys())) > 0
- @patch('shapash.explainer.smart_explainer.SmartExplainer.filter')
- @patch('shapash.explainer.smart_plotter.SmartPlotter.local_pred')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.filter")
+ @patch("shapash.explainer.smart_plotter.SmartPlotter.local_pred")
def test_local_plot_1(self, local_pred, filter):
"""
Unit test Local plot 1
@@ -158,33 +134,22 @@ def test_local_plot_1(self, local_pred, filter):
local_pred.return_value = 12.88
filter.return_value = None
self.smart_explainer._case = "regression"
- output = self.smart_explainer.plot.local_plot(index='person_B')
+ output = self.smart_explainer.plot.local_plot(index="person_B")
output_data = output.data
- feature_values = ['Age :
27', 'Education :
Master']
+ feature_values = ["Age :
27", "Education :
Master"]
contributions = [3.6, 1.2]
bars = []
- bars.append(go.Bar(
- x=[contributions[1]],
- y=[feature_values[1]],
- orientation='h'
- ))
- bars.append(go.Bar(
- x=[contributions[0]],
- y=[feature_values[0]],
- orientation='h'
- ))
- expected_output = go.Figure(
- data=bars,
- layout=go.Layout(yaxis=dict(type='category')))
+ bars.append(go.Bar(x=[contributions[1]], y=[feature_values[1]], orientation="h"))
+ bars.append(go.Bar(x=[contributions[0]], y=[feature_values[0]], orientation="h"))
+ expected_output = go.Figure(data=bars, layout=go.Layout(yaxis=dict(type="category")))
for part in list(zip(output_data, expected_output.data)):
assert part[0].x == part[1].x
assert part[0].y == part[1].y
-
- @patch('shapash.explainer.smart_plotter.select_lines')
+ @patch("shapash.explainer.smart_plotter.select_lines")
def test_local_plot_2(self, select_lines):
"""
Unit test local plot 2
@@ -197,18 +162,14 @@ def test_local_plot_2(self, select_lines):
self.smart_explainer._case = "regression"
with self.assertRaises(ValueError):
- condition = ''
+ condition = ""
output = self.smart_explainer.plot.local_plot(query=condition)
expected_output = go.Figure(
- data=go.Bar(
- x=[],
- y=[],
- orientation='h'),
- layout=go.Layout(yaxis=dict(type='category')))
+ data=go.Bar(x=[], y=[], orientation="h"), layout=go.Layout(yaxis=dict(type="category"))
+ )
assert output == expected_output
-
- @patch('shapash.explainer.smart_plotter.select_lines')
+ @patch("shapash.explainer.smart_plotter.select_lines")
def test_local_plot_3(self, select_lines):
"""
Unit test local plot 3
@@ -218,15 +179,17 @@ def test_local_plot_3(self, select_lines):
[description]
"""
select_lines.return_value = []
- condition = ''
+ condition = ""
output = self.smart_explainer.plot.local_plot(query=condition)
expected_output = go.Figure()
assert output.data == expected_output.data
- assert output.layout.annotations[0].text == "Select a valid single sample to display
Local Explanation plot."
+ assert (
+ output.layout.annotations[0].text == "Select a valid single sample to display
Local Explanation plot."
+ )
- @patch('shapash.explainer.smart_explainer.SmartExplainer.filter')
- @patch('shapash.explainer.smart_plotter.select_lines')
- @patch('shapash.explainer.smart_plotter.SmartPlotter.local_pred')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.filter")
+ @patch("shapash.explainer.smart_plotter.select_lines")
+ @patch("shapash.explainer.smart_plotter.SmartPlotter.local_pred")
def test_local_plot_4(self, local_pred, select_lines, filter):
"""
Unit test local plot 4
@@ -237,85 +200,36 @@ def test_local_plot_4(self, local_pred, select_lines, filter):
"""
filter.return_value = None
local_pred.return_value = 0.58
- select_lines.return_value = ['B']
- index = ['A', 'B']
- x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
+ select_lines.return_value = ["B"]
+ index = ["A", "B"]
+ x_init = pd.DataFrame(data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=index)
contrib_sorted1 = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78],
- [3.6, 1.2]]
- ),
- columns=['contrib_0', 'contrib_1'],
- index=index
+ data=np.array([[-3.4, 0.78], [3.6, 1.2]]), columns=["contrib_0", "contrib_1"], index=index
)
contrib_sorted2 = pd.DataFrame(
- data=np.array(
- [[-0.4, 0.78],
- [0.6, 0.2]]
- ),
- columns=['contrib_0', 'contrib_1'],
- index=index
+ data=np.array([[-0.4, 0.78], [0.6, 0.2]]), columns=["contrib_0", "contrib_1"], index=index
)
x_sorted1 = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- [27, 'Master']]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
+ data=np.array([["PhD", 34], [27, "Master"]]), columns=["feature_0", "feature_1"], index=index
)
x_sorted2 = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- [27, 'Master']]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
- )
- var_dict1 = pd.DataFrame(
- data=np.array(
- [[0, 1],
- [1, 0]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
- )
- var_dict2 = pd.DataFrame(
- data=np.array(
- [[0, 1],
- [1, 0]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
+ data=np.array([["PhD", 34], [27, "Master"]]), columns=["feature_0", "feature_1"], index=index
)
+ var_dict1 = pd.DataFrame(data=np.array([[0, 1], [1, 0]]), columns=["feature_0", "feature_1"], index=index)
+ var_dict2 = pd.DataFrame(data=np.array([[0, 1], [1, 0]]), columns=["feature_0", "feature_1"], index=index)
mask1 = pd.DataFrame(
- data=np.array(
- [[True, False],
- [True, True]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index)
+ data=np.array([[True, False], [True, True]]), columns=["feature_0", "feature_1"], index=index
+ )
mask2 = pd.DataFrame(
- data=np.array(
- [[True, True],
- [True, True]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index)
- feature_dictionary = {'X1': 'Education', 'X2': 'Age'}
+ data=np.array([[True, True], [True, True]]), columns=["feature_0", "feature_1"], index=index
+ )
+ feature_dictionary = {"X1": "Education", "X2": "Age"}
smart_explainer_mi = SmartExplainer(model=self.model, features_dict=feature_dictionary)
smart_explainer_mi.data = dict()
smart_explainer_mi.contributions = [contrib_sorted1, contrib_sorted2]
- smart_explainer_mi.data['contrib_sorted'] = [contrib_sorted1, contrib_sorted2]
- smart_explainer_mi.data['x_sorted'] = [x_sorted1, x_sorted2]
- smart_explainer_mi.data['var_dict'] = [var_dict1, var_dict2]
+ smart_explainer_mi.data["contrib_sorted"] = [contrib_sorted1, contrib_sorted2]
+ smart_explainer_mi.data["x_sorted"] = [x_sorted1, x_sorted2]
+ smart_explainer_mi.data["var_dict"] = [var_dict1, var_dict2]
smart_explainer_mi.x_init = x_init
smart_explainer_mi.columns_dict = {i: col for i, col in enumerate(smart_explainer_mi.x_init.columns)}
smart_explainer_mi.mask = [mask1, mask2]
@@ -324,31 +238,21 @@ def test_local_plot_4(self, local_pred, select_lines, filter):
smart_explainer_mi.state = MultiDecorator(SmartState())
condition = "index == 'B'"
output = smart_explainer_mi.plot.local_plot(query=condition)
- feature_values = ['Age :
27', 'Education :
Master']
+ feature_values = ["Age :
27", "Education :
Master"]
contributions = [0.6, 0.2]
bars = []
- bars.append(go.Bar(
- x=[contributions[1]],
- y=[feature_values[1]],
- orientation='h'
- ))
- bars.append(go.Bar(
- x=[contributions[0]],
- y=[feature_values[0]],
- orientation='h'
- ))
- expected_output = go.Figure(
- data=bars,
- layout=go.Layout(yaxis=dict(type='category')))
+ bars.append(go.Bar(x=[contributions[1]], y=[feature_values[1]], orientation="h"))
+ bars.append(go.Bar(x=[contributions[0]], y=[feature_values[0]], orientation="h"))
+ expected_output = go.Figure(data=bars, layout=go.Layout(yaxis=dict(type="category")))
for part in list(zip(output.data, expected_output.data)):
assert part[0].x == part[1].x
assert part[0].y == part[1].y
tit = "Local Explanation - Id: B
Response: 1 - Proba: 0.5800"
assert output.layout.title.text == tit
- @patch('shapash.explainer.smart_explainer.SmartExplainer.filter')
- @patch('shapash.explainer.smart_plotter.select_lines')
- @patch('shapash.explainer.smart_plotter.SmartPlotter.local_pred')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.filter")
+ @patch("shapash.explainer.smart_plotter.select_lines")
+ @patch("shapash.explainer.smart_plotter.SmartPlotter.local_pred")
def test_local_plot_5(self, local_pred, select_lines, filter):
"""
Unit test local plot 5
@@ -358,128 +262,66 @@ def test_local_plot_5(self, local_pred, select_lines, filter):
[description]
"""
local_pred.return_value = 0.58
- select_lines.return_value = ['B']
+ select_lines.return_value = ["B"]
filter.return_value = None
- index = ['A', 'B']
- x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
+ index = ["A", "B"]
+ x_init = pd.DataFrame(data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=index)
contrib_sorted1 = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78],
- [3.6, 1.2]]
- ),
- columns=['contrib_0', 'contrib_1'],
- index=index
+ data=np.array([[-3.4, 0.78], [3.6, 1.2]]), columns=["contrib_0", "contrib_1"], index=index
)
contrib_sorted2 = pd.DataFrame(
- data=np.array(
- [[-0.4, 0.78],
- [0.6, 0.2]]
- ),
- columns=['contrib_0', 'contrib_1'],
- index=index
+ data=np.array([[-0.4, 0.78], [0.6, 0.2]]), columns=["contrib_0", "contrib_1"], index=index
)
x_sorted1 = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- [27, 'Master']]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
+ data=np.array([["PhD", 34], [27, "Master"]]), columns=["feature_0", "feature_1"], index=index
)
x_sorted2 = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- [27, 'Master']]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
- )
- var_dict1 = pd.DataFrame(
- data=np.array(
- [[0, 1],
- [1, 0]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
- )
- var_dict2 = pd.DataFrame(
- data=np.array(
- [[0, 1],
- [1, 0]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
+ data=np.array([["PhD", 34], [27, "Master"]]), columns=["feature_0", "feature_1"], index=index
)
+ var_dict1 = pd.DataFrame(data=np.array([[0, 1], [1, 0]]), columns=["feature_0", "feature_1"], index=index)
+ var_dict2 = pd.DataFrame(data=np.array([[0, 1], [1, 0]]), columns=["feature_0", "feature_1"], index=index)
mask1 = pd.DataFrame(
- data=np.array(
- [[True, False],
- [True, True]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index)
+ data=np.array([[True, False], [True, True]]), columns=["feature_0", "feature_1"], index=index
+ )
mask2 = pd.DataFrame(
- data=np.array(
- [[False, True],
- [False, True]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index)
+ data=np.array([[False, True], [False, True]]), columns=["feature_0", "feature_1"], index=index
+ )
mask_contrib1 = pd.DataFrame(
- data=np.array(
- [[0.0, 0.78],
- [0.0, 1.20]]
- ),
- columns=['masked_neg', 'masked_pos'],
- index=index)
+ data=np.array([[0.0, 0.78], [0.0, 1.20]]), columns=["masked_neg", "masked_pos"], index=index
+ )
mask_contrib2 = pd.DataFrame(
- data=np.array(
- [[0.0, 0.78],
- [0.0, 0.20]]
- ),
- columns=['masked_neg', 'masked_pos'],
- index=index)
-
- feature_dictionary = {'X1': 'Education', 'X2': 'Age'}
+ data=np.array([[0.0, 0.78], [0.0, 0.20]]), columns=["masked_neg", "masked_pos"], index=index
+ )
+
+ feature_dictionary = {"X1": "Education", "X2": "Age"}
smart_explainer_mi = SmartExplainer(model=self.model, features_dict=feature_dictionary)
smart_explainer_mi.data = dict()
smart_explainer_mi.contributions = [contrib_sorted1, contrib_sorted2]
- smart_explainer_mi.data['contrib_sorted'] = [contrib_sorted1, contrib_sorted2]
- smart_explainer_mi.data['x_sorted'] = [x_sorted1, x_sorted2]
- smart_explainer_mi.data['var_dict'] = [var_dict1, var_dict2]
+ smart_explainer_mi.data["contrib_sorted"] = [contrib_sorted1, contrib_sorted2]
+ smart_explainer_mi.data["x_sorted"] = [x_sorted1, x_sorted2]
+ smart_explainer_mi.data["var_dict"] = [var_dict1, var_dict2]
smart_explainer_mi.x_init = x_init
smart_explainer_mi.columns_dict = {i: col for i, col in enumerate(smart_explainer_mi.x_init.columns)}
smart_explainer_mi.mask = [mask1, mask2]
smart_explainer_mi.masked_contributions = [mask_contrib1, mask_contrib2]
- smart_explainer_mi.mask_params = {'features_to_hide': None,
- 'threshold': None,
- 'positive': None,
- 'max_contrib': 1}
+ smart_explainer_mi.mask_params = {
+ "features_to_hide": None,
+ "threshold": None,
+ "positive": None,
+ "max_contrib": 1,
+ }
smart_explainer_mi._case = "classification"
smart_explainer_mi._classes = [0, 1]
smart_explainer_mi.state = MultiDecorator(SmartState())
condition = "index == 'B'"
output = smart_explainer_mi.plot.local_plot(query=condition)
- feature_values = ['Hidden Positive Contributions',
- 'Education :
Master']
+ feature_values = ["Hidden Positive Contributions", "Education :
Master"]
contributions = [0.2, 0.2]
bars = []
for elem in list(zip(feature_values, contributions)):
- bars.append(go.Bar(
- x=[elem[1]],
- y=[elem[0]],
- orientation='h'
- ))
- expected_output = go.Figure(
- data=bars,
- layout=go.Layout(yaxis=dict(type='category')))
+ bars.append(go.Bar(x=[elem[1]], y=[elem[0]], orientation="h"))
+ expected_output = go.Figure(data=bars, layout=go.Layout(yaxis=dict(type="category")))
assert len(expected_output.data) == len(output.data)
for part in list(zip(output.data, expected_output.data)):
@@ -491,18 +333,20 @@ def test_local_plot_5(self, local_pred, select_lines, filter):
output2 = smart_explainer_mi.plot.local_plot(query=condition, show_masked=False)
assert len(output2.data) == 1
assert expected_output.data[-1].x == output2.data[0].x
- smart_explainer_mi.mask_params = {'features_to_hide': None,
- 'threshold': None,
- 'positive': True,
- 'max_contrib': 1}
+ smart_explainer_mi.mask_params = {
+ "features_to_hide": None,
+ "threshold": None,
+ "positive": True,
+ "max_contrib": 1,
+ }
output3 = smart_explainer_mi.plot.local_plot(row_num=1)
assert len(output3.data) == 2
assert expected_output.data[-1].x == output3.data[-1].x
assert expected_output.data[-2].x == output3.data[-2].x
- @patch('shapash.explainer.smart_explainer.SmartExplainer.filter')
- @patch('shapash.explainer.smart_plotter.select_lines')
- @patch('shapash.explainer.smart_plotter.SmartPlotter.local_pred')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.filter")
+ @patch("shapash.explainer.smart_plotter.select_lines")
+ @patch("shapash.explainer.smart_plotter.SmartPlotter.local_pred")
def test_local_plot_groups_features(self, local_pred, select_lines, filter):
"""
Unit test local plot 6 for groups of features
@@ -511,132 +355,141 @@ def test_local_plot_groups_features(self, local_pred, select_lines, filter):
select_lines.return_value = [10]
filter.return_value = None
index = [3, 10]
- x_init = pd.DataFrame({
- 'X1': {3: 1, 10: 5},
- 'X2': {3: 42, 10: 4},
- 'X3': {3: 9, 10: 1},
- 'X4': {3: 2008, 10: 2008}
- })
- x_init_groups = pd.DataFrame({
- 'X2': {3: 42, 10: 4},
- 'X4': {3: 2008, 10: 2008},
- 'group1': {3: -2361.80078125, 10: 2361.80078125}
- })
- contrib_sorted1 = pd.DataFrame({
- 'contribution_0': {3: 0.15, 10: 0.22},
- 'contribution_1': {3: -0.13, 10: -0.12},
- 'contribution_2': {3: -0.03, 10: 0.02},
- 'contribution_3': {3: 0.0, 10: 0.01}
- })
- contrib_sorted2 = pd.DataFrame({
- 'contribution_0': {3: -0.15, 10: -0.22},
- 'contribution_1': {3: 0.13, 10: 0.12},
- 'contribution_2': {3: 0.03, 10: -0.02},
- 'contribution_3': {3: -0.0, 10: -0.01}
- })
- x_sorted1 = pd.DataFrame({
- 'feature_0': {3: 9, 10: 5},
- 'feature_1': {3: 1, 10: 1},
- 'feature_2': {3: 2008, 10: 2008},
- 'feature_3': {3: 42, 10: 4}
- })
- x_sorted2 = pd.DataFrame({
- 'feature_0': {3: 9, 10: 5},
- 'feature_1': {3: 1, 10: 1},
- 'feature_2': {3: 2008, 10: 2008},
- 'feature_3': {3: 42, 10: 4}
- })
- var_dict1 = pd.DataFrame({
- 'feature_0': {3: 2, 10: 0},
- 'feature_1': {3: 0, 10: 2},
- 'feature_2': {3: 3, 10: 3},
- 'feature_3': {3: 1, 10: 1}
- })
- var_dict2 = pd.DataFrame({
- 'feature_0': {3: 2, 10: 0},
- 'feature_1': {3: 0, 10: 2},
- 'feature_2': {3: 3, 10: 3},
- 'feature_3': {3: 1, 10: 1}
- })
-
- contrib_groups_sorted1 = pd.DataFrame({
- 'contribution_0': {3: 0.03, 10: 0.09},
- 'contribution_1': {3: -0.03, 10: 0.02},
- 'contribution_2': {3: 0.0, 10: 0.01}
- })
- contrib_groups_sorted2 = pd.DataFrame({
- 'contribution_0': {3: -0.03, 10: -0.09},
- 'contribution_1': {3: 0.03, 10: -0.02},
- 'contribution_2': {3: -0.0, 10: -0.01}
- })
- x_groups_sorted1 = pd.DataFrame({
- 'feature_0': {3: -2361.8, 10: 2361.8},
- 'feature_1': {3: 2008.0, 10: 2008.0},
- 'feature_2': {3: 42.0, 10: 4.0}
- })
- x_groups_sorted2 = pd.DataFrame({
- 'feature_0': {3: -2361.8, 10: 2361.8},
- 'feature_1': {3: 2008.0, 10: 2008.0},
- 'feature_2': {3: 42.0, 10: 4.0}
- })
- groups_var_dict1 = pd.DataFrame({
- 'feature_0': {3: 2, 10: 2},
- 'feature_1': {3: 1, 10: 1},
- 'feature_2': {3: 0, 10: 0}
- })
- groups_var_dict2 = pd.DataFrame({
- 'feature_0': {3: 2, 10: 2},
- 'feature_1': {3: 1, 10: 1},
- 'feature_2': {3: 0, 10: 0}
- })
+ x_init = pd.DataFrame(
+ {"X1": {3: 1, 10: 5}, "X2": {3: 42, 10: 4}, "X3": {3: 9, 10: 1}, "X4": {3: 2008, 10: 2008}}
+ )
+ x_init_groups = pd.DataFrame(
+ {"X2": {3: 42, 10: 4}, "X4": {3: 2008, 10: 2008}, "group1": {3: -2361.80078125, 10: 2361.80078125}}
+ )
+ contrib_sorted1 = pd.DataFrame(
+ {
+ "contribution_0": {3: 0.15, 10: 0.22},
+ "contribution_1": {3: -0.13, 10: -0.12},
+ "contribution_2": {3: -0.03, 10: 0.02},
+ "contribution_3": {3: 0.0, 10: 0.01},
+ }
+ )
+ contrib_sorted2 = pd.DataFrame(
+ {
+ "contribution_0": {3: -0.15, 10: -0.22},
+ "contribution_1": {3: 0.13, 10: 0.12},
+ "contribution_2": {3: 0.03, 10: -0.02},
+ "contribution_3": {3: -0.0, 10: -0.01},
+ }
+ )
+ x_sorted1 = pd.DataFrame(
+ {
+ "feature_0": {3: 9, 10: 5},
+ "feature_1": {3: 1, 10: 1},
+ "feature_2": {3: 2008, 10: 2008},
+ "feature_3": {3: 42, 10: 4},
+ }
+ )
+ x_sorted2 = pd.DataFrame(
+ {
+ "feature_0": {3: 9, 10: 5},
+ "feature_1": {3: 1, 10: 1},
+ "feature_2": {3: 2008, 10: 2008},
+ "feature_3": {3: 42, 10: 4},
+ }
+ )
+ var_dict1 = pd.DataFrame(
+ {
+ "feature_0": {3: 2, 10: 0},
+ "feature_1": {3: 0, 10: 2},
+ "feature_2": {3: 3, 10: 3},
+ "feature_3": {3: 1, 10: 1},
+ }
+ )
+ var_dict2 = pd.DataFrame(
+ {
+ "feature_0": {3: 2, 10: 0},
+ "feature_1": {3: 0, 10: 2},
+ "feature_2": {3: 3, 10: 3},
+ "feature_3": {3: 1, 10: 1},
+ }
+ )
+
+ contrib_groups_sorted1 = pd.DataFrame(
+ {
+ "contribution_0": {3: 0.03, 10: 0.09},
+ "contribution_1": {3: -0.03, 10: 0.02},
+ "contribution_2": {3: 0.0, 10: 0.01},
+ }
+ )
+ contrib_groups_sorted2 = pd.DataFrame(
+ {
+ "contribution_0": {3: -0.03, 10: -0.09},
+ "contribution_1": {3: 0.03, 10: -0.02},
+ "contribution_2": {3: -0.0, 10: -0.01},
+ }
+ )
+ x_groups_sorted1 = pd.DataFrame(
+ {
+ "feature_0": {3: -2361.8, 10: 2361.8},
+ "feature_1": {3: 2008.0, 10: 2008.0},
+ "feature_2": {3: 42.0, 10: 4.0},
+ }
+ )
+ x_groups_sorted2 = pd.DataFrame(
+ {
+ "feature_0": {3: -2361.8, 10: 2361.8},
+ "feature_1": {3: 2008.0, 10: 2008.0},
+ "feature_2": {3: 42.0, 10: 4.0},
+ }
+ )
+ groups_var_dict1 = pd.DataFrame(
+ {"feature_0": {3: 2, 10: 2}, "feature_1": {3: 1, 10: 1}, "feature_2": {3: 0, 10: 0}}
+ )
+ groups_var_dict2 = pd.DataFrame(
+ {"feature_0": {3: 2, 10: 2}, "feature_1": {3: 1, 10: 1}, "feature_2": {3: 0, 10: 0}}
+ )
mask1 = pd.DataFrame(
- data=np.array(
- [[True, True, False],
- [True, True, False]]
- ),
- columns=['contrib_0', 'contrib_1', 'contrib_2'],
- index=index)
+ data=np.array([[True, True, False], [True, True, False]]),
+ columns=["contrib_0", "contrib_1", "contrib_2"],
+ index=index,
+ )
mask2 = pd.DataFrame(
- data=np.array(
- [[True, True, False],
- [True, True, False]]
- ),
- columns=['contrib_0', 'contrib_1', 'contrib_2'],
- index=index)
+ data=np.array([[True, True, False], [True, True, False]]),
+ columns=["contrib_0", "contrib_1", "contrib_2"],
+ index=index,
+ )
mask_contrib1 = pd.DataFrame(
- data=np.array(
- [[-0.002, 0.0],
- [-0.024, 0.0]]
- ),
- columns=['masked_neg', 'masked_pos'],
- index=index)
+ data=np.array([[-0.002, 0.0], [-0.024, 0.0]]), columns=["masked_neg", "masked_pos"], index=index
+ )
mask_contrib2 = pd.DataFrame(
- data=np.array(
- [[0.0, 0.002],
- [0.0, 0.024]]
- ),
- columns=['masked_neg', 'masked_pos'],
- index=index)
-
- feature_dictionary = {'X1': 'X1_label', 'X2': 'X2_label', 'X3': 'X3_label', 'X4': 'X4_label',
- 'group1': 'group1_label'}
+ data=np.array([[0.0, 0.002], [0.0, 0.024]]), columns=["masked_neg", "masked_pos"], index=index
+ )
+
+ feature_dictionary = {
+ "X1": "X1_label",
+ "X2": "X2_label",
+ "X3": "X3_label",
+ "X4": "X4_label",
+ "group1": "group1_label",
+ }
smart_explainer_mi = SmartExplainer(model=self.model, features_dict=feature_dictionary)
- smart_explainer_mi.features_groups = {'group1': ['X1', 'X3']}
- smart_explainer_mi.inv_features_dict = {'X1_label': 'X1', 'X2_label': 'X2', 'X3_label': 'X3',
- 'X4_label': 'X4', 'group1_label': 'group1'}
+ smart_explainer_mi.features_groups = {"group1": ["X1", "X3"]}
+ smart_explainer_mi.inv_features_dict = {
+ "X1_label": "X1",
+ "X2_label": "X2",
+ "X3_label": "X3",
+ "X4_label": "X4",
+ "group1_label": "group1",
+ }
smart_explainer_mi.data = dict()
smart_explainer_mi.contributions = [contrib_sorted1, contrib_sorted2]
- smart_explainer_mi.data['contrib_sorted'] = [contrib_sorted1, contrib_sorted2]
- smart_explainer_mi.data['x_sorted'] = [x_sorted1, x_sorted2]
- smart_explainer_mi.data['var_dict'] = [var_dict1, var_dict2]
+ smart_explainer_mi.data["contrib_sorted"] = [contrib_sorted1, contrib_sorted2]
+ smart_explainer_mi.data["x_sorted"] = [x_sorted1, x_sorted2]
+ smart_explainer_mi.data["var_dict"] = [var_dict1, var_dict2]
smart_explainer_mi.data_groups = dict()
- smart_explainer_mi.data_groups['contrib_sorted'] = [contrib_groups_sorted1, contrib_groups_sorted2]
- smart_explainer_mi.data_groups['x_sorted'] = [x_groups_sorted1, x_groups_sorted2]
- smart_explainer_mi.data_groups['var_dict'] = [groups_var_dict1, groups_var_dict2]
+ smart_explainer_mi.data_groups["contrib_sorted"] = [contrib_groups_sorted1, contrib_groups_sorted2]
+ smart_explainer_mi.data_groups["x_sorted"] = [x_groups_sorted1, x_groups_sorted2]
+ smart_explainer_mi.data_groups["var_dict"] = [groups_var_dict1, groups_var_dict2]
smart_explainer_mi.x_init = x_init
smart_explainer_mi.x_init_groups = x_init_groups
@@ -644,10 +497,12 @@ def test_local_plot_groups_features(self, local_pred, select_lines, filter):
smart_explainer_mi.columns_dict = {i: col for i, col in enumerate(smart_explainer_mi.x_init.columns)}
smart_explainer_mi.mask = [mask1, mask2]
smart_explainer_mi.masked_contributions = [mask_contrib1, mask_contrib2]
- smart_explainer_mi.mask_params = {'features_to_hide': None,
- 'threshold': None,
- 'positive': None,
- 'max_contrib': 2}
+ smart_explainer_mi.mask_params = {
+ "features_to_hide": None,
+ "threshold": None,
+ "positive": None,
+ "max_contrib": 2,
+ }
smart_explainer_mi._case = "classification"
smart_explainer_mi._classes = [0, 1]
@@ -657,9 +512,9 @@ def test_local_plot_groups_features(self, local_pred, select_lines, filter):
assert len(output_fig.data) == 3
- @patch('shapash.explainer.smart_explainer.SmartExplainer.filter')
- @patch('shapash.explainer.smart_plotter.select_lines')
- @patch('shapash.explainer.smart_plotter.SmartPlotter.local_pred')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.filter")
+ @patch("shapash.explainer.smart_plotter.select_lines")
+ @patch("shapash.explainer.smart_plotter.SmartPlotter.local_pred")
def test_local_plot_multi_index(self, local_pred, select_lines, filter):
"""
Unit test local plot multi index
@@ -669,65 +524,38 @@ def test_local_plot_multi_index(self, local_pred, select_lines, filter):
[description]
"""
local_pred.return_value = 12.78
- select_lines.return_value = [('C', 'A')]
+ select_lines.return_value = [("C", "A")]
filter.return_value = None
- index = pd.MultiIndex.from_tuples(
- [('A', 'A'), ('C', 'A')],
- names=('col1', 'col2')
- )
+ index = pd.MultiIndex.from_tuples([("A", "A"), ("C", "A")], names=("col1", "col2"))
x_init_multi_index = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]
- ),
- columns=['X1', 'X2'],
- index=index
+ data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=index
)
contrib_sorted_multi_index = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78],
- [3.6, 1.2]]
- ),
- columns=['contrib_0', 'contrib_1'],
- index=index
+ data=np.array([[-3.4, 0.78], [3.6, 1.2]]), columns=["contrib_0", "contrib_1"], index=index
)
x_sorted_multi_index = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- [27, 'Master']]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
+ data=np.array([["PhD", 34], [27, "Master"]]), columns=["feature_0", "feature_1"], index=index
)
var_dict_multi_index = pd.DataFrame(
- data=np.array(
- [[0, 1],
- [1, 0]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index
+ data=np.array([[0, 1], [1, 0]]), columns=["feature_0", "feature_1"], index=index
)
mask_multi_index = pd.DataFrame(
- data=np.array(
- [[True, False],
- [True, True]]
- ),
- columns=['feature_0', 'feature_1'],
- index=index)
+ data=np.array([[True, False], [True, True]]), columns=["feature_0", "feature_1"], index=index
+ )
- feature_dictionary = {'X1': 'Education', 'X2': 'Age'}
+ feature_dictionary = {"X1": "Education", "X2": "Age"}
smart_explainer_mi = SmartExplainer(model=self.model, features_dict=feature_dictionary)
smart_explainer_mi.data = dict()
smart_explainer_mi.contributions = contrib_sorted_multi_index
- smart_explainer_mi.data['contrib_sorted'] = contrib_sorted_multi_index
- smart_explainer_mi.data['x_sorted'] = x_sorted_multi_index
- smart_explainer_mi.data['var_dict'] = var_dict_multi_index
+ smart_explainer_mi.data["contrib_sorted"] = contrib_sorted_multi_index
+ smart_explainer_mi.data["x_sorted"] = x_sorted_multi_index
+ smart_explainer_mi.data["var_dict"] = var_dict_multi_index
smart_explainer_mi.x_init = x_init_multi_index
smart_explainer_mi.columns_dict = {i: col for i, col in enumerate(smart_explainer_mi.x_init.columns)}
smart_explainer_mi.mask = mask_multi_index
@@ -739,23 +567,13 @@ def test_local_plot_multi_index(self, local_pred, select_lines, filter):
output = smart_explainer_mi.plot.local_plot(query=condition)
- feature_values = ['Age :
27', 'Education :
Master']
+ feature_values = ["Age :
27", "Education :
Master"]
contributions = [3.6, 1.2]
bars = []
- bars.append(go.Bar(
- x=[contributions[1]],
- y=[feature_values[1]],
- orientation='h'
- ))
- bars.append(go.Bar(
- x=[contributions[0]],
- y=[feature_values[0]],
- orientation='h'
- ))
- expected_output = go.Figure(
- data=bars,
- layout=go.Layout(yaxis=dict(type='category')))
+ bars.append(go.Bar(x=[contributions[1]], y=[feature_values[1]], orientation="h"))
+ bars.append(go.Bar(x=[contributions[0]], y=[feature_values[0]], orientation="h"))
+ expected_output = go.Figure(data=bars, layout=go.Layout(yaxis=dict(type="category")))
for part in list(zip(output.data, expected_output.data)):
assert part[0].x == part[1].x
assert part[0].y == part[1].y
@@ -764,14 +582,9 @@ def test_get_selection(self):
"""
Unit test get selection
"""
- line = ['person_A']
- output = self.smart_explainer.plot.get_selection(
- line,
- self.var_dict,
- self.x_sorted,
- self.contrib_sorted
- )
- expected_output = np.array([0, 1]), np.array(['PhD', 34]), np.array([-3.4, 0.78])
+ line = ["person_A"]
+ output = self.smart_explainer.plot.get_selection(line, self.var_dict, self.x_sorted, self.contrib_sorted)
+ expected_output = np.array([0, 1]), np.array(["PhD", 34]), np.array([-3.4, 0.78])
assert len(output) == 3
assert np.array_equal(output[0], expected_output[0])
assert np.array_equal(output[1], expected_output[1])
@@ -781,17 +594,12 @@ def test_apply_mask_one_line(self):
"""
Unit test apply mask one line
"""
- line = ['person_A']
+ line = ["person_A"]
var_dict = np.array([0, 1])
- x_sorted = np.array(['PhD', 34])
+ x_sorted = np.array(["PhD", 34])
contrib_sorted = np.array([-3.4, 0.78])
- output = self.smart_explainer.plot.apply_mask_one_line(
- line,
- var_dict,
- x_sorted,
- contrib_sorted
- )
- expected_output = np.array([0]), np.array(['PhD']), np.array([-3.4])
+ output = self.smart_explainer.plot.apply_mask_one_line(line, var_dict, x_sorted, contrib_sorted)
+ expected_output = np.array([0]), np.array(["PhD"]), np.array([-3.4])
assert len(output) == 3
assert np.array_equal(output[0], expected_output[0])
assert np.array_equal(output[1], expected_output[1])
@@ -801,15 +609,15 @@ def test_check_masked_contributions_1(self):
"""
Unit test check masked contributions 1
"""
- line = ['person_A']
- var_dict = ['X1', 'X2']
- x_val = ['PhD', 34]
+ line = ["person_A"]
+ var_dict = ["X1", "X2"]
+ x_val = ["PhD", 34]
contrib = [-3.4, 0.78]
var_dict, x_val, contrib = self.smart_explainer.plot.check_masked_contributions(line, var_dict, x_val, contrib)
- expected_var_dict = ['X1', 'X2']
- expected_x_val = ['PhD', 34]
+ expected_var_dict = ["X1", "X2"]
+ expected_x_val = ["PhD", 34]
expected_contrib = [-3.4, 0.78]
- self.assertListEqual(var_dict,expected_var_dict)
+ self.assertListEqual(var_dict, expected_var_dict)
self.assertListEqual(x_val, expected_x_val)
self.assertListEqual(contrib, expected_contrib)
@@ -817,20 +625,18 @@ def test_check_masked_contributions_2(self):
"""
Unit test check masked contributions 2
"""
- line = ['person_A']
- var_dict = ['X1', 'X2']
- x_val = ['PhD', 34]
+ line = ["person_A"]
+ var_dict = ["X1", "X2"]
+ x_val = ["PhD", 34]
contrib = [-3.4, 0.78]
self.smart_explainer.masked_contributions = pd.DataFrame(
- data=[[0.0, 2.5], [0.0, 1.6]],
- columns=['masked_neg', 'masked_pos'],
- index=['person_A', 'person_B']
+ data=[[0.0, 2.5], [0.0, 1.6]], columns=["masked_neg", "masked_pos"], index=["person_A", "person_B"]
)
var_dict, x_val, contrib = self.smart_explainer.plot.check_masked_contributions(line, var_dict, x_val, contrib)
- expected_var_dict = ['X1', 'X2', 'Hidden Positive Contributions']
- expected_x_val = ['PhD', 34, '' ]
+ expected_var_dict = ["X1", "X2", "Hidden Positive Contributions"]
+ expected_x_val = ["PhD", 34, ""]
expected_contrib = [-3.4, 0.78, 2.5]
- self.assertListEqual(var_dict,expected_var_dict)
+ self.assertListEqual(var_dict, expected_var_dict)
self.assertListEqual(x_val, expected_x_val)
self.assertListEqual(contrib, expected_contrib)
@@ -838,21 +644,17 @@ def test_plot_bar_chart_1(self):
"""
Unit test plot bar chart 1
"""
- var_dict = ['X1', 'X2']
- x_val = ['PhD', 34]
+ var_dict = ["X1", "X2"]
+ x_val = ["PhD", 34]
contributions = [-3.4, 0.78]
bars = []
for num, elem in enumerate(var_dict):
- bars.append(go.Bar(
- x=[contributions[num]],
- y=['{} :
{}'.format(elem, x_val[num])],
- orientation='h'
- ))
- expected_output_fig = go.Figure(
- data=bars,
- layout=go.Layout(yaxis=dict(type='category')))
+ bars.append(
+ go.Bar(x=[contributions[num]], y=["{} :
{}".format(elem, x_val[num])], orientation="h")
+ )
+ expected_output_fig = go.Figure(data=bars, layout=go.Layout(yaxis=dict(type="category")))
self.smart_explainer._case = "regression"
- fig_output = self.smart_explainer.plot.plot_bar_chart('ind', var_dict, x_val, contributions)
+ fig_output = self.smart_explainer.plot.plot_bar_chart("ind", var_dict, x_val, contributions)
for part in list(zip(fig_output.data, expected_output_fig.data)):
assert part[0].x == part[1].x
assert part[0].y == part[1].y
@@ -861,27 +663,21 @@ def test_plot_bar_chart_2(self):
"""
Unit test plot bar chart 2
"""
- var_dict = ['X1', 'X2', 'Hidden Positive Contributions']
- x_val = ['PhD', 34, '']
+ var_dict = ["X1", "X2", "Hidden Positive Contributions"]
+ x_val = ["PhD", 34, ""]
order = [3, 1, 2]
contributions = [-3.4, 0.78, 2.5]
- ylabel = ['X1 :
PhD', 'X2 :
34', 'Hidden Positive Contributions']
+ ylabel = ["X1 :
PhD", "X2 :
34", "Hidden Positive Contributions"]
self.smart_explainer.masked_contributions = pd.DataFrame()
bars = []
comblist = list(zip(order, contributions, ylabel))
comblist.sort(reverse=True)
for elem in comblist:
- bars.append(go.Bar(
- x=[elem[1]],
- y=[elem[2]],
- orientation='h'
- ))
- expected_output_fig = go.Figure(
- data=bars,
- layout=go.Layout(yaxis=dict(type='category')))
+ bars.append(go.Bar(x=[elem[1]], y=[elem[2]], orientation="h"))
+ expected_output_fig = go.Figure(data=bars, layout=go.Layout(yaxis=dict(type="category")))
self.smart_explainer._case = "regression"
- fig_output = self.smart_explainer.plot.plot_bar_chart('ind', var_dict, x_val, contributions)
+ fig_output = self.smart_explainer.plot.plot_bar_chart("ind", var_dict, x_val, contributions)
for part in list(zip(fig_output.data, expected_output_fig.data)):
assert part[0].x == part[1].x
assert part[0].y == part[1].y
@@ -890,12 +686,14 @@ def test_contribution_plot_1(self):
"""
Classification
"""
- col = 'X1'
+ col = "X1"
output = self.smart_explainer.plot.contribution_plot(col, violin_maxf=0, proba=False)
- expected_output = go.Scatter(x=self.smart_explainer.x_init[col],
- y=self.smart_explainer.contributions[-1][col],
- mode='markers',
- hovertext=[f"Id: {x}
" for x in self.smart_explainer.x_init.index])
+ expected_output = go.Scatter(
+ x=self.smart_explainer.x_init[col],
+ y=self.smart_explainer.contributions[-1][col],
+ mode="markers",
+ hovertext=[f"Id: {x}
" for x in self.smart_explainer.x_init.index],
+ )
assert np.array_equal(output.data[0].x, expected_output.x)
assert np.array_equal(output.data[0].y, expected_output.y)
assert len(np.unique(output.data[0].marker.color)) == 1
@@ -905,16 +703,18 @@ def test_contribution_plot_2(self):
"""
Regression
"""
- col = 'X2'
+ col = "X2"
xpl = self.smart_explainer
xpl.contributions = self.contrib1
xpl._case = "regression"
xpl.state = SmartState()
output = xpl.plot.contribution_plot(col, violin_maxf=0)
- expected_output = go.Scatter(x=xpl.x_init[col],
- y=xpl.contributions[col],
- mode='markers',
- hovertext=[f"Id: {x}" for x in xpl.x_init.index])
+ expected_output = go.Scatter(
+ x=xpl.x_init[col],
+ y=xpl.contributions[col],
+ mode="markers",
+ hovertext=[f"Id: {x}" for x in xpl.x_init.index],
+ )
assert np.array_equal(output.data[0].x, expected_output.x)
assert np.array_equal(output.data[0].y, expected_output.y)
@@ -926,15 +726,17 @@ def test_contribution_plot_3(self):
"""
Color Plot classification
"""
- col = 'X2'
+ col = "X2"
xpl = self.smart_explainer
- xpl.y_pred = pd.DataFrame([0, 1], columns=['pred'], index=xpl.x_init.index)
+ xpl.y_pred = pd.DataFrame([0, 1], columns=["pred"], index=xpl.x_init.index)
xpl._classes = [0, 1]
output = xpl.plot.contribution_plot(col, violin_maxf=0, proba=False)
- expected_output = go.Scatter(x=xpl.x_init[col],
- y=xpl.contributions[-1][col],
- mode='markers',
- hovertext=[f"Id: {x}
Predict: {y}" for x, y in zip(xpl.x_init.index, xpl.y_pred.iloc[:, 0].tolist())])
+ expected_output = go.Scatter(
+ x=xpl.x_init[col],
+ y=xpl.contributions[-1][col],
+ mode="markers",
+ hovertext=[f"Id: {x}
Predict: {y}" for x, y in zip(xpl.x_init.index, xpl.y_pred.iloc[:, 0].tolist())],
+ )
assert np.array_equal(output.data[0].x, expected_output.x)
assert np.array_equal(output.data[0].y, expected_output.y)
@@ -946,17 +748,21 @@ def test_contribution_plot_4(self):
"""
Regression Color Plot
"""
- col = 'X2'
+ col = "X2"
xpl = self.smart_explainer
xpl.contributions = self.contrib1
xpl._case = "regression"
xpl.state = SmartState()
- xpl.y_pred = pd.DataFrame([0.46989877093, 12.749302948], columns=['pred'], index=xpl.x_init.index)
+ xpl.y_pred = pd.DataFrame([0.46989877093, 12.749302948], columns=["pred"], index=xpl.x_init.index)
output = xpl.plot.contribution_plot(col, violin_maxf=0)
- expected_output = go.Scatter(x=xpl.x_init[col],
- y=xpl.contributions[col],
- mode='markers',
- hovertext=[f"Id: {x}
Predict: {round(y,3)}" for x, y in zip(xpl.x_init.index, xpl.y_pred.iloc[:, 0].tolist())])
+ expected_output = go.Scatter(
+ x=xpl.x_init[col],
+ y=xpl.contributions[col],
+ mode="markers",
+ hovertext=[
+ f"Id: {x}
Predict: {round(y,3)}" for x, y in zip(xpl.x_init.index, xpl.y_pred.iloc[:, 0].tolist())
+ ],
+ )
assert np.array_equal(output.data[0].x, expected_output.x)
assert np.array_equal(output.data[0].y, expected_output.y)
@@ -968,19 +774,21 @@ def test_contribution_plot_5(self):
"""
Regression Color Plot with pred
"""
- col = 'X2'
+ col = "X2"
xpl = self.smart_explainer
- xpl.contributions = pd.concat([self.contrib1]*10, ignore_index=True)
+ xpl.contributions = pd.concat([self.contrib1] * 10, ignore_index=True)
xpl._case = "regression"
xpl.state = SmartState()
- xpl.x_init = pd.concat([xpl.x_init]*10, ignore_index=True)
+ xpl.x_init = pd.concat([xpl.x_init] * 10, ignore_index=True)
xpl.postprocessing_modifications = False
- xpl.y_pred = pd.concat([pd.DataFrame([0.46989877093, 12.749302948])]*10, ignore_index=True)
+ xpl.y_pred = pd.concat([pd.DataFrame([0.46989877093, 12.749302948])] * 10, ignore_index=True)
output = xpl.plot.contribution_plot(col)
- np_hv = np.array([f"Id: {x}
Predict: {round(y,2)}" for x, y in zip(xpl.x_init.index, xpl.y_pred.iloc[:, 0].tolist())])
+ np_hv = np.array(
+ [f"Id: {x}
Predict: {round(y,2)}" for x, y in zip(xpl.x_init.index, xpl.y_pred.iloc[:, 0].tolist())]
+ )
assert len(output.data) == 3
- assert output.data[0].type == 'violin'
- assert output.data[-1].type == 'scatter'
+ assert output.data[0].type == "violin"
+ assert output.data[-1].type == "scatter"
assert len(np.unique(output.data[-1].marker.color)) >= 2
assert np.array_equal(output.data[-1].hovertext, np_hv)
assert output.layout.xaxis.title.text == xpl.features_dict[col]
@@ -989,12 +797,12 @@ def test_contribution_plot_6(self):
"""
Regression without pred
"""
- col = 'X2'
+ col = "X2"
xpl = self.smart_explainer
- xpl.contributions = pd.concat([self.contrib1]*10, ignore_index=True)
+ xpl.contributions = pd.concat([self.contrib1] * 10, ignore_index=True)
xpl._case = "regression"
xpl.state = SmartState()
- xpl.x_init = pd.concat([xpl.x_init]*10, ignore_index=True)
+ xpl.x_init = pd.concat([xpl.x_init] * 10, ignore_index=True)
xpl.postprocessing_modifications = False
output = xpl.plot.contribution_plot(col)
np_hv = [f"Id: {x}" for x in xpl.x_init.index]
@@ -1005,7 +813,7 @@ def test_contribution_plot_6(self):
annot_list.sort()
assert len(output.data) == 2
for elem in output.data:
- assert elem.type == 'violin'
+ assert elem.type == "violin"
assert output.data[-1].marker.color == output.data[-2].marker.color
self.assertListEqual(annot_list, np_hv)
assert output.layout.xaxis.title.text == xpl.features_dict[col]
@@ -1014,7 +822,7 @@ def test_contribution_plot_7(self):
"""
Classification without pred
"""
- col = 'X1'
+ col = "X1"
xpl = self.smart_explainer
xpl.contributions[0] = pd.concat([xpl.contributions[0]] * 10, ignore_index=True)
xpl.contributions[1] = pd.concat([xpl.contributions[1]] * 10, ignore_index=True)
@@ -1029,7 +837,7 @@ def test_contribution_plot_7(self):
annot_list.sort()
assert len(output.data) == 2
for elem in output.data:
- assert elem.type == 'violin'
+ assert elem.type == "violin"
assert output.data[-1].marker.color == output.data[-2].marker.color
self.assertListEqual(annot_list, np_hv)
assert output.layout.xaxis.title.text == xpl.features_dict[col]
@@ -1038,7 +846,7 @@ def test_contribution_plot_8(self):
"""
Classification with pred
"""
- col = 'X1'
+ col = "X1"
xpl = self.smart_explainer
xpl.x_init = pd.concat([xpl.x_init] * 10, ignore_index=True)
xpl.x_init.index = [i for i in range(xpl.x_init.shape[0])]
@@ -1047,8 +855,9 @@ def test_contribution_plot_8(self):
xpl.contributions[1] = pd.concat([xpl.contributions[1]] * 10, ignore_index=True)
xpl.contributions[0].index = xpl.x_init.index
xpl.contributions[1].index = xpl.x_init.index
- xpl.y_pred = pd.DataFrame([3, 1, 1, 3, 3, 3, 1, 3, 1, 1, 1, 3, 3, 1, 1, 1, 1, 3, 3, 3],
- columns=['pred'], index=xpl.x_init.index)
+ xpl.y_pred = pd.DataFrame(
+ [3, 1, 1, 3, 3, 3, 1, 3, 1, 1, 1, 3, 3, 1, 1, 1, 1, 3, 3, 3], columns=["pred"], index=xpl.x_init.index
+ )
model = lambda: None
model.classes_ = np.array([0, 1])
xpl.model = model
@@ -1061,9 +870,9 @@ def test_contribution_plot_8(self):
annot_list.sort()
assert len(output.data) == 4
for elem in output.data:
- assert elem.type == 'violin'
- assert output.data[0].side == 'negative'
- assert output.data[1].side == 'positive'
+ assert elem.type == "violin"
+ assert output.data[0].side == "negative"
+ assert output.data[1].side == "positive"
assert output.data[-1].line.color == output.data[-3].line.color
assert output.data[-1].line.color != output.data[-2].line.color
self.assertListEqual(annot_list, np_hv)
@@ -1073,7 +882,7 @@ def test_contribution_plot_9(self):
"""
Classification with pred and sampling
"""
- col = 'X1'
+ col = "X1"
xpl = self.smart_explainer
xpl.x_init = pd.concat([xpl.x_init] * 20, ignore_index=True)
xpl.x_init.index = [i for i in range(xpl.x_init.shape[0])]
@@ -1082,17 +891,16 @@ def test_contribution_plot_9(self):
xpl.contributions[1] = pd.concat([xpl.contributions[1]] * 20, ignore_index=True)
xpl.contributions[0].index = xpl.x_init.index
xpl.contributions[1].index = xpl.x_init.index
- xpl.y_pred = pd.DataFrame([3, 1, 1, 3, 3]*8,
- columns=['pred'], index=xpl.x_init.index)
+ xpl.y_pred = pd.DataFrame([3, 1, 1, 3, 3] * 8, columns=["pred"], index=xpl.x_init.index)
model = lambda: None
model.classes_ = np.array([0, 1])
xpl.model = model
output = xpl.plot.contribution_plot(col, max_points=39)
assert len(output.data) == 4
for elem in output.data:
- assert elem.type == 'violin'
- assert output.data[0].side == 'negative'
- assert output.data[1].side == 'positive'
+ assert elem.type == "violin"
+ assert output.data[0].side == "negative"
+ assert output.data[1].side == "positive"
assert output.data[-1].line.color == output.data[-3].line.color
assert output.data[-1].line.color != output.data[-2].line.color
assert output.layout.xaxis.title.text == xpl.features_dict[col]
@@ -1101,13 +909,13 @@ def test_contribution_plot_9(self):
total_row = total_row + data.x.shape[0]
assert total_row == 39
expected_title = "Education - Feature Contribution
Response: 1 - Length of random Subset: 39 (98%)"
- assert output.layout.title['text'] == expected_title
+ assert output.layout.title["text"] == expected_title
def test_contribution_plot_10(self):
"""
Regression with pred and subset
"""
- col = 'X2'
+ col = "X2"
xpl = self.smart_explainer
xpl.x_init = pd.concat([xpl.x_init] * 4, ignore_index=True)
xpl.x_init.index = [i for i in range(xpl.x_init.shape[0])]
@@ -1115,15 +923,18 @@ def test_contribution_plot_10(self):
xpl.contributions = pd.concat([self.contrib1] * 4, ignore_index=True)
xpl._case = "regression"
xpl.state = SmartState()
- xpl.y_pred = pd.DataFrame([0.46989877093, 12.749302948]*4, columns=['pred'], index=xpl.x_init.index)
+ xpl.y_pred = pd.DataFrame([0.46989877093, 12.749302948] * 4, columns=["pred"], index=xpl.x_init.index)
subset = [1, 2, 6, 7]
output = xpl.plot.contribution_plot(col, selection=subset, violin_maxf=0)
- expected_output = go.Scatter(x=xpl.x_init[col].loc[subset],
- y=xpl.contributions[col].loc[subset],
- mode='markers',
- hovertext=[f"Id: {x}
Predict: {y:.2f}"
- for x, y in zip(xpl.x_init.loc[subset].index,
- xpl.y_pred.loc[subset].iloc[:, 0].tolist())])
+ expected_output = go.Scatter(
+ x=xpl.x_init[col].loc[subset],
+ y=xpl.contributions[col].loc[subset],
+ mode="markers",
+ hovertext=[
+ f"Id: {x}
Predict: {y:.2f}"
+ for x, y in zip(xpl.x_init.loc[subset].index, xpl.y_pred.loc[subset].iloc[:, 0].tolist())
+ ],
+ )
assert np.array_equal(output.data[0].x, expected_output.x)
assert np.array_equal(output.data[0].y, expected_output.y)
@@ -1131,39 +942,32 @@ def test_contribution_plot_10(self):
assert np.array_equal(output.data[0].hovertext, expected_output.hovertext)
assert output.layout.xaxis.title.text == self.smart_explainer.features_dict[col]
expected_title = "Age - Feature Contribution
Length of user-defined Subset: 4 (50%)"
- assert output.layout.title['text'] == expected_title
+ assert output.layout.title["text"] == expected_title
def test_contribution_plot_11(self):
"""
classification with proba
"""
- col = 'X1'
+ col = "X1"
xpl = self.smart_explainer
xpl.proba_values = pd.DataFrame(
- data=np.array(
- [[0.4, 0.6],
- [0.3, 0.7]]),
- columns=['class_1', 'class_2'],
- index=xpl.x_encoded.index.values)
+ data=np.array([[0.4, 0.6], [0.3, 0.7]]), columns=["class_1", "class_2"], index=xpl.x_encoded.index.values
+ )
output = self.smart_explainer.plot.contribution_plot(col)
assert str(type(output.data[-1])) == ""
- self.assertListEqual(list(output.data[-1]['marker']['color']), [0.6, 0.7])
- self.assertListEqual(list(output.data[-1]['y']), [-0.3, 4.7])
+ self.assertListEqual(list(output.data[-1]["marker"]["color"]), [0.6, 0.7])
+ self.assertListEqual(list(output.data[-1]["y"]), [-0.3, 4.7])
def test_contribution_plot_12(self):
"""
contribution plot with groups of features for classification case
"""
x_encoded = pd.DataFrame(
- data=np.array(
- [[0, 34],
- [1, 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[0, 34], [1, 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
xpl = self.smart_explainer
xpl.inv_features_dict = {}
- col = 'group1'
+ col = "group1"
xpl.x_encoded = x_encoded
xpl.contributions[0] = pd.concat([xpl.contributions[0]] * 10, ignore_index=True)
xpl.contributions[1] = pd.concat([xpl.contributions[1]] * 10, ignore_index=True)
@@ -1172,7 +976,7 @@ def test_contribution_plot_12(self):
xpl.postprocessing_modifications = False
xpl.preprocessing = None
# Creates a group of features named group1
- xpl.features_groups = {'group1': ['X1', 'X2']}
+ xpl.features_groups = {"group1": ["X1", "X2"]}
xpl.contributions_groups = xpl.state.compute_grouped_contributions(xpl.contributions, xpl.features_groups)
xpl.features_imp_groups = None
xpl._update_features_dict_with_groups(features_groups=xpl.features_groups)
@@ -1180,7 +984,7 @@ def test_contribution_plot_12(self):
output = xpl.plot.contribution_plot(col, proba=False)
assert len(output.data) == 1
- assert output.data[0].type == 'scatter'
+ assert output.data[0].type == "scatter"
self.setUp()
def test_contribution_plot_13(self):
@@ -1188,15 +992,11 @@ def test_contribution_plot_13(self):
contribution plot with groups of features for regression case
"""
x_encoded = pd.DataFrame(
- data=np.array(
- [[0, 34],
- [1, 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[0, 34], [1, 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
xpl = self.smart_explainer
xpl.inv_features_dict = {}
- col = 'group1'
+ col = "group1"
xpl.x_encoded = x_encoded
xpl.contributions = pd.concat([self.contrib1] * 10, ignore_index=True)
xpl._case = "regression"
@@ -1207,7 +1007,7 @@ def test_contribution_plot_13(self):
xpl.postprocessing_modifications = False
xpl.preprocessing = None
# Creates a group of features named group1
- xpl.features_groups = {'group1': ['X1', 'X2']}
+ xpl.features_groups = {"group1": ["X1", "X2"]}
xpl.contributions_groups = xpl.state.compute_grouped_contributions(xpl.contributions, xpl.features_groups)
xpl.features_imp_groups = None
xpl._update_features_dict_with_groups(features_groups=xpl.features_groups)
@@ -1215,7 +1015,7 @@ def test_contribution_plot_13(self):
output = xpl.plot.contribution_plot(col, proba=False)
assert len(output.data) == 1
- assert output.data[0].type == 'scatter'
+ assert output.data[0].type == "scatter"
self.setUp()
def test_contribution_plot_14(self):
@@ -1223,15 +1023,11 @@ def test_contribution_plot_14(self):
contribution plot with groups of features for classification case and subset
"""
x_encoded = pd.DataFrame(
- data=np.array(
- [[0, 34],
- [1, 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[0, 34], [1, 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
xpl = self.smart_explainer
xpl.inv_features_dict = {}
- col = 'group1'
+ col = "group1"
xpl.x_encoded = x_encoded
xpl.contributions[0] = pd.concat([xpl.contributions[0]] * 10, ignore_index=True)
xpl.contributions[1] = pd.concat([xpl.contributions[1]] * 10, ignore_index=True)
@@ -1240,7 +1036,7 @@ def test_contribution_plot_14(self):
xpl.postprocessing_modifications = False
xpl.preprocessing = None
# Creates a group of features named group1
- xpl.features_groups = {'group1': ['X1', 'X2']}
+ xpl.features_groups = {"group1": ["X1", "X2"]}
xpl.contributions_groups = xpl.state.compute_grouped_contributions(xpl.contributions, xpl.features_groups)
xpl.features_imp_groups = None
xpl._update_features_dict_with_groups(features_groups=xpl.features_groups)
@@ -1249,7 +1045,7 @@ def test_contribution_plot_14(self):
output = xpl.plot.contribution_plot(col, proba=False, selection=subset)
assert len(output.data) == 1
- assert output.data[0].type == 'scatter'
+ assert output.data[0].type == "scatter"
assert len(output.data[0].x) == 10
self.setUp()
@@ -1258,15 +1054,11 @@ def test_contribution_plot_15(self):
contribution plot with groups of features for regression case with subset
"""
x_encoded = pd.DataFrame(
- data=np.array(
- [[0, 34],
- [1, 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[0, 34], [1, 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
xpl = self.smart_explainer
xpl.inv_features_dict = {}
- col = 'group1'
+ col = "group1"
xpl.x_encoded = x_encoded
xpl.contributions = pd.concat([self.contrib1] * 10, ignore_index=True)
xpl._case = "regression"
@@ -1277,7 +1069,7 @@ def test_contribution_plot_15(self):
xpl.postprocessing_modifications = False
xpl.preprocessing = None
# Creates a group of features named group1
- xpl.features_groups = {'group1': ['X1', 'X2']}
+ xpl.features_groups = {"group1": ["X1", "X2"]}
xpl.contributions_groups = xpl.state.compute_grouped_contributions(xpl.contributions, xpl.features_groups)
xpl.features_imp_groups = None
xpl._update_features_dict_with_groups(features_groups=xpl.features_groups)
@@ -1286,7 +1078,7 @@ def test_contribution_plot_15(self):
output = xpl.plot.contribution_plot(col, proba=False, selection=subset)
assert len(output.data) == 1
- assert output.data[0].type == 'scatter'
+ assert output.data[0].type == "scatter"
assert len(output.data[0].x) == 10
self.setUp()
@@ -1294,14 +1086,9 @@ def test_plot_features_import_1(self):
"""
Unit test plot features import 1
"""
- serie1 = pd.Series([0.131, 0.51], index=['col1', 'col2'])
+ serie1 = pd.Series([0.131, 0.51], index=["col1", "col2"])
output = self.smart_explainer.plot.plot_features_import(serie1)
- data = go.Bar(
- x=serie1,
- y=serie1.index,
- name='Global',
- orientation='h'
- )
+ data = go.Bar(x=serie1, y=serie1.index, name="Global", orientation="h")
expected_output = go.Figure(data=data)
assert np.array_equal(output.data[0].x, expected_output.data[0].x)
@@ -1313,21 +1100,11 @@ def test_plot_features_import_2(self):
"""
Unit test plot features import 2
"""
- serie1 = pd.Series([0.131, 0.51], index=['col1', 'col2'])
- serie2 = pd.Series([0.33, 0.11], index=['col1', 'col2'])
+ serie1 = pd.Series([0.131, 0.51], index=["col1", "col2"])
+ serie2 = pd.Series([0.33, 0.11], index=["col1", "col2"])
output = self.smart_explainer.plot.plot_features_import(serie1, serie2)
- data1 = go.Bar(
- x=serie1,
- y=serie1.index,
- name='Global',
- orientation='h'
- )
- data2 = go.Bar(
- x=serie2,
- y=serie2.index,
- name='Subset',
- orientation='h'
- )
+ data1 = go.Bar(x=serie1, y=serie1.index, name="Global", orientation="h")
+ data2 = go.Bar(x=serie2, y=serie2.index, name="Subset", orientation="h")
expected_output = go.Figure(data=[data2, data1])
assert np.array_equal(output.data[0].x, expected_output.data[0].x)
assert np.array_equal(output.data[0].y, expected_output.data[0].y)
@@ -1344,19 +1121,11 @@ def test_features_importance_1(self):
"""
xpl = self.smart_explainer
xpl.explain_data = None
- output = xpl.plot.features_importance(selection=['person_A', 'person_B'])
+ output = xpl.plot.features_importance(selection=["person_A", "person_B"])
- data1 = go.Bar(
- x=np.array([0.2296, 0.7704]),
- y=np.array(['Age', 'Education']),
- name='Subset',
- orientation='h')
+ data1 = go.Bar(x=np.array([0.2296, 0.7704]), y=np.array(["Age", "Education"]), name="Subset", orientation="h")
- data2 = go.Bar(
- x=np.array([0.2296, 0.7704]),
- y=np.array(['Age', 'Education']),
- name='Global',
- orientation='h')
+ data2 = go.Bar(x=np.array([0.2296, 0.7704]), y=np.array(["Age", "Education"]), name="Global", orientation="h")
expected_output = go.Figure(data=[data1, data2])
@@ -1374,25 +1143,17 @@ def test_features_importance_2(self):
Unit test features importance 2
"""
xpl = self.smart_explainer
- #regression
+ # regression
xpl.contributions = self.contrib1
xpl.backend.state = SmartState()
xpl.explain_data = None
xpl._case = "regression"
xpl.state = SmartState()
- output = xpl.plot.features_importance(selection=['person_A', 'person_B'])
+ output = xpl.plot.features_importance(selection=["person_A", "person_B"])
- data1 = go.Bar(
- x=np.array([0.2296, 0.7704]),
- y=np.array(['Age', 'Education']),
- name='Subset',
- orientation='h')
+ data1 = go.Bar(x=np.array([0.2296, 0.7704]), y=np.array(["Age", "Education"]), name="Subset", orientation="h")
- data2 = go.Bar(
- x=np.array([0.2296, 0.7704]),
- y=np.array(['Age', 'Education']),
- name='Global',
- orientation='h')
+ data2 = go.Bar(x=np.array([0.2296, 0.7704]), y=np.array(["Age", "Education"]), name="Global", orientation="h")
expected_output = go.Figure(data=[data1, data2])
@@ -1410,19 +1171,15 @@ def test_features_importance_3(self):
Unit test features importance for groups of features
"""
x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34, 1],
- ['Master', 27, 0]]),
- columns=['X1', 'X2', 'X3'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34, 1], ["Master", 27, 0]]),
+ columns=["X1", "X2", "X3"],
+ index=["person_A", "person_B"],
)
contrib = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78, 1.2],
- [1.2, 3.6, -0.3]]),
- columns=['X1', 'X2', 'X3'],
- index=['person_A', 'person_B']
+ data=np.array([[-3.4, 0.78, 1.2], [1.2, 3.6, -0.3]]),
+ columns=["X1", "X2", "X3"],
+ index=["person_A", "person_B"],
)
smart_explainer = SmartExplainer(model=self.model)
@@ -1431,10 +1188,10 @@ def test_features_importance_3(self):
smart_explainer.postprocessing_modifications = False
smart_explainer.features_imp_groups = None
smart_explainer.features_imp = None
- smart_explainer.features_groups = {'group0': ['X1', 'X2']}
+ smart_explainer.features_groups = {"group0": ["X1", "X2"]}
smart_explainer.contributions = [contrib, -contrib]
- smart_explainer.features_dict = {'X1': 'X1', 'X2': 'X2', 'X3': 'X3', 'group0': 'group0'}
- smart_explainer.inv_features_dict = {'X1': 'X1', 'X2': 'X2', 'X3': 'X3', 'group0': 'group0'}
+ smart_explainer.features_dict = {"X1": "X1", "X2": "X2", "X3": "X3", "group0": "group0"}
+ smart_explainer.inv_features_dict = {"X1": "X1", "X2": "X2", "X3": "X3", "group0": "group0"}
smart_explainer.model = self.smart_explainer.model
smart_explainer._case, smart_explainer._classes = check_model(self.smart_explainer.model)
smart_explainer.backend = ShapBackend(model=self.smart_explainer.model)
@@ -1442,17 +1199,15 @@ def test_features_importance_3(self):
smart_explainer.explain_data = None
smart_explainer.state = MultiDecorator(SmartState())
smart_explainer.contributions_groups = smart_explainer.state.compute_grouped_contributions(
- smart_explainer.contributions, smart_explainer.features_groups)
+ smart_explainer.contributions, smart_explainer.features_groups
+ )
smart_explainer.features_imp_groups = smart_explainer.state.compute_features_import(
- smart_explainer.contributions_groups)
+ smart_explainer.contributions_groups
+ )
output = smart_explainer.plot.features_importance()
- data1 = go.Bar(
- x=np.array([0.1682, 0.8318]),
- y=np.array(['X3', 'group0']),
- name='Global',
- orientation='h')
+ data1 = go.Bar(x=np.array([0.1682, 0.8318]), y=np.array(["X3", "group0"]), name="Global", orientation="h")
expected_output = go.Figure(data=[data1])
assert np.array_equal(output.data[0].x, expected_output.data[0].x)
@@ -1465,19 +1220,15 @@ def test_features_importance_4(self):
Unit test features importance for groups of features when displaying a group
"""
x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34, 1],
- ['Master', 27, 0]]),
- columns=['X1', 'X2', 'X3'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34, 1], ["Master", 27, 0]]),
+ columns=["X1", "X2", "X3"],
+ index=["person_A", "person_B"],
)
contrib = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78, 1.2],
- [1.2, 3.6, -0.3]]),
- columns=['X1', 'X2', 'X3'],
- index=['person_A', 'person_B']
+ data=np.array([[-3.4, 0.78, 1.2], [1.2, 3.6, -0.3]]),
+ columns=["X1", "X2", "X3"],
+ index=["person_A", "person_B"],
)
smart_explainer = SmartExplainer(model=self.model)
@@ -1486,10 +1237,10 @@ def test_features_importance_4(self):
smart_explainer.postprocessing_modifications = False
smart_explainer.features_imp_groups = None
smart_explainer.features_imp = None
- smart_explainer.features_groups = {'group0': ['X1', 'X2']}
+ smart_explainer.features_groups = {"group0": ["X1", "X2"]}
smart_explainer.contributions = [contrib, -contrib]
- smart_explainer.features_dict = {'X1': 'X1', 'X2': 'X2', 'X3': 'X3', 'group0': 'group0'}
- smart_explainer.inv_features_dict = {'X1': 'X1', 'X2': 'X2', 'X3': 'X3', 'group0': 'group0'}
+ smart_explainer.features_dict = {"X1": "X1", "X2": "X2", "X3": "X3", "group0": "group0"}
+ smart_explainer.inv_features_dict = {"X1": "X1", "X2": "X2", "X3": "X3", "group0": "group0"}
smart_explainer.model = self.smart_explainer.model
smart_explainer.backend = ShapBackend(model=self.smart_explainer.model)
smart_explainer.backend.state = MultiDecorator(SmartState())
@@ -1497,17 +1248,15 @@ def test_features_importance_4(self):
smart_explainer._case, smart_explainer._classes = check_model(self.smart_explainer.model)
smart_explainer.state = smart_explainer.backend.state
smart_explainer.contributions_groups = smart_explainer.state.compute_grouped_contributions(
- smart_explainer.contributions, smart_explainer.features_groups)
+ smart_explainer.contributions, smart_explainer.features_groups
+ )
smart_explainer.features_imp_groups = smart_explainer.state.compute_features_import(
- smart_explainer.contributions_groups)
+ smart_explainer.contributions_groups
+ )
- output = smart_explainer.plot.features_importance(group_name='group0')
+ output = smart_explainer.plot.features_importance(group_name="group0")
- data1 = go.Bar(
- x=np.array([0.4179, 0.4389]),
- y=np.array(['X2', 'X1']),
- name='Global',
- orientation='h')
+ data1 = go.Bar(x=np.array([0.4179, 0.4389]), y=np.array(["X2", "X1"]), name="Global", orientation="h")
expected_output = go.Figure(data=[data1])
assert np.array_equal(output.data[0].x, expected_output.data[0].x)
@@ -1517,7 +1266,7 @@ def test_features_importance_4(self):
def test_local_pred_1(self):
xpl = self.smart_explainer
- output = xpl.plot.local_pred('person_A',label=0)
+ output = xpl.plot.local_pred("person_A", label=0)
assert isinstance(output, float)
def test_plot_line_comparison_1(self):
@@ -1525,45 +1274,41 @@ def test_plot_line_comparison_1(self):
Unit test 1 for plot_line_comparison
"""
xpl = self.smart_explainer
- index = ['person_A', 'person_B']
- data = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
- features_dict = {'X1': 'X1', 'X2': 'X2'}
- xpl.inv_features_dict = {'X1': 'X1', 'X2': 'X2'}
- colors = ['rgba(244, 192, 0, 1.0)', 'rgba(74, 99, 138, 0.7)']
- var_dict = ['X1', 'X2']
+ index = ["person_A", "person_B"]
+ data = pd.DataFrame(data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=index)
+ features_dict = {"X1": "X1", "X2": "X2"}
+ xpl.inv_features_dict = {"X1": "X1", "X2": "X2"}
+ colors = ["rgba(244, 192, 0, 1.0)", "rgba(74, 99, 138, 0.7)"]
+ var_dict = ["X1", "X2"]
contributions = [[-3.4, 0.78], [1.2, 3.6]]
- title = 'Compare plot - index : person_A ; person_B'
+ title = "Compare plot - index : person_A ; person_B"
predictions = [data.loc[id] for id in index]
fig = list()
for i in range(2):
- fig.append(go.Scatter(
- x=[contributions[0][i], contributions[1][i]],
- y=['' + feat + '' for feat in var_dict],
- mode='lines+markers',
- name=f'Id: {index[i]}',
- hovertext=[f'Id: {index[i]}
X1
Contribution: {contributions[0][i]:.4f}
'
- + f'Value: {data.iloc[i][0]}',
- f'Id: {index[i]}
X2
Contribution: {contributions[1][i]:.4f}
'
- + f'Value: {data.iloc[i][1]}'
- ],
- marker={'color': colors[i]}
+ fig.append(
+ go.Scatter(
+ x=[contributions[0][i], contributions[1][i]],
+ y=["" + feat + "" for feat in var_dict],
+ mode="lines+markers",
+ name=f"Id: {index[i]}",
+ hovertext=[
+ f"Id: {index[i]}
X1
Contribution: {contributions[0][i]:.4f}
"
+ + f"Value: {data.iloc[i][0]}",
+ f"Id: {index[i]}
X2
Contribution: {contributions[1][i]:.4f}
"
+ + f"Value: {data.iloc[i][1]}",
+ ],
+ marker={"color": colors[i]},
)
)
expected_output = go.Figure(data=fig)
- output = xpl.plot.plot_line_comparison(['person_A', 'person_B'], var_dict, contributions,
- predictions=predictions, dict_features=features_dict)
+ output = xpl.plot.plot_line_comparison(
+ ["person_A", "person_B"], var_dict, contributions, predictions=predictions, dict_features=features_dict
+ )
for i in range(2):
- assert np.array_equal(output.data[i]['x'], expected_output.data[i]['x'])
- assert np.array_equal(output.data[i]['y'], expected_output.data[i]['y'])
+ assert np.array_equal(output.data[i]["x"], expected_output.data[i]["x"])
+ assert np.array_equal(output.data[i]["y"], expected_output.data[i]["y"])
assert output.data[i].name == expected_output.data[i].name
assert output.data[i].hovertext == expected_output.data[i].hovertext
assert output.data[i].marker == expected_output.data[i].marker
@@ -1574,26 +1319,26 @@ def test_plot_line_comparison_2(self):
Unit test 2 for plot_line_comparison
"""
xpl = self.smart_explainer
- index = ['person_A', 'person_B']
- data = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
- xpl.inv_features_dict = {'X1': 'X1', 'X2': 'X2'}
- var_dict = ['X1', 'X2']
+ index = ["person_A", "person_B"]
+ data = pd.DataFrame(data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=index)
+ xpl.inv_features_dict = {"X1": "X1", "X2": "X2"}
+ var_dict = ["X1", "X2"]
contributions = [[-3.4, 0.78], [1.2, 3.6]]
- subtitle = 'This is a good test'
- title = 'Compare plot - index : person_A ; person_B' \
- + "
This is a good test"
+ subtitle = "This is a good test"
+ title = (
+ "Compare plot - index : person_A ; person_B"
+ + "
This is a good test"
+ )
predictions = [data.loc[id] for id in index]
- output = xpl.plot.plot_line_comparison(index, var_dict, contributions,
- subtitle=subtitle, predictions=predictions,
- dict_features=xpl.inv_features_dict)
+ output = xpl.plot.plot_line_comparison(
+ index,
+ var_dict,
+ contributions,
+ subtitle=subtitle,
+ predictions=predictions,
+ dict_features=xpl.inv_features_dict,
+ )
assert title == output.layout.title.text
@@ -1603,20 +1348,21 @@ def test_compare_plot_1(self):
"""
xpl = self.smart_explainer
xpl.contributions = pd.DataFrame(
- data=[[-3.4, 0.78], [1.2, 3.6]],
- index=['person_A', 'person_B'],
- columns=['X1', 'X2']
+ data=[[-3.4, 0.78], [1.2, 3.6]], index=["person_A", "person_B"], columns=["X1", "X2"]
)
- xpl.inv_features_dict = {'Education': 'X1', 'Age': 'X2'}
+ xpl.inv_features_dict = {"Education": "X1", "Age": "X2"}
xpl._case = "regression"
output = xpl.plot.compare_plot(row_num=[1], show_predict=False)
title = "Compare plot - index : person_B
"
- data = [go.Scatter(
- x=[1.2, 3.6],
- y=['Education', 'Age'],
- name='Id: person_B',
- hovertext=['Id: person_B
Education
Contribution: 1.2000
Value: Master',
- 'Id: person_B
Age
Contribution: 3.6000
Value: 27']
+ data = [
+ go.Scatter(
+ x=[1.2, 3.6],
+ y=["Education", "Age"],
+ name="Id: person_B",
+ hovertext=[
+ "Id: person_B
Education
Contribution: 1.2000
Value: Master",
+ "Id: person_B
Age
Contribution: 3.6000
Value: 27",
+ ],
)
]
expected_output = go.Figure(data=data)
@@ -1631,36 +1377,32 @@ def test_compare_plot_2(self):
Unit test 2 for compare_plot
"""
xpl = self.smart_explainer
- xpl.inv_features_dict = {'Education': 'X1', 'Age': 'X2'}
- index = ['person_A', 'person_B']
+ xpl.inv_features_dict = {"Education": "X1", "Age": "X2"}
+ index = ["person_A", "person_B"]
contributions = [[-3.4, 0.78], [1.2, 3.6]]
- xpl.contributions = pd.DataFrame(
- data=contributions,
- index=index,
- columns=['X1', 'X2']
- )
- data = np.array(
- [['PhD', 34],
- ['Master', 27]]
- )
+ xpl.contributions = pd.DataFrame(data=contributions, index=index, columns=["X1", "X2"])
+ data = np.array([["PhD", 34], ["Master", 27]])
xpl._case = "regression"
output = xpl.plot.compare_plot(index=index, show_predict=True)
- title_and_subtitle = "Compare plot - index : person_A ;" \
- " person_B
" \
- "Predictions: person_A: 0 ; person_B: 1"
+ title_and_subtitle = (
+ "Compare plot - index : person_A ;"
+ " person_B
"
+ "Predictions: person_A: 0 ; person_B: 1"
+ )
fig = list()
for i in range(2):
- fig.append(go.Scatter(
- x=contributions[i][::-1],
- y=['Age', 'Education'],
- name=f'Id: {index[i]}',
- hovertext=[
- f'Id: {index[i]}
Age
Contribution: {contributions[i][1]:.4f}'
- f'
Value: {data[i][1]}',
- f'Id: {index[i]}
Education
Contribution: {contributions[i][0]:.4f}'
- f'
Value: {data[i][0]}'
- ]
- )
+ fig.append(
+ go.Scatter(
+ x=contributions[i][::-1],
+ y=["Age", "Education"],
+ name=f"Id: {index[i]}",
+ hovertext=[
+ f"Id: {index[i]}
Age
Contribution: {contributions[i][1]:.4f}"
+ f"
Value: {data[i][1]}",
+ f"Id: {index[i]}
Education
Contribution: {contributions[i][0]:.4f}"
+ f"
Value: {data[i][0]}",
+ ],
+ )
)
expected_output = go.Figure(data=fig)
@@ -1675,55 +1417,26 @@ def test_compare_plot_3(self):
"""
Unit test 3 for compare_plot classification
"""
- index = ['A', 'B']
- x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
- contributions1 = pd.DataFrame(
- data=np.array(
- [[-3.4, 0.78],
- [1.2, 3.6]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
- contributions2 = pd.DataFrame(
- data=np.array(
- [[-0.4, 0.78],
- [0.2, 0.6]]
- ),
- columns=['X1', 'X2'],
- index=index
- )
- feature_dictionary = {'X1': 'Education', 'X2': 'Age'}
+ index = ["A", "B"]
+ x_init = pd.DataFrame(data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=index)
+ contributions1 = pd.DataFrame(data=np.array([[-3.4, 0.78], [1.2, 3.6]]), columns=["X1", "X2"], index=index)
+ contributions2 = pd.DataFrame(data=np.array([[-0.4, 0.78], [0.2, 0.6]]), columns=["X1", "X2"], index=index)
+ feature_dictionary = {"X1": "Education", "X2": "Age"}
smart_explainer_mi = SmartExplainer(model=self.model, features_dict=feature_dictionary)
smart_explainer_mi.contributions = [
- pd.DataFrame(
- data=contributions1,
- index=index,
- columns=['X1', 'X2']
- ),
- pd.DataFrame(
- data=contributions2,
- index=index,
- columns=['X1', 'X2']
- )
+ pd.DataFrame(data=contributions1, index=index, columns=["X1", "X2"]),
+ pd.DataFrame(data=contributions2, index=index, columns=["X1", "X2"]),
]
- smart_explainer_mi.inv_features_dict = {'Education': 'X1', 'Age': 'X2'}
+ smart_explainer_mi.inv_features_dict = {"Education": "X1", "Age": "X2"}
smart_explainer_mi.data = dict()
smart_explainer_mi.x_init = x_init
smart_explainer_mi.columns_dict = {i: col for i, col in enumerate(smart_explainer_mi.x_init.columns)}
smart_explainer_mi._case = "classification"
smart_explainer_mi._classes = [0, 1]
- smart_explainer_mi.model = 'predict_proba'
+ smart_explainer_mi.model = "predict_proba"
- output_label0 = smart_explainer_mi.plot.compare_plot(index=['A', 'B'], label=0, show_predict=False)
- output_label1 = smart_explainer_mi.plot.compare_plot(index=['A', 'B'], show_predict=False)
+ output_label0 = smart_explainer_mi.plot.compare_plot(index=["A", "B"], label=0, show_predict=False)
+ output_label1 = smart_explainer_mi.plot.compare_plot(index=["A", "B"], show_predict=False)
title_0 = "Compare plot - index : A ; B
"
title_1 = "Compare plot - index : A ; B
"
@@ -1732,16 +1445,17 @@ def test_compare_plot_3(self):
x0 = contributions1.to_numpy()
x0.sort(axis=1)
for i in range(2):
- fig_0.append(go.Scatter(
- x=x0[i][::-1],
- y=['Age', 'Education'],
- name=f'Id: {index[i]}',
- hovertext=[
- f'Id: {index[i]}
Age
Contribution: {x0[i][1]:.4f}'
- f'
Value: {x_init.to_numpy()[i][1]}',
- f'Id: {index[i]}
Education
Contribution: {x0[i][0]:.4f}'
- f'
Value: {x_init.to_numpy()[i][0]}'
- ]
+ fig_0.append(
+ go.Scatter(
+ x=x0[i][::-1],
+ y=["Age", "Education"],
+ name=f"Id: {index[i]}",
+ hovertext=[
+ f"Id: {index[i]}
Age
Contribution: {x0[i][1]:.4f}"
+ f"
Value: {x_init.to_numpy()[i][1]}",
+ f"Id: {index[i]}
Education
Contribution: {x0[i][0]:.4f}"
+ f"
Value: {x_init.to_numpy()[i][0]}",
+ ],
)
)
@@ -1749,16 +1463,17 @@ def test_compare_plot_3(self):
x1 = contributions2.to_numpy()
x1.sort(axis=1)
for i in range(2):
- fig_1.append(go.Scatter(
- x=x1[i][::-1],
- y=['Age', 'Education'],
- name=f'Id: {index[i]}',
- hovertext=[
- f'Id: {index[i]}
Age
Contribution: {x1[i][1]:.4f}'
- f'
Value: {x_init.to_numpy()[i][1]}',
- f'Id: {index[i]}
Education
Contribution: {x1[i][0]:.4f}'
- f'
Value: {x_init.to_numpy()[i][0]}'
- ]
+ fig_1.append(
+ go.Scatter(
+ x=x1[i][::-1],
+ y=["Age", "Education"],
+ name=f"Id: {index[i]}",
+ hovertext=[
+ f"Id: {index[i]}
Age
Contribution: {x1[i][1]:.4f}"
+ f"
Value: {x_init.to_numpy()[i][1]}",
+ f"Id: {index[i]}
Education
Contribution: {x1[i][0]:.4f}"
+ f"
Value: {x_init.to_numpy()[i][0]}",
+ ],
)
)
@@ -1781,21 +1496,18 @@ def test_interactions_plot_1(self):
Unit test 1 for test interaction plot : scatter plot for categorical x categorical features
"""
- col1 = 'X1'
- col2 = 'X2'
+ col1 = "X1"
+ col2 = "X2"
- interaction_values = np.array([
- [[0.1, -0.7], [-0.6, 0.3]],
- [[0.2, -0.1], [-0.2, 0.1]]
- ])
+ interaction_values = np.array([[[0.1, -0.7], [-0.6, 0.3]], [[0.2, -0.1], [-0.2, 0.1]]])
self.smart_explainer.interaction_values = interaction_values
self.smart_explainer.x_interaction = self.smart_explainer.x_encoded
output = self.smart_explainer.plot.interactions_plot(col1, col2, violin_maxf=0)
- expected_output = px.scatter(x=self.x_init[col1],
- y=self.smart_explainer.interaction_values[:, 0, 1],
- color=self.x_init[col1])
+ expected_output = px.scatter(
+ x=self.x_init[col1], y=self.smart_explainer.interaction_values[:, 0, 1], color=self.x_init[col1]
+ )
assert np.array_equal(output.data[0].x, expected_output.data[0].x)
assert np.array_equal(output.data[1].y, expected_output.data[1].y * 2)
@@ -1806,31 +1518,24 @@ def test_interactions_plot_2(self):
"""
Unit test 1 for test interaction plot : scatter plot for categorical x numeric features
"""
- col1 = 'X1'
- col2 = 'X2'
+ col1 = "X1"
+ col2 = "X2"
smart_explainer = self.smart_explainer
smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
- smart_explainer.x_encoded['X2'] = smart_explainer.x_encoded['X2'].astype(float)
+ smart_explainer.x_encoded["X2"] = smart_explainer.x_encoded["X2"].astype(float)
- interaction_values = np.array([
- [[0.1, -0.7], [-0.7, 0.3]],
- [[0.2, -0.1], [-0.1, 0.1]]
- ])
+ interaction_values = np.array([[[0.1, -0.7], [-0.7, 0.3]], [[0.2, -0.1], [-0.1, 0.1]]])
smart_explainer.interaction_values = interaction_values
smart_explainer.x_interaction = smart_explainer.x_encoded
output = smart_explainer.plot.interactions_plot(col1, col2, violin_maxf=0)
- assert np.array_equal(output.data[0].x, ['PhD', 'Master'])
+ assert np.array_equal(output.data[0].x, ["PhD", "Master"])
assert np.array_equal(output.data[0].y, [-1.4, -0.2])
- assert np.array_equal(output.data[0].marker.color, [34., 27.])
+ assert np.array_equal(output.data[0].marker.color, [34.0, 27.0])
assert len(output.data) == 1
self.setUp()
@@ -1839,35 +1544,28 @@ def test_interactions_plot_3(self):
"""
Unit test 1 for test interaction plot : scatter plot for numeric x categorical features
"""
- col1 = 'X1'
- col2 = 'X2'
+ col1 = "X1"
+ col2 = "X2"
smart_explainer = self.smart_explainer
smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
- smart_explainer.x_encoded['X2'] = smart_explainer.x_encoded['X2'].astype(float)
+ smart_explainer.x_encoded["X2"] = smart_explainer.x_encoded["X2"].astype(float)
- interaction_values = np.array([
- [[0.1, -0.7], [-0.7, 0.3]],
- [[0.2, -0.1], [-0.1, 0.1]]
- ])
+ interaction_values = np.array([[[0.1, -0.7], [-0.7, 0.3]], [[0.2, -0.1], [-0.1, 0.1]]])
smart_explainer.interaction_values = interaction_values
smart_explainer.x_interaction = smart_explainer.x_encoded
output = smart_explainer.plot.interactions_plot(col2, col1, violin_maxf=0)
- assert np.array_equal(output.data[0].x, [34.])
+ assert np.array_equal(output.data[0].x, [34.0])
assert np.array_equal(output.data[0].y, [-1.4])
- assert output.data[0].name == 'PhD'
+ assert output.data[0].name == "PhD"
- assert np.array_equal(output.data[1].x, [27.])
+ assert np.array_equal(output.data[1].x, [27.0])
assert np.array_equal(output.data[1].y, [-0.2])
- assert output.data[1].name == 'Master'
+ assert output.data[1].name == "Master"
assert len(output.data) == 2
@@ -1877,24 +1575,17 @@ def test_interactions_plot_4(self):
"""
Unit test 1 for test interaction plot : scatter plot for numeric x numeric features
"""
- col1 = 'X1'
- col2 = 'X2'
+ col1 = "X1"
+ col2 = "X2"
smart_explainer = self.smart_explainer
smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
- data=np.array(
- [[520, 34],
- [12800, 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([[520, 34], [12800, 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
- smart_explainer.x_encoded['X1'] = smart_explainer.x_encoded['X1'].astype(float)
- smart_explainer.x_encoded['X2'] = smart_explainer.x_encoded['X2'].astype(float)
+ smart_explainer.x_encoded["X1"] = smart_explainer.x_encoded["X1"].astype(float)
+ smart_explainer.x_encoded["X2"] = smart_explainer.x_encoded["X2"].astype(float)
- interaction_values = np.array([
- [[0.1, -0.7], [-0.7, 0.3]],
- [[0.2, -0.1], [-0.1, 0.1]]
- ])
+ interaction_values = np.array([[[0.1, -0.7], [-0.7, 0.3]], [[0.2, -0.1], [-0.1, 0.1]]])
smart_explainer.interaction_values = interaction_values
smart_explainer.x_interaction = smart_explainer.x_encoded
@@ -1903,7 +1594,7 @@ def test_interactions_plot_4(self):
assert np.array_equal(output.data[0].x, [520, 12800])
assert np.array_equal(output.data[0].y, [-1.4, -0.2])
- assert np.array_equal(output.data[0].marker.color, [34., 27.])
+ assert np.array_equal(output.data[0].marker.color, [34.0, 27.0])
assert len(output.data) == 1
@@ -1913,22 +1604,15 @@ def test_interactions_plot_5(self):
"""
Unit test 1 for test interaction plot : violin plot for categorical x numeric features
"""
- col1 = 'X1'
- col2 = 'X2'
+ col1 = "X1"
+ col2 = "X2"
smart_explainer = self.smart_explainer
smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34],
- ['Master', 27]]),
- columns=['X1', 'X2'],
- index=['person_A', 'person_B']
+ data=np.array([["PhD", 34], ["Master", 27]]), columns=["X1", "X2"], index=["person_A", "person_B"]
)
- smart_explainer.x_encoded['X2'] = smart_explainer.x_encoded['X2'].astype(float)
+ smart_explainer.x_encoded["X2"] = smart_explainer.x_encoded["X2"].astype(float)
- interaction_values = np.array([
- [[0.1, -0.7], [-0.7, 0.3]],
- [[0.2, -0.1], [-0.1, 0.1]]
- ])
+ interaction_values = np.array([[[0.1, -0.7], [-0.7, 0.3]], [[0.2, -0.1], [-0.1, 0.1]]])
smart_explainer.interaction_values = interaction_values
smart_explainer.x_interaction = smart_explainer.x_encoded
@@ -1937,13 +1621,13 @@ def test_interactions_plot_5(self):
assert len(output.data) == 3
- assert output.data[0].type == 'violin'
- assert output.data[1].type == 'violin'
- assert output.data[2].type == 'scatter'
+ assert output.data[0].type == "violin"
+ assert output.data[1].type == "violin"
+ assert output.data[2].type == "scatter"
- assert np.array_equal(output.data[2].x, ['PhD', 'Master'])
+ assert np.array_equal(output.data[2].x, ["PhD", "Master"])
assert np.array_equal(output.data[2].y, [-1.4, -0.2])
- assert np.array_equal(output.data[2].marker.color, [34., 27.])
+ assert np.array_equal(output.data[2].marker.color, [34.0, 27.0])
self.setUp()
@@ -1953,22 +1637,30 @@ def test_top_interactions_plot_1(self):
"""
smart_explainer = self.smart_explainer
smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34, 16, 0.2, 12],
- ['Master', 27, -10, 0.65, 18]]),
- columns=['X1', 'X2', 'X3', 'X4', 'X5'],
- index=['person_A', 'person_B']
- ).astype({'X1': str, 'X2': float, 'X3': float, 'X4': float, 'X5': float})
+ data=np.array([["PhD", 34, 16, 0.2, 12], ["Master", 27, -10, 0.65, 18]]),
+ columns=["X1", "X2", "X3", "X4", "X5"],
+ index=["person_A", "person_B"],
+ ).astype({"X1": str, "X2": float, "X3": float, "X4": float, "X5": float})
smart_explainer.features_desc = dict(smart_explainer.x_init.nunique())
smart_explainer.columns_dict = {i: col for i, col in enumerate(smart_explainer.x_init.columns)}
- interaction_values = np.array([
- [[0.1, -0.7, 0.01, -0.9, 0.6], [-0.1, 0.8, 0.02, 0.7, -0.5], [0.2, 0.5, 0.04, -0.88, 0.7],
- [0.15, 0.6, -0.2, 0.5, 0.3]],
- [[0.2, -0.1, 0.2, 0.8, 0.55], [-0.2, 0.6, 0.02, -0.67, -0.6], [0.1, -0.5, 0.05, 1, 0.5],
- [0.3, 0.6, 0.02, -0.9, 0.4]]
- ])
+ interaction_values = np.array(
+ [
+ [
+ [0.1, -0.7, 0.01, -0.9, 0.6],
+ [-0.1, 0.8, 0.02, 0.7, -0.5],
+ [0.2, 0.5, 0.04, -0.88, 0.7],
+ [0.15, 0.6, -0.2, 0.5, 0.3],
+ ],
+ [
+ [0.2, -0.1, 0.2, 0.8, 0.55],
+ [-0.2, 0.6, 0.02, -0.67, -0.6],
+ [0.1, -0.5, 0.05, 1, 0.5],
+ [0.3, 0.6, 0.02, -0.9, 0.4],
+ ],
+ ]
+ )
smart_explainer.interaction_values = interaction_values
smart_explainer.x_interaction = smart_explainer.x_encoded
@@ -1976,46 +1668,54 @@ def test_top_interactions_plot_1(self):
output = smart_explainer.plot.top_interactions_plot(nb_top_interactions=5, violin_maxf=0)
assert len(output.layout.updatemenus[0].buttons) == 5
- assert isinstance(output.layout.updatemenus[0].buttons[0].args[0]['visible'], list)
- assert len(output.layout.updatemenus[0].buttons[0].args[0]['visible']) >= 5
- assert True in output.layout.updatemenus[0].buttons[0].args[0]['visible']
+ assert isinstance(output.layout.updatemenus[0].buttons[0].args[0]["visible"], list)
+ assert len(output.layout.updatemenus[0].buttons[0].args[0]["visible"]) >= 5
+ assert True in output.layout.updatemenus[0].buttons[0].args[0]["visible"]
self.setUp()
def test_top_interactions_plot_2(self):
- """
- Test top interactions plot with violin and scatter plots
- """
- smart_explainer = self.smart_explainer
- smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
- data=np.array(
- [['PhD', 34, 16, 0.2, 12],
- ['Master', 27, -10, 0.65, 18]]),
- columns=['X1', 'X2', 'X3', 'X4', 'X5'],
- index=['person_A', 'person_B']
- ).astype({'X1': str, 'X2': float, 'X3': float, 'X4': float, 'X5': float})
-
- smart_explainer.features_desc = dict(smart_explainer.x_init.nunique())
- smart_explainer.columns_dict = {i: col for i, col in enumerate(smart_explainer.x_init.columns)}
-
- interaction_values = np.array([
- [[0.1, -0.7, 0.01, -0.9, 0.6], [-0.1, 0.8, 0.02, 0.7, -0.5], [0.2, 0.5, 0.04, -0.88, 0.7],
- [0.15, 0.6, -0.2, 0.5, 0.3]],
- [[0.2, -0.1, 0.2, 0.8, 0.55], [-0.2, 0.6, 0.02, -0.67, -0.6], [0.1, -0.5, 0.05, 1, 0.5],
- [0.3, 0.6, 0.02, -0.9, 0.4]]
- ])
-
- smart_explainer.interaction_values = interaction_values
- smart_explainer.x_interaction = smart_explainer.x_encoded
-
- output = smart_explainer.plot.top_interactions_plot(nb_top_interactions=4)
-
- assert len(output.layout.updatemenus[0].buttons) == 4
- assert isinstance(output.layout.updatemenus[0].buttons[0].args[0]['visible'], list)
- assert len(output.layout.updatemenus[0].buttons[0].args[0]['visible']) >= 4
- assert True in output.layout.updatemenus[0].buttons[0].args[0]['visible']
-
- self.setUp()
+ """
+ Test top interactions plot with violin and scatter plots
+ """
+ smart_explainer = self.smart_explainer
+ smart_explainer.x_encoded = smart_explainer.x_init = pd.DataFrame(
+ data=np.array([["PhD", 34, 16, 0.2, 12], ["Master", 27, -10, 0.65, 18]]),
+ columns=["X1", "X2", "X3", "X4", "X5"],
+ index=["person_A", "person_B"],
+ ).astype({"X1": str, "X2": float, "X3": float, "X4": float, "X5": float})
+
+ smart_explainer.features_desc = dict(smart_explainer.x_init.nunique())
+ smart_explainer.columns_dict = {i: col for i, col in enumerate(smart_explainer.x_init.columns)}
+
+ interaction_values = np.array(
+ [
+ [
+ [0.1, -0.7, 0.01, -0.9, 0.6],
+ [-0.1, 0.8, 0.02, 0.7, -0.5],
+ [0.2, 0.5, 0.04, -0.88, 0.7],
+ [0.15, 0.6, -0.2, 0.5, 0.3],
+ ],
+ [
+ [0.2, -0.1, 0.2, 0.8, 0.55],
+ [-0.2, 0.6, 0.02, -0.67, -0.6],
+ [0.1, -0.5, 0.05, 1, 0.5],
+ [0.3, 0.6, 0.02, -0.9, 0.4],
+ ],
+ ]
+ )
+
+ smart_explainer.interaction_values = interaction_values
+ smart_explainer.x_interaction = smart_explainer.x_encoded
+
+ output = smart_explainer.plot.top_interactions_plot(nb_top_interactions=4)
+
+ assert len(output.layout.updatemenus[0].buttons) == 4
+ assert isinstance(output.layout.updatemenus[0].buttons[0].args[0]["visible"], list)
+ assert len(output.layout.updatemenus[0].buttons[0].args[0]["visible"]) >= 4
+ assert True in output.layout.updatemenus[0].buttons[0].args[0]["visible"]
+
+ self.setUp()
def test_correlations_1(self):
"""
@@ -2023,12 +1723,10 @@ def test_correlations_1(self):
"""
smart_explainer = self.smart_explainer
- df = pd.DataFrame({
- "A": [8, 90, 10, 110],
- "B": [4.3, 7.4, 10.2, 15.7],
- "C": ["C8", "C8", "C9", "C9"],
- "D": [1, -3, -5, -10]
- }, index=[8, 9, 10, 11])
+ df = pd.DataFrame(
+ {"A": [8, 90, 10, 110], "B": [4.3, 7.4, 10.2, 15.7], "C": ["C8", "C8", "C9", "C9"], "D": [1, -3, -5, -10]},
+ index=[8, 9, 10, 11],
+ )
output = smart_explainer.plot.correlations(df, max_features=3)
@@ -2043,14 +1741,12 @@ def test_correlations_2(self):
"""
smart_explainer = self.smart_explainer
- df = pd.DataFrame({
- "A": [8, 90, 10, 110],
- "B": [4.3, 7.4, 10.2, 15.7],
- "C": ["C8", "C8", "C9", "C9"],
- "D": [1, -3, -5, -10]
- }, index=[8, 9, 10, 11])
+ df = pd.DataFrame(
+ {"A": [8, 90, 10, 110], "B": [4.3, 7.4, 10.2, 15.7], "C": ["C8", "C8", "C9", "C9"], "D": [1, -3, -5, -10]},
+ index=[8, 9, 10, 11],
+ )
- output = smart_explainer.plot.correlations(df, max_features=3, facet_col='C')
+ output = smart_explainer.plot.correlations(df, max_features=3, facet_col="C")
assert len(output.data) == 2
assert len(output.data[0].x) == 3
@@ -2059,7 +1755,7 @@ def test_correlations_2(self):
def test_stability_plot_1(self):
np.random.seed(42)
- df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
model = DecisionTreeRegressor().fit(X, y)
@@ -2076,7 +1772,7 @@ def test_stability_plot_1(self):
def test_stability_plot_2(self):
np.random.seed(42)
- df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
model = DecisionTreeRegressor().fit(X, y)
@@ -2097,7 +1793,7 @@ def test_stability_plot_2(self):
def test_stability_plot_3(self):
np.random.seed(79)
- df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
model = DecisionTreeRegressor().fit(X, y)
@@ -2117,7 +1813,7 @@ def test_stability_plot_3(self):
def test_stability_plot_4(self):
np.random.seed(79)
- df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
model = DecisionTreeRegressor().fit(X, y)
@@ -2138,7 +1834,7 @@ def test_stability_plot_4(self):
def test_stability_plot_5(self):
np.random.seed(79)
- df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
model = DecisionTreeRegressor().fit(X, y)
@@ -2157,7 +1853,7 @@ def test_stability_plot_5(self):
assert np.array(list(output.data[0].x)).dtype == "float"
def test_local_neighbors_plot(self):
- df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list("ABCD"))
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
model = DecisionTreeRegressor().fit(X, y)
@@ -2173,11 +1869,11 @@ def test_local_neighbors_plot(self):
assert actual_shape == expected_shape
assert np.array(list(output.data[0].x)).dtype == "float"
- @patch('shapash.explainer.smart_explainer.SmartExplainer.compute_features_compacity')
+ @patch("shapash.explainer.smart_explainer.SmartExplainer.compute_features_compacity")
def test_compacity_plot(self, compute_features_compacity):
compute_features_compacity.return_value = None
- selection = ['person_A', 'person_B']
+ selection = ["person_A", "person_B"]
approx = 0.9
nb_features = 5
@@ -2192,10 +1888,10 @@ def test_scatter_plot_prediction_1(self):
"""
Regression
"""
- df_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X_train = df_train.iloc[:, :-1]
y_train = df_train.iloc[:, -1]
- df_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X_test = df_test.iloc[:, :-1]
y_test = df_test.iloc[:, -1]
model = DecisionTreeRegressor().fit(X_train, y_train)
@@ -2216,10 +1912,10 @@ def test_scatter_plot_prediction_2(self):
"""
Regression
"""
- df_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X_train = df_train.iloc[:, :-1]
y_train = df_train.iloc[:, -1]
- df_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list('ABCD'))
+ df_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 4)), columns=list("ABCD"))
X_test = df_test.iloc[:, :-1]
y_test = df_test.iloc[:, -1]
model = DecisionTreeRegressor().fit(X_train, y_train)
@@ -2240,9 +1936,9 @@ def test_scatter_plot_prediction_3(self):
"""
Classification
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 2, size=(50, 1)))
- X_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list('ABC'))
+ X_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list("ABC"))
y_test = pd.DataFrame(np.random.randint(0, 2, size=(50, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2263,9 +1959,9 @@ def test_scatter_plot_prediction_4(self):
"""
Classification
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 2, size=(50, 1)))
- X_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list('ABC'))
+ X_test = pd.DataFrame(np.random.randint(0, 100, size=(50, 3)), columns=list("ABC"))
y_test = pd.DataFrame(np.random.randint(0, 2, size=(50, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2288,9 +1984,9 @@ def test_scatter_plot_prediction_5(self):
"""
Classification Multiclass
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(100, 1)))
- X_test = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list('ABC'))
+ X_test = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list("ABC"))
y_test = pd.DataFrame(np.random.randint(0, 3, size=(100, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2312,9 +2008,9 @@ def test_scatter_plot_prediction_6(self):
"""
Classification Multiclass
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(100, 1)))
- X_test = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list('ABC'))
+ X_test = pd.DataFrame(np.random.randint(0, 100, size=(100, 3)), columns=list("ABC"))
y_test = pd.DataFrame(np.random.randint(0, 3, size=(100, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2337,20 +2033,20 @@ def test_subset_sampling_1(self):
"""
test _subset_sampling
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(30, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
xpl.compile(x=X_train, y_target=y_train)
list_ind, addnote = xpl.plot._subset_sampling(max_points=10)
assert len(list_ind) == 10
- assert addnote == 'Length of random Subset: 10 (33%)'
+ assert addnote == "Length of random Subset: 10 (33%)"
def test_subset_sampling_2(self):
"""
test _subset_sampling
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(30, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2363,7 +2059,7 @@ def test_subset_sampling_3(self):
"""
test _subset_sampling
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(30, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2371,14 +2067,14 @@ def test_subset_sampling_3(self):
selection = list(range(10, 20))
list_ind, addnote = xpl.plot._subset_sampling(selection=selection)
assert len(list_ind) == 10
- assert addnote == 'Length of user-defined Subset: 10 (33%)'
+ assert addnote == "Length of user-defined Subset: 10 (33%)"
assert list_ind == selection
def test_subset_sampling_4(self):
"""
test _subset_sampling
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(30, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2386,14 +2082,14 @@ def test_subset_sampling_4(self):
selection = list(range(10, 20))
list_ind, addnote = xpl.plot._subset_sampling(selection=selection, max_points=50)
assert len(list_ind) == 10
- assert addnote == 'Length of user-defined Subset: 10 (33%)'
+ assert addnote == "Length of user-defined Subset: 10 (33%)"
assert list_ind == selection
def test_subset_sampling_5(self):
"""
test _subset_sampling
"""
- X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list('ABC'))
+ X_train = pd.DataFrame(np.random.randint(0, 100, size=(30, 3)), columns=list("ABC"))
y_train = pd.DataFrame(np.random.randint(0, 3, size=(30, 1)))
model = DecisionTreeClassifier().fit(X_train, y_train)
xpl = SmartExplainer(model=model)
@@ -2401,4 +2097,3 @@ def test_subset_sampling_5(self):
selection = np.array(list(range(10, 20)))
with self.assertRaises(ValueError):
list_ind, addnote = xpl.plot._subset_sampling(selection=selection, max_points=50)
-
diff --git a/tests/unit_tests/explainer/test_smart_predictor.py b/tests/unit_tests/explainer/test_smart_predictor.py
index f6a07bf8..83f97047 100644
--- a/tests/unit_tests/explainer/test_smart_predictor.py
+++ b/tests/unit_tests/explainer/test_smart_predictor.py
@@ -2,25 +2,28 @@
Unit test smart predictor
"""
-import unittest
-from shapash.explainer.smart_predictor import SmartPredictor
-from shapash import SmartExplainer
-from shapash.explainer.smart_state import SmartState
-from shapash.explainer.multi_decorator import MultiDecorator
-from shapash.backend import ShapBackend
import os
+import types
+import unittest
from os import path
from pathlib import Path
-import pandas as pd
-import numpy as np
+from unittest.mock import patch
+
import catboost as cb
-from catboost import Pool
import category_encoders as ce
-from unittest.mock import patch
-import types
-from sklearn.compose import ColumnTransformer
-import sklearn.preprocessing as skp
+import numpy as np
+import pandas as pd
import shap
+import sklearn.preprocessing as skp
+from catboost import Pool
+from sklearn.compose import ColumnTransformer
+
+from shapash import SmartExplainer
+from shapash.backend import ShapBackend
+from shapash.explainer.multi_decorator import MultiDecorator
+from shapash.explainer.smart_predictor import SmartPredictor
+from shapash.explainer.smart_state import SmartState
+
def init_sme_to_pickle_test():
"""
@@ -32,8 +35,8 @@ def init_sme_to_pickle_test():
[description]
"""
current = Path(path.abspath(__file__)).parent.parent.parent
- pkl_file = path.join(current, 'data/predictor.pkl')
- y_pred = pd.DataFrame(data=np.array([1, 2]), columns=['pred'])
+ pkl_file = path.join(current, "data/predictor.pkl")
+ y_pred = pd.DataFrame(data=np.array([1, 2]), columns=["pred"])
dataframe_x = pd.DataFrame([[1, 2, 4], [1, 2, 3]])
clf = cb.CatBoostClassifier(n_estimators=1).fit(dataframe_x, y_pred)
xpl = SmartExplainer(model=clf, features_dict={})
@@ -41,31 +44,31 @@ def init_sme_to_pickle_test():
predictor = xpl.to_smartpredictor()
return pkl_file, predictor
+
class TestSmartPredictor(unittest.TestCase):
"""
Unit test Smart Predictor class
"""
+
def setUp(self):
- df = pd.DataFrame(range(0, 5), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 2 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = ["S", "M", "S", "D", "M"]
- df = df.set_index('id')
+ df = pd.DataFrame(range(0, 5), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 2 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = ["S", "M", "S", "D", "M"]
+ df = df.set_index("id")
encoder = ce.OrdinalEncoder(cols=["x2"], handle_unknown="None")
- encoder_fitted = encoder.fit(df[['x1', 'x2']])
- df_encoded = encoder_fitted.transform(df[['x1', 'x2']])
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[['x1', 'x2']], df['y'])
+ encoder_fitted = encoder.fit(df[["x1", "x2"]])
+ df_encoded = encoder_fitted.transform(df[["x1", "x2"]])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df_encoded[["x1", "x2"]], df["y"])
backend = ShapBackend(model=clf)
columns_dict = {0: "x1", 1: "x2"}
label_dict = {0: "Yes", 1: "No"}
- postprocessing = {"x2": {
- "type": "transcoding",
- "rule": {"S": "single", "M": "married", "D": "divorced"}}}
+ postprocessing = {"x2": {"type": "transcoding", "rule": {"S": "single", "M": "married", "D": "divorced"}}}
features_dict = {"x1": "age", "x2": "family_situation"}
- features_types = {features: str(df[features].dtypes) for features in df[['x1', 'x2']]}
+ features_types = {features: str(df[features].dtypes) for features in df[["x1", "x2"]]}
self.df_1 = df.copy()
self.preprocessing_1 = encoder_fitted
@@ -78,22 +81,30 @@ def setUp(self):
self.features_dict_1 = features_dict
self.features_types_1 = features_types
- self.predictor_1 = SmartPredictor(features_dict, clf,
- columns_dict, backend, features_types, label_dict,
- encoder_fitted, postprocessing)
+ self.predictor_1 = SmartPredictor(
+ features_dict, clf, columns_dict, backend, features_types, label_dict, encoder_fitted, postprocessing
+ )
- self.features_groups = {'group1': ['x1', 'x2']}
+ self.features_groups = {"group1": ["x1", "x2"]}
self.features_dict_w_groups = {"x1": "age", "x2": "weight", "group1": "group1"}
- self.predictor_1_w_groups = SmartPredictor(self.features_dict_w_groups, clf,
- columns_dict, backend, features_types, label_dict,
- encoder_fitted, postprocessing, self.features_groups)
+ self.predictor_1_w_groups = SmartPredictor(
+ self.features_dict_w_groups,
+ clf,
+ columns_dict,
+ backend,
+ features_types,
+ label_dict,
+ encoder_fitted,
+ postprocessing,
+ self.features_groups,
+ )
self.predictor_1.backend.state = SmartState()
- df['x2'] = np.random.randint(1, 100, df.shape[0])
+ df["x2"] = np.random.randint(1, 100, df.shape[0])
encoder = ce.OrdinalEncoder(cols=["x2"], handle_unknown="None")
encoder_fitted = encoder.fit(df[["x1", "x2"]])
df_encoded = encoder_fitted.transform(df[["x1", "x2"]])
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
backend = ShapBackend(model=clf)
features_dict = {"x1": "age", "x2": "weight"}
features_types = {features: str(df[features].dtypes) for features in df[["x1", "x2"]].columns}
@@ -109,21 +120,21 @@ def setUp(self):
self.features_dict_2 = features_dict
self.features_types_2 = features_types
- self.predictor_2 = SmartPredictor(features_dict, clf,
- columns_dict, backend, features_types, label_dict,
- encoder_fitted, postprocessing)
+ self.predictor_2 = SmartPredictor(
+ features_dict, clf, columns_dict, backend, features_types, label_dict, encoder_fitted, postprocessing
+ )
self.predictor_2.backend.state = SmartState()
- df['x1'] = [25, 39, 50, 43, 67]
- df['x2'] = [90, 78, 84, 85, 53]
+ df["x1"] = [25, 39, 50, 43, 67]
+ df["x2"] = [90, 78, 84, 85, 53]
columns_dict = {0: "x1", 1: "x2"}
label_dict = {0: "No", 1: "Yes"}
features_dict = {"x1": "age", "x2": "weight"}
- features_types = {features: str(df[features].dtypes) for features in df[['x1', 'x2']].columns}
+ features_types = {features: str(df[features].dtypes) for features in df[["x1", "x2"]].columns}
- clf = cb.CatBoostRegressor(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ clf = cb.CatBoostRegressor(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
backend_3 = ShapBackend(model=clf)
self.df_3 = df.copy()
@@ -137,72 +148,62 @@ def setUp(self):
self.features_dict_3 = features_dict
self.features_types_3 = features_types
- self.predictor_3 = SmartPredictor(features_dict, clf,
- columns_dict, backend,
- features_types, label_dict)
+ self.predictor_3 = SmartPredictor(features_dict, clf, columns_dict, backend, features_types, label_dict)
self.predictor_3.backend.state = SmartState()
def predict_proba(self, arg1, arg2):
"""
predict_proba method
"""
- matrx = np.array(
- [[0.2, 0.8],
- [0.3, 0.7],
- [0.4, 0.6]]
- )
+ matrx = np.array([[0.2, 0.8], [0.3, 0.7], [0.4, 0.6]])
return matrx
def predict(self, arg1, arg2):
"""
predict method
"""
- matrx = np.array(
- [12, 3, 7]
- )
+ matrx = np.array([12, 3, 7])
return matrx
def test_init_1(self):
"""
Test init smart predictor
"""
- predictor_1 = SmartPredictor(self.features_dict_1,
- self.clf_1,
- self.columns_dict_1,
- self.backend_1,
- self.features_types_1,
- self.label_dict_1,
- self.preprocessing_1,
- self.postprocessing_1)
-
- assert hasattr(predictor_1, 'model')
- assert hasattr(predictor_1, 'backend')
- assert hasattr(predictor_1, 'features_dict')
- assert hasattr(predictor_1, 'label_dict')
- assert hasattr(predictor_1, '_case')
- assert hasattr(predictor_1, '_classes')
- assert hasattr(predictor_1, 'columns_dict')
- assert hasattr(predictor_1, 'features_types')
- assert hasattr(predictor_1, 'preprocessing')
- assert hasattr(predictor_1, 'postprocessing')
- assert hasattr(predictor_1, 'mask_params')
- assert hasattr(predictor_1, 'features_groups')
+ predictor_1 = SmartPredictor(
+ self.features_dict_1,
+ self.clf_1,
+ self.columns_dict_1,
+ self.backend_1,
+ self.features_types_1,
+ self.label_dict_1,
+ self.preprocessing_1,
+ self.postprocessing_1,
+ )
+
+ assert hasattr(predictor_1, "model")
+ assert hasattr(predictor_1, "backend")
+ assert hasattr(predictor_1, "features_dict")
+ assert hasattr(predictor_1, "label_dict")
+ assert hasattr(predictor_1, "_case")
+ assert hasattr(predictor_1, "_classes")
+ assert hasattr(predictor_1, "columns_dict")
+ assert hasattr(predictor_1, "features_types")
+ assert hasattr(predictor_1, "preprocessing")
+ assert hasattr(predictor_1, "postprocessing")
+ assert hasattr(predictor_1, "mask_params")
+ assert hasattr(predictor_1, "features_groups")
assert predictor_1.model == self.clf_1
assert predictor_1.backend == self.backend_1
assert predictor_1.features_dict == self.features_dict_1
assert predictor_1.label_dict == self.label_dict_1
assert predictor_1._case == "classification"
- assert predictor_1._classes == [0,1]
+ assert predictor_1._classes == [0, 1]
assert predictor_1.columns_dict == self.columns_dict_1
assert predictor_1.preprocessing == self.preprocessing_1
assert predictor_1.postprocessing == self.postprocessing_1
- mask_params = {'features_to_hide': None,
- 'threshold': None,
- 'positive': True,
- 'max_contrib': 1
- }
+ mask_params = {"features_to_hide": None, "threshold": None, "positive": True, "max_contrib": 1}
predictor_1.mask_params = mask_params
assert predictor_1.mask_params == mask_params
@@ -210,28 +211,30 @@ def test_init_2(self):
"""
Test init smart predictor with groups of features
"""
- predictor_1 = SmartPredictor(self.features_dict_w_groups,
- self.clf_1,
- self.columns_dict_1,
- self.backend_1,
- self.features_types_1,
- self.label_dict_1,
- self.preprocessing_1,
- self.postprocessing_1,
- self.features_groups)
-
- assert hasattr(predictor_1, 'model')
- assert hasattr(predictor_1, 'backend')
- assert hasattr(predictor_1, 'features_dict')
- assert hasattr(predictor_1, 'label_dict')
- assert hasattr(predictor_1, '_case')
- assert hasattr(predictor_1, '_classes')
- assert hasattr(predictor_1, 'columns_dict')
- assert hasattr(predictor_1, 'features_types')
- assert hasattr(predictor_1, 'preprocessing')
- assert hasattr(predictor_1, 'postprocessing')
- assert hasattr(predictor_1, 'mask_params')
- assert hasattr(predictor_1, 'features_groups')
+ predictor_1 = SmartPredictor(
+ self.features_dict_w_groups,
+ self.clf_1,
+ self.columns_dict_1,
+ self.backend_1,
+ self.features_types_1,
+ self.label_dict_1,
+ self.preprocessing_1,
+ self.postprocessing_1,
+ self.features_groups,
+ )
+
+ assert hasattr(predictor_1, "model")
+ assert hasattr(predictor_1, "backend")
+ assert hasattr(predictor_1, "features_dict")
+ assert hasattr(predictor_1, "label_dict")
+ assert hasattr(predictor_1, "_case")
+ assert hasattr(predictor_1, "_classes")
+ assert hasattr(predictor_1, "columns_dict")
+ assert hasattr(predictor_1, "features_types")
+ assert hasattr(predictor_1, "preprocessing")
+ assert hasattr(predictor_1, "postprocessing")
+ assert hasattr(predictor_1, "mask_params")
+ assert hasattr(predictor_1, "features_groups")
assert predictor_1.model == self.clf_1
assert predictor_1.backend == self.backend_1
@@ -243,12 +246,7 @@ def test_init_2(self):
assert predictor_1.preprocessing == self.preprocessing_1
assert predictor_1.postprocessing == self.postprocessing_1
- mask_params = {
- 'features_to_hide': None,
- 'threshold': None,
- 'positive': True,
- 'max_contrib': 1
- }
+ mask_params = {"features_to_hide": None, "threshold": None, "positive": True, "max_contrib": 1}
predictor_1.mask_params = mask_params
assert predictor_1.mask_params == mask_params
@@ -256,18 +254,18 @@ def test_add_input_1(self):
"""
Test add_input method from smart predictor
"""
- ypred = self.df_1['y']
+ ypred = self.df_1["y"]
shap_values = self.clf_1.get_feature_importance(Pool(self.df_encoded_1), type="ShapValues")
predictor_1 = self.predictor_1
predictor_1.add_input(x=self.df_1[["x1", "x2"]], contributions=shap_values[:, :-1])
predictor_1_contrib = predictor_1.data["contributions"]
- assert all(attribute in predictor_1.data.keys()
- for attribute in ["x", "x_preprocessed", "contributions", "ypred"])
+ assert all(
+ attribute in predictor_1.data.keys() for attribute in ["x", "x_preprocessed", "contributions", "ypred"]
+ )
assert predictor_1.data["x"].shape == predictor_1.data["x_preprocessed"].shape
- assert all(feature in predictor_1.data["x"].columns
- for feature in predictor_1.data["x_preprocessed"].columns)
+ assert all(feature in predictor_1.data["x"].columns for feature in predictor_1.data["x_preprocessed"].columns)
assert predictor_1_contrib.shape == predictor_1.data["x"].shape
predictor_1.add_input(ypred=ypred)
@@ -280,23 +278,25 @@ def test_add_input_2(self):
"""
Test add_input method from smart predictor with groups of features
"""
- ypred = self.df_1['y']
+ ypred = self.df_1["y"]
shap_values = self.clf_1.get_feature_importance(Pool(self.df_encoded_1), type="ShapValues")
predictor_1 = self.predictor_1_w_groups
predictor_1.add_input(x=self.df_1[["x1", "x2"]], contributions=shap_values[:, :-1])
predictor_1_contrib = predictor_1.data["contributions"]
- assert all(attribute in predictor_1.data.keys()
- for attribute in ["x", "x_preprocessed", "x_postprocessed", "contributions", "ypred"])
+ assert all(
+ attribute in predictor_1.data.keys()
+ for attribute in ["x", "x_preprocessed", "x_postprocessed", "contributions", "ypred"]
+ )
- assert hasattr(predictor_1, 'data_groups')
- assert all(attribute in predictor_1.data_groups.keys()
- for attribute in ["x_postprocessed", "contributions", "ypred"])
+ assert hasattr(predictor_1, "data_groups")
+ assert all(
+ attribute in predictor_1.data_groups.keys() for attribute in ["x_postprocessed", "contributions", "ypred"]
+ )
assert predictor_1.data["x"].shape == predictor_1.data["x_preprocessed"].shape
- assert all(feature in predictor_1.data["x"].columns
- for feature in predictor_1.data["x_preprocessed"].columns)
+ assert all(feature in predictor_1.data["x"].columns for feature in predictor_1.data["x_preprocessed"].columns)
assert predictor_1_contrib.shape == predictor_1.data["x"].shape
predictor_1.add_input(ypred=ypred)
@@ -307,9 +307,11 @@ def test_add_input_2(self):
print(predictor_1.data["contributions"].sum(axis=1).values)
- print(predictor_1.data_groups['contributions'])
+ print(predictor_1.data_groups["contributions"])
- assert all(predictor_1.data_groups['contributions']['group1'].values == predictor_1.data["contributions"].sum(axis=1))
+ assert all(
+ predictor_1.data_groups["contributions"]["group1"].values == predictor_1.data["contributions"].sum(axis=1)
+ )
def test_check_contributions(self):
"""
@@ -325,16 +327,26 @@ def test_check_contributions(self):
predictor_1.data["x_preprocessed"] = self.df_2[["x1", "x2"]]
predictor_1.data["ypred"] = self.df_2["y"]
- adapt_contrib = [np.array([[-0.04395604, 0.13186813],
- [-0.04395604, 0.13186813],
- [-0.0021978, 0.01318681],
- [-0.0021978, 0.01318681],
- [-0.04395604, 0.13186813]]),
- np.array([[0.04395604, -0.13186813],
- [0.04395604, -0.13186813],
- [0.0021978, -0.01318681],
- [0.0021978, -0.01318681],
- [0.04395604, -0.13186813]])]
+ adapt_contrib = [
+ np.array(
+ [
+ [-0.04395604, 0.13186813],
+ [-0.04395604, 0.13186813],
+ [-0.0021978, 0.01318681],
+ [-0.0021978, 0.01318681],
+ [-0.04395604, 0.13186813],
+ ]
+ ),
+ np.array(
+ [
+ [0.04395604, -0.13186813],
+ [0.04395604, -0.13186813],
+ [0.0021978, -0.01318681],
+ [0.0021978, -0.01318681],
+ [0.04395604, -0.13186813],
+ ]
+ ),
+ ]
contributions = list()
for element in adapt_contrib:
contributions.append(pd.DataFrame(element, columns=["x1", "x2"]))
@@ -357,7 +369,7 @@ def test_check_model_1(self):
predictor_1.model = model
_case, _classes = predictor_1.check_model()
- assert _case == 'regression'
+ assert _case == "regression"
assert _classes is None
def test_check_model_2(self):
@@ -375,37 +387,58 @@ def test_check_model_2(self):
predictor_1.model = model
_case, _classes = predictor_1.check_model()
- assert _case == 'classification'
+ assert _case == "classification"
self.assertListEqual(_classes, [1, 2])
- @patch('shapash.explainer.smart_predictor.SmartPredictor.check_model')
- @patch('shapash.utils.check.check_preprocessing_options')
- @patch('shapash.utils.check.check_consistency_model_features')
- @patch('shapash.utils.check.check_consistency_model_label')
- def test_check_preprocessing_1(self, check_consistency_model_label,
- check_consistency_model_features,
- check_preprocessing_options,
- check_model):
+ @patch("shapash.explainer.smart_predictor.SmartPredictor.check_model")
+ @patch("shapash.utils.check.check_preprocessing_options")
+ @patch("shapash.utils.check.check_consistency_model_features")
+ @patch("shapash.utils.check.check_consistency_model_label")
+ def test_check_preprocessing_1(
+ self, check_consistency_model_label, check_consistency_model_features, check_preprocessing_options, check_model
+ ):
"""
Test check preprocessing on multiple preprocessing
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
features_dict = None
- columns_dict = {i:features for i,features in enumerate(train.columns)}
+ columns_dict = {i: features for i, features in enumerate(train.columns)}
features_types = {features: str(train[features].dtypes) for features in train.columns}
label_dict = None
- enc_ordinal_all = ce.OrdinalEncoder(cols=['Onehot1', 'Onehot2', 'Binary1', 'Binary2', 'Ordinal1', 'Ordinal2',
- 'BaseN1', 'BaseN2', 'Target1', 'Target2', 'other']).fit(train)
- train_ordinal_all = enc_ordinal_all.transform(train)
-
- y = pd.DataFrame({'y_class': [0, 0, 0, 1]})
+ enc_ordinal_all = ce.OrdinalEncoder(
+ cols=[
+ "Onehot1",
+ "Onehot2",
+ "Binary1",
+ "Binary2",
+ "Ordinal1",
+ "Ordinal2",
+ "BaseN1",
+ "BaseN2",
+ "Target1",
+ "Target2",
+ "other",
+ ]
+ ).fit(train)
+ train_ordinal_all = enc_ordinal_all.transform(train)
+
+ y = pd.DataFrame({"y_class": [0, 0, 0, 1]})
model = cb.CatBoostClassifier(n_estimators=1).fit(train_ordinal_all, y)
backend = ShapBackend(model=model)
@@ -415,59 +448,50 @@ def test_check_preprocessing_1(self, check_consistency_model_label,
check_consistency_model_label.return_value = True
check_model.return_value = "classification", [0, 1]
- predictor_1 = SmartPredictor(features_dict, model,
- columns_dict, backend, features_types, label_dict)
-
+ predictor_1 = SmartPredictor(features_dict, model, columns_dict, backend, features_types, label_dict)
- y = pd.DataFrame(data=[0, 1, 0, 0], columns=['y'])
+ y = pd.DataFrame(data=[0, 1, 0, 0], columns=["y"])
- enc_onehot = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2']).fit(train)
+ enc_onehot = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"]).fit(train)
train_onehot = enc_onehot.transform(train)
- enc_binary = ce.BinaryEncoder(cols=['Binary1', 'Binary2']).fit(train_onehot)
+ enc_binary = ce.BinaryEncoder(cols=["Binary1", "Binary2"]).fit(train_onehot)
train_binary = enc_binary.transform(train_onehot)
- enc_ordinal = ce.OrdinalEncoder(cols=['Ordinal1', 'Ordinal2']).fit(train_binary)
+ enc_ordinal = ce.OrdinalEncoder(cols=["Ordinal1", "Ordinal2"]).fit(train_binary)
train_ordinal = enc_ordinal.transform(train_binary)
- enc_basen = ce.BaseNEncoder(cols=['BaseN1', 'BaseN2']).fit(train_ordinal)
+ enc_basen = ce.BaseNEncoder(cols=["BaseN1", "BaseN2"]).fit(train_ordinal)
train_basen = enc_basen.transform(train_ordinal)
- enc_target = ce.TargetEncoder(cols=['Target1', 'Target2']).fit(train_basen, y)
+ enc_target = ce.TargetEncoder(cols=["Target1", "Target2"]).fit(train_basen, y)
input_dict1 = dict()
- input_dict1['col'] = 'Onehot2'
- input_dict1['mapping'] = pd.Series(data=['C', 'D', np.nan], index=['C', 'D', 'missing'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "Onehot2"
+ input_dict1["mapping"] = pd.Series(data=["C", "D", np.nan], index=["C", "D", "missing"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'Binary2'
- input_dict2['mapping'] = pd.Series(data=['G', 'H', np.nan], index=['G', 'H', 'missing'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "Binary2"
+ input_dict2["mapping"] = pd.Series(data=["G", "H", np.nan], index=["G", "H", "missing"])
+ input_dict2["data_type"] = "object"
input_dict = dict()
- input_dict['col'] = 'state'
- input_dict['mapping'] = pd.Series(data=['US', 'FR-1', 'FR-2'], index=['US', 'FR', 'FR'])
- input_dict['data_type'] = 'object'
+ input_dict["col"] = "state"
+ input_dict["mapping"] = pd.Series(data=["US", "FR-1", "FR-2"], index=["US", "FR", "FR"])
+ input_dict["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'Ordinal2'
- input_dict3['mapping'] = pd.Series(data=['K', 'L', np.nan], index=['K', 'L', 'missing'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "Ordinal2"
+ input_dict3["mapping"] = pd.Series(data=["K", "L", np.nan], index=["K", "L", "missing"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
- enc = ColumnTransformer(
- transformers=[
- ('onehot', skp.OneHotEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
+ enc = ColumnTransformer(transformers=[("onehot", skp.OneHotEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
wrong_prepro = skp.OneHotEncoder().fit(train, y)
- predictor_1.preprocessing = [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1,
- list_dict]
+ predictor_1.preprocessing = [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1, list_dict]
predictor_1.check_preprocessing()
for preprocessing in [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target]:
@@ -502,37 +526,58 @@ def test_check_label_dict_2(self):
predictor_1 = self.predictor_1
predictor_1.label_dict = None
- predictor_1._case = 'regression'
+ predictor_1._case = "regression"
predictor_1.check_label_dict()
- @patch('shapash.explainer.smart_predictor.SmartPredictor.check_model')
- @patch('shapash.utils.check.check_preprocessing_options')
- @patch('shapash.utils.check.check_consistency_model_features')
- @patch('shapash.utils.check.check_consistency_model_label')
- def test_check_mask_params(self, check_consistency_model_label,
- check_consistency_model_features,
- check_preprocessing_options,
- check_model):
+ @patch("shapash.explainer.smart_predictor.SmartPredictor.check_model")
+ @patch("shapash.utils.check.check_preprocessing_options")
+ @patch("shapash.utils.check.check_consistency_model_features")
+ @patch("shapash.utils.check.check_consistency_model_label")
+ def test_check_mask_params(
+ self, check_consistency_model_label, check_consistency_model_features, check_preprocessing_options, check_model
+ ):
"""
Unit test check mask params
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
features_dict = None
columns_dict = {i: features for i, features in enumerate(train.columns)}
features_types = {features: str(train[features].dtypes) for features in train.columns}
label_dict = None
- enc_ordinal = ce.OrdinalEncoder(cols=['Onehot1', 'Onehot2', 'Binary1', 'Binary2', 'Ordinal1', 'Ordinal2',
- 'BaseN1', 'BaseN2', 'Target1', 'Target2', 'other']).fit(train)
+ enc_ordinal = ce.OrdinalEncoder(
+ cols=[
+ "Onehot1",
+ "Onehot2",
+ "Binary1",
+ "Binary2",
+ "Ordinal1",
+ "Ordinal2",
+ "BaseN1",
+ "BaseN2",
+ "Target1",
+ "Target2",
+ "other",
+ ]
+ ).fit(train)
train_ordinal = enc_ordinal.transform(train)
- y = pd.DataFrame({'y_class': [0, 0, 0, 1]})
+ y = pd.DataFrame({"y_class": [0, 0, 0, 1]})
model = cb.CatBoostClassifier(n_estimators=1).fit(train_ordinal, y)
backend = ShapBackend(model=model)
@@ -544,31 +589,22 @@ def test_check_mask_params(self, check_consistency_model_label,
wrong_mask_params_1 = list()
wrong_mask_params_2 = None
- wrong_mask_params_3 = {
- "features_to_hide": None,
- "threshold": None,
- "positive": None
- }
- wright_mask_params = {
- "features_to_hide": None,
- "threshold": None,
- "positive": True,
- "max_contrib": 5
- }
+ wrong_mask_params_3 = {"features_to_hide": None, "threshold": None, "positive": None}
+ wright_mask_params = {"features_to_hide": None, "threshold": None, "positive": True, "max_contrib": 5}
with self.assertRaises(ValueError):
- predictor_1 = SmartPredictor(features_dict, model,
- columns_dict, backend, features_types, label_dict,
- mask_params=wrong_mask_params_1)
- predictor_1 = SmartPredictor(features_dict, model,
- columns_dict, backend, features_types, label_dict,
- mask_params=wrong_mask_params_2)
- predictor_1 = SmartPredictor(features_dict, model,
- columns_dict, backend, features_types, label_dict,
- mask_params=wrong_mask_params_3)
-
- predictor_1 = SmartPredictor(features_dict, model,
- columns_dict, backend, features_types, label_dict,
- mask_params=wright_mask_params)
+ predictor_1 = SmartPredictor(
+ features_dict, model, columns_dict, backend, features_types, label_dict, mask_params=wrong_mask_params_1
+ )
+ predictor_1 = SmartPredictor(
+ features_dict, model, columns_dict, backend, features_types, label_dict, mask_params=wrong_mask_params_2
+ )
+ predictor_1 = SmartPredictor(
+ features_dict, model, columns_dict, backend, features_types, label_dict, mask_params=wrong_mask_params_3
+ )
+
+ predictor_1 = SmartPredictor(
+ features_dict, model, columns_dict, backend, features_types, label_dict, mask_params=wright_mask_params
+ )
def test_check_ypred_1(self):
"""
@@ -576,9 +612,8 @@ def test_check_ypred_1(self):
"""
predictor_1 = self.predictor_1
-
predictor_1.data = {"x": None, "ypred": None, "contributions": None}
- predictor_1.data["x"] = self.df_1[["x1","x2"]]
+ predictor_1.data["x"] = self.df_1[["x1", "x2"]]
y_pred = None
predictor_1.check_ypred(ypred=y_pred)
@@ -586,10 +621,7 @@ def test_check_ypred_2(self):
"""
Unit test check y pred 2
"""
- y_pred = pd.DataFrame(
- data=np.array(['1', 0, 0, 1, 0]),
- columns=['Y']
- )
+ y_pred = pd.DataFrame(data=np.array(["1", 0, 0, 1, 0]), columns=["Y"])
predictor_1 = self.predictor_1
predictor_1.data = {"x": None, "ypred": None, "contributions": None}
predictor_1.data["x"] = self.df_1
@@ -604,11 +636,8 @@ def test_check_ypred_3(self):
predictor_1 = self.predictor_1
predictor_1.data = {"x": None, "ypred": None, "contributions": None}
- predictor_1.data["x"] = self.df_1[["x1","x2"]]
- y_pred = pd.DataFrame(
- data=np.array([0]),
- columns=['Y']
- )
+ predictor_1.data["x"] = self.df_1[["x1", "x2"]]
+ y_pred = pd.DataFrame(data=np.array([0]), columns=["Y"])
with self.assertRaises(ValueError):
predictor_1.check_ypred(y_pred)
@@ -629,11 +658,9 @@ def test_check_ypred_5(self):
"""
predictor_1 = self.predictor_1
predictor_1.data = {"x": None, "ypred": None, "contributions": None}
- predictor_1.data["x"] = self.df_1[["x1","x2"]]
+ predictor_1.data["x"] = self.df_1[["x1", "x2"]]
- y_pred = pd.Series(
- data=np.array(['0'])
- )
+ y_pred = pd.Series(data=np.array(["0"]))
with self.assertRaises(ValueError):
predictor_1.check_ypred(y_pred)
@@ -643,7 +670,7 @@ def test_predict_proba_1(self):
"""
predictor_1 = self.predictor_1
- clf = cb.CatBoostRegressor(n_estimators=1).fit(self.df_encoded_1[['x1', 'x2']], self.df_1['y'])
+ clf = cb.CatBoostRegressor(n_estimators=1).fit(self.df_encoded_1[["x1", "x2"]], self.df_1["y"])
backend = ShapBackend(model=clf)
predictor_1.model = clf
predictor_1.backend = backend
@@ -667,15 +694,14 @@ def test_predict_proba_2(self):
"""
Unit test of predict_proba method.
"""
- clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_2[['x1', 'x2']], self.df_2['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_2[["x1", "x2"]], self.df_2["y"])
predictor_1 = self.predictor_2
predictor_1.model = clf
predictor_1.backend = ShapBackend(model=clf)
predictor_1.preprocessing = None
-
- predictor_1.data = {"x": None, "ypred": None, "contributions": None, "x_preprocessed":None}
+ predictor_1.data = {"x": None, "ypred": None, "contributions": None, "x_preprocessed": None}
predictor_1.data["x"] = self.df_2[["x1", "x2"]]
predictor_1.data["x_preprocessed"] = self.df_2[["x1", "x2"]]
@@ -710,13 +736,13 @@ def test_detail_contributions_2(self):
"""
Unit test 2 of detail_contributions method.
"""
- clf = cb.CatBoostRegressor(n_estimators=1).fit(self.df_2[['x1', 'x2']], self.df_2['y'])
+ clf = cb.CatBoostRegressor(n_estimators=1).fit(self.df_2[["x1", "x2"]], self.df_2["y"])
predictor_1 = self.predictor_1
predictor_1.model = clf
predictor_1.backend = ShapBackend(model=clf)
predictor_1.preprocessing = None
- predictor_1.backend._case = 'regression'
+ predictor_1.backend._case = "regression"
predictor_1._case = "regression"
predictor_1._classes = None
@@ -731,7 +757,7 @@ def test_detail_contributions_2(self):
assert all(contributions.index == predictor_1.data["x"].index)
assert contributions.shape[1] == predictor_1.data["x"].shape[1] + 1
- clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_2[['x1', 'x2']], self.df_2['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_2[["x1", "x2"]], self.df_2["y"])
backend = ShapBackend(model=clf)
predictor_1 = self.predictor_2
@@ -767,22 +793,21 @@ def test_save_1(self):
assert path.exists(pkl_file)
os.remove(pkl_file)
- @patch('shapash.explainer.smart_predictor.SmartPredictor.check_model')
- @patch('shapash.utils.check.check_preprocessing_options')
- @patch('shapash.utils.check.check_consistency_model_features')
- @patch('shapash.utils.check.check_consistency_model_label')
- def test_apply_preprocessing_1(self,check_consistency_model_label,
- check_consistency_model_features,
- check_preprocessing_options,
- check_model ):
+ @patch("shapash.explainer.smart_predictor.SmartPredictor.check_model")
+ @patch("shapash.utils.check.check_preprocessing_options")
+ @patch("shapash.utils.check.check_consistency_model_features")
+ @patch("shapash.utils.check.check_consistency_model_label")
+ def test_apply_preprocessing_1(
+ self, check_consistency_model_label, check_consistency_model_features, check_preprocessing_options, check_model
+ ):
"""
Unit test for apply preprocessing method
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2]})
- enc = ColumnTransformer(transformers=[('power', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='passthrough')
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2]})
+ enc = ColumnTransformer(
+ transformers=[("power", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="passthrough"
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train), index=train.index)
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
@@ -798,10 +823,8 @@ def test_apply_preprocessing_1(self,check_consistency_model_label,
check_consistency_model_label.return_value = True
check_model.return_value = "classification", [0, 1]
- predictor_1 = SmartPredictor(features_dict, clf,
- columns_dict, backend,
- features_types, label_dict, enc)
- predictor_1.data = {"x":None}
+ predictor_1 = SmartPredictor(features_dict, clf, columns_dict, backend, features_types, label_dict, enc)
+ predictor_1.data = {"x": None}
predictor_1.data["x"] = train
predictor_1.data["x_preprocessed"] = predictor_1.apply_preprocessing()
@@ -809,47 +832,55 @@ def test_apply_preprocessing_1(self,check_consistency_model_label,
assert output_preprocessed.shape == train_preprocessed.shape
assert [column in clf.feature_names_ for column in output_preprocessed.columns]
assert all(train.index == output_preprocessed.index)
- assert all([str(type_result) == str(train_preprocessed.dtypes[index])
- for index, type_result in enumerate(output_preprocessed.dtypes)])
+ assert all(
+ [
+ str(type_result) == str(train_preprocessed.dtypes[index])
+ for index, type_result in enumerate(output_preprocessed.dtypes)
+ ]
+ )
def test_summarize_1(self):
"""
Unit test 1 summarize method
"""
- clf = cb.CatBoostRegressor(n_estimators=1).fit(self.df_3[['x1', 'x2']], self.df_3['y'])
+ clf = cb.CatBoostRegressor(n_estimators=1).fit(self.df_3[["x1", "x2"]], self.df_3["y"])
backend = ShapBackend(model=clf)
predictor_1 = self.predictor_3
predictor_1.model = clf
predictor_1.backend = backend
- predictor_1.data = {"x": None,
- "x_preprocessed": None,
- "x_postprocessed": None,
- "ypred": None,
- "contributions": None}
-
- predictor_1.data["x"] = self.df_3[['x1', 'x2']]
- predictor_1.data["x_postprocessed"] = self.df_3[['x1', 'x2']]
- predictor_1.data["x_preprocessed"] = self.df_3[['x1', 'x2']]
+ predictor_1.data = {
+ "x": None,
+ "x_preprocessed": None,
+ "x_postprocessed": None,
+ "ypred": None,
+ "contributions": None,
+ }
+
+ predictor_1.data["x"] = self.df_3[["x1", "x2"]]
+ predictor_1.data["x_postprocessed"] = self.df_3[["x1", "x2"]]
+ predictor_1.data["x_preprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["ypred"] = self.df_3["y"]
- contribution = pd.DataFrame([[0.0, 0.094286],
- [0.0, -0.023571],
- [0.0, -0.023571],
- [0.0, -0.023571],
- [0.0, -0.023571]], columns=["x1", "x2"])
+ contribution = pd.DataFrame(
+ [[0.0, 0.094286], [0.0, -0.023571], [0.0, -0.023571], [0.0, -0.023571], [0.0, -0.023571]],
+ columns=["x1", "x2"],
+ )
predictor_1.data["contributions"] = contribution
output = predictor_1.summarize()
print(output)
- expected_output = pd.DataFrame({
- "y": [1, 1, 0, 0, 0],
- "feature_1": ["weight", "weight", "weight", "weight", "weight"],
- "value_1": ["90", "78", "84", "85", "53"],
- "contribution_1": ["0.0942857", "-0.0235714", "-0.0235714", "-0.0235714", "-0.0235714"],
- "feature_2": ["age", "age", "age", "age", "age"],
- "value_2": ["25", "39", "50", "43", "67"],
- "contribution_2": ["0", "0", "0", "0", "0"]
- }, dtype=object)
+ expected_output = pd.DataFrame(
+ {
+ "y": [1, 1, 0, 0, 0],
+ "feature_1": ["weight", "weight", "weight", "weight", "weight"],
+ "value_1": ["90", "78", "84", "85", "53"],
+ "contribution_1": ["0.0942857", "-0.0235714", "-0.0235714", "-0.0235714", "-0.0235714"],
+ "feature_2": ["age", "age", "age", "age", "age"],
+ "value_2": ["25", "39", "50", "43", "67"],
+ "contribution_2": ["0", "0", "0", "0", "0"],
+ },
+ dtype=object,
+ )
expected_output["y"] = expected_output["y"].astype(int)
feature_expected = [column for column in expected_output.columns if column.startswith("feature_")]
@@ -873,7 +904,7 @@ def test_summarize_2(self):
predictor_1 = self.predictor_3
predictor_1._case = "classification"
predictor_1._classes = [0, 1]
- clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_3[['x1', 'x2']], self.df_3['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_3[["x1", "x2"]], self.df_3["y"])
backend = ShapBackend(model=clf)
predictor_1.model = clf
predictor_1.backend = backend
@@ -881,36 +912,39 @@ def test_summarize_2(self):
with self.assertRaises(ValueError):
predictor_1.summarize()
- predictor_1.data = {"x": None,
- "x_preprocessed": None,
- "x_postprocessed": None,
- "ypred": None,
- "contributions": None}
+ predictor_1.data = {
+ "x": None,
+ "x_preprocessed": None,
+ "x_postprocessed": None,
+ "ypred": None,
+ "contributions": None,
+ }
predictor_1.data["x"] = self.df_3[["x1", "x2"]]
predictor_1.data["x_preprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["x_postprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["ypred"] = pd.DataFrame(
- {"y": ["Yes", "Yes", "No", "No", "No"],
- "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209]}
- )
+ {"y": ["Yes", "Yes", "No", "No", "No"], "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209]}
+ )
predictor_1.data["contributions"] = pd.DataFrame(
- {"x1": [0, 0, -0, -0, -0],
- "x2": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
- )
+ {"x1": [0, 0, -0, -0, -0], "x2": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
+ )
output = predictor_1.summarize()
- expected_output = pd.DataFrame({
- "y": ["Yes", "Yes", "No", "No", "No"],
- "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209],
- "feature_1": ["weight", "weight", "weight", "weight", "weight"],
- "value_1": ["90", "78", "84", "85", "53"],
- "contribution_1": ["0.161538", "-0.0403846", "0.0403846", "0.0403846", "0.0403846"],
- "feature_2": ["age", "age", "age", "age", "age"],
- "value_2": ["25", "39", "50", "43", "67"],
- "contribution_2": ["0", "0", "0", "0", "0"]
- }, dtype=object)
+ expected_output = pd.DataFrame(
+ {
+ "y": ["Yes", "Yes", "No", "No", "No"],
+ "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209],
+ "feature_1": ["weight", "weight", "weight", "weight", "weight"],
+ "value_1": ["90", "78", "84", "85", "53"],
+ "contribution_1": ["0.161538", "-0.0403846", "0.0403846", "0.0403846", "0.0403846"],
+ "feature_2": ["age", "age", "age", "age", "age"],
+ "value_2": ["25", "39", "50", "43", "67"],
+ "contribution_2": ["0", "0", "0", "0", "0"],
+ },
+ dtype=object,
+ )
expected_output["proba"] = expected_output["proba"].astype(float)
feature_expected = [column for column in expected_output.columns if column.startswith("feature_")]
@@ -933,41 +967,40 @@ def test_summarize_3(self):
Unit test 3 summarize method
"""
predictor_1 = self.predictor_3
- predictor_1.mask_params = {"features_to_hide": None,
- "threshold": None,
- "positive": None,
- "max_contrib": 1
- }
-
- predictor_1.data = {"x": None,
- "x_preprocessed": None,
- "x_postprocessed": None,
- "ypred": None,
- "contributions": None}
+ predictor_1.mask_params = {"features_to_hide": None, "threshold": None, "positive": None, "max_contrib": 1}
+
+ predictor_1.data = {
+ "x": None,
+ "x_preprocessed": None,
+ "x_postprocessed": None,
+ "ypred": None,
+ "contributions": None,
+ }
predictor_1.data["x"] = self.df_3[["x1", "x2"]]
predictor_1.data["x_preprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["x_postprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["ypred"] = pd.DataFrame(
- {"y": ["Yes", "Yes", "No", "No", "No"],
- "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209]}
- )
+ {"y": ["Yes", "Yes", "No", "No", "No"], "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209]}
+ )
predictor_1.data["contributions"] = pd.DataFrame(
- {"x1": [0, 0, -0, -0, -0],
- "x2": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
- )
+ {"x1": [0, 0, -0, -0, -0], "x2": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
+ )
output = predictor_1.summarize()
- expected_output = pd.DataFrame({
- "y": ["Yes", "Yes", "No", "No", "No"],
- "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209],
- "feature_1": ["weight", "weight", "weight", "weight", "weight"],
- "value_1": ["90", "78", "84", "85", "53"],
- "contribution_1": ["0.161538", "-0.0403846", "0.0403846", "0.0403846", "0.0403846"],
- "feature_2": ["age", "age", "age", "age", "age"],
- "value_2": ["25", "39", "50", "43", "67"],
- "contribution_2": ["0", "0", "0", "0", "0"]
- }, dtype=object)
+ expected_output = pd.DataFrame(
+ {
+ "y": ["Yes", "Yes", "No", "No", "No"],
+ "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209],
+ "feature_1": ["weight", "weight", "weight", "weight", "weight"],
+ "value_1": ["90", "78", "84", "85", "53"],
+ "contribution_1": ["0.161538", "-0.0403846", "0.0403846", "0.0403846", "0.0403846"],
+ "feature_2": ["age", "age", "age", "age", "age"],
+ "value_2": ["25", "39", "50", "43", "67"],
+ "contribution_2": ["0", "0", "0", "0", "0"],
+ },
+ dtype=object,
+ )
expected_output["proba"] = expected_output["proba"].astype(float)
feature_expected = [column for column in expected_output.columns if column.startswith("feature_")]
@@ -985,10 +1018,7 @@ def test_summarize_3(self):
assert not len(contribution_expected) == len(contribution_output)
assert not len(output.columns) == len(expected_output.columns)
- predictor_1.mask_params = {"features_to_hide": None,
- "threshold": None,
- "positive": None,
- "max_contrib": None}
+ predictor_1.mask_params = {"features_to_hide": None, "threshold": None, "positive": None, "max_contrib": None}
def test_summarize_4(self):
"""
@@ -997,7 +1027,7 @@ def test_summarize_4(self):
predictor_1 = self.predictor_1_w_groups
predictor_1._case = "classification"
predictor_1._classes = [0, 1]
- clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_3[['x1', 'x2']], self.df_3['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df_3[["x1", "x2"]], self.df_3["y"])
backend = ShapBackend(model=clf)
predictor_1.model = clf
predictor_1.backend = backend
@@ -1010,41 +1040,38 @@ def test_summarize_4(self):
"x_preprocessed": None,
"x_postprocessed": None,
"ypred": None,
- "contributions": None
+ "contributions": None,
}
- predictor_1.data_groups = {
- "x_postprocessed": None,
- "ypred": None,
- "contributions": None
- }
+ predictor_1.data_groups = {"x_postprocessed": None, "ypred": None, "contributions": None}
predictor_1.data["x"] = self.df_3[["x1", "x2"]]
predictor_1.data["x_preprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["x_postprocessed"] = self.df_3[["x1", "x2"]]
predictor_1.data["ypred"] = pd.DataFrame(
- {"y": ["Yes", "Yes", "No", "No", "No"],
- "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209]}
- )
- predictor_1.data_groups["x_postprocessed"] = self.df_3[["x1"]].rename(columns={'x1': 'group1'})
+ {"y": ["Yes", "Yes", "No", "No", "No"], "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209]}
+ )
+ predictor_1.data_groups["x_postprocessed"] = self.df_3[["x1"]].rename(columns={"x1": "group1"})
predictor_1.data_groups["ypred"] = predictor_1.data["ypred"]
predictor_1.data["contributions"] = pd.DataFrame(
- {"x1": [0, 0, -0, -0, -0],
- "x2": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
- )
+ {"x1": [0, 0, -0, -0, -0], "x2": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
+ )
predictor_1.data_groups["contributions"] = pd.DataFrame(
{"group1": [0.161538, -0.0403846, 0.0403846, 0.0403846, 0.0403846]}
)
output = predictor_1.summarize()
- expected_output = pd.DataFrame({
- "y": ["Yes", "Yes", "No", "No", "No"],
- "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209],
- "feature_1": ["weight", "weight", "weight", "weight", "weight"],
- "value_1": ["90", "78", "84", "85", "53"],
- "contribution_1": ["0.161538", "-0.0403846", "0.0403846", "0.0403846", "0.0403846"]
- }, dtype=object)
+ expected_output = pd.DataFrame(
+ {
+ "y": ["Yes", "Yes", "No", "No", "No"],
+ "proba": [0.519221, 0.468791, 0.531209, 0.531209, 0.531209],
+ "feature_1": ["weight", "weight", "weight", "weight", "weight"],
+ "value_1": ["90", "78", "84", "85", "53"],
+ "contribution_1": ["0.161538", "-0.0403846", "0.0403846", "0.0403846", "0.0403846"],
+ },
+ dtype=object,
+ )
expected_output["proba"] = expected_output["proba"].astype(float)
feature_expected = [column for column in expected_output.columns if column.startswith("feature_")]
@@ -1084,36 +1111,22 @@ def test_apply_postprocessing_1(self):
"""
predictor_1 = self.predictor_3
predictor_1.data = {"x": None, "ypred": None, "contributions": None}
- predictor_1.data["x"] = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
+ predictor_1.data["x"] = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
assert np.array_equal(predictor_1.data["x"], predictor_1.apply_postprocessing())
def test_apply_postprocessing_2(self):
"""
Unit test apply_postprocessing 2
"""
- postprocessing = {'x1': {'type': 'suffix', 'rule': ' t'},
- 'x2': {'type': 'prefix', 'rule': 'test'}}
+ postprocessing = {"x1": {"type": "suffix", "rule": " t"}, "x2": {"type": "prefix", "rule": "test"}}
predictor_1 = self.predictor_3
predictor_1.postprocessing = postprocessing
predictor_1.data = {"x": None, "ypred": None, "contributions": None}
- predictor_1.data["x"] = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['x1', 'x2'],
- index=['Id1', 'Id2']
- )
+ predictor_1.data["x"] = pd.DataFrame([[1, 2], [3, 4]], columns=["x1", "x2"], index=["Id1", "Id2"])
expected_output = pd.DataFrame(
- data=[['1 t', 'test2'],
- ['3 t', 'test4']],
- columns=['x1', 'x2'],
- index=['Id1', 'Id2']
+ data=[["1 t", "test2"], ["3 t", "test4"]], columns=["x1", "x2"], index=["Id1", "Id2"]
)
output = predictor_1.apply_postprocessing()
assert np.array_equal(output, expected_output)
@@ -1126,14 +1139,18 @@ def test_convert_dict_dataset(self):
x = predictor_1.convert_dict_dataset(x={"x1": 1, "x2": "M"})
- assert all([str(x[feature].dtypes) == predictor_1.features_types[feature]
- for feature in predictor_1.features_types.keys()])
+ assert all(
+ [
+ str(x[feature].dtypes) == predictor_1.features_types[feature]
+ for feature in predictor_1.features_types.keys()
+ ]
+ )
with self.assertRaises(ValueError):
- predictor_1.convert_dict_dataset(x={"x1":"M", "x2":"M"})
+ predictor_1.convert_dict_dataset(x={"x1": "M", "x2": "M"})
predictor_1.convert_dict_dataset(x={"x1": 1, "x2": "M", "x3": "M"})
- @patch('shapash.explainer.smart_predictor.SmartPredictor.convert_dict_dataset')
+ @patch("shapash.explainer.smart_predictor.SmartPredictor.convert_dict_dataset")
def test_check_dataset_type(self, convert_dict_dataset):
"""
Unit test check_dataset_type
@@ -1163,8 +1180,12 @@ def test_check_dataset_features(self):
predictor_1.check_dataset_features(x=pd.DataFrame({"x1": ["M"], "x2": ["M"]}))
x = predictor_1.check_dataset_features(x=pd.DataFrame({"x2": ["M"], "x1": [1]}))
- assert all([str(x[feature].dtypes) == predictor_1.features_types[feature]
- for feature in predictor_1.features_types.keys()])
+ assert all(
+ [
+ str(x[feature].dtypes) == predictor_1.features_types[feature]
+ for feature in predictor_1.features_types.keys()
+ ]
+ )
features_order = []
for order in range(min(predictor_1.columns_dict.keys()), max(predictor_1.columns_dict.keys()) + 1):
@@ -1186,9 +1207,7 @@ def test_to_smartexplainer(self):
data_ypred_init = pd.DataFrame({"ypred": [1, 0, 0, 0, 0]})
data_ypred_init.index = data_x.index
- predictor_1.data = {"x": data_x,
- "ypred_init": data_ypred_init,
- "x_preprocessed": data_x_preprocessed}
+ predictor_1.data = {"x": data_x, "ypred_init": data_ypred_init, "x_preprocessed": data_x_preprocessed}
xpl = predictor_1.to_smartexplainer()
@@ -1206,9 +1225,10 @@ def test_to_smartexplainer(self):
ct = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OrdinalEncoder(), ['x2']),
+ ("onehot_ce", ce.OrdinalEncoder(), ["x2"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
ct.fit(data_x)
predictor_1.preprocessing = ct
diff --git a/tests/unit_tests/explainer/test_smart_state.py b/tests/unit_tests/explainer/test_smart_state.py
index 264d6cac..4132c5c6 100644
--- a/tests/unit_tests/explainer/test_smart_state.py
+++ b/tests/unit_tests/explainer/test_smart_state.py
@@ -2,9 +2,11 @@
Unit test smart state
"""
import unittest
-from unittest.mock import patch, Mock
-import pandas as pd
+from unittest.mock import Mock, patch
+
import numpy as np
+import pandas as pd
+
from shapash.explainer.smart_state import SmartState
@@ -13,6 +15,7 @@ class TestSmartState(unittest.TestCase):
Unit test Smart State Class
TODO: Docstring
"""
+
def test_validate_contributions_1(self):
"""
Unit test validate contributions
@@ -23,12 +26,7 @@ def test_validate_contributions_1(self):
"""
state = SmartState()
x_init = Mock()
- contributions = pd.DataFrame(
- [[2, 1],
- [8, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
+ contributions = pd.DataFrame([[2, 1], [8, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
expected_output = contributions
output = state.validate_contributions(contributions, x_init)
assert not pd.testing.assert_frame_equal(expected_output, output)
@@ -39,22 +37,12 @@ def test_validate_contributions_2(self):
"""
state = SmartState()
contributions = np.array([[2, 1], [8, 4]])
- x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
- expected_output = pd.DataFrame(
- [[2, 1],
- [8, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
+ x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
+ expected_output = pd.DataFrame([[2, 1], [8, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
output = state.validate_contributions(contributions, x_init)
assert not pd.testing.assert_frame_equal(expected_output, output)
- @patch('shapash.explainer.smart_state.inverse_transform_contributions')
+ @patch("shapash.explainer.smart_state.inverse_transform_contributions")
def test_inverse_transform_contributions(self, mock_inverse_transform_contributions):
"""
Unit test inverse transform contributions
@@ -73,13 +61,10 @@ def test_check_contributions_1(self):
"""
state = SmartState()
contributions = pd.DataFrame(
- [[-0.2, 0.1],
- [0.8, -0.4],
- [0.5, -0.7]],
+ [[-0.2, 0.1], [0.8, -0.4], [0.5, -0.7]],
)
x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
+ [[1, 2], [3, 4]],
)
assert not state.check_contributions(contributions, x_init)
@@ -88,15 +73,8 @@ def test_check_contributions_2(self):
Unit test check contributions 2
"""
state = SmartState()
- contributions = pd.DataFrame(
- [[-0.2, 0.1],
- [0.8, -0.4]],
- index=['row_1', 'row_2']
- )
- x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]]
- )
+ contributions = pd.DataFrame([[-0.2, 0.1], [0.8, -0.4]], index=["row_1", "row_2"])
+ x_init = pd.DataFrame([[1, 2], [3, 4]])
assert not state.check_contributions(contributions, x_init)
def test_check_contributions_3(self):
@@ -105,13 +83,11 @@ def test_check_contributions_3(self):
"""
state = SmartState()
contributions = pd.DataFrame(
- [[-0.2, 0.1],
- [0.8, -0.4]],
- columns=['col_1', 'col_2'],
+ [[-0.2, 0.1], [0.8, -0.4]],
+ columns=["col_1", "col_2"],
)
x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
+ [[1, 2], [3, 4]],
)
assert not state.check_contributions(contributions, x_init)
@@ -120,21 +96,11 @@ def test_check_contributions_4(self):
Unit test check contributions 4
"""
state = SmartState()
- contributions = pd.DataFrame(
- [[-0.2, 0.1],
- [0.8, -0.4]],
- columns=['col_1', 'col_2'],
- index=['row_1', 'row_2']
- )
- x_init = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['col_1', 'col_2'],
- index=['row_1', 'row_2']
- )
+ contributions = pd.DataFrame([[-0.2, 0.1], [0.8, -0.4]], columns=["col_1", "col_2"], index=["row_1", "row_2"])
+ x_init = pd.DataFrame([[1, 2], [3, 4]], columns=["col_1", "col_2"], index=["row_1", "row_2"])
assert state.check_contributions(contributions, x_init)
- @patch('shapash.explainer.smart_state.rank_contributions')
+ @patch("shapash.explainer.smart_state.rank_contributions")
def test_rank_contributions(self, mock_rank_contributions):
"""
Unit test rank contributions
@@ -153,11 +119,7 @@ def test_assign_contributions_1(self):
"""
state = SmartState()
output = state.assign_contributions([1, 2, 3])
- expected = {
- 'contrib_sorted': 1,
- 'x_sorted': 2,
- 'var_dict': 3
- }
+ expected = {"contrib_sorted": 1, "x_sorted": 2, "var_dict": 3}
self.assertDictEqual(output, expected)
def test_assign_contributions_2(self):
@@ -169,7 +131,7 @@ def test_assign_contributions_2(self):
with self.assertRaises(ValueError):
state.assign_contributions(ranked)
- @patch('shapash.explainer.smart_state.cap_contributions')
+ @patch("shapash.explainer.smart_state.cap_contributions")
def test_cap_contributions(self, mock_cap_contributions):
"""
Unit test cap contributions
@@ -182,7 +144,7 @@ def test_cap_contributions(self, mock_cap_contributions):
state.cap_contributions(Mock(), Mock())
mock_cap_contributions.assert_called()
- @patch('shapash.explainer.smart_state.hide_contributions')
+ @patch("shapash.explainer.smart_state.hide_contributions")
def test_hide_contributions(self, mock_hide_contributions):
"""
Unit test hide contributions
@@ -195,7 +157,7 @@ def test_hide_contributions(self, mock_hide_contributions):
state.hide_contributions(Mock(), Mock())
mock_hide_contributions.assert_called()
- @patch('shapash.explainer.smart_state.sign_contributions')
+ @patch("shapash.explainer.smart_state.sign_contributions")
def test_sign_contributions(self, mock_sign_contributions):
"""
Unit test sign contributions
@@ -208,7 +170,7 @@ def test_sign_contributions(self, mock_sign_contributions):
state.sign_contributions(Mock(), Mock())
mock_sign_contributions.assert_called()
- @patch('shapash.explainer.smart_state.cutoff_contributions')
+ @patch("shapash.explainer.smart_state.cutoff_contributions")
def test_cutoff_contributions(self, mock_cutoff_contributions):
"""
Unit test cutoff contributions
@@ -221,7 +183,7 @@ def test_cutoff_contributions(self, mock_cutoff_contributions):
state.cutoff_contributions(Mock(), Mock())
mock_cutoff_contributions.assert_called()
- @patch('shapash.explainer.smart_state.combine_masks')
+ @patch("shapash.explainer.smart_state.combine_masks")
def test_combine_masks(self, mock_combine_masks):
"""
Unit test combine masks
@@ -234,7 +196,7 @@ def test_combine_masks(self, mock_combine_masks):
state.combine_masks(Mock())
mock_combine_masks.assert_called()
- @patch('shapash.explainer.smart_state.compute_masked_contributions')
+ @patch("shapash.explainer.smart_state.compute_masked_contributions")
def test_compute_masked_contributions(self, mock_compute_masked_contributions):
"""
Unit test compute masked contributions
@@ -247,7 +209,7 @@ def test_compute_masked_contributions(self, mock_compute_masked_contributions):
state.compute_masked_contributions(Mock(), Mock())
mock_compute_masked_contributions.assert_called()
- @patch('shapash.explainer.smart_state.init_mask')
+ @patch("shapash.explainer.smart_state.init_mask")
def test_init_mask(self, mock_init_mask):
"""
Unit test init mask
@@ -266,48 +228,45 @@ def test_summarize_1(self):
"""
state = SmartState()
contrib_sorted = pd.DataFrame(
- [[0.32230754, 0.1550689, 0.10183475, 0.05471339],
- [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
- [-0.48666675, 0.25507156, -0.16968889, 0.0757443]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ [
+ [0.32230754, 0.1550689, 0.10183475, 0.05471339],
+ [-0.58547512, -0.37050409, -0.07249285, 0.00171975],
+ [-0.48666675, 0.25507156, -0.16968889, 0.0757443],
+ ],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
var_dict = pd.DataFrame(
- [[1, 0, 2, 3],
- [1, 0, 3, 2],
- [1, 0, 2, 3]],
- columns=['feature_0', 'feature_1', 'feature_2', 'feature_3'],
- index=[0, 1, 2]
+ [[1, 0, 2, 3], [1, 0, 3, 2], [1, 0, 2, 3]],
+ columns=["feature_0", "feature_1", "feature_2", "feature_3"],
+ index=[0, 1, 2],
)
x_sorted = pd.DataFrame(
- [[1., 3., 22., 1.],
- [2., 1., 2., 38.],
- [2., 3., 26., 1.]],
- columns=['feature_0', 'feature_1', 'feature_2', 'feature_3'],
- index=[0, 1, 2]
+ [[1.0, 3.0, 22.0, 1.0], [2.0, 1.0, 2.0, 38.0], [2.0, 3.0, 26.0, 1.0]],
+ columns=["feature_0", "feature_1", "feature_2", "feature_3"],
+ index=[0, 1, 2],
)
mask = pd.DataFrame(
- [[True, True, False, False],
- [True, True, False, False],
- [True, True, False, False]],
- columns=['contribution_0', 'contribution_1', 'contribution_2', 'contribution_3'],
- index=[0, 1, 2]
+ [[True, True, False, False], [True, True, False, False], [True, True, False, False]],
+ columns=["contribution_0", "contribution_1", "contribution_2", "contribution_3"],
+ index=[0, 1, 2],
)
- columns_dict = {0: 'Pclass', 1: 'Sex', 2: 'Age', 3: 'Embarked'}
- features_dict = {'Pclass': 'Pclass', 'Sex': 'Sex', 'Age': 'Age', 'Embarked': 'Embarked'}
+ columns_dict = {0: "Pclass", 1: "Sex", 2: "Age", 3: "Embarked"}
+ features_dict = {"Pclass": "Pclass", "Sex": "Sex", "Age": "Age", "Embarked": "Embarked"}
output = state.summarize(contrib_sorted, var_dict, x_sorted, mask, columns_dict, features_dict)
expected = pd.DataFrame(
- [['Sex', 1.0, 0.32230754, 'Pclass', 3.0, 0.1550689],
- ['Sex', 2.0, -0.58547512, 'Pclass', 1.0, -0.37050409],
- ['Sex', 2.0, -0.48666675, 'Pclass', 3.0, 0.25507156]],
- columns=['feature_1', 'value_1', 'contribution_1', 'feature_2', 'value_2',
- 'contribution_2'],
+ [
+ ["Sex", 1.0, 0.32230754, "Pclass", 3.0, 0.1550689],
+ ["Sex", 2.0, -0.58547512, "Pclass", 1.0, -0.37050409],
+ ["Sex", 2.0, -0.48666675, "Pclass", 3.0, 0.25507156],
+ ],
+ columns=["feature_1", "value_1", "contribution_1", "feature_2", "value_2", "contribution_2"],
index=[0, 1, 2],
- dtype=object
+ dtype=object,
)
assert not pd.testing.assert_frame_equal(expected, output)
- @patch('shapash.explainer.smart_state.compute_features_import')
+ @patch("shapash.explainer.smart_state.compute_features_import")
def test_compute_features_import(self, mock_compute_features_import):
"""
Unit test compute features import
@@ -316,7 +275,7 @@ def test_compute_features_import(self, mock_compute_features_import):
state.compute_features_import(Mock())
mock_compute_features_import.assert_called()
- @patch('shapash.explainer.smart_state.group_contributions')
+ @patch("shapash.explainer.smart_state.group_contributions")
def test_compute_grouped_contributions(self, mock_group_contributions):
"""
Unit test compute features groups contributions
diff --git a/tests/unit_tests/manipulation/test_filters.py b/tests/unit_tests/manipulation/test_filters.py
index 1e7f3da9..c51eeb38 100644
--- a/tests/unit_tests/manipulation/test_filters.py
+++ b/tests/unit_tests/manipulation/test_filters.py
@@ -2,13 +2,18 @@
Unit test for filters
"""
import unittest
+
import numpy as np
import pandas as pd
-from shapash.manipulation.filters import cutoff_contributions, cutoff_contributions_old
-from shapash.manipulation.filters import cap_contributions
-from shapash.manipulation.filters import hide_contributions
-from shapash.manipulation.filters import sign_contributions
-from shapash.manipulation.filters import combine_masks
+
+from shapash.manipulation.filters import (
+ cap_contributions,
+ combine_masks,
+ cutoff_contributions,
+ cutoff_contributions_old,
+ hide_contributions,
+ sign_contributions,
+)
class TestFilter(unittest.TestCase):
@@ -16,22 +21,15 @@ class TestFilter(unittest.TestCase):
Unit test class for filters
TODO: Docstring
"""
+
def test_hide_contributions_1(self):
"""
Unit test hide contributions 1
"""
- dataframe = pd.DataFrame(
- [[2, 0, 1],
- [0, 1, 2],
- [2, 3, 1]],
- columns=['col1', 'col2', 'col3']
- )
+ dataframe = pd.DataFrame([[2, 0, 1], [0, 1, 2], [2, 3, 1]], columns=["col1", "col2", "col3"])
output = hide_contributions(dataframe, [0, 2])
expected = pd.DataFrame(
- [[False, False, True],
- [False, True, False],
- [False, True, True]],
- columns=['col1', 'col2', 'col3']
+ [[False, False, True], [False, True, False], [False, True, True]], columns=["col1", "col2", "col3"]
)
pd.testing.assert_frame_equal(output, expected)
@@ -39,18 +37,10 @@ def test_hide_contributions_2(self):
"""
Unit test hide contributions 2
"""
- dataframe = pd.DataFrame(
- [[2, 0, 1],
- [0, 1, 2],
- [2, 3, 1]],
- columns=['col1', 'col2', 'col3']
- )
+ dataframe = pd.DataFrame([[2, 0, 1], [0, 1, 2], [2, 3, 1]], columns=["col1", "col2", "col3"])
output = hide_contributions(dataframe, [-5])
expected = pd.DataFrame(
- [[True, True, True],
- [True, True, True],
- [True, True, True]],
- columns=['col1', 'col2', 'col3']
+ [[True, True, True], [True, True, True], [True, True, True]], columns=["col1", "col2", "col3"]
)
pd.testing.assert_frame_equal(output, expected)
@@ -58,18 +48,10 @@ def test_hide_contributions_3(self):
"""
Unit test hide contributions 3
"""
- dataframe = pd.DataFrame(
- [[2, 0, 1],
- [0, 1, 2],
- [2, 3, 1]],
- columns=['col1', 'col2', 'col3']
- )
+ dataframe = pd.DataFrame([[2, 0, 1], [0, 1, 2], [2, 3, 1]], columns=["col1", "col2", "col3"])
output = hide_contributions(dataframe, [])
expected = pd.DataFrame(
- [[True, True, True],
- [True, True, True],
- [True, True, True]],
- columns=['col1', 'col2', 'col3']
+ [[True, True, True], [True, True, True], [True, True, True]], columns=["col1", "col2", "col3"]
)
pd.testing.assert_frame_equal(output, expected)
@@ -78,17 +60,11 @@ def test_cap_contributions(self):
Unit test cap contributions
"""
xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=['c1', 'c2', 'c3']
+ [[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=["c1", "c2", "c3"]
)
thresholdvalue = 0.3
result = pd.DataFrame(
- [[False, True, False],
- [True, False, True],
- [True, False, True]],
- columns=['c1', 'c2', 'c3']
+ [[False, True, False], [True, False, True], [True, False, True]], columns=["c1", "c2", "c3"]
)
output = cap_contributions(xmatr, thresholdvalue)
assert xmatr.shape == output.shape
@@ -98,42 +74,18 @@ def test_sign_contributions_1(self):
"""
Unit test sign contributions 1
"""
- dataframe = pd.DataFrame(
- {
- 'val1': [1, -1],
- 'val2': [-2, -2],
- 'val3': [0.5, -2]
- }
- )
+ dataframe = pd.DataFrame({"val1": [1, -1], "val2": [-2, -2], "val3": [0.5, -2]})
output = sign_contributions(dataframe, positive=True)
- expected = pd.DataFrame(
- {
- 'val1': [True, False],
- 'val2': [False, False],
- 'val3': [True, False]
- }
- )
+ expected = pd.DataFrame({"val1": [True, False], "val2": [False, False], "val3": [True, False]})
pd.testing.assert_frame_equal(output, expected)
def test_sign_contributions_2(self):
"""
Unit test sign contributions 2
"""
- dataframe = pd.DataFrame(
- {
- 'val1': [1, -1],
- 'val2': [-2, -2],
- 'val3': [0.5, -2]
- }
- )
+ dataframe = pd.DataFrame({"val1": [1, -1], "val2": [-2, -2], "val3": [0.5, -2]})
output = sign_contributions(dataframe, positive=False)
- expected = pd.DataFrame(
- {
- 'val1': [False, True],
- 'val2': [True, True],
- 'val3': [False, True]
- }
- )
+ expected = pd.DataFrame({"val1": [False, True], "val2": [True, True], "val3": [False, True]})
pd.testing.assert_frame_equal(output, expected)
def test_cutoff_contributions_old(self):
@@ -141,15 +93,10 @@ def test_cutoff_contributions_old(self):
Unit test cutoff contributions old
"""
dataframe = pd.DataFrame(np.tile(np.array([1, 2, 3, 4]), (4, 2)))
- dataframe.columns = ['col_{}'.format(col) for col in dataframe.columns]
+ dataframe.columns = [f"col_{col}" for col in dataframe.columns]
output = cutoff_contributions_old(dataframe, max_contrib=4)
- expected = pd.DataFrame(
- np.tile(
- np.array([True, True, True, True, False, False, False, False]),
- (4, 1)
- )
- )
- expected.columns = ['col_{}'.format(col) for col in expected.columns]
+ expected = pd.DataFrame(np.tile(np.array([True, True, True, True, False, False, False, False]), (4, 1)))
+ expected.columns = [f"col_{col}" for col in expected.columns]
assert output.equals(expected)
def test_cutoff_contributions_0(self):
@@ -158,19 +105,19 @@ def test_cutoff_contributions_0(self):
"""
dataframe = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, True]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, True],
}
)
output = cutoff_contributions(dataframe, 0)
expected = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, False],
- 'val3': [False, False, False],
- 'val4': [False, False, False]
+ "val1": [False, False, False],
+ "val2": [False, False, False],
+ "val3": [False, False, False],
+ "val4": [False, False, False],
}
)
pd.testing.assert_frame_equal(output, expected)
@@ -181,19 +128,19 @@ def test_cutoff_contributions_1(self):
"""
dataframe = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, True]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, True],
}
)
output = cutoff_contributions(dataframe, 1)
expected = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, False]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, False],
}
)
pd.testing.assert_frame_equal(output, expected)
@@ -204,19 +151,19 @@ def test_cutoff_contributions_2(self):
"""
dataframe = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, True]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, True],
}
)
output = cutoff_contributions(dataframe, 2)
expected = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, True]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, True],
}
)
pd.testing.assert_frame_equal(output, expected)
@@ -227,19 +174,19 @@ def test_cutoff_contributions_3(self):
"""
dataframe = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, True]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, True],
}
)
output = cutoff_contributions(dataframe)
expected = pd.DataFrame(
{
- 'val1': [False, False, False],
- 'val2': [False, False, True],
- 'val3': [False, True, False],
- 'val4': [False, False, True]
+ "val1": [False, False, False],
+ "val2": [False, False, True],
+ "val3": [False, True, False],
+ "val4": [False, False, True],
}
)
pd.testing.assert_frame_equal(output, expected)
@@ -249,22 +196,10 @@ def test_combine_masks_1(self):
Unit test combine mask 1
"""
df1 = pd.DataFrame(
- [[True, False, True],
- [True, True, True],
- [False, False, False]],
- columns=['col1', 'col2', 'col3']
- )
- df2 = pd.DataFrame(
- [[True, False, True],
- [True, False, False]],
- columns=['col1', 'col2', 'col3']
- )
- df3 = pd.DataFrame(
- [[False, False],
- [True, False],
- [True, False]],
- columns=['col1', 'col2']
+ [[True, False, True], [True, True, True], [False, False, False]], columns=["col1", "col2", "col3"]
)
+ df2 = pd.DataFrame([[True, False, True], [True, False, False]], columns=["col1", "col2", "col3"])
+ df3 = pd.DataFrame([[False, False], [True, False], [True, False]], columns=["col1", "col2"])
self.assertRaises(ValueError, combine_masks, [df1, df2])
self.assertRaises(ValueError, combine_masks, [df1, df3])
@@ -273,22 +208,15 @@ def test_combine_masks_2(self):
Unit test combine masks 2
"""
df1 = pd.DataFrame(
- [[True, False, True],
- [True, True, True],
- [False, False, False]],
- columns=['col1', 'col2', 'col3']
+ [[True, False, True], [True, True, True], [False, False, False]], columns=["col1", "col2", "col3"]
)
df2 = pd.DataFrame(
- [[False, False, True],
- [True, False, True],
- [True, False, False]],
- columns=['contrib_1', 'contrib_2', 'contrib_3']
+ [[False, False, True], [True, False, True], [True, False, False]],
+ columns=["contrib_1", "contrib_2", "contrib_3"],
)
output = combine_masks([df1, df2])
expected_output = pd.DataFrame(
- [[False, False, True],
- [True, False, True],
- [False, False, False]],
- columns=['contrib_1', 'contrib_2', 'contrib_3']
+ [[False, False, True], [True, False, True], [False, False, False]],
+ columns=["contrib_1", "contrib_2", "contrib_3"],
)
pd.testing.assert_frame_equal(output, expected_output)
diff --git a/tests/unit_tests/manipulation/test_mask.py b/tests/unit_tests/manipulation/test_mask.py
index 4c96bdfc..50c4fd09 100644
--- a/tests/unit_tests/manipulation/test_mask.py
+++ b/tests/unit_tests/manipulation/test_mask.py
@@ -2,7 +2,9 @@
Unit test of mask
"""
import unittest
+
import pandas as pd
+
from shapash.manipulation.mask import compute_masked_contributions, init_mask
@@ -11,30 +13,18 @@ class TestMask(unittest.TestCase):
Class of Unit test for Mask
TODO: Docstring
"""
+
def test_compute_masked_contributions_1(self):
"""
test of compute masked contributions 1
"""
- column_name = ['col1', 'col2', 'col3']
- xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=column_name
- )
+ column_name = ["col1", "col2", "col3"]
+ xmatr = pd.DataFrame([[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=column_name)
masktest = pd.DataFrame(
- [[True, False, False],
- [False, True, False],
- [False, False, False]],
- columns=column_name
+ [[True, False, False], [False, True, False], [False, False, False]], columns=column_name
)
output = compute_masked_contributions(xmatr, masktest)
- expected = pd.DataFrame(
- [[-0.02, 0.43],
- [-1.08, 0.0],
- [-0.008, 1.02]],
- columns=['masked_neg', 'masked_pos']
- )
+ expected = pd.DataFrame([[-0.02, 0.43], [-1.08, 0.0], [-0.008, 1.02]], columns=["masked_neg", "masked_pos"])
assert (xmatr.shape[0], 2) == output.shape
assert output.equals(expected)
@@ -42,26 +32,11 @@ def test_compute_masked_contributions_2(self):
"""
test of compute masked contributions 2
"""
- column_name = ['col1', 'col2', 'col3']
- xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=column_name
- )
- masktest = pd.DataFrame(
- [[True, False, False],
- [True, True, True],
- [False, False, False]],
- columns=column_name
- )
+ column_name = ["col1", "col2", "col3"]
+ xmatr = pd.DataFrame([[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=column_name)
+ masktest = pd.DataFrame([[True, False, False], [True, True, True], [False, False, False]], columns=column_name)
output = compute_masked_contributions(xmatr, masktest)
- expected = pd.DataFrame(
- [[-0.02, 0.43],
- [0.0, 0.0],
- [-0.008, 1.02]],
- columns=['masked_neg', 'masked_pos']
- )
+ expected = pd.DataFrame([[-0.02, 0.43], [0.0, 0.0], [-0.008, 1.02]], columns=["masked_neg", "masked_pos"])
assert (xmatr.shape[0], 2) == output.shape
assert output.equals(expected)
@@ -69,26 +44,11 @@ def test_compute_masked_contributions_3(self):
"""
test of compute masked contributions 3
"""
- column_name = ['col1', 'col2', 'col3']
- xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=column_name
- )
- masktest = pd.DataFrame(
- [[True, True, True],
- [True, True, True],
- [True, True, True]],
- columns=column_name
- )
+ column_name = ["col1", "col2", "col3"]
+ xmatr = pd.DataFrame([[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=column_name)
+ masktest = pd.DataFrame([[True, True, True], [True, True, True], [True, True, True]], columns=column_name)
output = compute_masked_contributions(xmatr, masktest)
- expected = pd.DataFrame(
- [[0.0, 0.0],
- [0.0, 0.0],
- [0.0, 0.0]],
- columns=['masked_neg', 'masked_pos']
- )
+ expected = pd.DataFrame([[0.0, 0.0], [0.0, 0.0], [0.0, 0.0]], columns=["masked_neg", "masked_pos"])
assert (xmatr.shape[0], 2) == output.shape
assert output.equals(expected)
@@ -96,18 +56,8 @@ def test_init_mask(self):
"""
test of initialization of mask
"""
- column_name = ['col1', 'col2']
- s_ord = pd.DataFrame(
- [[0.1, 0.43],
- [-0.78, 0.002],
- [0.62, -0.008]],
- columns=column_name
- )
- expected = pd.DataFrame(
- [[True, True],
- [True, True],
- [True, True]],
- columns=column_name
- )
+ column_name = ["col1", "col2"]
+ s_ord = pd.DataFrame([[0.1, 0.43], [-0.78, 0.002], [0.62, -0.008]], columns=column_name)
+ expected = pd.DataFrame([[True, True], [True, True], [True, True]], columns=column_name)
output = init_mask(s_ord)
assert output.equals(expected)
diff --git a/tests/unit_tests/manipulation/test_select_lines.py b/tests/unit_tests/manipulation/test_select_lines.py
index 15ad9594..d0962685 100644
--- a/tests/unit_tests/manipulation/test_select_lines.py
+++ b/tests/unit_tests/manipulation/test_select_lines.py
@@ -2,16 +2,15 @@
Unit test for select lines
"""
import unittest
+
import pandas as pd
+
from shapash.manipulation.select_lines import select_lines
DF = pd.DataFrame(
- [['A', 'A', -16.4, 12],
- ['C', 'A', 3.4, 0],
- ['C', 'B', 8.4, 9],
- ['C', 'B', -9, -5]],
- columns=['col1', 'col2', 'col3', 'col4']
- )
+ [["A", "A", -16.4, 12], ["C", "A", 3.4, 0], ["C", "B", 8.4, 9], ["C", "B", -9, -5]],
+ columns=["col1", "col2", "col3", "col4"],
+)
class TestSelectLines(unittest.TestCase):
@@ -19,18 +18,19 @@ class TestSelectLines(unittest.TestCase):
test select lines unit test
TODO: Docstring
"""
+
def test_compute_select_no_lines(self):
"""
test of compute select no lines
"""
- output = select_lines(DF, 'col3 < -3000')
+ output = select_lines(DF, "col3 < -3000")
assert len(output) == 0
def test_compute_select_simple_index_lines(self):
"""
test of compute select simple index lines
"""
- output = select_lines(DF, 'col3 < col4')
+ output = select_lines(DF, "col3 < col4")
expected = [0, 2, 3]
assert output == expected
@@ -38,7 +38,7 @@ def test_compute_select_multiple_index_lines(self):
"""
test of compute select multiple index lines
"""
- dataframe = DF.set_index(['col1', 'col2'])
- output = select_lines(dataframe, 'col3 < 0')
- expected = [('A', 'A'), ('C', 'B')]
+ dataframe = DF.set_index(["col1", "col2"])
+ output = select_lines(dataframe, "col3 < 0")
+ expected = [("A", "A"), ("C", "B")]
assert output == expected
diff --git a/tests/unit_tests/manipulation/test_summarize.py b/tests/unit_tests/manipulation/test_summarize.py
index 55172dcc..a0f91252 100644
--- a/tests/unit_tests/manipulation/test_summarize.py
+++ b/tests/unit_tests/manipulation/test_summarize.py
@@ -2,10 +2,12 @@
Unit test of summarize
"""
import unittest
+
+import numpy as np
import pandas as pd
from pandas.testing import assert_frame_equal
-import numpy as np
-from shapash.manipulation.summarize import summarize_el, compute_features_import, group_contributions
+
+from shapash.manipulation.summarize import compute_features_import, group_contributions, summarize_el
class TestSummarize(unittest.TestCase):
@@ -13,28 +15,17 @@ class TestSummarize(unittest.TestCase):
Unit test for summarize
TODO: Docstring
"""
+
def test_summarize_el_1(self):
"""
Test summarize el 1
"""
- column_name = ['col1', 'col2', 'col3']
- xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=column_name
- )
- masktest = pd.DataFrame(
- [[True, False, False],
- [False, True, False],
- [False, True, True]],
- columns=column_name
- )
+ column_name = ["col1", "col2", "col3"]
+ xmatr = pd.DataFrame([[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=column_name)
+ masktest = pd.DataFrame([[True, False, False], [False, True, False], [False, True, True]], columns=column_name)
output = summarize_el(xmatr, masktest, "feat")
expected = pd.DataFrame(
- [[0.1, np.nan], [0.002, np.nan], [-0.008, 0.4]],
- columns=["feat1", "feat2"],
- dtype=object
+ [[0.1, np.nan], [0.002, np.nan], [-0.008, 0.4]], columns=["feat1", "feat2"], dtype=object
)
assert xmatr.shape[0] == output.shape[0]
assert output.equals(expected)
@@ -43,25 +34,11 @@ def test_summarize_el_2(self):
"""
Test summarize el 2
"""
- column_name = ['col1', 'col2', 'col3']
- xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=column_name
- )
- masktest = pd.DataFrame(
- [[True, False, False],
- [False, True, False],
- [False, False, True]],
- columns=column_name
- )
+ column_name = ["col1", "col2", "col3"]
+ xmatr = pd.DataFrame([[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=column_name)
+ masktest = pd.DataFrame([[True, False, False], [False, True, False], [False, False, True]], columns=column_name)
output = summarize_el(xmatr, masktest, "feat")
- expected = pd.DataFrame(
- [[0.1], [0.002], [0.4]],
- columns=["feat1"],
- dtype=object
- )
+ expected = pd.DataFrame([[0.1], [0.002], [0.4]], columns=["feat1"], dtype=object)
assert xmatr.shape[0] == output.shape[0]
assert output.equals(expected)
@@ -69,24 +46,14 @@ def test_summarize_el_3(self):
"""
Test summarize el 3
"""
- column_name = ['col1', 'col2', 'col3']
+ column_name = ["col1", "col2", "col3"]
xmatr = pd.DataFrame(
- [["dfkj", "nfk", "bla"],
- ["Buble", "blue", "cool"],
- ["angry", "peace", "deep"]],
- columns=column_name
- )
- masktest = pd.DataFrame(
- [[True, False, False],
- [False, True, False],
- [False, True, True]],
- columns=column_name
+ [["dfkj", "nfk", "bla"], ["Buble", "blue", "cool"], ["angry", "peace", "deep"]], columns=column_name
)
+ masktest = pd.DataFrame([[True, False, False], [False, True, False], [False, True, True]], columns=column_name)
output = summarize_el(xmatr, masktest, "temp")
expected = pd.DataFrame(
- [['dfkj', np.nan], ['blue', np.nan], ['peace', 'deep']],
- columns=["temp1", "temp2"],
- dtype=object
+ [["dfkj", np.nan], ["blue", np.nan], ["peace", "deep"]], columns=["temp1", "temp2"], dtype=object
)
assert xmatr.shape[0] == output.shape[0]
assert output.equals(expected)
@@ -95,27 +62,15 @@ def test_summarize_el_4(self):
"""
Test summarize el 4
"""
- column_name = ['col1', 'col2', 'col3']
- index_list = ['A', 'B', 'C']
- xmatr = pd.DataFrame(
- [[0.1, 0.43, -0.02],
- [-0.78, 0.002, -0.3],
- [0.62, -0.008, 0.4]],
- columns=column_name
- )
- masktest = pd.DataFrame(
- [[True, False, False],
- [False, True, False],
- [False, True, True]],
- columns=column_name
- )
+ column_name = ["col1", "col2", "col3"]
+ index_list = ["A", "B", "C"]
+ xmatr = pd.DataFrame([[0.1, 0.43, -0.02], [-0.78, 0.002, -0.3], [0.62, -0.008, 0.4]], columns=column_name)
+ masktest = pd.DataFrame([[True, False, False], [False, True, False], [False, True, True]], columns=column_name)
xmatr.index = index_list
masktest.index = index_list
output = summarize_el(xmatr, masktest, "temp")
expected = pd.DataFrame(
- [[0.1, np.nan], [0.002, np.nan], [-0.008, 0.4]],
- columns=["temp1", "temp2"],
- dtype=object
+ [[0.1, np.nan], [0.002, np.nan], [-0.008, 0.4]], columns=["temp1", "temp2"], dtype=object
)
expected.index = index_list
assert xmatr.shape[0] == output.shape[0]
@@ -125,20 +80,13 @@ def test_compute_features_import_1(self):
"""
Test compute features import 1
"""
- column_name = ['col1', 'col2', 'col3']
- index_list = ['A', 'B', 'C']
+ column_name = ["col1", "col2", "col3"]
+ index_list = ["A", "B", "C"]
xmatr = pd.DataFrame(
- [[0.1, 0.4, -0.02],
- [-0.1, 0.2, -0.03],
- [0.2, -0.8, 0.4]],
- columns=column_name,
- index=index_list
+ [[0.1, 0.4, -0.02], [-0.1, 0.2, -0.03], [0.2, -0.8, 0.4]], columns=column_name, index=index_list
)
output = compute_features_import(xmatr)
- expected = pd.Series(
- [0.4, 1.4, 0.45],
- index=column_name
- )
+ expected = pd.Series([0.4, 1.4, 0.45], index=column_name)
expected = expected / expected.sum()
expected = expected.sort_values(ascending=True)
assert output.equals(expected)
@@ -147,24 +95,16 @@ def test_group_contributions_1(self):
"""
Test compute contributions groups
"""
- column_name = ['col1', 'col2', 'col3']
- index_list = ['A', 'B', 'C']
+ column_name = ["col1", "col2", "col3"]
+ index_list = ["A", "B", "C"]
xmatr = pd.DataFrame(
- [[0.1, 0.4, -0.02],
- [-0.1, 0.2, -0.03],
- [0.2, -0.8, 0.4]],
- columns=column_name,
- index=index_list
+ [[0.1, 0.4, -0.02], [-0.1, 0.2, -0.03], [0.2, -0.8, 0.4]], columns=column_name, index=index_list
)
- features_groups = {'group1': ['col1', 'col2']}
+ features_groups = {"group1": ["col1", "col2"]}
output = group_contributions(xmatr, features_groups)
expected = pd.DataFrame(
- [[-0.02, 0.5],
- [-0.03, 0.1],
- [0.40, -0.6]],
- columns=['col3', 'group1'],
- index=index_list
+ [[-0.02, 0.5], [-0.03, 0.1], [0.40, -0.6]], columns=["col3", "group1"], index=index_list
)
assert_frame_equal(output, expected)
@@ -172,23 +112,15 @@ def test_group_contributions_2(self):
"""
Test compute contributions groups
"""
- column_name = ['col1', 'col2', 'col3']
- index_list = ['A', 'B', 'C']
+ column_name = ["col1", "col2", "col3"]
+ index_list = ["A", "B", "C"]
xmatr = pd.DataFrame(
- [[0.1, 0.4, -0.02],
- [-0.1, 0.2, -0.03],
- [0.2, -0.8, 0.4]],
- columns=column_name,
- index=index_list
+ [[0.1, 0.4, -0.02], [-0.1, 0.2, -0.03], [0.2, -0.8, 0.4]], columns=column_name, index=index_list
)
- features_groups = {'group1': ['col1', 'col2'], 'group2': ['col3']}
+ features_groups = {"group1": ["col1", "col2"], "group2": ["col3"]}
output = group_contributions(xmatr, features_groups)
expected = pd.DataFrame(
- [[0.5, -0.02],
- [0.1, -0.03],
- [-0.6, 0.4]],
- columns=['group1', 'group2'],
- index=index_list
+ [[0.5, -0.02], [0.1, -0.03], [-0.6, 0.4]], columns=["group1", "group2"], index=index_list
)
assert_frame_equal(output, expected)
diff --git a/tests/unit_tests/report/test_common.py b/tests/unit_tests/report/test_common.py
index 66dfabcb..e8039b6a 100644
--- a/tests/unit_tests/report/test_common.py
+++ b/tests/unit_tests/report/test_common.py
@@ -1,13 +1,19 @@
import unittest
+
import numpy as np
import pandas as pd
-from shapash.report.common import VarType, series_dtype, numeric_is_continuous, get_callable, \
- display_value, replace_dict_values
+from shapash.report.common import (
+ VarType,
+ display_value,
+ get_callable,
+ numeric_is_continuous,
+ replace_dict_values,
+ series_dtype,
+)
class TestCommon(unittest.TestCase):
-
def test_series_dtype_1(self):
"""
Test string series
@@ -89,9 +95,10 @@ def test_numeric_is_continuous_4(self):
assert numeric_is_continuous(s) is False
def test_get_callable(self):
- fn = get_callable('sklearn.metrics.accuracy_score')
+ fn = get_callable("sklearn.metrics.accuracy_score")
from sklearn.metrics import accuracy_score
+
y_true = [1, 1, 0, 1, 0]
y_pred = [1, 1, 1, 0, 0]
@@ -99,22 +106,22 @@ def test_get_callable(self):
def test_display_value_1(self):
value = 123456.789
- expected_str = '123,456.789'
- assert display_value(value, ',', '.') == expected_str
+ expected_str = "123,456.789"
+ assert display_value(value, ",", ".") == expected_str
def test_display_value_2(self):
value = 123456.789
- expected_str = '123 456,789'
- assert display_value(value, ' ', ',') == expected_str
+ expected_str = "123 456,789"
+ assert display_value(value, " ", ",") == expected_str
def test_display_value_3(self):
value = 123456.789
- expected_str = '123.456,789'
- assert display_value(value, '.', ',') == expected_str
+ expected_str = "123.456,789"
+ assert display_value(value, ".", ",") == expected_str
def test_replace_dict_values_1(self):
- d = {'a': 1234, 'b': 0.1234, 'c': {'d': 123.456, 'e': 1234, 'f': {'g': 1234}}}
- expected_d = {'a': '1,234', 'b': '0.1234', 'c': {'d': '123.456', 'e': '1,234', 'f': {'g': '1,234'}}}
- res_d = replace_dict_values(d, display_value, ',', '.')
+ d = {"a": 1234, "b": 0.1234, "c": {"d": 123.456, "e": 1234, "f": {"g": 1234}}}
+ expected_d = {"a": "1,234", "b": "0.1234", "c": {"d": "123.456", "e": "1,234", "f": {"g": "1,234"}}}
+ res_d = replace_dict_values(d, display_value, ",", ".")
assert d == expected_d
assert res_d == expected_d # The replace_dict_values function modify inplace but also returns the result dict
diff --git a/tests/unit_tests/report/test_data_analysis.py b/tests/unit_tests/report/test_data_analysis.py
index 6a57108c..1bf3abba 100644
--- a/tests/unit_tests/report/test_data_analysis.py
+++ b/tests/unit_tests/report/test_data_analysis.py
@@ -1,27 +1,29 @@
import unittest
from unittest import TestCase
-import pandas as pd
+
import numpy as np
+import pandas as pd
-from shapash.report.data_analysis import perform_global_dataframe_analysis, perform_univariate_dataframe_analysis
from shapash.report.common import compute_col_types
+from shapash.report.data_analysis import perform_global_dataframe_analysis, perform_univariate_dataframe_analysis
class TestDataAnalysis(unittest.TestCase):
-
def test_perform_global_dataframe_analysis_1(self):
- df = pd.DataFrame({
- "string_data": ["a", "b", "c", "d", "e", "f"],
- "bool_data": [True, True, False, False, False, False],
- "int_data": [10, 20, 30, 40, 50, 0],
- })
+ df = pd.DataFrame(
+ {
+ "string_data": ["a", "b", "c", "d", "e", "f"],
+ "bool_data": [True, True, False, False, False, False],
+ "int_data": [10, 20, 30, 40, 50, 0],
+ }
+ )
d = perform_global_dataframe_analysis(df)
expected_d = {
- 'number of features': '3',
- 'number of observations': '6',
- 'missing values': '0',
- '% missing values': '0',
+ "number of features": "3",
+ "number of observations": "6",
+ "missing values": "0",
+ "% missing values": "0",
}
TestCase().assertDictEqual(d, expected_d)
@@ -31,52 +33,42 @@ def test_perform_global_dataframe_analysis_2(self):
assert d == {}
def test_perform_univariate_dataframe_analysis_1(self):
- df = pd.DataFrame({
- "string_data": ["a", "b", "c", "d", "e", "f"]*10,
- "bool_data": [True, True, False, False, False, True]*10,
- "int_continuous_data": list(range(60)),
- "float_continuous_data": np.linspace(0, 2, 60),
- "int_cat_data": [1, 1, 1, 2, 2, 2]*10,
- "float_cat_data": [0.2, 0.2, 0.2, 0.6, 0.6, 0.6]*10
- })
+ df = pd.DataFrame(
+ {
+ "string_data": ["a", "b", "c", "d", "e", "f"] * 10,
+ "bool_data": [True, True, False, False, False, True] * 10,
+ "int_continuous_data": list(range(60)),
+ "float_continuous_data": np.linspace(0, 2, 60),
+ "int_cat_data": [1, 1, 1, 2, 2, 2] * 10,
+ "float_cat_data": [0.2, 0.2, 0.2, 0.6, 0.6, 0.6] * 10,
+ }
+ )
d = perform_univariate_dataframe_analysis(df, col_types=compute_col_types(df))
expected_d = {
- 'int_continuous_data': {
- 'count': '60',
- 'mean': '29.5',
- 'std': '17.5',
- 'min': '0',
- '25%': '14.8',
- '50%': '29.5',
- '75%': '44.2',
- 'max': '59'
- },
- 'float_continuous_data': {
- 'count': '60',
- 'mean': '1',
- 'std': '0.592',
- 'min': '0',
- '25%': '0.5',
- '50%': '1',
- '75%': '1.5',
- 'max': '2'
+ "int_continuous_data": {
+ "count": "60",
+ "mean": "29.5",
+ "std": "17.5",
+ "min": "0",
+ "25%": "14.8",
+ "50%": "29.5",
+ "75%": "44.2",
+ "max": "59",
},
- 'int_cat_data': {
- 'distinct values': '2',
- 'missing values': '0'
+ "float_continuous_data": {
+ "count": "60",
+ "mean": "1",
+ "std": "0.592",
+ "min": "0",
+ "25%": "0.5",
+ "50%": "1",
+ "75%": "1.5",
+ "max": "2",
},
- 'float_cat_data': {
- 'distinct values': '2',
- 'missing values': '0'
- },
- 'string_data': {
- 'distinct values': '6',
- 'missing values': '0'
- },
- 'bool_data': {
- 'distinct values': '2',
- 'missing values': '0'
- }
+ "int_cat_data": {"distinct values": "2", "missing values": "0"},
+ "float_cat_data": {"distinct values": "2", "missing values": "0"},
+ "string_data": {"distinct values": "6", "missing values": "0"},
+ "bool_data": {"distinct values": "2", "missing values": "0"},
}
TestCase().assertDictEqual(d, expected_d)
diff --git a/tests/unit_tests/report/test_plots.py b/tests/unit_tests/report/test_plots.py
index 9087d493..6dd764b2 100644
--- a/tests/unit_tests/report/test_plots.py
+++ b/tests/unit_tests/report/test_plots.py
@@ -1,77 +1,83 @@
import unittest
from unittest.mock import patch
-import pandas as pd
-import numpy as np
+
import matplotlib.pyplot as plt
+import numpy as np
+import pandas as pd
-from shapash.report.plots import generate_fig_univariate, generate_fig_univariate_continuous, \
- generate_fig_univariate_categorical
from shapash.report.common import VarType
+from shapash.report.plots import (
+ generate_fig_univariate,
+ generate_fig_univariate_categorical,
+ generate_fig_univariate_continuous,
+)
class TestPlots(unittest.TestCase):
-
- @patch('shapash.report.plots.generate_fig_univariate_continuous')
- @patch('shapash.report.plots.generate_fig_univariate_categorical')
+ @patch("shapash.report.plots.generate_fig_univariate_continuous")
+ @patch("shapash.report.plots.generate_fig_univariate_categorical")
def test_generate_fig_univariate_1(self, mock_plot_cat, mock_plot_cont):
- df = pd.DataFrame({
- "string_data": ["a", "b", "c", "d", "e", np.nan],
- "data_train_test": ['train', 'train', 'train', 'train', 'test', 'test']
- })
-
- generate_fig_univariate(df, 'string_data', 'data_train_test', type=VarType.TYPE_CAT)
+ df = pd.DataFrame(
+ {
+ "string_data": ["a", "b", "c", "d", "e", np.nan],
+ "data_train_test": ["train", "train", "train", "train", "test", "test"],
+ }
+ )
+
+ generate_fig_univariate(df, "string_data", "data_train_test", type=VarType.TYPE_CAT)
mock_plot_cat.assert_called_once()
self.assertEqual(mock_plot_cont.call_count, 0)
- @patch('shapash.report.plots.generate_fig_univariate_continuous')
- @patch('shapash.report.plots.generate_fig_univariate_categorical')
+ @patch("shapash.report.plots.generate_fig_univariate_continuous")
+ @patch("shapash.report.plots.generate_fig_univariate_categorical")
def test_generate_fig_univariate_2(self, mock_plot_cat, mock_plot_cont):
- df = pd.DataFrame({
- "int_data": list(range(50)),
- "data_train_test": ['train', 'train', 'train', 'train', 'test']*10
- })
+ df = pd.DataFrame(
+ {"int_data": list(range(50)), "data_train_test": ["train", "train", "train", "train", "test"] * 10}
+ )
- generate_fig_univariate(df, 'int_data', 'data_train_test', type=VarType.TYPE_NUM)
+ generate_fig_univariate(df, "int_data", "data_train_test", type=VarType.TYPE_NUM)
mock_plot_cont.assert_called_once()
self.assertEqual(mock_plot_cat.call_count, 0)
- @patch('shapash.report.plots.generate_fig_univariate_continuous')
- @patch('shapash.report.plots.generate_fig_univariate_categorical')
+ @patch("shapash.report.plots.generate_fig_univariate_continuous")
+ @patch("shapash.report.plots.generate_fig_univariate_categorical")
def test_generate_fig_univariate_3(self, mock_plot_cat, mock_plot_cont):
- df = pd.DataFrame({
- "int_cat_data": [10, 10, 20, 20, 20, 10],
- "data_train_test": ['train', 'train', 'train', 'train', 'test', 'test']
- })
-
- generate_fig_univariate(df, 'int_cat_data', 'data_train_test', type=VarType.TYPE_CAT)
+ df = pd.DataFrame(
+ {
+ "int_cat_data": [10, 10, 20, 20, 20, 10],
+ "data_train_test": ["train", "train", "train", "train", "test", "test"],
+ }
+ )
+
+ generate_fig_univariate(df, "int_cat_data", "data_train_test", type=VarType.TYPE_CAT)
mock_plot_cat.assert_called_once()
self.assertEqual(mock_plot_cont.call_count, 0)
def test_generate_fig_univariate_continuous(self):
- df = pd.DataFrame({
- "int_data": [10, 20, 30, 40, 50, 0],
- "data_train_test": ['train', 'train', 'train', 'train', 'test', 'test']
- })
- fig = generate_fig_univariate_continuous(df, 'int_data', 'data_train_test')
+ df = pd.DataFrame(
+ {
+ "int_data": [10, 20, 30, 40, 50, 0],
+ "data_train_test": ["train", "train", "train", "train", "test", "test"],
+ }
+ )
+ fig = generate_fig_univariate_continuous(df, "int_data", "data_train_test")
assert isinstance(fig, plt.Figure)
def test_generate_fig_univariate_categorical_1(self):
- df = pd.DataFrame({
- "int_data": [0, 0, 0, 1, 1, 0],
- "data_train_test": ['train', 'train', 'train', 'train', 'test', 'test']
- })
+ df = pd.DataFrame(
+ {"int_data": [0, 0, 0, 1, 1, 0], "data_train_test": ["train", "train", "train", "train", "test", "test"]}
+ )
- fig = generate_fig_univariate_categorical(df, 'int_data', 'data_train_test')
+ fig = generate_fig_univariate_categorical(df, "int_data", "data_train_test")
assert len(fig.axes[0].patches) == 4 # Number of bars
def test_generate_fig_univariate_categorical_2(self):
- df = pd.DataFrame({
- "int_data": [0, 0, 0, 1, 1, 0],
- "data_train_test": ['train', 'train', 'train', 'train', 'train', 'train']
- })
+ df = pd.DataFrame(
+ {"int_data": [0, 0, 0, 1, 1, 0], "data_train_test": ["train", "train", "train", "train", "train", "train"]}
+ )
- fig = generate_fig_univariate_categorical(df, 'int_data', 'data_train_test')
+ fig = generate_fig_univariate_categorical(df, "int_data", "data_train_test")
assert len(fig.axes[0].patches) == 2 # Number of bars
@@ -79,12 +85,45 @@ def test_generate_fig_univariate_categorical_3(self):
"""
Test merging small categories into 'other' category
"""
- df = pd.DataFrame({
- "int_data": [0, 0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3, 4, 4, 4, 5, 5, 5, 6, 6, 6, 7, 7, 7, 8, 8, 8, 9, 10, 11],
- "data_train_test": ['train'] * 30
- })
-
- fig = generate_fig_univariate_categorical(df, 'int_data', 'data_train_test', nb_cat_max=7)
+ df = pd.DataFrame(
+ {
+ "int_data": [
+ 0,
+ 0,
+ 0,
+ 1,
+ 1,
+ 1,
+ 2,
+ 2,
+ 2,
+ 3,
+ 3,
+ 3,
+ 4,
+ 4,
+ 4,
+ 5,
+ 5,
+ 5,
+ 6,
+ 6,
+ 6,
+ 7,
+ 7,
+ 7,
+ 8,
+ 8,
+ 8,
+ 9,
+ 10,
+ 11,
+ ],
+ "data_train_test": ["train"] * 30,
+ }
+ )
+
+ fig = generate_fig_univariate_categorical(df, "int_data", "data_train_test", nb_cat_max=7)
assert len(fig.axes[0].patches) == 7 # Number of bars
@@ -92,26 +131,56 @@ def test_generate_fig_univariate_categorical_4(self):
"""
Test merging small categories into 'other' category
"""
- df = pd.DataFrame({
- "int_data": [0, 0, 0, 1, 1, 1, 2, 2, 2, 3, 3, 3, 4, 4, 4, 5, 5, 5, 6, 6, 6, 7, 7, 7, 8, 8, 8, 9, 10, 11],
- "data_train_test": ['train'] * 20 + ['test']*10
- })
-
- fig = generate_fig_univariate_categorical(df, 'int_data', 'data_train_test', nb_cat_max=7)
+ df = pd.DataFrame(
+ {
+ "int_data": [
+ 0,
+ 0,
+ 0,
+ 1,
+ 1,
+ 1,
+ 2,
+ 2,
+ 2,
+ 3,
+ 3,
+ 3,
+ 4,
+ 4,
+ 4,
+ 5,
+ 5,
+ 5,
+ 6,
+ 6,
+ 6,
+ 7,
+ 7,
+ 7,
+ 8,
+ 8,
+ 8,
+ 9,
+ 10,
+ 11,
+ ],
+ "data_train_test": ["train"] * 20 + ["test"] * 10,
+ }
+ )
+
+ fig = generate_fig_univariate_categorical(df, "int_data", "data_train_test", nb_cat_max=7)
# Number of bars (multiplied by two as we have train + test for each cat)
- assert len(fig.axes[0].patches) == 7*2
+ assert len(fig.axes[0].patches) == 7 * 2
def test_generate_fig_univariate_categorical_5(self):
"""
Test merging small categories into 'other' category
"""
- df = pd.DataFrame({
- "int_data": [k for k in range(10) for _ in range(k)],
- "data_train_test": ['train'] * 45
- })
+ df = pd.DataFrame({"int_data": [k for k in range(10) for _ in range(k)], "data_train_test": ["train"] * 45})
- fig = generate_fig_univariate_categorical(df, 'int_data', 'data_train_test', nb_cat_max=7)
+ fig = generate_fig_univariate_categorical(df, "int_data", "data_train_test", nb_cat_max=7)
assert len(fig.axes[0].patches) == 7 # Number of bars
@@ -119,14 +188,16 @@ def test_generate_fig_univariate_categorical_6(self):
"""
Test merging small categories into 'other' category
"""
- df = pd.DataFrame({
- "int_data": [k for k in range(10) for _ in range(k)],
- "data_train_test": ['train'] * 10 + ['test'] * 10 + ['train'] * 25
- })
+ df = pd.DataFrame(
+ {
+ "int_data": [k for k in range(10) for _ in range(k)],
+ "data_train_test": ["train"] * 10 + ["test"] * 10 + ["train"] * 25,
+ }
+ )
- fig = generate_fig_univariate_categorical(df, 'int_data', 'data_train_test', nb_cat_max=7)
+ fig = generate_fig_univariate_categorical(df, "int_data", "data_train_test", nb_cat_max=7)
print(len(fig.axes[0].patches))
# Number of bars (multiplied by two as we have train + test for each cat)
- assert len(fig.axes[0].patches) == 7*2
+ assert len(fig.axes[0].patches) == 7 * 2
diff --git a/tests/unit_tests/report/test_project_report.py b/tests/unit_tests/report/test_project_report.py
index d50ecff9..4a66020f 100644
--- a/tests/unit_tests/report/test_project_report.py
+++ b/tests/unit_tests/report/test_project_report.py
@@ -1,45 +1,55 @@
+import os
import unittest
from unittest.mock import patch
-import os
+
+import catboost as cb
import numpy as np
import pandas as pd
-import catboost as cb
from category_encoders import OrdinalEncoder
from shapash import SmartExplainer
from shapash.report.project_report import ProjectReport
-expected_attrs = ['explainer', 'metadata', 'x_train_init', 'y_test', 'x_init', 'config',
- 'col_names', 'df_train_test', 'title_story', 'title_description']
+expected_attrs = [
+ "explainer",
+ "metadata",
+ "x_train_init",
+ "y_test",
+ "x_init",
+ "config",
+ "col_names",
+ "df_train_test",
+ "title_story",
+ "title_description",
+]
current_path = os.path.dirname(os.path.abspath(__file__))
class TestProjectReport(unittest.TestCase):
-
def setUp(self):
- self.df = pd.DataFrame(range(0, 21), columns=['id'])
- self.df['y'] = self.df['id'].apply(lambda x: 1 if x < 10 else 0)
- self.df['x1'] = np.random.randint(1, 123, self.df.shape[0])
- self.df['x2'] = np.random.randint(1, 3, self.df.shape[0])
- self.df = self.df.set_index('id')
- self.clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df[['x1', 'x2']], self.df['y'])
+ self.df = pd.DataFrame(range(0, 21), columns=["id"])
+ self.df["y"] = self.df["id"].apply(lambda x: 1 if x < 10 else 0)
+ self.df["x1"] = np.random.randint(1, 123, self.df.shape[0])
+ self.df["x2"] = np.random.randint(1, 3, self.df.shape[0])
+ self.df = self.df.set_index("id")
+ self.clf = cb.CatBoostClassifier(n_estimators=1).fit(self.df[["x1", "x2"]], self.df["y"])
self.xpl = SmartExplainer(model=self.clf)
- self.xpl.compile(x=self.df[['x1', 'x2']])
+ self.xpl.compile(x=self.df[["x1", "x2"]])
self.report1 = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
)
self.report2 = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
)
def test_init_1(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
)
for attr in expected_attrs:
assert hasattr(report, attr)
@@ -47,8 +57,8 @@ def test_init_1(self):
def test_init_2(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
)
for attr in expected_attrs:
assert hasattr(report, attr)
@@ -56,9 +66,9 @@ def test_init_2(self):
def test_init_3(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
- y_test=self.df['y']
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
+ y_test=self.df["y"],
)
for attr in expected_attrs:
assert hasattr(report, attr)
@@ -66,10 +76,10 @@ def test_init_3(self):
def test_init_4(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
- y_test=self.df['y'],
- config={}
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
+ y_test=self.df["y"],
+ config={},
)
for attr in expected_attrs:
assert hasattr(report, attr)
@@ -77,53 +87,55 @@ def test_init_4(self):
def test_init_5(self):
ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
- y_test=self.df['y'],
- config={'metrics': [{'path': 'sklearn.metrics.mean_squared_error'}]}
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
+ y_test=self.df["y"],
+ config={"metrics": [{"path": "sklearn.metrics.mean_squared_error"}]},
)
def test_init_6(self):
- self.assertRaises(ValueError, ProjectReport,
+ self.assertRaises(
+ ValueError,
+ ProjectReport,
self.xpl,
- os.path.join(current_path, '../../data/metadata.yaml'),
- self.df[['x1', 'x2']],
- self.df['y'],
- {'metrics': ['sklearn.metrics.mean_squared_error']}
+ os.path.join(current_path, "../../data/metadata.yaml"),
+ self.df[["x1", "x2"]],
+ self.df["y"],
+ {"metrics": ["sklearn.metrics.mean_squared_error"]},
)
- @patch('shapash.report.project_report.print_html')
+ @patch("shapash.report.project_report.print_html")
def test_display_title_description_1(self, mock_print_html):
self.report1.display_title_description()
mock_print_html.assert_called_once()
- @patch('shapash.report.project_report.print_html')
+ @patch("shapash.report.project_report.print_html")
def test_display_title_description_2(self, mock_print_html):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
- y_test=self.df['y'],
- config={'title_story': "My project report",
- 'title_description': """This document is a data science project report."""}
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
+ y_test=self.df["y"],
+ config={
+ "title_story": "My project report",
+ "title_description": """This document is a data science project report.""",
+ },
)
report.display_title_description()
self.assertEqual(mock_print_html.call_count, 2)
- @patch('shapash.report.project_report.print_md')
+ @patch("shapash.report.project_report.print_md")
def test_display_general_information_1(self, mock_print_html):
report = ProjectReport(
- explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml')
+ explainer=self.xpl, project_info_file=os.path.join(current_path, "../../data/metadata.yaml")
)
report.display_project_information()
self.assertTrue(mock_print_html.called)
- @patch('shapash.report.project_report.print_md')
+ @patch("shapash.report.project_report.print_md")
def test_display_model_information_1(self, mock_print_md):
report = ProjectReport(
- explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml')
+ explainer=self.xpl, project_info_file=os.path.join(current_path, "../../data/metadata.yaml")
)
report.display_model_analysis()
self.assertTrue(mock_print_md.called)
@@ -131,15 +143,15 @@ def test_display_model_information_1(self, mock_print_md):
def test_display_dataset_analysis_1(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=self.df[['x1', 'x2']],
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=self.df[["x1", "x2"]],
)
report.display_dataset_analysis()
def test_display_dataset_analysis_2(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
)
report.display_dataset_analysis()
@@ -148,22 +160,19 @@ def test_display_dataset_analysis_3(self):
Test we don't have a problem when only categorical features
"""
df = self.df.copy()
- df['x1'] = 'a'
- df['x2'] = df['x2'].astype(str)
- encoder = OrdinalEncoder(
- cols=['x1', 'x2'],
- handle_unknown='ignore',
- return_df=True).fit(df)
+ df["x1"] = "a"
+ df["x2"] = df["x2"].astype(str)
+ encoder = OrdinalEncoder(cols=["x1", "x2"], handle_unknown="ignore", return_df=True).fit(df)
df = encoder.transform(df)
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
xpl = SmartExplainer(model=clf)
- xpl.compile(x=df[['x1', 'x2']])
+ xpl.compile(x=df[["x1", "x2"]])
report = ProjectReport(
explainer=xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- x_train=df[['x1', 'x2']],
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ x_train=df[["x1", "x2"]],
)
report.display_dataset_analysis()
@@ -171,7 +180,7 @@ def test_display_dataset_analysis_3(self):
def test_display_model_explainability_1(self):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
)
report.display_model_explainability()
@@ -179,67 +188,63 @@ def test_display_model_explainability_2(self):
"""
Tests multiclass case
"""
- df = pd.DataFrame(range(0, 21), columns=['id'])
- df['y'] = df['id'].apply(
- lambda x: 0 if x < 5 else 1 if (5 <= x < 10) else 2 if (10 <= x < 15) else 3)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df['x2'] = np.random.randint(1, 3, df.shape[0])
- df = df.set_index('id')
- clf = cb.CatBoostClassifier(n_estimators=1).fit(df[['x1', 'x2']], df['y'])
+ df = pd.DataFrame(range(0, 21), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 0 if x < 5 else 1 if (5 <= x < 10) else 2 if (10 <= x < 15) else 3)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df["x2"] = np.random.randint(1, 3, df.shape[0])
+ df = df.set_index("id")
+ clf = cb.CatBoostClassifier(n_estimators=1).fit(df[["x1", "x2"]], df["y"])
xpl = SmartExplainer(model=clf)
- xpl.compile(x=df[['x1', 'x2']])
- report = ProjectReport(
- explainer=xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml')
- )
+ xpl.compile(x=df[["x1", "x2"]])
+ report = ProjectReport(explainer=xpl, project_info_file=os.path.join(current_path, "../../data/metadata.yaml"))
report.display_model_explainability()
- @patch('shapash.report.project_report.logging')
+ @patch("shapash.report.project_report.logging")
def test_display_model_performance_1(self, mock_logging):
"""
No y_test given
"""
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
)
report.display_model_performance()
mock_logging.info.assert_called_once()
- @patch('shapash.report.project_report.logging')
+ @patch("shapash.report.project_report.logging")
def test_display_model_performance_2(self, mock_logging):
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- y_test=self.df['y'],
- config=dict(metrics=[{'path': 'sklearn.metrics.mean_squared_error'}])
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ y_test=self.df["y"],
+ config=dict(metrics=[{"path": "sklearn.metrics.mean_squared_error"}]),
)
report.display_model_performance()
self.assertEqual(mock_logging.call_count, 0)
- @patch('shapash.report.project_report.logging')
+ @patch("shapash.report.project_report.logging")
def test_display_model_performance_3(self, mock_logging):
"""
No metrics given in ProjectReport
"""
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- y_test=self.df['y'],
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ y_test=self.df["y"],
)
report.display_model_performance()
mock_logging.info.assert_called_once()
- @patch('shapash.report.project_report.logging')
+ @patch("shapash.report.project_report.logging")
def test_display_model_performance_4(self, mock_logging):
"""
Test use of proba values.
"""
report = ProjectReport(
explainer=self.xpl,
- project_info_file=os.path.join(current_path, '../../data/metadata.yaml'),
- y_test=self.df['y'],
- config=dict(metrics=[{'path': 'sklearn.metrics.log_loss', 'use_proba_values': True}])
+ project_info_file=os.path.join(current_path, "../../data/metadata.yaml"),
+ y_test=self.df["y"],
+ config=dict(metrics=[{"path": "sklearn.metrics.log_loss", "use_proba_values": True}]),
)
report.display_model_performance()
self.assertEqual(mock_logging.call_count, 0)
diff --git a/tests/unit_tests/style/test_style_utils.py b/tests/unit_tests/style/test_style_utils.py
index 15b20a0c..157ee565 100644
--- a/tests/unit_tests/style/test_style_utils.py
+++ b/tests/unit_tests/style/test_style_utils.py
@@ -1,14 +1,15 @@
"""
Unit test of style_utils
"""
-import unittest
import re
+import unittest
+
from shapash.style.style_utils import (
colors_loading,
- select_palette,
- define_style,
convert_str_color_to_plt_format,
- get_pyplot_color
+ define_style,
+ get_pyplot_color,
+ select_palette,
)
@@ -25,7 +26,7 @@ def rgb_string_detector(self, string_val):
"""
check rgb() or rgba() format of a str variable
"""
- matching = re.match(r"^rgba?\((\d+),\s*(\d+),\s*(\d+)(?:,\s*(\d+(?:\.\d+)?))?\)$",string_val)
+ matching = re.match(r"^rgba?\((\d+),\s*(\d+),\s*(\d+)(?:,\s*(\d+(?:\.\d+)?))?\)$", string_val)
matching = False if matching is None else True
return matching
@@ -71,9 +72,9 @@ def test_define_style(self):
def test_get_pyplot_color(self):
available_palettes = colors_loading()
for palette_name in available_palettes.keys():
- palette = colors_loading()[palette_name]['report_feature_distribution']
+ palette = colors_loading()[palette_name]["report_feature_distribution"]
colors = get_pyplot_color(colors=palette)
assert isinstance(colors, dict)
- col1 = colors_loading()[palette_name]['title_color']
+ col1 = colors_loading()[palette_name]["title_color"]
color = get_pyplot_color(col1)
assert isinstance(color, list)
diff --git a/tests/unit_tests/utils/test_category_encoders_backend.py b/tests/unit_tests/utils/test_category_encoders_backend.py
index 553857b1..a85df3f4 100644
--- a/tests/unit_tests/utils/test_category_encoders_backend.py
+++ b/tests/unit_tests/utils/test_category_encoders_backend.py
@@ -2,23 +2,24 @@
Unit test of Inverse Transform
"""
import unittest
-import pandas as pd
-import numpy as np
-import category_encoders as ce
+
import catboost as cb
+import category_encoders as ce
import lightgbm
+import numpy as np
+import pandas as pd
import xgboost
-from shapash.utils.transform import inverse_transform, apply_preprocessing, get_col_mapping_ce
from sklearn.ensemble import GradientBoostingClassifier
+from shapash.utils.transform import apply_preprocessing, get_col_mapping_ce, inverse_transform
+
class TestInverseTransformCaterogyEncoder(unittest.TestCase):
def test_inverse_transform_1(self):
"""
Test no preprocessing
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"]})
original = inverse_transform(train)
pd.testing.assert_frame_equal(original, train)
@@ -26,53 +27,80 @@ def test_inverse_transform_2(self):
"""
Test multiple preprocessing
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
-
- test = pd.DataFrame({'Onehot1': ['A', 'B', 'A'], 'Onehot2': ['C', 'D', 'ZZ'],
- 'Binary1': ['E', 'F', 'F'], 'Binary2': ['G', 'H', 'ZZ'],
- 'Ordinal1': ['I', 'J', 'J'], 'Ordinal2': ['K', 'L', 'ZZ'],
- 'BaseN1': ['M', 'N', 'N'], 'BaseN2': ['O', 'P', 'ZZ'],
- 'Target1': ['Q', 'R', 'R'], 'Target2': ['S', 'T', 'ZZ'],
- 'other': ['other', '123', np.nan]})
-
- expected = pd.DataFrame({'Onehot1': ['A', 'B', 'A'], 'Onehot2': ['C', 'D', 'missing'],
- 'Binary1': ['E', 'F', 'F'], 'Binary2': ['G', 'H', 'missing'],
- 'Ordinal1': ['I', 'J', 'J'], 'Ordinal2': ['K', 'L', 'missing'],
- 'BaseN1': ['M', 'N', 'N'], 'BaseN2': ['O', 'P', np.nan],
- 'Target1': ['Q', 'R', 'R'], 'Target2': ['S', 'T', 'NaN'],
- 'other': ['other', '123', np.nan]})
-
- y = pd.DataFrame(data=[0, 1, 0, 0], columns=['y'])
-
- enc_onehot = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2']).fit(train)
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
+
+ test = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A"],
+ "Onehot2": ["C", "D", "ZZ"],
+ "Binary1": ["E", "F", "F"],
+ "Binary2": ["G", "H", "ZZ"],
+ "Ordinal1": ["I", "J", "J"],
+ "Ordinal2": ["K", "L", "ZZ"],
+ "BaseN1": ["M", "N", "N"],
+ "BaseN2": ["O", "P", "ZZ"],
+ "Target1": ["Q", "R", "R"],
+ "Target2": ["S", "T", "ZZ"],
+ "other": ["other", "123", np.nan],
+ }
+ )
+
+ expected = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A"],
+ "Onehot2": ["C", "D", "missing"],
+ "Binary1": ["E", "F", "F"],
+ "Binary2": ["G", "H", "missing"],
+ "Ordinal1": ["I", "J", "J"],
+ "Ordinal2": ["K", "L", "missing"],
+ "BaseN1": ["M", "N", "N"],
+ "BaseN2": ["O", "P", np.nan],
+ "Target1": ["Q", "R", "R"],
+ "Target2": ["S", "T", "NaN"],
+ "other": ["other", "123", np.nan],
+ }
+ )
+
+ y = pd.DataFrame(data=[0, 1, 0, 0], columns=["y"])
+
+ enc_onehot = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"]).fit(train)
train_onehot = enc_onehot.transform(train)
- enc_binary = ce.BinaryEncoder(cols=['Binary1', 'Binary2']).fit(train_onehot)
+ enc_binary = ce.BinaryEncoder(cols=["Binary1", "Binary2"]).fit(train_onehot)
train_binary = enc_binary.transform(train_onehot)
- enc_ordinal = ce.OrdinalEncoder(cols=['Ordinal1', 'Ordinal2']).fit(train_binary)
+ enc_ordinal = ce.OrdinalEncoder(cols=["Ordinal1", "Ordinal2"]).fit(train_binary)
train_ordinal = enc_ordinal.transform(train_binary)
- enc_basen = ce.BaseNEncoder(cols=['BaseN1', 'BaseN2']).fit(train_ordinal)
+ enc_basen = ce.BaseNEncoder(cols=["BaseN1", "BaseN2"]).fit(train_ordinal)
train_basen = enc_basen.transform(train_ordinal)
- enc_target = ce.TargetEncoder(cols=['Target1', 'Target2']).fit(train_basen, y)
+ enc_target = ce.TargetEncoder(cols=["Target1", "Target2"]).fit(train_basen, y)
input_dict1 = dict()
- input_dict1['col'] = 'Onehot2'
- input_dict1['mapping'] = pd.Series(data=['C', 'D', np.nan], index=['C', 'D', 'missing'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "Onehot2"
+ input_dict1["mapping"] = pd.Series(data=["C", "D", np.nan], index=["C", "D", "missing"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'Binary2'
- input_dict2['mapping'] = pd.Series(data=['G', 'H', np.nan], index=['G', 'H', 'missing'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "Binary2"
+ input_dict2["mapping"] = pd.Series(data=["G", "H", np.nan], index=["G", "H", "missing"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'Ordinal2'
- input_dict3['mapping'] = pd.Series(data=['K', 'L', np.nan], index=['K', 'L', 'missing'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "Ordinal2"
+ input_dict3["mapping"] = pd.Series(data=["K", "L", np.nan], index=["K", "L", "missing"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
result1 = enc_onehot.transform(test)
@@ -81,8 +109,9 @@ def test_inverse_transform_2(self):
result4 = enc_basen.transform(result3)
result5 = enc_target.transform(result4)
- original = inverse_transform(result5, [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1,
- list_dict])
+ original = inverse_transform(
+ result5, [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1, list_dict]
+ )
pd.testing.assert_frame_equal(expected, original)
@@ -90,18 +119,22 @@ def test_inverse_transform_3(self):
"""
Test target encoding
"""
- train = pd.DataFrame({'city': ['chicago', 'paris', 'paris', 'chicago', 'chicago'],
- 'state': ['US', 'FR', 'FR', 'US', 'US'],
- 'other': ['A', 'A', np.nan, 'B', 'B']})
- test = pd.DataFrame({'city': ['chicago', 'paris', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- expected = pd.DataFrame({'city': ['chicago', 'paris', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- y = pd.DataFrame(data=[0, 1, 1, 0, 1], columns=['y'])
-
- enc = ce.TargetEncoder(cols=['city', 'state']).fit(train, y)
+ train = pd.DataFrame(
+ {
+ "city": ["chicago", "paris", "paris", "chicago", "chicago"],
+ "state": ["US", "FR", "FR", "US", "US"],
+ "other": ["A", "A", np.nan, "B", "B"],
+ }
+ )
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "paris"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ expected = pd.DataFrame(
+ {"city": ["chicago", "paris", "paris"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1, 0, 1], columns=["y"])
+
+ enc = ce.TargetEncoder(cols=["city", "state"]).fit(train, y)
result = enc.transform(test)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(expected, original)
@@ -110,10 +143,10 @@ def test_inverse_transform_4(self):
"""
Test ordinal encoding
"""
- train = pd.DataFrame({'city': ['chicago', 'st louis']})
- test = pd.DataFrame({'city': ['chicago', 'los angeles']})
- expected = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.OrdinalEncoder(handle_missing='value', handle_unknown='value')
+ train = pd.DataFrame({"city": ["chicago", "st louis"]})
+ test = pd.DataFrame({"city": ["chicago", "los angeles"]})
+ expected = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.OrdinalEncoder(handle_missing="value", handle_unknown="value")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -123,8 +156,8 @@ def test_inverse_transform_5(self):
"""
Test inverse_transform having Nan in train and handle missing value expect returned with nan_Ordinal
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.OrdinalEncoder(handle_missing='value', handle_unknown='value')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.OrdinalEncoder(handle_missing="value", handle_unknown="value")
result = enc.fit_transform(train)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(train, original)
@@ -133,8 +166,8 @@ def test_inverse_transform_6(self):
"""
test inverse_transform having Nan in train and handle missing return Nan expect returned with nan_Ordinal
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.OrdinalEncoder(handle_missing='return_nan', handle_unknown='value')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.OrdinalEncoder(handle_missing="return_nan", handle_unknown="value")
result = enc.fit_transform(train)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(train, original)
@@ -143,9 +176,9 @@ def test_inverse_transform_7(self):
"""
test inverse_transform both fields are return Nan with Nan Expect ValueError Ordinal
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', 'los angeles']})
- enc = ce.OrdinalEncoder(handle_missing='return_nan', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", "los angeles"]})
+ enc = ce.OrdinalEncoder(handle_missing="return_nan", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -155,9 +188,9 @@ def test_inverse_transform_8(self):
"""
test inverse_transform having missing and no Uknown expect inversed ordinal
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', 'los angeles']})
- enc = ce.OrdinalEncoder(handle_missing='value', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", "los angeles"]})
+ enc = ce.OrdinalEncoder(handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -167,10 +200,10 @@ def test_inverse_transform_9(self):
"""
test inverse_transform having handle missing value and handle unknown return Nan expect best inverse ordinal
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', np.nan, 'los angeles']})
- expected = pd.DataFrame({'city': ['chicago', np.nan, np.nan]})
- enc = ce.OrdinalEncoder(handle_missing='value', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", np.nan, "los angeles"]})
+ expected = pd.DataFrame({"city": ["chicago", np.nan, np.nan]})
+ enc = ce.OrdinalEncoder(handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = enc.inverse_transform(result)
@@ -180,16 +213,12 @@ def test_inverse_transform_10(self):
"""
test inverse_transform with multiple ordinal
"""
- data = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['a', 'b']})
- test = pd.DataFrame({'city': [1, 2, 2],
- 'state': [1, 2, 2],
- 'other': ['a', 'b', 'a']})
- expected = pd.DataFrame({'city': ['chicago', 'paris', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['a', 'b', 'a']})
- enc = ce.OrdinalEncoder(cols=['city', 'state'])
+ data = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["a", "b"]})
+ test = pd.DataFrame({"city": [1, 2, 2], "state": [1, 2, 2], "other": ["a", "b", "a"]})
+ expected = pd.DataFrame(
+ {"city": ["chicago", "paris", "paris"], "state": ["US", "FR", "FR"], "other": ["a", "b", "a"]}
+ )
+ enc = ce.OrdinalEncoder(cols=["city", "state"])
enc.fit(data)
original = inverse_transform(test, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -198,19 +227,17 @@ def test_inverse_transform_11(self):
"""
Test binary encoding
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', np.nan]})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", np.nan]})
- test = pd.DataFrame({'city': ['chicago', 'paris', 'monaco'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, 'B']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "monaco"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, "B"]}
+ )
- expected = pd.DataFrame({'city': ['chicago', 'paris', np.nan],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, 'B']})
+ expected = pd.DataFrame(
+ {"city": ["chicago", "paris", np.nan], "state": ["US", "FR", "FR"], "other": ["A", np.nan, "B"]}
+ )
- enc = ce.BinaryEncoder(cols=['city', 'state']).fit(train)
+ enc = ce.BinaryEncoder(cols=["city", "state"]).fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -219,7 +246,7 @@ def test_inverse_transform_12(self):
"""
test inverse_transform having data expecting a returned result
"""
- train = pd.Series(list('abcd')).to_frame('letter')
+ train = pd.Series(list("abcd")).to_frame("letter")
enc = ce.BaseNEncoder(base=2)
result = enc.fit_transform(train)
inversed_result = inverse_transform(result, enc)
@@ -229,8 +256,8 @@ def test_inverse_transform_13(self):
"""
Test basen encoding
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.BaseNEncoder(handle_missing='value', handle_unknown='value')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.BaseNEncoder(handle_missing="value", handle_unknown="value")
result = enc.fit_transform(train)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(train, original)
@@ -239,9 +266,9 @@ def test_inverse_transform_14(self):
"""
test inverse_transform having Nan in train and handle missing expected a result with Nan
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
- enc = ce.BaseNEncoder(handle_missing='return_nan', handle_unknown='value')
+ enc = ce.BaseNEncoder(handle_missing="return_nan", handle_unknown="value")
result = enc.fit_transform(train)
original = inverse_transform(result, enc)
@@ -251,10 +278,10 @@ def test_inverse_transform_15(self):
"""
test inverse_transform having missing and no unknown
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', 'los angeles']})
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", "los angeles"]})
- enc = ce.BaseNEncoder(handle_missing='value', handle_unknown='return_nan')
+ enc = ce.BaseNEncoder(handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -265,10 +292,10 @@ def test_inverse_transform_16(self):
"""
test inverse_transform having handle missing value and Unknown
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', np.nan, 'los angeles']})
- expected = pd.DataFrame({'city': ['chicago', np.nan, np.nan]})
- enc = ce.BaseNEncoder(handle_missing='value', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", np.nan, "los angeles"]})
+ expected = pd.DataFrame({"city": ["chicago", np.nan, np.nan]})
+ enc = ce.BaseNEncoder(handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -278,13 +305,9 @@ def test_inverse_transform_17(self):
"""
test inverse_transform with multiple baseN
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR']})
- test = pd.DataFrame({'city_0': [0, 1],
- 'city_1': [1, 0],
- 'state_0': [0, 1],
- 'state_1': [1, 0]})
- enc = ce.BaseNEncoder(cols=['city', 'state'], handle_missing='value', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"]})
+ test = pd.DataFrame({"city_0": [0, 1], "city_1": [1, 0], "state_0": [0, 1], "state_1": [1, 0]})
+ enc = ce.BaseNEncoder(cols=["city", "state"], handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
original = inverse_transform(test, enc)
pd.testing.assert_frame_equal(original, train)
@@ -293,8 +316,8 @@ def test_inverse_transform_18(self):
"""
Test Onehot encoding
"""
- encoder = ce.OneHotEncoder(cols=['match', 'match_box'], use_cat_names=True)
- value = pd.DataFrame({'match': pd.Series('box_-1'), 'match_box': pd.Series(-1)})
+ encoder = ce.OneHotEncoder(cols=["match", "match_box"], use_cat_names=True)
+ value = pd.DataFrame({"match": pd.Series("box_-1"), "match_box": pd.Series(-1)})
transformed = encoder.fit_transform(value)
inversed_result = inverse_transform(transformed, encoder)
pd.testing.assert_frame_equal(value, inversed_result)
@@ -303,8 +326,8 @@ def test_inverse_transform_19(self):
"""
test inverse_transform having no categories names
"""
- encoder = ce.OneHotEncoder(cols=['match', 'match_box'], use_cat_names=False)
- value = pd.DataFrame({'match': pd.Series('box_-1'), 'match_box': pd.Series(-1)})
+ encoder = ce.OneHotEncoder(cols=["match", "match_box"], use_cat_names=False)
+ value = pd.DataFrame({"match": pd.Series("box_-1"), "match_box": pd.Series(-1)})
transformed = encoder.fit_transform(value)
inversed_result = inverse_transform(transformed, encoder)
pd.testing.assert_frame_equal(value, inversed_result)
@@ -313,8 +336,8 @@ def test_inverse_transform_20(self):
"""
test inverse_transform with Nan in training expecting Nan_Onehot returned result
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.OneHotEncoder(handle_missing='value', handle_unknown='value')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.OneHotEncoder(handle_missing="value", handle_unknown="value")
result = enc.fit_transform(train)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(train, original)
@@ -323,8 +346,8 @@ def test_inverse_transform_21(self):
"""
test inverse_transform with Nan in training expecting Nan_Onehot returned result
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.OneHotEncoder(handle_missing='return_nan', handle_unknown='value')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.OneHotEncoder(handle_missing="return_nan", handle_unknown="value")
result = enc.fit_transform(train)
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(train, original)
@@ -333,10 +356,10 @@ def test_inverse_transform_22(self):
"""
test inverse_transform with Both fields return_nan
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', 'los angeles']})
- expected = pd.DataFrame({'city': ['chicago', np.nan]})
- enc = ce.OneHotEncoder(handle_missing='return_nan', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", "los angeles"]})
+ expected = pd.DataFrame({"city": ["chicago", np.nan]})
+ enc = ce.OneHotEncoder(handle_missing="return_nan", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -346,9 +369,9 @@ def test_inverse_transform_23(self):
"""
test inverse_transform having missing and No Unknown
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', 'los angeles']})
- enc = ce.OneHotEncoder(handle_missing='value', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", "los angeles"]})
+ enc = ce.OneHotEncoder(handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -358,10 +381,10 @@ def test_inverse_transform_24(self):
"""
test inverse_transform having handle missing value and Handle Unknown
"""
- train = pd.DataFrame({'city': ['chicago', np.nan]})
- test = pd.DataFrame({'city': ['chicago', np.nan, 'los angeles']})
- expected = pd.DataFrame({'city': ['chicago', np.nan, np.nan]})
- enc = ce.OneHotEncoder(handle_missing='value', handle_unknown='return_nan')
+ train = pd.DataFrame({"city": ["chicago", np.nan]})
+ test = pd.DataFrame({"city": ["chicago", np.nan, "los angeles"]})
+ expected = pd.DataFrame({"city": ["chicago", np.nan, np.nan]})
+ enc = ce.OneHotEncoder(handle_missing="value", handle_unknown="return_nan")
enc.fit(train)
result = enc.transform(test)
original = inverse_transform(result, enc)
@@ -371,17 +394,17 @@ def test_inverse_transform_25(self):
"""
Test dict encoding
"""
- data = pd.DataFrame({'city': ['chicago', 'paris-1', 'paris-2'],
- 'state': ['US', 'FR-1', 'FR-2'],
- 'other': ['A', 'B', np.nan]})
+ data = pd.DataFrame(
+ {"city": ["chicago", "paris-1", "paris-2"], "state": ["US", "FR-1", "FR-2"], "other": ["A", "B", np.nan]}
+ )
- expected = pd.DataFrame({'city': ['chicago', 'paris-1', 'paris-2'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', np.nan]})
+ expected = pd.DataFrame(
+ {"city": ["chicago", "paris-1", "paris-2"], "state": ["US", "FR", "FR"], "other": ["A", "B", np.nan]}
+ )
input_dict = dict()
- input_dict['col'] = 'state'
- input_dict['mapping'] = pd.Series(data=['US', 'FR-1', 'FR-2'], index=['US', 'FR', 'FR'])
- input_dict['data_type'] = 'object'
+ input_dict["col"] = "state"
+ input_dict["mapping"] = pd.Series(data=["US", "FR-1", "FR-2"], index=["US", "FR", "FR"])
+ input_dict["data_type"] = "object"
result = inverse_transform(data, input_dict)
pd.testing.assert_frame_equal(result, expected)
@@ -389,55 +412,82 @@ def test_inverse_transform_26(self):
"""
Test multiple dict encoding
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
-
- test = pd.DataFrame({'Onehot1': ['A', 'B', 'A'], 'Onehot2': ['C', 'D', 'ZZ'],
- 'Binary1': ['E', 'F', 'F'], 'Binary2': ['G', 'H', 'ZZ'],
- 'Ordinal1': ['I', 'J', 'J'], 'Ordinal2': ['K', 'L', 'ZZ'],
- 'BaseN1': ['M', 'N', 'N'], 'BaseN2': ['O', 'P', 'ZZ'],
- 'Target1': ['Q', 'R', 'R'], 'Target2': ['S', 'T', 'ZZ'],
- 'other': ['other', '123', np.nan]},
- index=['index1', 'index2', 'index3'])
-
- expected = pd.DataFrame({'Onehot1': ['A', 'B', 'A'], 'Onehot2': ['C', 'D', 'missing'],
- 'Binary1': ['E', 'F', 'F'], 'Binary2': ['G', 'H', 'missing'],
- 'Ordinal1': ['I', 'J', 'J'], 'Ordinal2': ['K', 'L', 'missing'],
- 'BaseN1': ['M', 'N', 'N'], 'BaseN2': ['O', 'P', np.nan],
- 'Target1': ['Q', 'R', 'R'], 'Target2': ['S', 'T', 'NaN'],
- 'other': ['other', '123', np.nan]},
- index=['index1', 'index2', 'index3'])
-
- y = pd.DataFrame(data=[0, 1, 0, 0], columns=['y'])
-
- enc_onehot = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2']).fit(train)
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
+
+ test = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A"],
+ "Onehot2": ["C", "D", "ZZ"],
+ "Binary1": ["E", "F", "F"],
+ "Binary2": ["G", "H", "ZZ"],
+ "Ordinal1": ["I", "J", "J"],
+ "Ordinal2": ["K", "L", "ZZ"],
+ "BaseN1": ["M", "N", "N"],
+ "BaseN2": ["O", "P", "ZZ"],
+ "Target1": ["Q", "R", "R"],
+ "Target2": ["S", "T", "ZZ"],
+ "other": ["other", "123", np.nan],
+ },
+ index=["index1", "index2", "index3"],
+ )
+
+ expected = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A"],
+ "Onehot2": ["C", "D", "missing"],
+ "Binary1": ["E", "F", "F"],
+ "Binary2": ["G", "H", "missing"],
+ "Ordinal1": ["I", "J", "J"],
+ "Ordinal2": ["K", "L", "missing"],
+ "BaseN1": ["M", "N", "N"],
+ "BaseN2": ["O", "P", np.nan],
+ "Target1": ["Q", "R", "R"],
+ "Target2": ["S", "T", "NaN"],
+ "other": ["other", "123", np.nan],
+ },
+ index=["index1", "index2", "index3"],
+ )
+
+ y = pd.DataFrame(data=[0, 1, 0, 0], columns=["y"])
+
+ enc_onehot = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"]).fit(train)
train_onehot = enc_onehot.transform(train)
- enc_binary = ce.BinaryEncoder(cols=['Binary1', 'Binary2']).fit(train_onehot)
+ enc_binary = ce.BinaryEncoder(cols=["Binary1", "Binary2"]).fit(train_onehot)
train_binary = enc_binary.transform(train_onehot)
- enc_ordinal = ce.OrdinalEncoder(cols=['Ordinal1', 'Ordinal2']).fit(train_binary)
+ enc_ordinal = ce.OrdinalEncoder(cols=["Ordinal1", "Ordinal2"]).fit(train_binary)
train_ordinal = enc_ordinal.transform(train_binary)
- enc_basen = ce.BaseNEncoder(cols=['BaseN1', 'BaseN2']).fit(train_ordinal)
+ enc_basen = ce.BaseNEncoder(cols=["BaseN1", "BaseN2"]).fit(train_ordinal)
train_basen = enc_basen.transform(train_ordinal)
- enc_target = ce.TargetEncoder(cols=['Target1', 'Target2']).fit(train_basen, y)
+ enc_target = ce.TargetEncoder(cols=["Target1", "Target2"]).fit(train_basen, y)
input_dict1 = dict()
- input_dict1['col'] = 'Onehot2'
- input_dict1['mapping'] = pd.Series(data=['C', 'D', np.nan], index=['C', 'D', 'missing'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "Onehot2"
+ input_dict1["mapping"] = pd.Series(data=["C", "D", np.nan], index=["C", "D", "missing"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'Binary2'
- input_dict2['mapping'] = pd.Series(data=['G', 'H', np.nan], index=['G', 'H', 'missing'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "Binary2"
+ input_dict2["mapping"] = pd.Series(data=["G", "H", np.nan], index=["G", "H", "missing"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'Ordinal2'
- input_dict3['mapping'] = pd.Series(data=['K', 'L', np.nan], index=['K', 'L', 'missing'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "Ordinal2"
+ input_dict3["mapping"] = pd.Series(data=["K", "L", np.nan], index=["K", "L", "missing"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
result1 = enc_onehot.transform(test)
@@ -446,8 +496,9 @@ def test_inverse_transform_26(self):
result4 = enc_basen.transform(result3)
result5 = enc_target.transform(result4)
- original = inverse_transform(result5, [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1,
- list_dict])
+ original = inverse_transform(
+ result5, [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1, list_dict]
+ )
pd.testing.assert_frame_equal(expected, original)
@@ -455,11 +506,9 @@ def test_transform_ce_1(self):
"""
Unit test for apply preprocessing on OneHotEncoder
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.one_hot.OneHotEncoder(cols=["num1", "num2"])
@@ -467,9 +516,7 @@ def test_transform_ce_1(self):
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -481,20 +528,16 @@ def test_transform_ce_2(self):
"""
Unit test for apply preprocessing on OrdinalEncoder
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.ordinal.OrdinalEncoder(cols=["num1", "num2"])
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -506,11 +549,9 @@ def test_transform_ce_3(self):
"""
Unit test for apply preprocessing on BaseNEncoder
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.basen.BaseNEncoder(cols=["num1", "num2"])
@@ -518,9 +559,7 @@ def test_transform_ce_3(self):
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -532,20 +571,16 @@ def test_transform_ce_4(self):
"""
Unit test for apply preprocessing on BinaryEncoder
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.binary.BinaryEncoder(cols=["num1", "num2"])
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -557,11 +592,9 @@ def test_transform_ce_5(self):
"""
Unit test for apply preprocessing with sklearn model
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.ordinal.OrdinalEncoder(cols=["num1", "num2"])
@@ -569,9 +602,7 @@ def test_transform_ce_5(self):
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = GradientBoostingClassifier().fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -582,11 +613,9 @@ def test_transform_ce_6(self):
"""
Unit test for apply preprocessing with catboost model
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.ordinal.OrdinalEncoder(cols=["num1", "num2"])
@@ -594,9 +623,7 @@ def test_transform_ce_6(self):
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -608,11 +635,9 @@ def test_transform_ce_7(self):
"""
Unit test for apply preprocessing with lightgbm model
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.ordinal.OrdinalEncoder(cols=["num1", "num2"])
@@ -620,9 +645,7 @@ def test_transform_ce_7(self):
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = lightgbm.sklearn.LGBMClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -634,11 +657,9 @@ def test_transform_ce_8(self):
"""
Unit test for apply preprocessing with xgboost model
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
enc = ce.ordinal.OrdinalEncoder(cols=["num1", "num2"])
@@ -646,9 +667,7 @@ def test_transform_ce_8(self):
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = xgboost.sklearn.XGBClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -660,16 +679,16 @@ def test_get_col_mapping_ce_1(self):
"""
Test test_get_col_mapping_ce with target encoding
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "paris"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
- enc = ce.TargetEncoder(cols=['city', 'state'])
+ enc = ce.TargetEncoder(cols=["city", "state"])
enc.fit(test, y)
mapping = get_col_mapping_ce(enc)
- expected_mapping = {'city': ['city'], 'state': ['state']}
+ expected_mapping = {"city": ["city"], "state": ["state"]}
self.assertDictEqual(mapping, expected_mapping)
@@ -677,16 +696,16 @@ def test_get_col_mapping_ce_2(self):
"""
Test test_get_col_mapping_ce with target OrdinalEncoder
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "paris"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
- enc = ce.OrdinalEncoder(handle_missing='value', handle_unknown='value')
+ enc = ce.OrdinalEncoder(handle_missing="value", handle_unknown="value")
enc.fit(test, y)
mapping = get_col_mapping_ce(enc)
- expected_mapping = {'city': ['city'], 'state': ['state'], 'other': ['other']}
+ expected_mapping = {"city": ["city"], "state": ["state"], "other": ["other"]}
self.assertDictEqual(mapping, expected_mapping)
@@ -694,16 +713,16 @@ def test_get_col_mapping_ce_3(self):
"""
Test test_get_col_mapping_ce with target BinaryEncoder
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "paris"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
- enc = ce.BinaryEncoder(cols=['city', 'state'])
+ enc = ce.BinaryEncoder(cols=["city", "state"])
enc.fit(test, y)
mapping = get_col_mapping_ce(enc)
- expected_mapping = {'city': ['city_0', 'city_1'], 'state': ['state_0', 'state_1']}
+ expected_mapping = {"city": ["city_0", "city_1"], "state": ["state_0", "state_1"]}
self.assertDictEqual(mapping, expected_mapping)
@@ -711,21 +730,27 @@ def test_get_col_mapping_ce_4(self):
"""
Test test_get_col_mapping_ce with target BaseNEncoder
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'new york'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "new york"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
enc = ce.BaseNEncoder(base=2)
enc.fit(test, y)
mapping = get_col_mapping_ce(enc)
- if ce.__version__ <= '2.2.2':
- expected_mapping = {'city': ['city_0', 'city_1', 'city_2'], 'state': ['state_0', 'state_1'],
- 'other': ['other_0', 'other_1']}
+ if ce.__version__ <= "2.2.2":
+ expected_mapping = {
+ "city": ["city_0", "city_1", "city_2"],
+ "state": ["state_0", "state_1"],
+ "other": ["other_0", "other_1"],
+ }
else:
- expected_mapping = {'city': ['city_0', 'city_1'], 'state': ['state_0', 'state_1'],
- 'other': ['other_0', 'other_1']}
+ expected_mapping = {
+ "city": ["city_0", "city_1"],
+ "state": ["state_0", "state_1"],
+ "other": ["other_0", "other_1"],
+ }
self.assertDictEqual(mapping, expected_mapping)
@@ -733,15 +758,15 @@ def test_get_col_mapping_ce_5(self):
"""
Test test_get_col_mapping_ce with target BaseNEncoder
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', np.nan, np.nan]})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "chicago"], "state": ["US", "FR", "FR"], "other": ["A", np.nan, np.nan]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
- enc = ce.OneHotEncoder(cols=['city', 'state'], use_cat_names=True)
+ enc = ce.OneHotEncoder(cols=["city", "state"], use_cat_names=True)
enc.fit(test, y)
mapping = get_col_mapping_ce(enc)
- expected_mapping = {'city': ['city_chicago', 'city_paris'], 'state': ['state_US', 'state_FR']}
+ expected_mapping = {"city": ["city_chicago", "city_paris"], "state": ["state_US", "state_FR"]}
self.assertDictEqual(mapping, expected_mapping)
diff --git a/tests/unit_tests/utils/test_check.py b/tests/unit_tests/utils/test_check.py
index c7f144b9..acdc96d4 100644
--- a/tests/unit_tests/utils/test_check.py
+++ b/tests/unit_tests/utils/test_check.py
@@ -1,101 +1,120 @@
"""
Unit test of Check
"""
+import types
import unittest
-import pytest
-import pandas as pd
-import numpy as np
+
+import catboost as cb
import category_encoders as ce
-from shapash.utils.check import check_preprocessing, check_model, check_label_dict,\
- check_mask_params, check_y, check_contribution_object,\
- check_preprocessing_options, check_postprocessing, \
- check_consistency_model_features, check_consistency_model_label, \
- check_additional_data
-from sklearn.compose import ColumnTransformer
-import sklearn.preprocessing as skp
-import types
+import lightgbm as lgb
+import numpy as np
+import pandas as pd
+import pytest
import sklearn.ensemble as ske
-import sklearn.svm as svm
import sklearn.linear_model as skl
+import sklearn.preprocessing as skp
+import sklearn.svm as svm
import xgboost as xgb
-import lightgbm as lgb
-import catboost as cb
+from sklearn.compose import ColumnTransformer
+from shapash.utils.check import (
+ check_additional_data,
+ check_consistency_model_features,
+ check_consistency_model_label,
+ check_contribution_object,
+ check_label_dict,
+ check_mask_params,
+ check_model,
+ check_postprocessing,
+ check_preprocessing,
+ check_preprocessing_options,
+ check_y,
+)
-class TestCheck(unittest.TestCase):
+class TestCheck(unittest.TestCase):
def setUp(self):
self.modellist = [
- lgb.LGBMRegressor(n_estimators=1), lgb.LGBMClassifier(n_estimators=1),
- xgb.XGBRegressor(n_estimators=1), xgb.XGBRegressor(n_estimators=1),
- cb.CatBoostRegressor(n_estimators=1), cb.CatBoostClassifier(n_estimators=1),
- ske.GradientBoostingRegressor(n_estimators=1), ske.GradientBoostingClassifier(n_estimators=1),
- ske.ExtraTreesRegressor(n_estimators=1), ske.ExtraTreesClassifier(n_estimators=1),
- ske.RandomForestRegressor(n_estimators=1), ske.RandomForestClassifier(n_estimators=1),
- skl.LogisticRegression(), skl.LinearRegression(),
- svm.SVR(kernel='linear'), svm.SVC(kernel='linear')
+ lgb.LGBMRegressor(n_estimators=1),
+ lgb.LGBMClassifier(n_estimators=1),
+ xgb.XGBRegressor(n_estimators=1),
+ xgb.XGBRegressor(n_estimators=1),
+ cb.CatBoostRegressor(n_estimators=1),
+ cb.CatBoostClassifier(n_estimators=1),
+ ske.GradientBoostingRegressor(n_estimators=1),
+ ske.GradientBoostingClassifier(n_estimators=1),
+ ske.ExtraTreesRegressor(n_estimators=1),
+ ske.ExtraTreesClassifier(n_estimators=1),
+ ske.RandomForestRegressor(n_estimators=1),
+ ske.RandomForestClassifier(n_estimators=1),
+ skl.LogisticRegression(),
+ skl.LinearRegression(),
+ svm.SVR(kernel="linear"),
+ svm.SVC(kernel="linear"),
]
def test_check_preprocessing_1(self):
"""
Test check preprocessing on multiple preprocessing
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
- y = pd.DataFrame(data=[0, 1, 0, 0], columns=['y'])
+ y = pd.DataFrame(data=[0, 1, 0, 0], columns=["y"])
- enc_onehot = ce.OneHotEncoder(cols=['Onehot1', 'Onehot2']).fit(train)
+ enc_onehot = ce.OneHotEncoder(cols=["Onehot1", "Onehot2"]).fit(train)
train_onehot = enc_onehot.transform(train)
- enc_binary = ce.BinaryEncoder(cols=['Binary1', 'Binary2']).fit(train_onehot)
+ enc_binary = ce.BinaryEncoder(cols=["Binary1", "Binary2"]).fit(train_onehot)
train_binary = enc_binary.transform(train_onehot)
- enc_ordinal = ce.OrdinalEncoder(cols=['Ordinal1', 'Ordinal2']).fit(train_binary)
+ enc_ordinal = ce.OrdinalEncoder(cols=["Ordinal1", "Ordinal2"]).fit(train_binary)
train_ordinal = enc_ordinal.transform(train_binary)
- enc_basen = ce.BaseNEncoder(cols=['BaseN1', 'BaseN2']).fit(train_ordinal)
+ enc_basen = ce.BaseNEncoder(cols=["BaseN1", "BaseN2"]).fit(train_ordinal)
train_basen = enc_basen.transform(train_ordinal)
- enc_target = ce.TargetEncoder(cols=['Target1', 'Target2']).fit(train_basen, y)
+ enc_target = ce.TargetEncoder(cols=["Target1", "Target2"]).fit(train_basen, y)
input_dict1 = dict()
- input_dict1['col'] = 'Onehot2'
- input_dict1['mapping'] = pd.Series(data=['C', 'D', np.nan], index=['C', 'D', 'missing'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "Onehot2"
+ input_dict1["mapping"] = pd.Series(data=["C", "D", np.nan], index=["C", "D", "missing"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'Binary2'
- input_dict2['mapping'] = pd.Series(data=['G', 'H', np.nan], index=['G', 'H', 'missing'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "Binary2"
+ input_dict2["mapping"] = pd.Series(data=["G", "H", np.nan], index=["G", "H", "missing"])
+ input_dict2["data_type"] = "object"
input_dict = dict()
- input_dict['col'] = 'state'
- input_dict['mapping'] = pd.Series(data=['US', 'FR-1', 'FR-2'], index=['US', 'FR', 'FR'])
- input_dict['data_type'] = 'object'
+ input_dict["col"] = "state"
+ input_dict["mapping"] = pd.Series(data=["US", "FR-1", "FR-2"], index=["US", "FR", "FR"])
+ input_dict["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'Ordinal2'
- input_dict3['mapping'] = pd.Series(data=['K', 'L', np.nan], index=['K', 'L', 'missing'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "Ordinal2"
+ input_dict3["mapping"] = pd.Series(data=["K", "L", np.nan], index=["K", "L", "missing"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
- enc = ColumnTransformer(
- transformers=[
- ('onehot', skp.OneHotEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
+ enc = ColumnTransformer(transformers=[("onehot", skp.OneHotEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
wrong_prepro = skp.OneHotEncoder().fit(train, y)
- check_preprocessing([enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1,
- list_dict])
+ check_preprocessing([enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target, input_dict1, list_dict])
for preprocessing in [enc_onehot, enc_binary, enc_ordinal, enc_basen, enc_target]:
check_preprocessing(preprocessing)
@@ -113,27 +132,21 @@ def test_check_model_1(self):
model = lambda: None
model.predict = types.MethodType(self.predict, model)
_case, _classes = check_model(model)
- assert _case == 'regression'
+ assert _case == "regression"
assert _classes is None
def predict_proba(self, arg1, arg2):
"""
predict_proba method
"""
- matrx = np.array(
- [[0.2, 0.8],
- [0.3, 0.7],
- [0.4, 0.6]]
- )
+ matrx = np.array([[0.2, 0.8], [0.3, 0.7], [0.4, 0.6]])
return matrx
def predict(self, arg1, arg2):
"""
predict method
"""
- matrx = np.array(
- [12, 3, 7]
- )
+ matrx = np.array([12, 3, 7])
return matrx
def test_check_model_2(self):
@@ -145,16 +158,16 @@ def test_check_model_2(self):
model.predict = types.MethodType(self.predict, model)
model.predict_proba = types.MethodType(self.predict_proba, model)
_case, _classes = check_model(model)
- assert _case == 'classification'
+ assert _case == "classification"
self.assertListEqual(_classes, [1, 2])
def test_check_label_dict_1(self):
"""
Unit test check label dict 1
"""
- label_dict={1: 'Yes', 0: 'No'}
+ label_dict = {1: "Yes", 0: "No"}
_classes = [0, 1]
- _case = 'classification'
+ _case = "classification"
check_label_dict(label_dict, _case, _classes)
def test_check_label_dict_2(self):
@@ -162,7 +175,7 @@ def test_check_label_dict_2(self):
Unit test check label dict 2
"""
label_dict = {}
- _case = 'regression'
+ _case = "regression"
check_label_dict(label_dict, _case)
def test_check_mask_params(self):
@@ -171,17 +184,8 @@ def test_check_mask_params(self):
"""
wrong_mask_params_1 = list()
wrong_mask_params_2 = None
- wrong_mask_params_3 = {
- "features_to_hide": None,
- "threshold": None,
- "positive": None
- }
- wright_mask_params = {
- "features_to_hide": None,
- "threshold": None,
- "positive": True,
- "max_contrib": 5
- }
+ wrong_mask_params_3 = {"features_to_hide": None, "threshold": None, "positive": None}
+ wright_mask_params = {"features_to_hide": None, "threshold": None, "positive": True, "max_contrib": 5}
with self.assertRaises(ValueError):
check_mask_params(wrong_mask_params_1)
check_mask_params(wrong_mask_params_2)
@@ -199,14 +203,8 @@ def test_check_y_2(self):
"""
Unit test check y 2
"""
- x_init = pd.DataFrame(
- data=np.array([[1, 2], [3, 4]]),
- columns=['Col1', 'Col2']
- )
- y_pred = pd.DataFrame(
- data=np.array(['1', 0]),
- columns=['Y']
- )
+ x_init = pd.DataFrame(data=np.array([[1, 2], [3, 4]]), columns=["Col1", "Col2"])
+ y_pred = pd.DataFrame(data=np.array(["1", 0]), columns=["Y"])
with self.assertRaises(ValueError):
check_y(x_init, y_pred)
@@ -214,14 +212,8 @@ def test_check_y_3(self):
"""
Unit test check y 3
"""
- x_init = pd.DataFrame(
- data=np.array([[1, 2], [3, 4]]),
- columns=['Col1', 'Col2']
- )
- y_pred = pd.DataFrame(
- data=np.array([0]),
- columns=['Y']
- )
+ x_init = pd.DataFrame(data=np.array([[1, 2], [3, 4]]), columns=["Col1", "Col2"])
+ y_pred = pd.DataFrame(data=np.array([0]), columns=["Y"])
with self.assertRaises(ValueError):
check_y(x_init, y_pred)
@@ -237,13 +229,8 @@ def test_check_y_5(self):
"""
Unit test check y 5
"""
- x_init = pd.DataFrame(
- data=np.array([[1, 2], [3, 4]]),
- columns=['Col1', 'Col2']
- )
- y_pred = pd.Series(
- data=np.array(['0'])
- )
+ x_init = pd.DataFrame(data=np.array([[1, 2], [3, 4]]), columns=["Col1", "Col2"])
+ y_pred = pd.Series(data=np.array(["0"]))
with self.assertRaises(ValueError):
check_y(x_init, y_pred)
@@ -251,25 +238,17 @@ def test_check_y_6(self):
"""
Unit test check y 6
"""
- x_init = pd.DataFrame(
- data=np.array([[1, 2], [3, 4]]),
- columns=['Col1', 'Col2']
- )
- y_pred = pd.Series(
- data=np.array(['0'])
- )
+ x_init = pd.DataFrame(data=np.array([[1, 2], [3, 4]]), columns=["Col1", "Col2"])
+ y_pred = pd.Series(data=np.array(["0"]))
with pytest.raises(Exception) as exc_info:
check_y(x_init, y_pred, y_name="y_pred")
- assert str(exc_info.value) == 'x and y_pred should have the same index.'
+ assert str(exc_info.value) == "x and y_pred should have the same index."
def test_check_contribution_object_1(self):
"""
Unit test check_contribution_object 1
"""
- contributions_1 = [
- np.array([[2, 1], [8, 4]]),
- np.array([[5, 5], [0, 0]])
- ]
+ contributions_1 = [np.array([[2, 1], [8, 4]]), np.array([[5, 5], [0, 0]])]
contributions_2 = np.array([[2, 1], [8, 4]])
model = lambda: None
@@ -294,83 +273,122 @@ def test_check_consistency_model_features_1(self):
"""
Test check_consistency_model_features 1
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
features_dict = None
- columns_dict = {i:features for i,features in enumerate(train.columns)}
+ columns_dict = {i: features for i, features in enumerate(train.columns)}
features_types = {features: str(train[features].dtypes) for features in train.columns}
label_dict = None
mask_params = None
- enc_ordinal_all = ce.OrdinalEncoder(cols=['Onehot1', 'Onehot2', 'Binary1', 'Binary2', 'Ordinal1', 'Ordinal2',
- 'BaseN1', 'BaseN2', 'Target1', 'Target2', 'other']).fit(train)
- train_ordinal_all = enc_ordinal_all.transform(train)
+ enc_ordinal_all = ce.OrdinalEncoder(
+ cols=[
+ "Onehot1",
+ "Onehot2",
+ "Binary1",
+ "Binary2",
+ "Ordinal1",
+ "Ordinal2",
+ "BaseN1",
+ "BaseN2",
+ "Target1",
+ "Target2",
+ "other",
+ ]
+ ).fit(train)
+ train_ordinal_all = enc_ordinal_all.transform(train)
preprocessing = enc_ordinal_all
- y = pd.DataFrame({'y_class': [0, 0, 0, 1]})
+ y = pd.DataFrame({"y_class": [0, 0, 0, 1]})
model = cb.CatBoostClassifier(n_estimators=1).fit(train_ordinal_all, y)
- check_consistency_model_features(features_dict, model, columns_dict,
- features_types, mask_params, preprocessing)
+ check_consistency_model_features(features_dict, model, columns_dict, features_types, mask_params, preprocessing)
def test_check_consistency_model_features_2(self):
"""
Test check_consistency_model_features 2
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
features_dict = None
columns_dict = {i: features for i, features in enumerate(train.columns)}
features_types = {features: str(train[features].dtypes) for features in train.columns}
- mask_params = {
- "features_to_hide": 'Binary3',
- "threshold": None,
- "positive": True,
- "max_contrib": 5
- }
-
- enc_ordinal_all = ce.OrdinalEncoder(cols=['Onehot1', 'Onehot2', 'Binary1', 'Binary2', 'Ordinal1', 'Ordinal2',
- 'BaseN1', 'BaseN2', 'Target1', 'Target2', 'other']).fit(train)
+ mask_params = {"features_to_hide": "Binary3", "threshold": None, "positive": True, "max_contrib": 5}
+
+ enc_ordinal_all = ce.OrdinalEncoder(
+ cols=[
+ "Onehot1",
+ "Onehot2",
+ "Binary1",
+ "Binary2",
+ "Ordinal1",
+ "Ordinal2",
+ "BaseN1",
+ "BaseN2",
+ "Target1",
+ "Target2",
+ "other",
+ ]
+ ).fit(train)
train_ordinal_all = enc_ordinal_all.transform(train)
preprocessing = enc_ordinal_all
- y = pd.DataFrame({'y_class': [0, 0, 0, 1]})
+ y = pd.DataFrame({"y_class": [0, 0, 0, 1]})
model = cb.CatBoostClassifier(n_estimators=1).fit(train_ordinal_all, y)
with self.assertRaises(ValueError):
- check_consistency_model_features(features_dict, model, columns_dict,
- features_types, mask_params, preprocessing)
+ check_consistency_model_features(
+ features_dict, model, columns_dict, features_types, mask_params, preprocessing
+ )
def test_check_preprocessing_options_1(self):
"""
Unit test 1 for check_preprocessing_options
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
- enc = ColumnTransformer(transformers=[('power', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='drop')
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
+ enc = ColumnTransformer(
+ transformers=[("power", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="drop"
+ )
enc.fit(train, y)
with self.assertRaises(ValueError):
check_preprocessing_options(enc)
- enc = ColumnTransformer(transformers=[('power', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[("power", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="passthrough"
+ )
enc.fit(train, y)
check_preprocessing_options(enc)
@@ -378,42 +396,69 @@ def test_check_consistency_model_features_4(self):
"""
Test check_consistency_model_features 1
"""
- train = pd.DataFrame({'Onehot1': ['A', 'B', 'A', 'B'], 'Onehot2': ['C', 'D', 'C', 'D'],
- 'Binary1': ['E', 'F', 'E', 'F'], 'Binary2': ['G', 'H', 'G', 'H'],
- 'Ordinal1': ['I', 'J', 'I', 'J'], 'Ordinal2': ['K', 'L', 'K', 'L'],
- 'BaseN1': ['M', 'N', 'M', 'N'], 'BaseN2': ['O', 'P', 'O', 'P'],
- 'Target1': ['Q', 'R', 'Q', 'R'], 'Target2': ['S', 'T', 'S', 'T'],
- 'other': ['other', np.nan, 'other', 'other']})
+ train = pd.DataFrame(
+ {
+ "Onehot1": ["A", "B", "A", "B"],
+ "Onehot2": ["C", "D", "C", "D"],
+ "Binary1": ["E", "F", "E", "F"],
+ "Binary2": ["G", "H", "G", "H"],
+ "Ordinal1": ["I", "J", "I", "J"],
+ "Ordinal2": ["K", "L", "K", "L"],
+ "BaseN1": ["M", "N", "M", "N"],
+ "BaseN2": ["O", "P", "O", "P"],
+ "Target1": ["Q", "R", "Q", "R"],
+ "Target2": ["S", "T", "S", "T"],
+ "other": ["other", np.nan, "other", "other"],
+ }
+ )
features_dict = None
- columns_dict = {i:features for i,features in enumerate(train.columns)}
+ columns_dict = {i: features for i, features in enumerate(train.columns)}
features_types = {features: str(train[features].dtypes) for features in train.columns}
label_dict = None
mask_params = None
- enc_ordinal_all = ce.OrdinalEncoder(cols=['Onehot1', 'Onehot2', 'Binary1', 'Binary2', 'Ordinal1', 'Ordinal2',
- 'BaseN1', 'BaseN2', 'Target1', 'Target2', 'other']).fit(train)
- train_ordinal_all = enc_ordinal_all.transform(train)
+ enc_ordinal_all = ce.OrdinalEncoder(
+ cols=[
+ "Onehot1",
+ "Onehot2",
+ "Binary1",
+ "Binary2",
+ "Ordinal1",
+ "Ordinal2",
+ "BaseN1",
+ "BaseN2",
+ "Target1",
+ "Target2",
+ "other",
+ ]
+ ).fit(train)
+ train_ordinal_all = enc_ordinal_all.transform(train)
preprocessing = enc_ordinal_all
- y = pd.DataFrame({'y_class': [0, 0, 0, 1]})
+ y = pd.DataFrame({"y_class": [0, 0, 0, 1]})
for model in self.modellist:
print(type(model))
model.fit(train_ordinal_all, y)
- check_consistency_model_features(features_dict, model, columns_dict,
- features_types, mask_params, preprocessing,
- list_preprocessing=[preprocessing])
+ check_consistency_model_features(
+ features_dict,
+ model,
+ columns_dict,
+ features_types,
+ mask_params,
+ preprocessing,
+ list_preprocessing=[preprocessing],
+ )
def test_check_consistency_model_features_5(self):
"""
Unit test check_consistency_model_features 5
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': [5, 10]},
- index=['index1', 'index2'])
+ train = pd.DataFrame(
+ {"city": ["chicago", "paris"], "state": ["US", "FR"], "other": [5, 10]}, index=["index1", "index2"]
+ )
features_dict = None
columns_dict = {i: features for i, features in enumerate(train.columns)}
@@ -422,71 +467,72 @@ def test_check_consistency_model_features_5(self):
enc = ColumnTransformer(
transformers=[
- ('Ordinal_ce', ce.OrdinalEncoder(), ['city', 'state']),
- ('Ordinal_skp', skp.OrdinalEncoder(), ['city', 'state'])
+ ("Ordinal_ce", ce.OrdinalEncoder(), ["city", "state"]),
+ ("Ordinal_skp", skp.OrdinalEncoder(), ["city", "state"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
enc_2 = ColumnTransformer(
transformers=[
- ('Ordinal_ce', ce.OrdinalEncoder(), ['city', 'state']),
- ('Ordinal_skp', skp.OrdinalEncoder(), ['city', 'state'])
+ ("Ordinal_ce", ce.OrdinalEncoder(), ["city", "state"]),
+ ("Ordinal_skp", skp.OrdinalEncoder(), ["city", "state"]),
],
- remainder='drop')
+ remainder="drop",
+ )
enc.fit(train)
train_1 = pd.DataFrame(enc.transform(train), columns=["city_ce", "state_ce", "city_skp", "state_skp", "other"])
train_1["y"] = np.array([1, 0])
- clf_1 = cb.CatBoostClassifier(n_estimators=1) \
- .fit(train_1[["city_ce", "state_ce", "city_skp", "state_skp", "other"]],
- train_1['y'])
+ clf_1 = cb.CatBoostClassifier(n_estimators=1).fit(
+ train_1[["city_ce", "state_ce", "city_skp", "state_skp", "other"]], train_1["y"]
+ )
enc_2.fit(train)
train_2 = pd.DataFrame(enc_2.transform(train), columns=["city_ce", "state_ce", "city_skp", "state_skp"])
train_2["y"] = np.array([1, 0])
- clf_2 = cb.CatBoostClassifier(n_estimators=1) \
- .fit(train_2[["city_ce", "state_ce", "city_skp", "state_skp"]],
- train_2['y'])
+ clf_2 = cb.CatBoostClassifier(n_estimators=1).fit(
+ train_2[["city_ce", "state_ce", "city_skp", "state_skp"]], train_2["y"]
+ )
- enc_3 = ce.OneHotEncoder(cols=['city', 'state'])
+ enc_3 = ce.OneHotEncoder(cols=["city", "state"])
enc_3.fit(train)
train_3 = enc_3.transform(train)
train_3["y"] = np.array([1, 0])
- clf_3 = cb.CatBoostClassifier(n_estimators=1) \
- .fit(train_3[["city_1", "city_2", "state_1", "state_2", "other"]],
- train_3['y'])
+ clf_3 = cb.CatBoostClassifier(n_estimators=1).fit(
+ train_3[["city_1", "city_2", "state_1", "state_2", "other"]], train_3["y"]
+ )
- dict_4 = {'col': 'state',
- 'mapping': pd.Series(data=[1, 2],
- index=['US', 'FR']),
- 'data_type': 'object'}
+ dict_4 = {"col": "state", "mapping": pd.Series(data=[1, 2], index=["US", "FR"]), "data_type": "object"}
- dict_5 = {'col': 'city',
- 'mapping': pd.Series(data=[1, 2],
- index=['chicago', 'paris']),
- 'data_type': 'object'}
+ dict_5 = {"col": "city", "mapping": pd.Series(data=[1, 2], index=["chicago", "paris"]), "data_type": "object"}
enc_4 = [enc_3, [dict_4]]
enc_5 = [enc_3, [dict_4, dict_5]]
- check_consistency_model_features(features_dict, clf_1, columns_dict,
- features_types, mask_params, enc, list_preprocessing=[enc])
+ check_consistency_model_features(
+ features_dict, clf_1, columns_dict, features_types, mask_params, enc, list_preprocessing=[enc]
+ )
- check_consistency_model_features(features_dict, clf_2, columns_dict,
- features_types, mask_params, enc_2, list_preprocessing=[enc_2])
+ check_consistency_model_features(
+ features_dict, clf_2, columns_dict, features_types, mask_params, enc_2, list_preprocessing=[enc_2]
+ )
- check_consistency_model_features(features_dict, clf_3, columns_dict,
- features_types, mask_params, enc_3, list_preprocessing=[enc_3])
+ check_consistency_model_features(
+ features_dict, clf_3, columns_dict, features_types, mask_params, enc_3, list_preprocessing=[enc_3]
+ )
- check_consistency_model_features(features_dict, clf_3, columns_dict,
- features_types, mask_params, enc_4, list_preprocessing=enc_4)
+ check_consistency_model_features(
+ features_dict, clf_3, columns_dict, features_types, mask_params, enc_4, list_preprocessing=enc_4
+ )
- check_consistency_model_features(features_dict, clf_3, columns_dict,
- features_types, mask_params, enc_5, list_preprocessing=enc_5)
+ check_consistency_model_features(
+ features_dict, clf_3, columns_dict, features_types, mask_params, enc_5, list_preprocessing=enc_5
+ )
def test_check_consistency_model_label_1(self):
"""
@@ -511,20 +557,15 @@ def test_check_postprocessing_1(self):
"""
Unit test check_consistency_postprocessing
"""
- x = pd.DataFrame(
- [[1, 2],
- [3, 4]],
- columns=['Col1', 'Col2'],
- index=['Id1', 'Id2']
- )
- columns_dict = {0: 'Col1', 1: 'Col2'}
+ x = pd.DataFrame([[1, 2], [3, 4]], columns=["Col1", "Col2"], index=["Id1", "Id2"])
+ columns_dict = {0: "Col1", 1: "Col2"}
features_types = {features: str(x[features].dtypes) for features in x.columns}
- postprocessing1 = {0: {'Error': 'suffix', 'rule': ' t'}}
- postprocessing2 = {0: {'type': 'Error', 'rule': ' t'}}
- postprocessing3 = {0: {'type': 'suffix', 'Error': ' t'}}
- postprocessing4 = {0: {'type': 'suffix', 'rule': ' '}}
- postprocessing5 = {0: {'type': 'case', 'rule': 'lower'}}
- postprocessing6 = {0: {'type': 'case', 'rule': 'Error'}}
+ postprocessing1 = {0: {"Error": "suffix", "rule": " t"}}
+ postprocessing2 = {0: {"type": "Error", "rule": " t"}}
+ postprocessing3 = {0: {"type": "suffix", "Error": " t"}}
+ postprocessing4 = {0: {"type": "suffix", "rule": " "}}
+ postprocessing5 = {0: {"type": "case", "rule": "lower"}}
+ postprocessing6 = {0: {"type": "case", "rule": "Error"}}
with self.assertRaises(ValueError):
check_postprocessing(features_types, postprocessing1)
check_postprocessing(features_types, postprocessing2)
@@ -537,30 +578,34 @@ def test_check_preprocessing_options_1(self):
"""
Unit test check_preprocessing_options 1
"""
- df = pd.DataFrame(range(0, 5), columns=['id'])
- df['y'] = df['id'].apply(lambda x: 1 if x < 2 else 0)
- df['x1'] = np.random.randint(1, 123, df.shape[0])
- df = df.set_index('id')
- df['x2'] = ["S", "M", "S", "D", "M"]
- df['x3'] = np.random.randint(1, 123, df.shape[0])
- df['x4'] = ["S", "M", "S", "D", "M"]
+ df = pd.DataFrame(range(0, 5), columns=["id"])
+ df["y"] = df["id"].apply(lambda x: 1 if x < 2 else 0)
+ df["x1"] = np.random.randint(1, 123, df.shape[0])
+ df = df.set_index("id")
+ df["x2"] = ["S", "M", "S", "D", "M"]
+ df["x3"] = np.random.randint(1, 123, df.shape[0])
+ df["x4"] = ["S", "M", "S", "D", "M"]
- features_dict = {"x1": "age", "x2": "weight", "x3": 'test', 'x4': "test2"}
+ features_dict = {"x1": "age", "x2": "weight", "x3": "test", "x4": "test2"}
columns_dict = {0: "x1", 1: "x2", 2: "x3", 3: "x4"}
- encoder = ColumnTransformer(transformers=[('onehot_ce_1', ce.OneHotEncoder(), ['x2']),
- ('onehot_ce_2', ce.OneHotEncoder(), ['x4'])],
- remainder='drop')
- encoder_fitted = encoder.fit(df[["x1", "x2", "x3", 'x4']])
+ encoder = ColumnTransformer(
+ transformers=[("onehot_ce_1", ce.OneHotEncoder(), ["x2"]), ("onehot_ce_2", ce.OneHotEncoder(), ["x4"])],
+ remainder="drop",
+ )
+ encoder_fitted = encoder.fit(df[["x1", "x2", "x3", "x4"]])
- encoder_2 = ColumnTransformer(transformers=[('onehot_ce_1', ce.OneHotEncoder(), ['x2']),
- ('onehot_ce_2', ce.OneHotEncoder(), ['x4'])],
- remainder='passthrough')
- encoder_fitted_2 = encoder_2.fit(df[["x1", "x2", "x3", 'x4']])
+ encoder_2 = ColumnTransformer(
+ transformers=[("onehot_ce_1", ce.OneHotEncoder(), ["x2"]), ("onehot_ce_2", ce.OneHotEncoder(), ["x4"])],
+ remainder="passthrough",
+ )
+ encoder_fitted_2 = encoder_2.fit(df[["x1", "x2", "x3", "x4"]])
- expected_dict = {'features_to_drop': ['x1', 'x3'],
- 'features_dict_op': {'x2': 'weight', 'x4': 'test2'},
- 'columns_dict_op': {0: 'x2', 1: 'x4'}}
+ expected_dict = {
+ "features_to_drop": ["x1", "x3"],
+ "features_dict_op": {"x2": "weight", "x4": "test2"},
+ "columns_dict_op": {0: "x2", 1: "x4"},
+ }
expected_dict_2 = None
@@ -570,28 +615,15 @@ def test_check_preprocessing_options_1(self):
assert drop_option_2 == expected_dict_2
def test_check_additional_data_raises_index(self):
- x_init = pd.DataFrame(
- data=np.array([[1, 2], [3, 4]]),
- columns=['Col1', 'Col2']
- )
- additional_data = pd.DataFrame(
- data=np.array([[5]]),
- columns=['Col3']
- )
+ x_init = pd.DataFrame(data=np.array([[1, 2], [3, 4]]), columns=["Col1", "Col2"])
+ additional_data = pd.DataFrame(data=np.array([[5]]), columns=["Col3"])
with pytest.raises(Exception) as exc_info:
check_additional_data(x_init, additional_data)
- assert str(exc_info.value) == 'x and additional_data should have the same index.'
+ assert str(exc_info.value) == "x and additional_data should have the same index."
def test_check_additional_data_raises_type(self):
- x_init = pd.DataFrame(
- data=np.array([[1, 2], [3, 4]]),
- columns=['Col1', 'Col2']
- )
- additional_data = pd.Series(
- data=np.array([5, 6]),
- name='Col3'
- )
+ x_init = pd.DataFrame(data=np.array([[1, 2], [3, 4]]), columns=["Col1", "Col2"])
+ additional_data = pd.Series(data=np.array([5, 6]), name="Col3")
with pytest.raises(Exception) as exc_info:
check_additional_data(x_init, additional_data)
- assert str(exc_info.value) == 'additional_data must be a pd.Dataframe.'
-
+ assert str(exc_info.value) == "additional_data must be a pd.Dataframe."
diff --git a/tests/unit_tests/utils/test_columntransformer_backend.py b/tests/unit_tests/utils/test_columntransformer_backend.py
index d0ba1785..6405fbda 100644
--- a/tests/unit_tests/utils/test_columntransformer_backend.py
+++ b/tests/unit_tests/utils/test_columntransformer_backend.py
@@ -2,57 +2,66 @@
Unit test of Inverse Transform
"""
import unittest
-import pandas as pd
-import numpy as np
-import category_encoders as ce
-from sklearn.compose import ColumnTransformer
-import sklearn.preprocessing as skp
+
import catboost as cb
-import sklearn as sk
+import category_encoders as ce
import lightgbm
+import numpy as np
+import pandas as pd
+import sklearn as sk
+import sklearn.preprocessing as skp
import xgboost
-from shapash.utils.transform import inverse_transform
-from shapash.utils.transform import apply_preprocessing
-from shapash.utils.columntransformer_backend import get_feature_names, get_names, get_list_features_names, \
- get_col_mapping_ct
+from sklearn.compose import ColumnTransformer
from sklearn.ensemble import GradientBoostingClassifier
+from shapash.utils.columntransformer_backend import (
+ get_col_mapping_ct,
+ get_feature_names,
+ get_list_features_names,
+ get_names,
+)
+from shapash.utils.transform import apply_preprocessing, inverse_transform
# TODO
# StandardScaler return object vs float vs int
# Target encoding return object vs float
+
class TestInverseTransformColumnsTransformer(unittest.TestCase):
def test_inv_transform_ct_1(self):
"""
test inv_transform_ct with multiple encoding and drop option
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']},
- index=['index1', 'index2'])
+ train = pd.DataFrame(
+ {"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]}, index=["index1", "index2"]
+ )
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='drop')
+ remainder="drop",
+ )
enc.fit(train)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
-
- expected = pd.DataFrame({'onehot_ce_city': ['chicago', 'chicago', 'paris'],
- 'onehot_ce_state': ['US', 'FR', 'FR'],
- 'onehot_skp_city': ['chicago', 'chicago', 'paris'],
- 'onehot_skp_state': ['US', 'FR', 'FR']},
- index=['index1', 'index2', 'index3'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
+
+ expected = pd.DataFrame(
+ {
+ "onehot_ce_city": ["chicago", "chicago", "paris"],
+ "onehot_ce_state": ["US", "FR", "FR"],
+ "onehot_skp_city": ["chicago", "chicago", "paris"],
+ "onehot_skp_state": ["US", "FR", "FR"],
+ },
+ index=["index1", "index2", "index3"],
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'col3_0', 'col3_1', 'col4_0', 'col4_1']
- result.index = ['index1', 'index2', 'index3']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "col3_0", "col3_1", "col4_0", "col4_1"]
+ result.index = ["index1", "index2", "index3"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -60,33 +69,37 @@ def test_inv_transform_ct_2(self):
"""
test inv_transform_ct with multiple encoding and passthrough option
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']},
- index=['index1', 'index2'])
+ train = pd.DataFrame(
+ {"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]}, index=["index1", "index2"]
+ )
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
enc.fit(train)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
-
- expected = pd.DataFrame({'onehot_ce_city': ['chicago', 'chicago', 'paris'],
- 'onehot_ce_state': ['US', 'FR', 'FR'],
- 'onehot_skp_city': ['chicago', 'chicago', 'paris'],
- 'onehot_skp_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
+
+ expected = pd.DataFrame(
+ {
+ "onehot_ce_city": ["chicago", "chicago", "paris"],
+ "onehot_ce_state": ["US", "FR", "FR"],
+ "onehot_skp_city": ["chicago", "chicago", "paris"],
+ "onehot_skp_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ },
+ index=["index1", "index2", "index3"],
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'col3_0', 'col3_1', 'col4_0', 'col4_1', 'other']
- result.index = ['index1', 'index2', 'index3']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "col3_0", "col3_1", "col4_0", "col4_1", "other"]
+ result.index = ["index1", "index2", "index3"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -94,48 +107,52 @@ def test_inv_transform_ct_3(self):
"""
test inv_transform_ct with multiple encoding and dictionnary
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']},
- index=['index1', 'index2'])
+ train = pd.DataFrame(
+ {"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]}, index=["index1", "index2"]
+ )
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
enc.fit(train)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
-
- expected = pd.DataFrame({'onehot_ce_city': ['CH', 'CH', 'PR'],
- 'onehot_ce_state': ['US-FR', 'US-FR', 'US-FR'],
- 'onehot_skp_city': ['chicago', 'chicago', 'paris'],
- 'onehot_skp_state': ['US', 'FR', 'FR'],
- 'other': ['A-B', 'A-B', 'C']},
- index=['index1', 'index2', 'index3'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
+
+ expected = pd.DataFrame(
+ {
+ "onehot_ce_city": ["CH", "CH", "PR"],
+ "onehot_ce_state": ["US-FR", "US-FR", "US-FR"],
+ "onehot_skp_city": ["chicago", "chicago", "paris"],
+ "onehot_skp_state": ["US", "FR", "FR"],
+ "other": ["A-B", "A-B", "C"],
+ },
+ index=["index1", "index2", "index3"],
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'col3_0', 'col3_1', 'col4_0', 'col4_1', 'other']
- result.index = ['index1', 'index2', 'index3']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "col3_0", "col3_1", "col4_0", "col4_1", "other"]
+ result.index = ["index1", "index2", "index3"]
input_dict1 = dict()
- input_dict1['col'] = 'onehot_ce_city'
- input_dict1['mapping'] = pd.Series(data=['chicago', 'paris'], index=['CH', 'PR'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "onehot_ce_city"
+ input_dict1["mapping"] = pd.Series(data=["chicago", "paris"], index=["CH", "PR"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'other'
- input_dict2['mapping'] = pd.Series(data=['A', 'B', 'C'], index=['A-B', 'A-B', 'C'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "other"
+ input_dict2["mapping"] = pd.Series(data=["A", "B", "C"], index=["A-B", "A-B", "C"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'onehot_ce_state'
- input_dict3['mapping'] = pd.Series(data=['US', 'FR'], index=['US-FR', 'US-FR'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "onehot_ce_state"
+ input_dict3["mapping"] = pd.Series(data=["US", "FR"], index=["US-FR", "US-FR"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
original = inverse_transform(result, [enc, input_dict1, list_dict])
@@ -145,30 +162,36 @@ def test_inv_transform_ct_4(self):
"""
test inv_transform_ct with single target category encoders and passthrough option
"""
- y = pd.DataFrame(data=[0, 1, 1, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1, 1, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR', 'US'],
- 'other': ['A', 'B', 'B', 'B']})
+ train = pd.DataFrame(
+ {
+ "city": ["chicago", "paris", "paris", "chicago"],
+ "state": ["US", "FR", "FR", "US"],
+ "other": ["A", "B", "B", "B"],
+ }
+ )
enc = ColumnTransformer(
- transformers=[
- ('target', ce.TargetEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
-
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
-
- expected = pd.DataFrame(data={'target_city': ['chicago', 'chicago', 'paris'],
- 'target_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- dtype=object)
+ transformers=[("target", ce.TargetEncoder(), ["city", "state"])], remainder="passthrough"
+ )
+
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
+
+ expected = pd.DataFrame(
+ data={
+ "target_city": ["chicago", "chicago", "paris"],
+ "target_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ },
+ dtype=object,
+ )
enc.fit(train, y)
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2', 'other']
+ result.columns = ["col1", "col2", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -176,28 +199,28 @@ def test_inv_transform_ct_5(self):
"""
test inv_transform_ct with single target category encoders and drop option
"""
- y = pd.DataFrame(data=[0, 1, 0, 0], columns=['y'])
+ y = pd.DataFrame(data=[0, 1, 0, 0], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'US', 'FR'],
- 'other': ['A', 'B', 'A', 'B']})
+ train = pd.DataFrame(
+ {
+ "city": ["chicago", "paris", "chicago", "paris"],
+ "state": ["US", "FR", "US", "FR"],
+ "other": ["A", "B", "A", "B"],
+ }
+ )
- enc = ColumnTransformer(
- transformers=[
- ('target', ce.TargetEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("target", ce.TargetEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame(data={
- 'target_city': ['chicago', 'chicago', 'paris'],
- 'target_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame(
+ data={"target_city": ["chicago", "chicago", "paris"], "target_state": ["US", "FR", "FR"]}
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2']
+ result.columns = ["col1", "col2"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -205,28 +228,21 @@ def test_inv_transform_ct_6(self):
"""
test inv_transform_ct with Ordinal Category Encoder and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('ordinal', ce.OrdinalEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("ordinal", ce.OrdinalEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'ordinal_city': ['chicago', 'chicago', 'paris'],
- 'ordinal_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame({"ordinal_city": ["chicago", "chicago", "paris"], "ordinal_state": ["US", "FR", "FR"]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2']
+ result.columns = ["col1", "col2"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -234,28 +250,28 @@ def test_inv_transform_ct_7(self):
"""
test inv_transform_ct with category Ordinal Encoder and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('ordinal', ce.OrdinalEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ transformers=[("ordinal", ce.OrdinalEncoder(), ["city", "state"])], remainder="passthrough"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
-
- expected = pd.DataFrame({'ordinal_city': ['chicago', 'chicago', 'paris'],
- 'ordinal_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
+
+ expected = pd.DataFrame(
+ {
+ "ordinal_city": ["chicago", "chicago", "paris"],
+ "ordinal_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ }
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2', 'other']
+ result.columns = ["col1", "col2", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -263,28 +279,21 @@ def test_inv_transform_ct_8(self):
"""
test inv_transform_ct with Binary encoder and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('binary', ce.BinaryEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("binary", ce.BinaryEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'binary_city': ['chicago', 'chicago', 'paris'],
- 'binary_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame({"binary_city": ["chicago", "chicago", "paris"], "binary_state": ["US", "FR", "FR"]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -292,28 +301,28 @@ def test_inv_transform_ct_9(self):
"""
test inv_transform_ct with Binary Encoder and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('binary', ce.BinaryEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ transformers=[("binary", ce.BinaryEncoder(), ["city", "state"])], remainder="passthrough"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
-
- expected = pd.DataFrame({'binary_city': ['chicago', 'chicago', 'paris'],
- 'binary_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
+
+ expected = pd.DataFrame(
+ {
+ "binary_city": ["chicago", "chicago", "paris"],
+ "binary_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ }
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'other']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -321,27 +330,20 @@ def test_inv_transform_ct_10(self):
"""
test inv_transform_ct with BaseN Encoder and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('basen', ce.BaseNEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("basen", ce.BaseNEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'basen_city': ['chicago', 'chicago', 'paris'],
- 'basen_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame({"basen_city": ["chicago", "chicago", "paris"], "basen_state": ["US", "FR", "FR"]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -349,28 +351,22 @@ def test_inv_transform_ct_11(self):
"""
test inv_transform_ct with BaseN Encoder and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('basen', ce.BaseNEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ enc = ColumnTransformer(transformers=[("basen", ce.BaseNEncoder(), ["city", "state"])], remainder="passthrough")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'basen_city': ['chicago', 'chicago', 'paris'],
- 'basen_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ expected = pd.DataFrame(
+ {"basen_city": ["chicago", "chicago", "paris"], "basen_state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'other']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -378,27 +374,20 @@ def test_inv_transform_ct_12(self):
"""
test inv_transform_ct with single OneHotEncoder and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('onehot', ce.OneHotEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("onehot", ce.OneHotEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'onehot_city': ['chicago', 'chicago', 'paris'],
- 'onehot_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame({"onehot_city": ["chicago", "chicago", "paris"], "onehot_state": ["US", "FR", "FR"]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -406,28 +395,28 @@ def test_inv_transform_ct_13(self):
"""
test inv_transform_ct with OneHotEncoder and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('onehot', ce.OneHotEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ transformers=[("onehot", ce.OneHotEncoder(), ["city", "state"])], remainder="passthrough"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
-
- expected = pd.DataFrame({'onehot_city': ['chicago', 'chicago', 'paris'],
- 'onehot_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
+
+ expected = pd.DataFrame(
+ {
+ "onehot_city": ["chicago", "chicago", "paris"],
+ "onehot_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ }
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'other']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -435,27 +424,20 @@ def test_inv_transform_ct_14(self):
"""
test inv_transform_ct with OneHotEncoder Sklearn and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('onehot', skp.OneHotEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("onehot", skp.OneHotEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'onehot_city': ['chicago', 'chicago', 'paris'],
- 'onehot_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame({"onehot_city": ["chicago", "chicago", "paris"], "onehot_state": ["US", "FR", "FR"]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -463,28 +445,28 @@ def test_inv_transform_ct_15(self):
"""
test inv_transform_ct with OneHotEncoder Sklearn and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('onehot', skp.OneHotEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ transformers=[("onehot", skp.OneHotEncoder(), ["city", "state"])], remainder="passthrough"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
-
- expected = pd.DataFrame({'onehot_city': ['chicago', 'chicago', 'paris'],
- 'onehot_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
+
+ expected = pd.DataFrame(
+ {
+ "onehot_city": ["chicago", "chicago", "paris"],
+ "onehot_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ }
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'other']
+ result.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -492,27 +474,20 @@ def test_inv_transform_ct_16(self):
"""
test inv_tranform_ct with ordinal Encoder sklearn and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('ordinal', skp.OrdinalEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("ordinal", skp.OrdinalEncoder(), ["city", "state"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
- expected = pd.DataFrame({'ordinal_city': ['chicago', 'chicago', 'paris'],
- 'ordinal_state': ['US', 'FR', 'FR']})
+ expected = pd.DataFrame({"ordinal_city": ["chicago", "chicago", "paris"], "ordinal_state": ["US", "FR", "FR"]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2']
+ result.columns = ["col1", "col2"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -520,28 +495,28 @@ def test_inv_transform_ct_17(self):
"""
test inv_transform_ct with OrdinalEncoder Sklearn and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"city": ["chicago", "paris"], "state": ["US", "FR"], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('ordinal', skp.OrdinalEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ transformers=[("ordinal", skp.OrdinalEncoder(), ["city", "state"])], remainder="passthrough"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
-
- expected = pd.DataFrame({'ordinal_city': ['chicago', 'chicago', 'paris'],
- 'ordinal_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]}
+ )
+
+ expected = pd.DataFrame(
+ {
+ "ordinal_city": ["chicago", "chicago", "paris"],
+ "ordinal_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ }
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2', 'other']
+ result.columns = ["col1", "col2", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -549,36 +524,26 @@ def test_inv_transform_ct_18(self):
"""
test inv_transform_ct with Standardscaler Encoder Sklearn and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('std', skp.StandardScaler(), ['num1', 'num2'])
- ],
- remainder='passthrough')
+ enc = ColumnTransformer(transformers=[("std", skp.StandardScaler(), ["num1", "num2"])], remainder="passthrough")
enc.fit(train, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']},
- )
- if sk.__version__ >="1.0.0":
- expected = pd.DataFrame({'std_num1': [0.0, 1.0, 1.0],
- 'std_num2': [0.0, 2.0, 3.0],
- 'other': ['A', 'B', 'C']},
- )
+ test = pd.DataFrame(
+ {"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]},
+ )
+ if sk.__version__ >= "1.0.0":
+ expected = pd.DataFrame(
+ {"std_num1": [0.0, 1.0, 1.0], "std_num2": [0.0, 2.0, 3.0], "other": ["A", "B", "C"]},
+ )
else:
- expected = pd.DataFrame({'std_num1': [0.0, 1.0, 1.0],
- 'std_num2': [0.0, 2.0, 3.0],
- 'other': ['A', 'B', 'C']},
- dtype=object
- )
+ expected = pd.DataFrame(
+ {"std_num1": [0.0, 1.0, 1.0], "std_num2": [0.0, 2.0, 3.0], "other": ["A", "B", "C"]}, dtype=object
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2', 'other']
+ result.columns = ["col1", "col2", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -586,27 +551,18 @@ def test_inv_transform_ct_19(self):
"""
test inv_transform_ct with StandarScaler Encoder Sklearn and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
- enc = ColumnTransformer(
- transformers=[
- ('std', skp.StandardScaler(), ['num1', 'num2'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("std", skp.StandardScaler(), ["num1", "num2"])], remainder="drop")
enc.fit(train, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
- expected = pd.DataFrame({'std_num1': [0.0, 1.0, 1.0],
- 'std_num2': [0.0, 2.0, 3.0]})
+ expected = pd.DataFrame({"std_num1": [0.0, 1.0, 1.0], "std_num2": [0.0, 2.0, 3.0]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['col1', 'col2']
+ result.columns = ["col1", "col2"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -614,28 +570,23 @@ def test_inv_transform_ct_20(self):
"""
test inv_transform_ct with QuantileTransformer Encoder Sklearn and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])
- ],
- remainder='passthrough')
+ transformers=[("quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])],
+ remainder="passthrough",
+ )
enc.fit(train, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
- expected = pd.DataFrame({'quantile_num1': [0.0, 1.0, 1.0],
- 'quantile_num2': [0.0, 2.0, 2.0],
- 'other': ['A', 'B', 'C']})
+ expected = pd.DataFrame(
+ {"quantile_num1": [0.0, 1.0, 1.0], "quantile_num2": [0.0, 2.0, 2.0], "other": ["A", "B", "C"]}
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['num1', 'num2', 'other']
+ result.columns = ["num1", "num2", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -643,27 +594,20 @@ def test_inv_transform_ct_21(self):
"""
test inv_transform_ct with QuandtileTransformer Encoder Sklearn and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])
- ],
- remainder='drop')
+ transformers=[("quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="drop"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
- expected = pd.DataFrame({'quantile_num1': [0.0, 1.0, 1.0],
- 'quantile_num2': [0.0, 2.0, 2.0]})
+ expected = pd.DataFrame({"quantile_num1": [0.0, 1.0, 1.0], "quantile_num2": [0.0, 2.0, 2.0]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['num1', 'num2']
+ result.columns = ["num1", "num2"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -671,28 +615,26 @@ def test_inv_transform_ct_22(self):
"""
test inv_transform_ct with PowerTransformer Encoder Sklearn and passthrough option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('power', skp.PowerTransformer(), ['num1', 'num2'])
- ],
- remainder='passthrough')
+ transformers=[("power", skp.PowerTransformer(), ["num1", "num2"])], remainder="passthrough"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
- expected = pd.DataFrame({'power_num1': [0.0, 1.0, 1.0],
- 'power_num2': [0.0, 1.9999999997665876, 3.000000000169985],
- 'other': ['A', 'B', 'C']})
+ expected = pd.DataFrame(
+ {
+ "power_num1": [0.0, 1.0, 1.0],
+ "power_num2": [0.0, 1.9999999997665876, 3.000000000169985],
+ "other": ["A", "B", "C"],
+ }
+ )
result = pd.DataFrame(enc.transform(test))
- result.columns = ['num1', 'num2', 'other']
+ result.columns = ["num1", "num2", "other"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -700,27 +642,20 @@ def test_inv_transform_ct_23(self):
"""
test inv_transform_ct with PowerTransformer Encoder Sklearn and drop option
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
enc = ColumnTransformer(
- transformers=[
- ('power', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])
- ],
- remainder='drop')
+ transformers=[("power", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="drop"
+ )
enc.fit(train, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
- expected = pd.DataFrame({'power_num1': [0.0, 1.0, 1.0],
- 'power_num2': [0.0, 2.0, 2.0]})
+ expected = pd.DataFrame({"power_num1": [0.0, 1.0, 1.0], "power_num2": [0.0, 2.0, 2.0]})
result = pd.DataFrame(enc.transform(test))
- result.columns = ['num1', 'num2']
+ result.columns = ["num1", "num2"]
original = inverse_transform(result, enc)
pd.testing.assert_frame_equal(original, expected)
@@ -728,80 +663,72 @@ def test_transform_ct_1(self):
"""
Unit test for apply_preprocessing on ColumnTransformer with drop option and sklearn encoder.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
- enc = ColumnTransformer(transformers=[('power', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='drop')
+ enc = ColumnTransformer(
+ transformers=[("power", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="drop"
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
expected = pd.DataFrame(enc.transform(test))
result = apply_preprocessing(test, clf, enc)
assert result.shape == expected.shape
assert [column in clf.feature_names_ for column in result.columns]
assert all(expected.index == result.index)
- assert all([str(type_result) == str(expected.dtypes[index])
- for index, type_result in enumerate(result.dtypes)])
+ assert all([str(type_result) == str(expected.dtypes[index]) for index, type_result in enumerate(result.dtypes)])
def test_transform_ct_2(self):
"""
Unit test for apply_preprocessing on ColumnTransformer with passthrough option and category encoder.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
- enc = ColumnTransformer(transformers=[('onehot_ce', ce.OneHotEncoder(), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[("onehot_ce", ce.OneHotEncoder(), ["num1", "num2"])], remainder="passthrough"
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': [1, 0, 3]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": [1, 0, 3]})
expected = pd.DataFrame(enc.transform(test))
result = apply_preprocessing(test, clf, enc)
assert result.shape == expected.shape
assert [column in clf.feature_names_ for column in result.columns]
assert all(expected.index == result.index)
- assert all([str(type_result) == str(expected.dtypes[index])
- for index, type_result in enumerate(result.dtypes)])
+ assert all([str(type_result) == str(expected.dtypes[index]) for index, type_result in enumerate(result.dtypes)])
def test_transform_ct_3(self):
"""
Unit test for apply_preprocessing on ColumnTransformer with sklearn encoder and category encoder.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
- enc = ColumnTransformer(transformers=[('onehot_ce', ce.OneHotEncoder(), ['num1', 'num2']),
- ('onehot_skp', skp.OneHotEncoder(), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[
+ ("onehot_ce", ce.OneHotEncoder(), ["num1", "num2"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["num1", "num2"]),
+ ],
+ remainder="passthrough",
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -813,47 +740,57 @@ def test_transform_ct_4(self):
"""
Unit test for apply_preprocessing on list of a dict, a list of dict and a ColumnTransformer.
"""
- train = pd.DataFrame({'city': ['CH', 'CH', 'PR'],
- 'state': ['US-FR', 'US-FR', 'US-FR'],
- 'other': ['A-B', 'A-B', 'C']},
- index=['index1', 'index2', 'index3'])
+ train = pd.DataFrame(
+ {"city": ["CH", "CH", "PR"], "state": ["US-FR", "US-FR", "US-FR"], "other": ["A-B", "A-B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
- y = pd.DataFrame(data=[0, 1, 0], columns=['y'], index=['index1', 'index2', 'index3'])
+ y = pd.DataFrame(data=[0, 1, 0], columns=["y"], index=["index1", "index2", "index3"])
train_preprocessed = train.copy()
input_dict1 = dict()
- input_dict1['col'] = 'city'
- input_dict1['mapping'] = pd.Series(data=['chicago', 'paris'], index=['CH', 'PR'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "city"
+ input_dict1["mapping"] = pd.Series(data=["chicago", "paris"], index=["CH", "PR"])
+ input_dict1["data_type"] = "object"
transform_input_1 = pd.Series(data=input_dict1.get("mapping").values, index=input_dict1.get("mapping").index)
- train_preprocessed[input_dict1.get("col")] = train_preprocessed[input_dict1.get("col")].map(
- transform_input_1).astype(input_dict1.get("mapping").values.dtype)
+ train_preprocessed[input_dict1.get("col")] = (
+ train_preprocessed[input_dict1.get("col")]
+ .map(transform_input_1)
+ .astype(input_dict1.get("mapping").values.dtype)
+ )
input_dict2 = dict()
- input_dict2['col'] = 'other'
- input_dict2['mapping'] = pd.Series(data=['A', 'C'], index=['A-B', 'C'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "other"
+ input_dict2["mapping"] = pd.Series(data=["A", "C"], index=["A-B", "C"])
+ input_dict2["data_type"] = "object"
transform_input_2 = pd.Series(data=input_dict2.get("mapping").values, index=input_dict2.get("mapping").index)
- train_preprocessed[input_dict2.get("col")] = train_preprocessed[input_dict2.get("col")].map(
- transform_input_2).astype(input_dict2.get("mapping").values.dtype)
+ train_preprocessed[input_dict2.get("col")] = (
+ train_preprocessed[input_dict2.get("col")]
+ .map(transform_input_2)
+ .astype(input_dict2.get("mapping").values.dtype)
+ )
input_dict3 = dict()
- input_dict3['col'] = 'state'
- input_dict3['mapping'] = pd.Series(data=['US FR'], index=['US-FR'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "state"
+ input_dict3["mapping"] = pd.Series(data=["US FR"], index=["US-FR"])
+ input_dict3["data_type"] = "object"
transform_input_3 = pd.Series(data=input_dict3.get("mapping").values, index=input_dict3.get("mapping").index)
- train_preprocessed[input_dict3.get("col")] = train_preprocessed[input_dict3.get("col")].map(
- transform_input_3).astype(input_dict3.get("mapping").values.dtype)
+ train_preprocessed[input_dict3.get("col")] = (
+ train_preprocessed[input_dict3.get("col")]
+ .map(transform_input_3)
+ .astype(input_dict3.get("mapping").values.dtype)
+ )
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['other'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["other"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
enc.fit(train_preprocessed)
train_preprocessed = pd.DataFrame(enc.transform(train_preprocessed), index=train.index)
@@ -870,22 +807,22 @@ def test_transform_ct_5(self):
"""
Unit test for apply_preprocessing with ColumnTransformer and sklearn model.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
- enc = ColumnTransformer(transformers=[('onehot_ce', ce.OneHotEncoder(), ['num1', 'num2']),
- ('onehot_skp', skp.OneHotEncoder(), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[
+ ("onehot_ce", ce.OneHotEncoder(), ["num1", "num2"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["num1", "num2"]),
+ ],
+ remainder="passthrough",
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = GradientBoostingClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -896,22 +833,22 @@ def test_transform_ct_6(self):
"""
Unit test for apply_preprocessing with ColumnTransformer and catboost model.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
- enc = ColumnTransformer(transformers=[('onehot_ce', ce.OneHotEncoder(), ['num1', 'num2']),
- ('onehot_skp', skp.OneHotEncoder(), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[
+ ("onehot_ce", ce.OneHotEncoder(), ["num1", "num2"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["num1", "num2"]),
+ ],
+ remainder="passthrough",
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = cb.CatBoostClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -923,22 +860,22 @@ def test_transform_ct_7(self):
"""
Unit test for apply_preprocessing with ColumnTransformer and lightgbm model.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
- enc = ColumnTransformer(transformers=[('onehot_ce', ce.OneHotEncoder(), ['num1', 'num2']),
- ('onehot_skp', skp.OneHotEncoder(), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[
+ ("onehot_ce", ce.OneHotEncoder(), ["num1", "num2"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["num1", "num2"]),
+ ],
+ remainder="passthrough",
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = lightgbm.sklearn.LGBMClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -950,22 +887,22 @@ def test_transform_ct_8(self):
"""
Unit test for apply_preprocessing with ColumnTransformer and xgboost model.
"""
- y = pd.DataFrame(data=[0, 1], columns=['y'])
+ y = pd.DataFrame(data=[0, 1], columns=["y"])
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': [1, 0]})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": [1, 0]})
- enc = ColumnTransformer(transformers=[('onehot_ce', ce.OneHotEncoder(), ['num1', 'num2']),
- ('onehot_skp', skp.OneHotEncoder(), ['num1', 'num2'])],
- remainder='passthrough')
+ enc = ColumnTransformer(
+ transformers=[
+ ("onehot_ce", ce.OneHotEncoder(), ["num1", "num2"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["num1", "num2"]),
+ ],
+ remainder="passthrough",
+ )
enc.fit(train, y)
train_preprocessed = pd.DataFrame(enc.transform(train))
clf = xgboost.sklearn.XGBClassifier(n_estimators=1).fit(train_preprocessed, y)
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 0],
- 'other': [1, 0, 0]})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 0], "other": [1, 0, 0]})
expected = pd.DataFrame(enc.transform(test), index=test.index)
result = apply_preprocessing(test, clf, enc)
@@ -977,15 +914,16 @@ def test_get_feature_names_1(self):
"""
Unit test get_feature_names 1
"""
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
- enc_1 = ColumnTransformer(transformers=[('Quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='drop')
+ enc_1 = ColumnTransformer(
+ transformers=[("Quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="drop"
+ )
- enc_2 = ColumnTransformer(transformers=[('Quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='passthrough')
+ enc_2 = ColumnTransformer(
+ transformers=[("Quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])],
+ remainder="passthrough",
+ )
enc_1.fit(train)
enc_2.fit(train)
@@ -1000,21 +938,20 @@ def test_get_names_1(self):
"""
Unit test get_names 1
"""
- train = pd.DataFrame({'num1': [0, 1],
- 'num2': [0, 2],
- 'other': ['A', 'B']})
+ train = pd.DataFrame({"num1": [0, 1], "num2": [0, 2], "other": ["A", "B"]})
- enc_1 = ColumnTransformer(transformers=[('quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='drop')
+ enc_1 = ColumnTransformer(
+ transformers=[("quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])], remainder="drop"
+ )
- enc_2 = ColumnTransformer(transformers=[('quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])],
- remainder='passthrough')
+ enc_2 = ColumnTransformer(
+ transformers=[("quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])],
+ remainder="passthrough",
+ )
- enc_3 = ColumnTransformer(transformers=[('onehot', skp.OneHotEncoder(), ['other'])],
- remainder='drop')
+ enc_3 = ColumnTransformer(transformers=[("onehot", skp.OneHotEncoder(), ["other"])], remainder="drop")
- enc_4 = ColumnTransformer(transformers=[('onehot', skp.OneHotEncoder(), ['other'])],
- remainder='passthrough')
+ enc_4 = ColumnTransformer(transformers=[("onehot", skp.OneHotEncoder(), ["other"])], remainder="passthrough")
enc_1.fit(train)
enc_2.fit(train)
@@ -1054,26 +991,27 @@ def test_get_list_features_names_1(self):
"""
Unit test get_list_features_names 1
"""
- train = pd.DataFrame({'city': ['chicago', 'paris'],
- 'state': ['US', 'FR'],
- 'other': [5, 10]},
- index=['index1', 'index2'])
+ train = pd.DataFrame(
+ {"city": ["chicago", "paris"], "state": ["US", "FR"], "other": [5, 10]}, index=["index1", "index2"]
+ )
columns_dict = {i: features for i, features in enumerate(train.columns)}
enc = ColumnTransformer(
transformers=[
- ('Ordinal_ce', ce.OrdinalEncoder(), ['city', 'state']),
- ('Ordinal_skp', skp.OrdinalEncoder(), ['city', 'state'])
+ ("Ordinal_ce", ce.OrdinalEncoder(), ["city", "state"]),
+ ("Ordinal_skp", skp.OrdinalEncoder(), ["city", "state"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
enc_2 = ColumnTransformer(
transformers=[
- ('Ordinal_ce', ce.OrdinalEncoder(), ['city', 'state']),
- ('Ordinal_skp', skp.OrdinalEncoder(), ['city', 'state'])
+ ("Ordinal_ce", ce.OrdinalEncoder(), ["city", "state"]),
+ ("Ordinal_skp", skp.OrdinalEncoder(), ["city", "state"]),
],
- remainder='drop')
+ remainder="drop",
+ )
enc.fit(train)
train_1 = pd.DataFrame(enc.transform(train), columns=["city_ce", "state_ce", "city_skp", "state_skp", "other"])
@@ -1083,20 +1021,14 @@ def test_get_list_features_names_1(self):
train_2 = pd.DataFrame(enc_2.transform(train), columns=["city_ce", "state_ce", "city_skp", "state_skp"])
train_2["y"] = np.array([1, 0])
- enc_3 = ce.OneHotEncoder(cols=['city', 'state'])
+ enc_3 = ce.OneHotEncoder(cols=["city", "state"])
enc_3.fit(train)
train_3 = enc_3.transform(train)
train_3["y"] = np.array([1, 0])
- dict_4 = {'col': 'state',
- 'mapping': pd.Series(data=[1, 2],
- index=['US', 'FR']),
- 'data_type': 'object'}
+ dict_4 = {"col": "state", "mapping": pd.Series(data=[1, 2], index=["US", "FR"]), "data_type": "object"}
- dict_5 = {'col': 'city',
- 'mapping': pd.Series(data=[1, 2],
- index=['chicago', 'paris']),
- 'data_type': 'object'}
+ dict_5 = {"col": "city", "mapping": pd.Series(data=[1, 2], index=["chicago", "paris"]), "data_type": "object"}
enc_4 = [enc_3, [dict_4]]
@@ -1120,21 +1052,26 @@ def test_get_col_mapping_ct_1(self):
"""
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='drop')
+ remainder="drop",
+ )
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'onehot_ce_city': [0, 1], 'onehot_ce_state': [
- 2, 3], 'onehot_skp_city': [4, 5], 'onehot_skp_state': [6, 7]}
+ expected_mapping = {
+ "onehot_ce_city": [0, 1],
+ "onehot_ce_state": [2, 3],
+ "onehot_skp_city": [4, 5],
+ "onehot_skp_state": [6, 7],
+ }
self.assertDictEqual(mapping, expected_mapping)
@@ -1144,21 +1081,27 @@ def test_get_col_mapping_ct_2(self):
"""
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='passthrough')
+ remainder="passthrough",
+ )
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'onehot_ce_city': [0, 1], 'onehot_ce_state': [2, 3], 'onehot_skp_city': [4, 5],
- 'onehot_skp_state': [6, 7], 'other': [8]}
+ expected_mapping = {
+ "onehot_ce_city": [0, 1],
+ "onehot_ce_state": [2, 3],
+ "onehot_skp_city": [4, 5],
+ "onehot_skp_state": [6, 7],
+ "other": [8],
+ }
self.assertDictEqual(mapping, expected_mapping)
@@ -1167,22 +1110,19 @@ def test_get_col_mapping_ct_3(self):
Test ColumnTransformer col mapping with target encoder
"""
enc = ColumnTransformer(
- transformers=[
- ('target', ce.TargetEncoder(), ['city', 'state'])
- ],
- remainder='passthrough')
+ transformers=[("target", ce.TargetEncoder(), ["city", "state"])], remainder="passthrough"
+ )
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'],
- index=['index1', 'index2', 'index3'])
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"], index=["index1", "index2", "index3"])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'other': [2], 'target_city': [0], 'target_state': [1]}
+ expected_mapping = {"other": [2], "target_city": [0], "target_state": [1]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1190,23 +1130,18 @@ def test_get_col_mapping_ct_4(self):
"""
Test ColumnTransformer col mapping with Ordinal Category Encoder and drop option
"""
- enc = ColumnTransformer(
- transformers=[
- ('ordinal', ce.OrdinalEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("ordinal", ce.OrdinalEncoder(), ["city", "state"])], remainder="drop")
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'],
- index=['index1', 'index2', 'index3'])
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"], index=["index1", "index2", "index3"])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'ordinal_city': [0], 'ordinal_state': [1]}
+ expected_mapping = {"ordinal_city": [0], "ordinal_state": [1]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1214,22 +1149,17 @@ def test_get_col_mapping_ct_5(self):
"""
test get_col_mapping_ct with Binary encoder and drop option
"""
- enc = ColumnTransformer(
- transformers=[
- ('binary', ce.BinaryEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("binary", ce.BinaryEncoder(), ["city", "state"])], remainder="drop")
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'],
- index=['index1', 'index2', 'index3'])
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"], index=["index1", "index2", "index3"])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'binary_city': [0, 1], 'binary_state': [2, 3]}
+ expected_mapping = {"binary_city": [0, 1], "binary_state": [2, 3]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1237,25 +1167,20 @@ def test_get_col_mapping_ct_6(self):
"""
test get_col_mapping_ct with BaseN Encoder and drop option
"""
- enc = ColumnTransformer(
- transformers=[
- ('basen', ce.BaseNEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("basen", ce.BaseNEncoder(), ["city", "state"])], remainder="drop")
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'],
- index=['index1', 'index2', 'index3'])
- test = pd.DataFrame({'city': ['chicago', 'new york', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"], index=["index1", "index2", "index3"])
+ test = pd.DataFrame(
+ {"city": ["chicago", "new york", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
mapping = get_col_mapping_ct(enc, test_encoded)
- if ce.__version__ <= '2.2.2':
- expected_mapping = {'basen_city': [0, 1, 2], 'basen_state': [3, 4]}
+ if ce.__version__ <= "2.2.2":
+ expected_mapping = {"basen_city": [0, 1, 2], "basen_state": [3, 4]}
else:
- expected_mapping = {'basen_city': [0, 1], 'basen_state': [2, 3]}
+ expected_mapping = {"basen_city": [0, 1], "basen_state": [2, 3]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1263,22 +1188,17 @@ def test_get_col_mapping_ct_7(self):
"""
test get_col_mapping_ct with ordinal Encoder sklearn and drop option
"""
- enc = ColumnTransformer(
- transformers=[
- ('ordinal', skp.OrdinalEncoder(), ['city', 'state'])
- ],
- remainder='drop')
+ enc = ColumnTransformer(transformers=[("ordinal", skp.OrdinalEncoder(), ["city", "state"])], remainder="drop")
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'],
- index=['index1', 'index2', 'index3'])
- test = pd.DataFrame({'city': ['chicago', 'new york', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"], index=["index1", "index2", "index3"])
+ test = pd.DataFrame(
+ {"city": ["chicago", "new york", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'ordinal_city': [0], 'ordinal_state': [1]}
+ expected_mapping = {"ordinal_city": [0], "ordinal_state": [1]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1286,18 +1206,12 @@ def test_get_col_mapping_ct_8(self):
"""
test get_col_mapping_ct with Standardscaler Encoder Sklearn and passthrough option
"""
- enc = ColumnTransformer(
- transformers=[
- ('std', skp.StandardScaler(), ['num1', 'num2'])
- ],
- remainder='passthrough')
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ enc = ColumnTransformer(transformers=[("std", skp.StandardScaler(), ["num1", "num2"])], remainder="passthrough")
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
test_encoded = pd.DataFrame(enc.fit_transform(test))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'std_num1': [0], 'std_num2': [1], 'other': [2]}
+ expected_mapping = {"std_num1": [0], "std_num2": [1], "other": [2]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1306,18 +1220,15 @@ def test_get_col_mapping_ct_9(self):
test get_col_mapping_ct with QuantileTransformer Encoder Sklearn and passthrough option
"""
enc = ColumnTransformer(
- transformers=[
- ('quantile', skp.QuantileTransformer(n_quantiles=2), ['num1', 'num2'])
- ],
- remainder='passthrough')
+ transformers=[("quantile", skp.QuantileTransformer(n_quantiles=2), ["num1", "num2"])],
+ remainder="passthrough",
+ )
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
test_encoded = pd.DataFrame(enc.fit_transform(test))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'quantile_num1': [0], 'quantile_num2': [1], 'other': [2]}
+ expected_mapping = {"quantile_num1": [0], "quantile_num2": [1], "other": [2]}
self.assertDictEqual(mapping, expected_mapping)
@@ -1326,17 +1237,13 @@ def test_get_col_mapping_ct_10(self):
test get_col_mapping_ct with PowerTransformer Encoder Sklearn and passthrough option
"""
enc = ColumnTransformer(
- transformers=[
- ('power', skp.PowerTransformer(), ['num1', 'num2'])
- ],
- remainder='passthrough')
+ transformers=[("power", skp.PowerTransformer(), ["num1", "num2"])], remainder="passthrough"
+ )
- test = pd.DataFrame({'num1': [0, 1, 1],
- 'num2': [0, 2, 3],
- 'other': ['A', 'B', 'C']})
+ test = pd.DataFrame({"num1": [0, 1, 1], "num2": [0, 2, 3], "other": ["A", "B", "C"]})
test_encoded = pd.DataFrame(enc.fit_transform(test))
mapping = get_col_mapping_ct(enc, test_encoded)
- expected_mapping = {'power_num1': [0], 'power_num2': [1], 'other': [2]}
+ expected_mapping = {"power_num1": [0], "power_num2": [1], "other": [2]}
self.assertDictEqual(mapping, expected_mapping)
diff --git a/tests/unit_tests/utils/test_explanation_metrics.py b/tests/unit_tests/utils/test_explanation_metrics.py
index f77f6c08..4fadc4e5 100644
--- a/tests/unit_tests/utils/test_explanation_metrics.py
+++ b/tests/unit_tests/utils/test_explanation_metrics.py
@@ -1,14 +1,22 @@
import unittest
+
import numpy as np
import pandas as pd
from sklearn.linear_model import LinearRegression
-from shapash.utils.explanation_metrics import _df_to_array, \
- _compute_distance, _compute_similarities, _get_radius, find_neighbors, \
- shap_neighbors, get_min_nb_features, get_distance
+from shapash.utils.explanation_metrics import (
+ _compute_distance,
+ _compute_similarities,
+ _df_to_array,
+ _get_radius,
+ find_neighbors,
+ get_distance,
+ get_min_nb_features,
+ shap_neighbors,
+)
-class TestExplanationMetrics(unittest.TestCase):
+class TestExplanationMetrics(unittest.TestCase):
def test_df_to_array(self):
df = pd.DataFrame([1, 2, 3], columns=["col"])
expected = np.array([[1], [2], [3]])
@@ -25,7 +33,7 @@ def test_compute_distance(self):
assert t == expected
def test_compute_similarities(self):
- df = pd.DataFrame(np.random.randint(0, 100, size=(5, 4)), columns=list('ABCD')).values
+ df = pd.DataFrame(np.random.randint(0, 100, size=(5, 4)), columns=list("ABCD")).values
instance = df[0, :]
expected_len = 5
expected_dist = 0
@@ -34,12 +42,12 @@ def test_compute_similarities(self):
assert t[0] == expected_dist
def test_get_radius(self):
- df = pd.DataFrame(np.random.randint(0, 100, size=(5, 4)), columns=list('ABCD')).values
+ df = pd.DataFrame(np.random.randint(0, 100, size=(5, 4)), columns=list("ABCD")).values
t = _get_radius(df, n_neighbors=3)
assert t > 0
def test_find_neighbors(self):
- df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list('ABCD'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list("ABCD"))
selection = [1, 3]
X = df.iloc[:, :-1]
y = df.iloc[:, -1]
@@ -50,8 +58,8 @@ def test_find_neighbors(self):
assert t[0].shape[1] == X.shape[1] + 2
def test_shap_neighbors(self):
- df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list('ABCD'))
- contrib = pd.DataFrame(np.random.randint(10, size=(15, 4)), columns=list('EFGH'))
+ df = pd.DataFrame(np.random.randint(0, 100, size=(15, 4)), columns=list("ABCD"))
+ contrib = pd.DataFrame(np.random.randint(10, size=(15, 4)), columns=list("EFGH"))
instance = df.values[:2, :]
extra_cols = np.repeat(np.array([0, 0]), 2).reshape(2, -1)
instance = np.append(instance, extra_cols, axis=1)
@@ -62,7 +70,7 @@ def test_shap_neighbors(self):
assert t[2].shape == (len(df.columns),)
def test_get_min_nb_features(self):
- contrib = pd.DataFrame(np.random.randint(10, size=(15, 4)), columns=list('ABCD'))
+ contrib = pd.DataFrame(np.random.randint(10, size=(15, 4)), columns=list("ABCD"))
selection = [1, 3]
distance = 0.1
mode = "regression"
@@ -72,7 +80,7 @@ def test_get_min_nb_features(self):
assert len(t) == len(selection)
def test_get_distance(self):
- contrib = pd.DataFrame(np.random.randint(10, size=(15, 4)), columns=list('ABCD'))
+ contrib = pd.DataFrame(np.random.randint(10, size=(15, 4)), columns=list("ABCD"))
selection = [1, 3]
nb_features = 2
mode = "regression"
diff --git a/tests/unit_tests/utils/test_load_smartpredictor.py b/tests/unit_tests/utils/test_load_smartpredictor.py
index d17e5768..4ce82b2e 100644
--- a/tests/unit_tests/utils/test_load_smartpredictor.py
+++ b/tests/unit_tests/utils/test_load_smartpredictor.py
@@ -1,22 +1,25 @@
"""
Unit test smart predictor
"""
-import unittest
import sys
+import unittest
from os import path
from pathlib import Path
-from shapash.utils.load_smartpredictor import load_smartpredictor
-from shapash import SmartExplainer
-import pandas as pd
-import numpy as np
+
import catboost as cb
+import numpy as np
+import pandas as pd
+
+from shapash import SmartExplainer
+from shapash.utils.load_smartpredictor import load_smartpredictor
+
class Test_load_smartpredictor(unittest.TestCase):
def test_load_smartpredictor_1(self):
"""
Unit test load_smartpredictor 1
"""
- y_pred = pd.DataFrame(data=np.array([1, 2]), columns=['pred'])
+ y_pred = pd.DataFrame(data=np.array([1, 2]), columns=["pred"])
dataframe_x = pd.DataFrame([[1, 2, 4], [1, 2, 3]])
clf = cb.CatBoostClassifier(n_estimators=1).fit(dataframe_x, y_pred)
xpl = SmartExplainer(model=clf, features_dict={})
@@ -24,14 +27,14 @@ def test_load_smartpredictor_1(self):
predictor = xpl.to_smartpredictor()
current = Path(path.abspath(__file__)).parent.parent.parent
- if str(sys.version)[0:4] == '3.10':
- pkl_file = path.join(current, 'data/predictor_to_load_310.pkl')
- elif str(sys.version)[0:3] == '3.8':
- pkl_file = path.join(current, 'data/predictor_to_load_38.pkl')
- elif str(sys.version)[0:3] == '3.9':
- pkl_file = path.join(current, 'data/predictor_to_load_39.pkl')
- elif str(sys.version)[0:4] == '3.11':
- pkl_file = path.join(current, 'data/predictor_to_load_311.pkl')
+ if str(sys.version)[0:4] == "3.10":
+ pkl_file = path.join(current, "data/predictor_to_load_310.pkl")
+ elif str(sys.version)[0:3] == "3.8":
+ pkl_file = path.join(current, "data/predictor_to_load_38.pkl")
+ elif str(sys.version)[0:3] == "3.9":
+ pkl_file = path.join(current, "data/predictor_to_load_39.pkl")
+ elif str(sys.version)[0:4] == "3.11":
+ pkl_file = path.join(current, "data/predictor_to_load_311.pkl")
else:
raise NotImplementedError
@@ -42,4 +45,4 @@ def test_load_smartpredictor_1(self):
attrib_predictor2 = [element for element in predictor2.__dict__.keys()]
assert all(attrib in attrib_predictor2 for attrib in attrib_predictor)
- assert all(attrib2 in attrib_predictor for attrib2 in attrib_predictor2)
\ No newline at end of file
+ assert all(attrib2 in attrib_predictor for attrib2 in attrib_predictor2)
diff --git a/tests/unit_tests/utils/test_model.py b/tests/unit_tests/utils/test_model.py
index 5439e00e..b99f91f4 100644
--- a/tests/unit_tests/utils/test_model.py
+++ b/tests/unit_tests/utils/test_model.py
@@ -1,24 +1,26 @@
-import pytest
import numpy as np
import pandas as pd
+import pytest
+
from shapash.utils.model import predict_error
-y1 = pd.DataFrame(data=np.array([1, 2, 3]), columns=['pred'])
-expected1 = pd.DataFrame(data=np.array([0., 0., 0.]), columns=['_error_'])
+y1 = pd.DataFrame(data=np.array([1, 2, 3]), columns=["pred"])
+expected1 = pd.DataFrame(data=np.array([0.0, 0.0, 0.0]), columns=["_error_"])
+
+y2 = pd.DataFrame(data=np.array([0, 2, 3]), columns=["pred"])
+expected2 = pd.DataFrame(data=np.array([1, 0, 0]), columns=["_error_"])
-y2 = pd.DataFrame(data=np.array([0, 2, 3]), columns=['pred'])
-expected2 = pd.DataFrame(data=np.array([1, 0, 0]), columns=['_error_'])
@pytest.mark.parametrize(
- "y_target, y_pred, case, expected",
- [
- (None, None, 'classification', None),
- (y1, y1, 'classification', None),
- (y1, None, 'regression', None),
- (None, y1, 'regression', None),
- (y1, y1, 'regression', expected1),
- (y2, y1, 'regression', expected2)
- ]
+ "y_target, y_pred, case, expected",
+ [
+ (None, None, "classification", None),
+ (y1, y1, "classification", None),
+ (y1, None, "regression", None),
+ (None, y1, "regression", None),
+ (y1, y1, "regression", expected1),
+ (y2, y1, "regression", expected2),
+ ],
)
def test_predict_error_works(y_target, y_pred, case, expected):
result = predict_error(y_target, y_pred, case)
diff --git a/tests/unit_tests/utils/test_transform.py b/tests/unit_tests/utils/test_transform.py
index 5ab34965..6611b30e 100644
--- a/tests/unit_tests/utils/test_transform.py
+++ b/tests/unit_tests/utils/test_transform.py
@@ -2,13 +2,19 @@
Unit test of transform module.
"""
import unittest
-import pandas as pd
+
+import category_encoders as ce
import numpy as np
-from sklearn.compose import ColumnTransformer
+import pandas as pd
import sklearn.preprocessing as skp
-import category_encoders as ce
-from shapash.utils.transform import get_preprocessing_mapping, get_features_transform_mapping, handle_categorical_missing
from pandas.testing import assert_frame_equal
+from sklearn.compose import ColumnTransformer
+
+from shapash.utils.transform import (
+ get_features_transform_mapping,
+ get_preprocessing_mapping,
+ handle_categorical_missing,
+)
class TestInverseTransformCaterogyEncoder(unittest.TestCase):
@@ -18,40 +24,45 @@ def test_get_preprocessing_mapping_1(self):
"""
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='drop')
+ remainder="drop",
+ )
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test))
input_dict1 = dict()
- input_dict1['col'] = 'onehot_ce_city'
- input_dict1['mapping'] = pd.Series(data=['chicago', 'paris'], index=['CH', 'PR'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "onehot_ce_city"
+ input_dict1["mapping"] = pd.Series(data=["chicago", "paris"], index=["CH", "PR"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'other'
- input_dict2['mapping'] = pd.Series(data=['A', 'B', 'C'], index=['A-B', 'A-B', 'C'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "other"
+ input_dict2["mapping"] = pd.Series(data=["A", "B", "C"], index=["A-B", "A-B", "C"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'onehot_ce_state'
- input_dict3['mapping'] = pd.Series(data=['US', 'FR'], index=['US-FR', 'US-FR'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "onehot_ce_state"
+ input_dict3["mapping"] = pd.Series(data=["US", "FR"], index=["US-FR", "US-FR"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
- test_encoded.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'col3_0', 'col3_1', 'col4_0', 'col4_1']
+ test_encoded.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "col3_0", "col3_1", "col4_0", "col4_1"]
mapping = get_preprocessing_mapping(test_encoded, [enc, input_dict1, list_dict])
- expected_mapping = {'onehot_ce_city': ['col1_0', 'col1_1'], 'onehot_ce_state': ['col2_0', 'col2_1'],
- 'onehot_skp_city': ['col3_0', 'col3_1'], 'onehot_skp_state': ['col4_0', 'col4_1']}
+ expected_mapping = {
+ "onehot_ce_city": ["col1_0", "col1_1"],
+ "onehot_ce_state": ["col2_0", "col2_1"],
+ "onehot_skp_city": ["col3_0", "col3_1"],
+ "onehot_skp_state": ["col4_0", "col4_1"],
+ }
self.assertDictEqual(mapping, expected_mapping)
def test_get_preprocessing_mapping_2(self):
@@ -60,22 +71,26 @@ def test_get_preprocessing_mapping_2(self):
"""
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('ordinal_skp', skp.OrdinalEncoder(), ['city'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("ordinal_skp", skp.OrdinalEncoder(), ["city"]),
],
- remainder='drop')
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C']},
- index=['index1', 'index2', 'index3'])
+ remainder="drop",
+ )
+ test = pd.DataFrame(
+ {"city": ["chicago", "chicago", "paris"], "state": ["US", "FR", "FR"], "other": ["A", "B", "C"]},
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test))
- test_encoded.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'col3']
+ test_encoded.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "col3"]
mapping = get_preprocessing_mapping(test_encoded, enc)
- expected_mapping = {'onehot_ce_city': ['col1_0', 'col1_1'], 'onehot_ce_state': ['col2_0', 'col2_1'],
- 'ordinal_skp_city': ['col3']}
+ expected_mapping = {
+ "onehot_ce_city": ["col1_0", "col1_1"],
+ "onehot_ce_state": ["col2_0", "col2_1"],
+ "ordinal_skp_city": ["col3"],
+ }
self.assertDictEqual(mapping, expected_mapping)
def test_get_features_transform_mapping_1(self):
@@ -84,50 +99,64 @@ def test_get_features_transform_mapping_1(self):
"""
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city', 'state']),
- ('onehot_skp', skp.OneHotEncoder(), ['city', 'state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city", "state"]),
+ ("onehot_skp", skp.OneHotEncoder(), ["city", "state"]),
],
- remainder='drop')
-
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C'],
- 'other2': ['D', 'E', 'F']},
- index=['index1', 'index2', 'index3'])
+ remainder="drop",
+ )
+
+ test = pd.DataFrame(
+ {
+ "city": ["chicago", "chicago", "paris"],
+ "state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ "other2": ["D", "E", "F"],
+ },
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test))
input_dict1 = dict()
- input_dict1['col'] = 'onehot_ce_city'
- input_dict1['mapping'] = pd.Series(data=['chicago', 'paris'], index=['CH', 'PR'])
- input_dict1['data_type'] = 'object'
+ input_dict1["col"] = "onehot_ce_city"
+ input_dict1["mapping"] = pd.Series(data=["chicago", "paris"], index=["CH", "PR"])
+ input_dict1["data_type"] = "object"
input_dict2 = dict()
- input_dict2['col'] = 'other'
- input_dict2['mapping'] = pd.Series(data=['A', 'B', 'C'], index=['A-B', 'A-B', 'C'])
- input_dict2['data_type'] = 'object'
+ input_dict2["col"] = "other"
+ input_dict2["mapping"] = pd.Series(data=["A", "B", "C"], index=["A-B", "A-B", "C"])
+ input_dict2["data_type"] = "object"
input_dict3 = dict()
- input_dict3['col'] = 'onehot_ce_state'
- input_dict3['mapping'] = pd.Series(data=['US', 'FR'], index=['US-FR', 'US-FR'])
- input_dict3['data_type'] = 'object'
+ input_dict3["col"] = "onehot_ce_state"
+ input_dict3["mapping"] = pd.Series(data=["US", "FR"], index=["US-FR", "US-FR"])
+ input_dict3["data_type"] = "object"
list_dict = [input_dict2, input_dict3]
- test_encoded.columns = ['col1_0', 'col1_1', 'col2_0', 'col2_1', 'col3_0', 'col3_1', 'col4_0', 'col4_1']
+ test_encoded.columns = ["col1_0", "col1_1", "col2_0", "col2_1", "col3_0", "col3_1", "col4_0", "col4_1"]
# Construct expected x_encoded that will be built from test dataframe
- x_encoded = pd.DataFrame({'onehot_ce_city': ['chicago', 'chicago', 'paris'],
- 'onehot_ce_state': ['US', 'FR', 'FR'],
- 'onehot_skp_city': ['chicago', 'chicago', 'paris'],
- 'onehot_skp_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C'],
- 'other2': ['D', 'E', 'F']},
- index=['index1', 'index2', 'index3'])
+ x_encoded = pd.DataFrame(
+ {
+ "onehot_ce_city": ["chicago", "chicago", "paris"],
+ "onehot_ce_state": ["US", "FR", "FR"],
+ "onehot_skp_city": ["chicago", "chicago", "paris"],
+ "onehot_skp_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ "other2": ["D", "E", "F"],
+ },
+ index=["index1", "index2", "index3"],
+ )
mapping = get_features_transform_mapping(x_encoded, test_encoded, [enc, input_dict1, list_dict])
- expected_mapping = {'onehot_ce_city': ['col1_0', 'col1_1'], 'onehot_ce_state': ['col2_0', 'col2_1'],
- 'onehot_skp_city': ['col3_0', 'col3_1'], 'onehot_skp_state': ['col4_0', 'col4_1'],
- 'other': ['other'], 'other2': ['other2']}
+ expected_mapping = {
+ "onehot_ce_city": ["col1_0", "col1_1"],
+ "onehot_ce_state": ["col2_0", "col2_1"],
+ "onehot_skp_city": ["col3_0", "col3_1"],
+ "onehot_skp_state": ["col4_0", "col4_1"],
+ "other": ["other"],
+ "other2": ["other2"],
+ }
self.assertDictEqual(mapping, expected_mapping)
def test_get_features_transform_mapping_2(self):
@@ -136,55 +165,69 @@ def test_get_features_transform_mapping_2(self):
"""
enc = ColumnTransformer(
transformers=[
- ('onehot_ce', ce.OneHotEncoder(), ['city']),
- ('ordinal_skp', skp.OrdinalEncoder(), ['state'])
+ ("onehot_ce", ce.OneHotEncoder(), ["city"]),
+ ("ordinal_skp", skp.OrdinalEncoder(), ["state"]),
],
- remainder='drop')
-
- test = pd.DataFrame({'city': ['chicago', 'chicago', 'paris'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C'],
- 'other2': ['D', 'E', 'F']},
- index=['index1', 'index2', 'index3'])
+ remainder="drop",
+ )
+
+ test = pd.DataFrame(
+ {
+ "city": ["chicago", "chicago", "paris"],
+ "state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ "other2": ["D", "E", "F"],
+ },
+ index=["index1", "index2", "index3"],
+ )
test_encoded = pd.DataFrame(enc.fit_transform(test))
- test_encoded.columns = ['col1_0', 'col1_1', 'col2']
+ test_encoded.columns = ["col1_0", "col1_1", "col2"]
# Construct expected x_encoded that will be built from test dataframe
- x_encoded = pd.DataFrame({'onehot_ce_city': ['chicago', 'chicago', 'paris'],
- 'ordinal_skp_state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'C'],
- 'other2': ['D', 'E', 'F']},
- index=['index1', 'index2', 'index3'])
+ x_encoded = pd.DataFrame(
+ {
+ "onehot_ce_city": ["chicago", "chicago", "paris"],
+ "ordinal_skp_state": ["US", "FR", "FR"],
+ "other": ["A", "B", "C"],
+ "other2": ["D", "E", "F"],
+ },
+ index=["index1", "index2", "index3"],
+ )
mapping = get_features_transform_mapping(x_encoded, test_encoded, enc)
print(mapping)
- expected_mapping = {'onehot_ce_city': ['col1_0', 'col1_1'],
- 'ordinal_skp_state': ['col2'],
- 'other': ['other'], 'other2': ['other2']}
+ expected_mapping = {
+ "onehot_ce_city": ["col1_0", "col1_1"],
+ "ordinal_skp_state": ["col2"],
+ "other": ["other"],
+ "other2": ["other2"],
+ }
self.assertDictEqual(mapping, expected_mapping)
def test_get_features_transform_mapping_3(self):
"""
test get_features_transform_mapping with category encoders
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'B']})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "chicago"], "state": ["US", "FR", "FR"], "other": ["A", "B", "B"]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
- enc = ce.OneHotEncoder(cols=['city', 'state'], use_cat_names=True)
+ enc = ce.OneHotEncoder(cols=["city", "state"], use_cat_names=True)
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
- x_encoded = pd.DataFrame({'city': ['chicago', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'B']})
+ x_encoded = pd.DataFrame(
+ {"city": ["chicago", "paris", "chicago"], "state": ["US", "FR", "FR"], "other": ["A", "B", "B"]}
+ )
mapping = get_features_transform_mapping(x_encoded, test_encoded, enc)
- expected_mapping = {'city': ['city_chicago', 'city_paris'],
- 'state': ['state_US', 'state_FR'],
- 'other': ['other']}
+ expected_mapping = {
+ "city": ["city_chicago", "city_paris"],
+ "state": ["state_US", "state_FR"],
+ "other": ["other"],
+ }
self.assertDictEqual(mapping, expected_mapping)
@@ -192,25 +235,23 @@ def test_get_features_transform_mapping_4(self):
"""
test get_features_transform_mapping with list of category encoders
"""
- test = pd.DataFrame({'city': ['chicago', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'B']})
- y = pd.DataFrame(data=[0, 1, 1], columns=['y'])
+ test = pd.DataFrame(
+ {"city": ["chicago", "paris", "chicago"], "state": ["US", "FR", "FR"], "other": ["A", "B", "B"]}
+ )
+ y = pd.DataFrame(data=[0, 1, 1], columns=["y"])
- enc = ce.OneHotEncoder(cols=['state'], use_cat_names=True)
+ enc = ce.OneHotEncoder(cols=["state"], use_cat_names=True)
test_encoded = pd.DataFrame(enc.fit_transform(test, y))
- enc2 = ce.OrdinalEncoder(cols=['city'])
+ enc2 = ce.OrdinalEncoder(cols=["city"])
test_encoded = enc2.fit_transform(test_encoded, y)
- x_encoded = pd.DataFrame({'city': ['chicago', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['A', 'B', 'B']})
+ x_encoded = pd.DataFrame(
+ {"city": ["chicago", "paris", "chicago"], "state": ["US", "FR", "FR"], "other": ["A", "B", "B"]}
+ )
mapping = get_features_transform_mapping(x_encoded, test_encoded, [enc, enc2])
- expected_mapping = {'city': ['city'],
- 'state': ['state_US', 'state_FR'],
- 'other': ['other']}
+ expected_mapping = {"city": ["city"], "state": ["state_US", "state_FR"], "other": ["other"]}
self.assertDictEqual(mapping, expected_mapping)
@@ -218,14 +259,14 @@ def handle_categorical_missing(self):
"""
test handle_categorical_missing
"""
- df_test = pd.DataFrame({'city': [np.nan, 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': [np.nan, 'B', 'B']})
+ df_test = pd.DataFrame(
+ {"city": [np.nan, "paris", "chicago"], "state": ["US", "FR", "FR"], "other": [np.nan, "B", "B"]}
+ )
df_test = handle_categorical_missing(df_test)
- df_expected = pd.DataFrame({'city': ['missing', 'paris', 'chicago'],
- 'state': ['US', 'FR', 'FR'],
- 'other': ['missing', 'B', 'B']})
+ df_expected = pd.DataFrame(
+ {"city": ["missing", "paris", "chicago"], "state": ["US", "FR", "FR"], "other": ["missing", "B", "B"]}
+ )
assert_frame_equal(df_test, df_expected)
diff --git a/tests/unit_tests/utils/test_translate.py b/tests/unit_tests/utils/test_translate.py
index 57e88388..9668defd 100644
--- a/tests/unit_tests/utils/test_translate.py
+++ b/tests/unit_tests/utils/test_translate.py
@@ -2,6 +2,7 @@
Unit test Translate
"""
import unittest
+
from shapash.utils.translate import translate
@@ -9,12 +10,13 @@ class TestTranslate(unittest.TestCase):
"""
Unit test Translate class
"""
+
def test_translate(self):
"""
Unit test translate
"""
- elements = ['X_1', 'X_2']
- mapping = {'X_1': 'âge', 'X_2': 'profession'}
+ elements = ["X_1", "X_2"]
+ mapping = {"X_1": "âge", "X_2": "profession"}
output = translate(elements, mapping)
- expected = ['âge', 'profession']
+ expected = ["âge", "profession"]
self.assertListEqual(output, expected)
diff --git a/tests/unit_tests/utils/test_utils.py b/tests/unit_tests/utils/test_utils.py
index 370b5456..91e5959f 100644
--- a/tests/unit_tests/utils/test_utils.py
+++ b/tests/unit_tests/utils/test_utils.py
@@ -1,13 +1,22 @@
import unittest
+
import numpy as np
import pandas as pd
-from shapash.utils.utils import inclusion, within_dict, is_nested_list,\
- compute_digit_number, truncate_str, add_line_break, maximum_difference_sort_value, \
- compute_sorted_variables_interactions_list_indices, compute_top_correlations_features
+from shapash.utils.utils import (
+ add_line_break,
+ compute_digit_number,
+ compute_sorted_variables_interactions_list_indices,
+ compute_top_correlations_features,
+ inclusion,
+ is_nested_list,
+ maximum_difference_sort_value,
+ truncate_str,
+ within_dict,
+)
-class TestUtils(unittest.TestCase):
+class TestUtils(unittest.TestCase):
def test_inclusion_1(self):
x1 = [1, 2, 3]
x2 = [1, 2, 3, 4]
@@ -65,47 +74,65 @@ def test_truncate_str_1(self):
assert t == 12
def test_truncate_str_2(self):
- t = truncate_str("this is a test",50)
+ t = truncate_str("this is a test", 50)
assert t == "this is a test"
def test_truncate_str_3(self):
- t = truncate_str("this is a test",10)
+ t = truncate_str("this is a test", 10)
assert t == "this is a..."
def test_add_line_break_1(self):
- t = add_line_break(3453,10)
+ t = add_line_break(3453, 10)
assert t == 3453
def test_add_line_break_2(self):
- t = add_line_break("this is a very long sentence in order to make a very great test",10)
- expected = 'this is a very
long sentence
in order to make
a very great
test'
+ t = add_line_break("this is a very long sentence in order to make a very great test", 10)
+ expected = "this is a very
long sentence
in order to make
a very great
test"
assert t == expected
def test_add_line_break_3(self):
- t = add_line_break("this is a very long sentence in order to make a very great test",15,maxlen=30)
- expected = 'this is a very long
sentence in order
to...'
+ t = add_line_break("this is a very long sentence in order to make a very great test", 15, maxlen=30)
+ expected = "this is a very long
sentence in order
to..."
assert t == expected
def test_maximum_difference_sort_value_1(self):
- t = maximum_difference_sort_value([[1, 20, 3], ['feat1', 'feat2', 'feat3']])
+ t = maximum_difference_sort_value([[1, 20, 3], ["feat1", "feat2", "feat3"]])
assert t == 19
def test_maximum_difference_sort_value_2(self):
- t = maximum_difference_sort_value([[100, 11, 281, 64, 6000], ['feat1', 'feat2']])
+ t = maximum_difference_sort_value([[100, 11, 281, 64, 6000], ["feat1", "feat2"]])
assert t == 5989
def test_maximum_difference_sort_value_3(self):
- t = maximum_difference_sort_value(([[1], ['feat1']]))
+ t = maximum_difference_sort_value([[1], ["feat1"]])
assert t == 1
def test_compute_sorted_variables_interactions_list_indices_1(self):
- interaction_values = np.array([
- [[0.1, -0.7, 0.01, -0.9], [-0.1, 0.8, 0.02, 0.7], [0.2, 0.5, 0.04, -0.88], [0.15, 0.6, -0.2, 0.5]],
- [[0.2, -0.1, 0.2, 0.8], [-0.2, 0.6, 0.02, -0.67], [0.1, -0.5, 0.05, 1], [0.3, 0.6, 0.02, -0.9]]
- ])
-
- expected_output = ([[3, 1], [2, 1], [3, 0], [2, 0], [1, 0], [3, 2], [3, 3], [2, 3], [2, 2], [1, 3],
- [1, 2], [1, 1], [0, 3], [0, 2], [0, 1], [0, 0]])
+ interaction_values = np.array(
+ [
+ [[0.1, -0.7, 0.01, -0.9], [-0.1, 0.8, 0.02, 0.7], [0.2, 0.5, 0.04, -0.88], [0.15, 0.6, -0.2, 0.5]],
+ [[0.2, -0.1, 0.2, 0.8], [-0.2, 0.6, 0.02, -0.67], [0.1, -0.5, 0.05, 1], [0.3, 0.6, 0.02, -0.9]],
+ ]
+ )
+
+ expected_output = [
+ [3, 1],
+ [2, 1],
+ [3, 0],
+ [2, 0],
+ [1, 0],
+ [3, 2],
+ [3, 3],
+ [2, 3],
+ [2, 2],
+ [1, 3],
+ [1, 2],
+ [1, 1],
+ [0, 3],
+ [0, 2],
+ [0, 1],
+ [0, 0],
+ ]
output = compute_sorted_variables_interactions_list_indices(interaction_values)
diff --git a/tests/unit_tests/webapp/test_webapp_settings.py b/tests/unit_tests/webapp/test_webapp_settings.py
index 72390490..f861a5d8 100644
--- a/tests/unit_tests/webapp/test_webapp_settings.py
+++ b/tests/unit_tests/webapp/test_webapp_settings.py
@@ -1,35 +1,36 @@
import unittest
-from shapash import SmartExplainer
-from sklearn.tree import DecisionTreeRegressor
-import pandas as pd
+
import numpy as np
+import pandas as pd
+from sklearn.tree import DecisionTreeRegressor
+
+from shapash import SmartExplainer
+
class TestWebappSettings(unittest.TestCase):
"""
Unit tests for webapp settings class
Checks that the webapp settings remain valid whether the user input is valid or not
"""
+
def __init__(self, *args, **kwargs):
"""
Constructor - loads a SmartExplainer object from the appropriate pickle
"""
contributions = pd.DataFrame([[-0.1, 0.2, -0.3], [0.1, -0.2, 0.3]])
- y_pred = pd.DataFrame(data=np.array([1, 2]), columns=['pred'])
+ y_pred = pd.DataFrame(data=np.array([1, 2]), columns=["pred"])
dataframe_x = pd.DataFrame([[1, 2, 3], [1, 2, 3]])
model = DecisionTreeRegressor().fit([[0]], [[0]])
self.xpl = SmartExplainer(model=model)
self.xpl.compile(contributions=contributions, x=dataframe_x, y_pred=y_pred)
self.xpl.filter(max_contrib=2)
- super(TestWebappSettings, self).__init__(*args, **kwargs)
+ super().__init__(*args, **kwargs)
def test_settings_types(self):
"""
Test settings dtypes (must be ints)
"""
- settings = {'rows': None,
- 'points': 5200.4,
- 'violin': -1,
- 'features': "oui"}
+ settings = {"rows": None, "points": 5200.4, "violin": -1, "features": "oui"}
self.xpl.init_app(settings)
print(self.xpl.smartapp.settings)
assert all(isinstance(attrib, int) for k, attrib in self.xpl.smartapp.settings.items())
@@ -38,10 +39,7 @@ def test_settings_values(self):
"""
Test settings values (must be >0)
"""
- settings = {'rows': 0,
- 'points': 5200.4,
- 'violin': -1,
- 'features': "oui"}
+ settings = {"rows": 0, "points": 5200.4, "violin": -1, "features": "oui"}
self.xpl.init_app(settings)
assert all(attrib > 0 for k, attrib in self.xpl.smartapp.settings.items())
@@ -49,8 +47,6 @@ def test_settings_keys(self):
"""
Test settings keys : the expected keys must be in the final settings dict, whatever the user input is
"""
- settings = {'oui': 1,
- 1: 2,
- "a": []}
+ settings = {"oui": 1, 1: 2, "a": []}
self.xpl.init_app(settings)
- assert all(k in ['rows', 'points', 'violin', 'features'] for k in self.xpl.smartapp.settings)
+ assert all(k in ["rows", "points", "violin", "features"] for k in self.xpl.smartapp.settings)
diff --git a/tests/unit_tests/webapp/utils/test_callbacks.py b/tests/unit_tests/webapp/utils/test_callbacks.py
index ec336843..e90fcbb4 100644
--- a/tests/unit_tests/webapp/utils/test_callbacks.py
+++ b/tests/unit_tests/webapp/utils/test_callbacks.py
@@ -1,220 +1,221 @@
-import unittest
import copy
+import unittest
+
import numpy as np
import pandas as pd
-from sklearn.tree import DecisionTreeClassifier
from dash import dcc
+from sklearn.tree import DecisionTreeClassifier
+
from shapash import SmartExplainer
from shapash.webapp.smart_app import SmartApp
from shapash.webapp.utils.callbacks import (
- select_data_from_prediction_picking,
- select_data_from_str_filters,
- select_data_from_bool_filters,
- select_data_from_numeric_filters,
- select_data_from_date_filters,
+ create_filter_modalities_selection,
+ create_id_card_data,
+ create_id_card_layout,
+ get_feature_contributions_sign_to_show,
+ get_feature_filter_options,
get_feature_from_clicked_data,
get_feature_from_features_groups,
+ get_figure_zoom,
get_group_name,
+ get_id_card_contrib,
+ get_id_card_features,
get_indexes_from_datatable,
+ select_data_from_bool_filters,
+ select_data_from_date_filters,
+ select_data_from_numeric_filters,
+ select_data_from_prediction_picking,
+ select_data_from_str_filters,
update_click_data_on_subset_changes,
- get_figure_zoom,
- get_feature_contributions_sign_to_show,
update_features_to_display,
- get_id_card_features,
- get_id_card_contrib,
- create_id_card_data,
- create_id_card_layout,
- get_feature_filter_options,
- create_filter_modalities_selection
)
class TestCallbacks(unittest.TestCase):
-
def __init__(self, *args, **kwargs):
data = {
- 'column1': [1, 2, 3, 4, 5],
- 'column2': ['a', 'b', 'c', 'd', 'e'],
- 'column3': [1.1, 3.3, 2.2, 4.4, 5.5],
- 'column4': [True, False, True, False, False],
- 'column5': pd.date_range('2023-01-01', periods=5),
+ "column1": [1, 2, 3, 4, 5],
+ "column2": ["a", "b", "c", "d", "e"],
+ "column3": [1.1, 3.3, 2.2, 4.4, 5.5],
+ "column4": [True, False, True, False, False],
+ "column5": pd.date_range("2023-01-01", periods=5),
}
df = pd.DataFrame(data)
self.df = df
- dataframe_x = df[['column1','column3']].copy()
- y_target = pd.DataFrame(data=np.array([0, 0, 0, 1, 1]), columns=['pred'])
+ dataframe_x = df[["column1", "column3"]].copy()
+ y_target = pd.DataFrame(data=np.array([0, 0, 0, 1, 1]), columns=["pred"])
model = DecisionTreeClassifier().fit(dataframe_x, y_target)
- features_dict = {'column3': 'Useless col'}
- additional_data = df[['column2','column4','column5']].copy()
- additional_features_dict = {'column2': 'Additional col'}
+ features_dict = {"column3": "Useless col"}
+ additional_data = df[["column2", "column4", "column5"]].copy()
+ additional_features_dict = {"column2": "Additional col"}
self.xpl = SmartExplainer(model=model, features_dict=features_dict)
self.xpl.compile(
- x=dataframe_x,
+ x=dataframe_x,
y_pred=y_target,
y_target=y_target,
- additional_data=additional_data,
- additional_features_dict=additional_features_dict
+ additional_data=additional_data,
+ additional_features_dict=additional_features_dict,
)
self.smart_app = SmartApp(self.xpl)
self.click_data = {
- 'points': [
+ "points": [
{
- 'curveNumber': 0,
- 'pointNumber': 3,
- 'pointIndex': 3,
- 'x': 0.4649,
- 'y': 'Sex',
- 'label': 'Sex',
- 'value': 0.4649,
- 'customdata': 'Sex',
- 'marker.color': 'rgba(244, 192, 0, 1.0)',
- 'bbox': {'x0': 717.3, 'x1': 717.3, 'y0': 82.97, 'y1': 130.78}
+ "curveNumber": 0,
+ "pointNumber": 3,
+ "pointIndex": 3,
+ "x": 0.4649,
+ "y": "Sex",
+ "label": "Sex",
+ "value": 0.4649,
+ "customdata": "Sex",
+ "marker.color": "rgba(244, 192, 0, 1.0)",
+ "bbox": {"x0": 717.3, "x1": 717.3, "y0": 82.97, "y1": 130.78},
}
]
}
- self.special_cols = ['_index_', '_predict_', '_target_']
+ self.special_cols = ["_index_", "_predict_", "_target_"]
- super(TestCallbacks, self).__init__(*args, **kwargs)
+ super().__init__(*args, **kwargs)
def test_default_init_data(self):
expected_result = pd.DataFrame(
{
- '_index_': [0, 1, 2, 3, 4],
- '_predict_': [0, 0, 0, 1, 1],
- '_target_': [0, 0, 0, 1, 1],
- 'column1': [1, 2, 3, 4, 5],
- 'column3': [1.1, 3.3, 2.2, 4.4, 5.5],
- '_column2': ['a', 'b', 'c', 'd', 'e'],
- '_column4': [True, False, True, False, False],
- '_column5': pd.date_range('2023-01-01', periods=5),
+ "_index_": [0, 1, 2, 3, 4],
+ "_predict_": [0, 0, 0, 1, 1],
+ "_target_": [0, 0, 0, 1, 1],
+ "column1": [1, 2, 3, 4, 5],
+ "column3": [1.1, 3.3, 2.2, 4.4, 5.5],
+ "_column2": ["a", "b", "c", "d", "e"],
+ "_column4": [True, False, True, False, False],
+ "_column5": pd.date_range("2023-01-01", periods=5),
},
)
self.smart_app.init_data()
pd.testing.assert_frame_equal(expected_result, self.smart_app.round_dataframe)
-
+
def test_limited_rows_init_data(self):
self.smart_app.init_data(3)
- assert len(self.smart_app.round_dataframe)==3
+ assert len(self.smart_app.round_dataframe) == 3
def test_select_data_from_prediction_picking(self):
- selected_data = {"points": [{"customdata":0}, {"customdata":2}]}
+ selected_data = {"points": [{"customdata": 0}, {"customdata": 2}]}
expected_result = pd.DataFrame(
{
- 'column1': [1, 3],
- 'column2': ['a', 'c'],
- 'column3': [1.1, 2.2],
- 'column4': [True, True],
- 'column5': [pd.Timestamp('2023-01-01'), pd.Timestamp('2023-01-03')],
+ "column1": [1, 3],
+ "column2": ["a", "c"],
+ "column3": [1.1, 2.2],
+ "column4": [True, True],
+ "column5": [pd.Timestamp("2023-01-01"), pd.Timestamp("2023-01-03")],
},
- index=[0, 2]
+ index=[0, 2],
)
result = select_data_from_prediction_picking(self.df, selected_data)
pd.testing.assert_frame_equal(expected_result, result)
-
+
def test_select_data_from_str_filters(self):
round_dataframe = self.df
- id_feature = [{'type': 'var_dropdown', 'index': 1}]
- feature_id = [id_feature[i]['index'] for i in range(len(id_feature))]
- id_str_modality = [{'type': 'dynamic-str', 'index': 1}]
- val_feature = ['column2']
- val_str_modality = [['a', 'c']]
+ id_feature = [{"type": "var_dropdown", "index": 1}]
+ feature_id = [id_feature[i]["index"] for i in range(len(id_feature))]
+ id_str_modality = [{"type": "dynamic-str", "index": 1}]
+ val_feature = ["column2"]
+ val_str_modality = [["a", "c"]]
expected_result = pd.DataFrame(
{
- 'column1': [1, 3],
- 'column2': ['a', 'c'],
- 'column3': [1.1, 2.2],
- 'column4': [True, True],
- 'column5': [pd.Timestamp('2023-01-01'), pd.Timestamp('2023-01-03')],
+ "column1": [1, 3],
+ "column2": ["a", "c"],
+ "column3": [1.1, 2.2],
+ "column4": [True, True],
+ "column5": [pd.Timestamp("2023-01-01"), pd.Timestamp("2023-01-03")],
},
- index=[0, 2]
+ index=[0, 2],
)
result = select_data_from_str_filters(
round_dataframe,
- feature_id,
- id_str_modality,
- val_feature,
+ feature_id,
+ id_str_modality,
+ val_feature,
val_str_modality,
)
pd.testing.assert_frame_equal(expected_result, result)
-
+
def test_select_data_from_bool_filters(self):
round_dataframe = self.df
- id_feature = [{'type': 'var_dropdown', 'index': 2}]
- feature_id = [id_feature[i]['index'] for i in range(len(id_feature))]
- id_bool_modality = [{'type': 'dynamic-bool', 'index': 2}]
- val_feature = ['column4']
+ id_feature = [{"type": "var_dropdown", "index": 2}]
+ feature_id = [id_feature[i]["index"] for i in range(len(id_feature))]
+ id_bool_modality = [{"type": "dynamic-bool", "index": 2}]
+ val_feature = ["column4"]
val_bool_modality = [True]
expected_result = pd.DataFrame(
{
- 'column1': [1, 3],
- 'column2': ['a', 'c'],
- 'column3': [1.1, 2.2],
- 'column4': [True, True],
- 'column5': [pd.Timestamp('2023-01-01'), pd.Timestamp('2023-01-03')],
+ "column1": [1, 3],
+ "column2": ["a", "c"],
+ "column3": [1.1, 2.2],
+ "column4": [True, True],
+ "column5": [pd.Timestamp("2023-01-01"), pd.Timestamp("2023-01-03")],
},
- index=[0, 2]
+ index=[0, 2],
)
result = select_data_from_bool_filters(
round_dataframe,
- feature_id,
+ feature_id,
id_bool_modality,
- val_feature,
+ val_feature,
val_bool_modality,
)
pd.testing.assert_frame_equal(expected_result, result)
-
+
def test_select_data_from_date_filters(self):
round_dataframe = self.df
- id_feature = [{'type': 'var_dropdown', 'index': 1}]
- feature_id = [id_feature[i]['index'] for i in range(len(id_feature))]
- id_date = [{'type': 'dynamic-date', 'index': 1}]
- val_feature = ['column5']
- start_date = [pd.Timestamp('2023-01-01')]
- end_date = [pd.Timestamp('2023-01-03')]
+ id_feature = [{"type": "var_dropdown", "index": 1}]
+ feature_id = [id_feature[i]["index"] for i in range(len(id_feature))]
+ id_date = [{"type": "dynamic-date", "index": 1}]
+ val_feature = ["column5"]
+ start_date = [pd.Timestamp("2023-01-01")]
+ end_date = [pd.Timestamp("2023-01-03")]
expected_result = pd.DataFrame(
{
- 'column1': [1, 2, 3],
- 'column2': ['a', 'b', 'c'],
- 'column3': [1.1, 3.3, 2.2],
- 'column4': [True, False, True],
- 'column5': [pd.Timestamp('2023-01-01'), pd.Timestamp('2023-01-02'), pd.Timestamp('2023-01-03')],
+ "column1": [1, 2, 3],
+ "column2": ["a", "b", "c"],
+ "column3": [1.1, 3.3, 2.2],
+ "column4": [True, False, True],
+ "column5": [pd.Timestamp("2023-01-01"), pd.Timestamp("2023-01-02"), pd.Timestamp("2023-01-03")],
},
- index=[0, 1, 2]
+ index=[0, 1, 2],
)
result = select_data_from_date_filters(
round_dataframe,
feature_id,
- id_date,
+ id_date,
val_feature,
start_date,
end_date,
)
pd.testing.assert_frame_equal(expected_result, result)
-
+
def test_select_data_from_numeric_filters(self):
round_dataframe = self.df
- id_feature = [{'type': 'var_dropdown', 'index': 1}, {'type': 'var_dropdown', 'index': 2}]
- feature_id = [id_feature[i]['index'] for i in range(len(id_feature))]
- id_lower_modality = [{'type': 'lower', 'index': 1}, {'type': 'lower', 'index': 2}]
- val_feature = ['column1', 'column3']
+ id_feature = [{"type": "var_dropdown", "index": 1}, {"type": "var_dropdown", "index": 2}]
+ feature_id = [id_feature[i]["index"] for i in range(len(id_feature))]
+ id_lower_modality = [{"type": "lower", "index": 1}, {"type": "lower", "index": 2}]
+ val_feature = ["column1", "column3"]
val_lower_modality = [0, 0]
val_upper_modality = [3, 3]
expected_result = pd.DataFrame(
{
- 'column1': [1, 3],
- 'column2': ['a', 'c'],
- 'column3': [1.1, 2.2],
- 'column4': [True, True],
- 'column5': [pd.Timestamp('2023-01-01'), pd.Timestamp('2023-01-03')],
+ "column1": [1, 3],
+ "column2": ["a", "c"],
+ "column3": [1.1, 2.2],
+ "column4": [True, True],
+ "column5": [pd.Timestamp("2023-01-01"), pd.Timestamp("2023-01-03")],
},
- index=[0, 2]
+ index=[0, 2],
)
result = select_data_from_numeric_filters(
round_dataframe,
@@ -225,11 +226,11 @@ def test_select_data_from_numeric_filters(self):
val_upper_modality,
)
pd.testing.assert_frame_equal(expected_result, result)
-
+
def test_get_feature_from_clicked_data(self):
feature = get_feature_from_clicked_data(self.click_data)
assert feature == "Sex"
-
+
def test_get_feature_from_features_groups(self):
features_groups = {"A": ["column1", "column3"]}
feature = get_feature_from_features_groups("column3", features_groups)
@@ -240,7 +241,7 @@ def test_get_feature_from_features_groups(self):
feature = get_feature_from_features_groups("column2", features_groups)
assert feature == "column2"
-
+
def test_get_group_name(self):
features_groups = {"A": ["column1", "column3"]}
feature = get_group_name("A", features_groups)
@@ -248,41 +249,41 @@ def test_get_group_name(self):
feature = get_group_name("column3", features_groups)
assert feature == None
-
+
def test_get_indexes_from_datatable(self):
- data = self.smart_app.components['table']['dataset'].data
+ data = self.smart_app.components["table"]["dataset"].data
subset = get_indexes_from_datatable(data)
assert subset == [0, 1, 2, 3, 4]
-
+
def test_get_indexes_from_datatable_no_subset(self):
- data = self.smart_app.components['table']['dataset'].data
+ data = self.smart_app.components["table"]["dataset"].data
subset = get_indexes_from_datatable(data, [0, 1, 2, 3, 4])
assert subset == None
-
+
def test_get_indexes_from_datatable_empty(self):
subset = get_indexes_from_datatable([], [0, 1, 2, 3, 4])
assert subset == None
-
+
def test_update_click_data_on_subset_changes(self):
click_data = {
- 'points': [
+ "points": [
{
- 'curveNumber': 1,
- 'pointNumber': 3,
- 'pointIndex': 3,
- 'x': 0.4649,
- 'y': 'Sex',
- 'label': 'Sex',
- 'value': 0.4649,
- 'customdata': 'Sex',
- 'marker.color': 'rgba(244, 192, 0, 1.0)',
- 'bbox': {'x0': 717.3, 'x1': 717.3, 'y0': 82.97, 'y1': 130.78}
+ "curveNumber": 1,
+ "pointNumber": 3,
+ "pointIndex": 3,
+ "x": 0.4649,
+ "y": "Sex",
+ "label": "Sex",
+ "value": 0.4649,
+ "customdata": "Sex",
+ "marker.color": "rgba(244, 192, 0, 1.0)",
+ "bbox": {"x0": 717.3, "x1": 717.3, "y0": 82.97, "y1": 130.78},
}
]
}
click_data = update_click_data_on_subset_changes(click_data)
assert click_data == self.click_data
-
+
def test_get_figure_zoom(self):
zoom_active = get_figure_zoom(None)
assert zoom_active == False
@@ -292,7 +293,7 @@ def test_get_figure_zoom(self):
zoom_active = get_figure_zoom(4)
assert zoom_active == False
-
+
def test_get_feature_contributions_sign_to_show(self):
sign = get_feature_contributions_sign_to_show([1], [1])
assert sign == None
@@ -308,126 +309,125 @@ def test_get_feature_contributions_sign_to_show(self):
def test_update_features_to_display(self):
value, max, marks = update_features_to_display(20, 40, 22)
- assert value==20
- assert max==20
- assert marks=={'1': '1', '5': '5', '10': '10', '15': '15', '20': '20'}
+ assert value == 20
+ assert max == 20
+ assert marks == {"1": "1", "5": "5", "10": "10", "15": "15", "20": "20"}
value, max, marks = update_features_to_display(7, 40, 6)
- assert value==6
- assert max==7
- assert marks=={'1': '1', '7': '7'}
-
+ assert value == 6
+ assert max == 7
+ assert marks == {"1": "1", "7": "7"}
+
def test_get_id_card_features(self):
- data = self.smart_app.components['table']['dataset'].data
+ data = self.smart_app.components["table"]["dataset"].data
features_dict = copy.deepcopy(self.xpl.features_dict)
features_dict.update(self.xpl.additional_features_dict)
selected_row = get_id_card_features(data, 3, self.special_cols, features_dict)
expected_result = pd.DataFrame(
{
- 'feature_value': [3, 1, 1, 4, 4.4, 'd', False, pd.Timestamp('2023-01-04')],
- 'feature_name': [
- '_index_',
- '_predict_',
- '_target_',
- 'column1',
- 'Useless col',
- '_Additional col',
- '_column4',
- '_column5',
+ "feature_value": [3, 1, 1, 4, 4.4, "d", False, pd.Timestamp("2023-01-04")],
+ "feature_name": [
+ "_index_",
+ "_predict_",
+ "_target_",
+ "column1",
+ "Useless col",
+ "_Additional col",
+ "_column4",
+ "_column5",
],
},
- index = ['_index_', '_predict_', '_target_', 'column1', 'column3', '_column2', '_column4', '_column5']
+ index=["_index_", "_predict_", "_target_", "column1", "column3", "_column2", "_column4", "_column5"],
)
pd.testing.assert_frame_equal(selected_row, expected_result)
-
+
def test_get_id_card_contrib(self):
data = self.xpl.data
selected_contrib = get_id_card_contrib(data, 3, self.xpl.features_dict, self.xpl.columns_dict, 0)
- assert set(selected_contrib['feature_name']) == {'Useless col', 'column1'}
- assert selected_contrib.columns.tolist() == ['feature_name', 'feature_contrib']
+ assert set(selected_contrib["feature_name"]) == {"Useless col", "column1"}
+ assert selected_contrib.columns.tolist() == ["feature_name", "feature_contrib"]
def test_create_id_card_data(self):
selected_row = pd.DataFrame(
{
- 'feature_value': [3, 1, 1, 4, 4.4, 'd', False, pd.Timestamp('2023-01-04')],
- 'feature_name': [
- '_index_',
- '_predict_',
- '_target_',
- 'column1',
- 'Useless col',
- '_Additional col',
- '_column4',
- '_column5',
+ "feature_value": [3, 1, 1, 4, 4.4, "d", False, pd.Timestamp("2023-01-04")],
+ "feature_name": [
+ "_index_",
+ "_predict_",
+ "_target_",
+ "column1",
+ "Useless col",
+ "_Additional col",
+ "_column4",
+ "_column5",
],
},
- index = ['_index_', '_predict_', '_target_', 'column1', 'column3', '_column2', '_column4', '_column5']
+ index=["_index_", "_predict_", "_target_", "column1", "column3", "_column2", "_column4", "_column5"],
)
selected_contrib = pd.DataFrame(
{
- 'feature_name': ['column1', 'Useless col'],
- 'feature_contrib': [-0.6, 0],
+ "feature_name": ["column1", "Useless col"],
+ "feature_contrib": [-0.6, 0],
}
)
selected_data = create_id_card_data(
- selected_row,
- selected_contrib,
- 'feature_name',
- True,
- self.special_cols,
- self.xpl.additional_features_dict
+ selected_row, selected_contrib, "feature_name", True, self.special_cols, self.xpl.additional_features_dict
)
expected_result = pd.DataFrame(
{
- 'feature_value': [3, 1, 1, 4.4, 4, 'd', False, pd.Timestamp('2023-01-04')],
- 'feature_name': [
- '_index_',
- '_predict_',
- '_target_',
- 'Useless col',
- 'column1',
- '_Additional col',
- '_column4',
- '_column5',
+ "feature_value": [3, 1, 1, 4.4, 4, "d", False, pd.Timestamp("2023-01-04")],
+ "feature_name": [
+ "_index_",
+ "_predict_",
+ "_target_",
+ "Useless col",
+ "column1",
+ "_Additional col",
+ "_column4",
+ "_column5",
],
- 'feature_contrib': [np.nan, np.nan, np.nan, 0.0, -0.6, np.nan, np.nan, np.nan]
+ "feature_contrib": [np.nan, np.nan, np.nan, 0.0, -0.6, np.nan, np.nan, np.nan],
},
- index = ['_index_', '_predict_', '_target_', 'column3', 'column1', '_column2', '_column4', '_column5']
+ index=["_index_", "_predict_", "_target_", "column3", "column1", "_column2", "_column4", "_column5"],
)
pd.testing.assert_frame_equal(selected_data, expected_result)
-
+
def test_create_id_card_layout(self):
selected_data = pd.DataFrame(
{
- 'feature_value': [3, 1, 1, 4.4, 4, 'd'],
- 'feature_name': ['_index_', '_predict_', '_target_', 'Useless col', 'column1', '_Additional col'],
- 'feature_contrib': [np.nan, np.nan, np.nan, 0.0, -0.6, np.nan]
+ "feature_value": [3, 1, 1, 4.4, 4, "d"],
+ "feature_name": ["_index_", "_predict_", "_target_", "Useless col", "column1", "_Additional col"],
+ "feature_contrib": [np.nan, np.nan, np.nan, 0.0, -0.6, np.nan],
},
- index = ['_index_', '_predict_', '_target_', 'column3', 'column1', '_column2']
+ index=["_index_", "_predict_", "_target_", "column3", "column1", "_column2"],
)
children = create_id_card_layout(selected_data, self.xpl.additional_features_dict)
- assert len(children)==6
-
+ assert len(children) == 6
+
def test_get_feature_filter_options(self):
features_dict = copy.deepcopy(self.xpl.features_dict)
features_dict.update(self.xpl.additional_features_dict)
options = get_feature_filter_options(self.smart_app.dataframe, features_dict, self.special_cols)
- assert [option["label"] for option in options]==[
- '_index_',
- '_predict_',
- '_target_',
- 'Useless col',
- '_Additional col',
- '_column4',
- '_column5',
- 'column1',
+ assert [option["label"] for option in options] == [
+ "_index_",
+ "_predict_",
+ "_target_",
+ "Useless col",
+ "_Additional col",
+ "_column4",
+ "_column5",
+ "column1",
]
-
+
def test_create_filter_modalities_selection(self):
- new_element = create_filter_modalities_selection("column3", {'type': 'var_dropdown', 'index': 1}, self.smart_app.round_dataframe)
- assert type(new_element.children[0])==dcc.Input
+ new_element = create_filter_modalities_selection(
+ "column3", {"type": "var_dropdown", "index": 1}, self.smart_app.round_dataframe
+ )
+ assert type(new_element.children[0]) == dcc.Input
- new_element = create_filter_modalities_selection("_column2", {'type': 'var_dropdown', 'index': 1}, self.smart_app.round_dataframe)
- assert type(new_element.children)==dcc.Dropdown
\ No newline at end of file
+ new_element = create_filter_modalities_selection(
+ "_column2", {"type": "var_dropdown", "index": 1}, self.smart_app.round_dataframe
+ )
+ assert type(new_element.children) == dcc.Dropdown
diff --git a/tests/unit_tests/webapp/utils/test_utils.py b/tests/unit_tests/webapp/utils/test_utils.py
index 8b25f9d4..4f75d6a0 100644
--- a/tests/unit_tests/webapp/utils/test_utils.py
+++ b/tests/unit_tests/webapp/utils/test_utils.py
@@ -1,9 +1,9 @@
import unittest
+
from shapash.webapp.utils.utils import round_to_k
class TestUtils(unittest.TestCase):
-
def test_round_to_k_1(self):
x = 123456789
expected_r_x = 123000000
diff --git a/tutorial/generate_report/output/.gitignore b/tutorial/generate_report/output/.gitignore
index 600a0007..bcff1706 100644
--- a/tutorial/generate_report/output/.gitignore
+++ b/tutorial/generate_report/output/.gitignore
@@ -1,3 +1,3 @@
# Ignore everything in this directory
*
-!.gitignore
\ No newline at end of file
+!.gitignore
diff --git a/tutorial/generate_report/shapash_report_example.py b/tutorial/generate_report/shapash_report_example.py
index 92420757..dfcac6e0 100644
--- a/tutorial/generate_report/shapash_report_example.py
+++ b/tutorial/generate_report/shapash_report_example.py
@@ -4,25 +4,23 @@
that generates the same report.
"""
import os
+
import pandas as pd
from category_encoders import OrdinalEncoder
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
-from shapash.data.data_loader import data_loading
from shapash import SmartExplainer
+from shapash.data.data_loader import data_loading
if __name__ == "__main__":
- house_df, house_dict = data_loading('house_prices')
- y_df = house_df['SalePrice']
- X_df = house_df[house_df.columns.difference(['SalePrice'])]
+ house_df, house_dict = data_loading("house_prices")
+ y_df = house_df["SalePrice"]
+ X_df = house_df[house_df.columns.difference(["SalePrice"])]
- categorical_features = [col for col in X_df.columns if X_df[col].dtype == 'object']
+ categorical_features = [col for col in X_df.columns if X_df[col].dtype == "object"]
- encoder = OrdinalEncoder(
- cols=categorical_features,
- handle_unknown='ignore',
- return_df=True).fit(X_df)
+ encoder = OrdinalEncoder(cols=categorical_features, handle_unknown="ignore", return_df=True).fit(X_df)
X_df = encoder.transform(X_df)
@@ -30,34 +28,34 @@
regressor = RandomForestRegressor(n_estimators=50).fit(Xtrain, ytrain)
- y_pred = pd.DataFrame(regressor.predict(Xtest), columns=['pred'], index=Xtest.index)
+ y_pred = pd.DataFrame(regressor.predict(Xtest), columns=["pred"], index=Xtest.index)
cur_dir = os.path.dirname(os.path.abspath(__file__))
xpl = SmartExplainer(
model=regressor,
preprocessing=encoder, # Optional: compile step can use inverse_transform method
- features_dict=house_dict
+ features_dict=house_dict,
)
xpl.compile(x=Xtest, y_pred=y_pred, y_target=ytest)
xpl.generate_report(
- output_file=os.path.join(cur_dir, 'output', 'report.html'),
- project_info_file=os.path.join(cur_dir, 'utils', 'project_info.yml'),
+ output_file=os.path.join(cur_dir, "output", "report.html"),
+ project_info_file=os.path.join(cur_dir, "utils", "project_info.yml"),
x_train=Xtrain,
y_train=ytrain,
y_test=ytest,
title_story="House prices report",
- title_description="""This document is a data science report of the kaggle house prices tutorial project.
+ title_description="""This document is a data science report of the kaggle house prices tutorial project.
It was generated using the Shapash library.""",
metrics=[
{
- 'path': 'sklearn.metrics.mean_absolute_error',
- 'name': 'Mean absolute error',
+ "path": "sklearn.metrics.mean_absolute_error",
+ "name": "Mean absolute error",
},
{
- 'path': 'sklearn.metrics.mean_squared_error',
- 'name': 'Mean squared error',
- }
- ]
+ "path": "sklearn.metrics.mean_squared_error",
+ "name": "Mean squared error",
+ },
+ ],
)
diff --git a/tutorial/generate_report/working/.gitignore b/tutorial/generate_report/working/.gitignore
index 600a0007..bcff1706 100644
--- a/tutorial/generate_report/working/.gitignore
+++ b/tutorial/generate_report/working/.gitignore
@@ -1,3 +1,3 @@
# Ignore everything in this directory
*
-!.gitignore
\ No newline at end of file
+!.gitignore

 -
-  +
+ 
 -As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
+As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
On the left side, you will find the official Shapash repository.
@@ -130,4 +130,4 @@ Your pull request is now ready to be submitted. A member of the Shapash team wil
You have contributed to an Open source project, thank you and congratulations ! 🥳
-Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
\ No newline at end of file
+Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
diff --git a/LICENSE b/LICENSE
index 4947287f..f433b1a5 100644
--- a/LICENSE
+++ b/LICENSE
@@ -174,4 +174,4 @@
incurred by, or claims asserted against, such Contributor by reason
of your accepting any such warranty or additional liability.
- END OF TERMS AND CONDITIONS
\ No newline at end of file
+ END OF TERMS AND CONDITIONS
diff --git a/Makefile b/Makefile
index 720b6909..1f65e00e 100644
--- a/Makefile
+++ b/Makefile
@@ -84,4 +84,4 @@ dist: clean ## builds source and wheel package
ls -l dist
install: clean ## install the package to the active Python's site-packages
- python setup.py install
\ No newline at end of file
+ python setup.py install
diff --git a/README.md b/README.md
index 8a159b3a..aee9e717 100644
--- a/README.md
+++ b/README.md
@@ -38,7 +38,7 @@ With Shapash, you can generate a **Webapp** that simplifies the comprehension of
Additionally, Shapash contributes to data science auditing by **presenting valuable information** about any model and data **in a comprehensive report**.
-Shapash is suitable for Regression, Binary Classification, and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
+Shapash is suitable for Regression, Binary Classification and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
> [!NOTE]
> If you want to give us feedback : [Feedback form](https://framaforms.org/shapash-collecting-your-feedback-and-use-cases-1687456776)
@@ -72,9 +72,9 @@ Shapash is suitable for Regression, Binary Classification, and Multiclass proble
| 2.2.x | Dataset Filter
-As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
+As you can see, you can select on the right side which branch of your forked repository you want to associate to the pull request.
On the left side, you will find the official Shapash repository.
@@ -130,4 +130,4 @@ Your pull request is now ready to be submitted. A member of the Shapash team wil
You have contributed to an Open source project, thank you and congratulations ! 🥳
-Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
\ No newline at end of file
+Show your contribution to Shapash in your curriculum, and share it on your social media. Be proud of yourself, you gave some code lines to the entire world !
diff --git a/LICENSE b/LICENSE
index 4947287f..f433b1a5 100644
--- a/LICENSE
+++ b/LICENSE
@@ -174,4 +174,4 @@
incurred by, or claims asserted against, such Contributor by reason
of your accepting any such warranty or additional liability.
- END OF TERMS AND CONDITIONS
\ No newline at end of file
+ END OF TERMS AND CONDITIONS
diff --git a/Makefile b/Makefile
index 720b6909..1f65e00e 100644
--- a/Makefile
+++ b/Makefile
@@ -84,4 +84,4 @@ dist: clean ## builds source and wheel package
ls -l dist
install: clean ## install the package to the active Python's site-packages
- python setup.py install
\ No newline at end of file
+ python setup.py install
diff --git a/README.md b/README.md
index 8a159b3a..aee9e717 100644
--- a/README.md
+++ b/README.md
@@ -38,7 +38,7 @@ With Shapash, you can generate a **Webapp** that simplifies the comprehension of
Additionally, Shapash contributes to data science auditing by **presenting valuable information** about any model and data **in a comprehensive report**.
-Shapash is suitable for Regression, Binary Classification, and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
+Shapash is suitable for Regression, Binary Classification and Multiclass problems. It is **compatible with numerous models**, including Catboost, Xgboost, LightGBM, Sklearn Ensemble, Linear models, and SVM. For other models, solutions to integrate Shapash are available; more details can be found [here](#how_shapash_works).
> [!NOTE]
> If you want to give us feedback : [Feedback form](https://framaforms.org/shapash-collecting-your-feedback-and-use-cases-1687456776)
@@ -72,9 +72,9 @@ Shapash is suitable for Regression, Binary Classification, and Multiclass proble
| 2.2.x | Dataset Filter  ](https://github.com/MAIF/shapash/blob/master/tutorial/tutorial01-Shapash-Overview-Launch-WebApp.ipynb)
| 2.0.x | Refactoring Shapash
](https://github.com/MAIF/shapash/blob/master/tutorial/tutorial01-Shapash-Overview-Launch-WebApp.ipynb)
| 2.0.x | Refactoring Shapash  ](https://github.com/MAIF/shapash/blob/master/tutorial/explainer_and_backend/tuto-expl06-Shapash-custom-backend.ipynb)
| 1.7.x | Variabilize Colors
](https://github.com/MAIF/shapash/blob/master/tutorial/explainer_and_backend/tuto-expl06-Shapash-custom-backend.ipynb)
| 1.7.x | Variabilize Colors  ](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common02-colors.ipynb)
-| 1.6.x | Explainability Quality Metrics
](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common02-colors.ipynb)
-| 1.6.x | Explainability Quality Metrics  ](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
-| 1.4.x | Groups of features
](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
-| 1.4.x | Groups of features  ](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
-| 1.3.x | Shapash Report
](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
-| 1.3.x | Shapash Report  ](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
+| 1.4.x | Groups of features
](https://github.com/MAIF/shapash/blob/master/tutorial/explainability_quality/tuto-quality01-Builing-confidence-explainability.ipynb)
+| 1.4.x | Groups of features  ](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
+| 1.3.x | Shapash Report
](https://github.com/MAIF/shapash/blob/master/tutorial/common/tuto-common01-groups_of_features.ipynb) |
+| 1.3.x | Shapash Report 
 -
-  +
+ 

 -
-  +
+ 

 -
-  +
+ 