diff --git a/_posts/2024-10-14-6th-issue-newsletter.md b/_posts/2024-10-14-6th-issue-newsletter.md
new file mode 100644
index 000000000..60f039958
--- /dev/null
+++ b/_posts/2024-10-14-6th-issue-newsletter.md
@@ -0,0 +1,234 @@
+---
+layout: post
+categories: newsletter
+title: "OBO Foundry Newsletter Issue 6"
+date: 2024-10-14
+author:
+ - OBO Operations Commitee
+---
+
+## OBO Foundry Newsletter Issue 6
+
+Welcome to the 6th issue of the OBO Foundry Newsletter! As always, we're excited to share the latest developments and achievements in the bio-ontology community.
+
+This edition continues our tradition of spotlighting contributors and ontologies. We're pleased to introduce two of our dedicated community members, as well as two ontologies that exemplify the principles and standards of the OBO Foundry.
+
+We will report on the progress of ongoing initiatives, including updates on ontology reviews, Principal standardization efforts, and new tools that are enhancing our collective work.
+
+This issue also features a recap of recent events and workshops that have fostered collaboration and knowledge-sharing within our community.
+
+As always, we invite you to share your thoughts, ideas, and suggestions with the OBO Operations Committee. Your input is invaluable in shaping the future of the OBO Foundry and advancing our shared mission of creating interoperable, high-quality ontologies for biomedical research.
+
+Best regards, [The OBO Foundry Operations Committee.](https://obofoundry.org/docs/OperationsCommittee.html)
+
+
+## Highlights
+
+
+#### New Dashboard release
+
+Our dashboard reflects the ongoing commitment of our community to harmonize metadata and increase interoperability across ontologies. We continue to invite the community to participate in the harmonization efforts, and get their ontologies to pass the dashboard. Ontologies that pass are highlighted on [https://obofoundry.org/](https://obofoundry.org/). The latest version of the dashboard can be accessed here: [https://dashboard.obofoundry.org/dashboard/index.html](https://dashboard.obofoundry.org/dashboard/index.html).
+
+
+---
+
+
+## Ongoing Discussions
+
+
+### [Principle 19](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/964) Development: Shaping Guidelines for Term Stability and Deprecation
+
+There is an ongoing discussion about the development and refinement of Principle 19. The discussion covers circumstances that require the obsoletion of terms and the stability of their meanings. The draft of [Principle 19](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/964) is currently in progress and focuses on term stability and deprecation.
+
+
+
+* Key points include:
+ * Defining circumstances for term deprecation
+ * Distinguishing between mandatory and recommended practices
+ * Exploring alternatives to the "obsolete class" method
+ * The committee discussed that all alternative identifiers (alt IDs) should be explicitly marked as obsolete. Furthermore, the OFOC decided that prepending "obsolete" to the label of an obsolete term is mandatory while prepending "obsolete" to the definition of an obsolete term is recommended but not mandatory.
+ * Documenting reasons for term obsoletion
+ * Ensuring timely communication about deprecated terms
+ * Automated compliance checks
+* The committee is considering:
+ * Using "must," "should," or "may" to indicate obligation levels
+ * Implementing automated checks for adherence to the principle
+ * Integrating the principle into ROBOT reports
+
+Discussions are ongoing, with the committee actively seeking feedback (which you can provide by commenting on the [issue](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/964)) and working towards a revised draft.
+
+
+---
+
+## Ontologies
+
+### New ontologies currently under review
+
+* [Exercise Medicine Ontology](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2615) (EXMO) is an ontology focused on exercise medicine. It is based on the Basic Formal Ontology (BFO), and includes terms related to physical activity, health status, and exercise prescriptions for various populations, including healthy individuals, those with chronic conditions, and people with disabilities. Developed to support personalized exercise prescriptions, EXMO bridges standardized guidelines with individualized approaches. It draws from guidelines, databases, and scientific literature and has been validated through expert evaluation and reasoners like ELK and JFact. Currently under review, EXMO aims to become a standard for personalized exercise prescriptions and a tool for databases and recommendation systems. [Join the discussion](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2615).
+* [Biomarker Ontology](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2604) (BMONT) is a comprehensive knowledge representation system that encompasses various medical and biological aspects. Developed in accordance with Basic Formal Ontology (BFO) and Open Biological and Biomedical Ontology (OBO) principles, BMONT builds upon a decade-old biomarker terminology created by Fraunhofer SCAI. It incorporates entities and definitions from this legacy system, as well as related terms gathered from scientific publications and books across diverse disease fields. The ontology aims to enhance biomarker identification tasks and serve as a supportive, integratable tool for advanced AI techniques such as Machine Learning (ML) and Large Learning Models (LLM). BMONT is under review, with contributors actively addressing feedback and making updates to improve its quality and alignment with OBO Foundry standards. [Join the discussion](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2604).
+* [Physiologically based pharmacokinetic modelling ontology](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2563) (PBPK) is a framework for representing the complex pharmacokinetic processes in living organisms. It includes classes like physiological parameters, PBPK types, biological compartments, and mathematical models, aiding in understanding drug behavior. This ontology provides a structured vocabulary for researchers, clinicians, and drug developers, promoting communication, data integration, and collaboration in pharmacokinetics and related life sciences. [ ](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2563)PBPK is currently under review - [join the discussion](https://github.com/OBOFoundry/OBOFoundry.github.io/issues/2563).
+
+
+### Spotlight on well-established OBO ontologies
+
+In this issue, we continue our ontology spotlight series, highlighting two ontologies from the OBO Foundry family, the [Medical Action Ontology (MAxO) ](https://obofoundry.org/ontology/maxo.html)and the [Drug Ontology (DrOn). ](https://obofoundry.org/ontology/dron.html)
+
+
+
+* The [Medical Action Ontology (MAxO) ](https://obofoundry.org/ontology/maxo.html)aims to provide a standardized framework for describing and organizing medical actions, particularly in the context of phenotypic annotations. This ontology plays a role in harmonizing the diverse terms used in clinical settings, making it easier for researchers and healthcare professionals to communicate effectively. A recent development in MAxO involves the integration of treatment annotations to phenotypes into the Human Phenotype Ontology (HPO) website ([example](https://hpo.jax.org/browse/term/HP:0000822), click on “Medical Action”). This update enhances the utility of HPO for researchers, allowing them to explore comprehensive annotations in one place, which improves disease diagnosis and phenotype-based research.
+* [The Drug Ontology (DrOn) ](https://obofoundry.org/ontology/dron.html)is a structured, formal representation of drug-related knowledge that organizes information about pharmaceuticals, their properties, ingredients, and relationships in a standardized format. It provides a common vocabulary and hierarchical classification system for drugs, containing aspects such as chemical structure, therapeutic uses, and mechanisms of action. Drug ontologies play a crucial role in various fields, including pharmacology, clinical research, and healthcare informatics, by facilitating data integration, enhancing interoperability between different systems, and supporting advanced applications like drug discovery, adverse event prediction, and personalized medicine. DrOn aims to provide a comprehensive classification of drug products and their ingredients, reusing the Chemical Entities of Biological Interest (ChEBI) ontology for compounds that are drug ingredients. It reuses other OBO ontologies as needed, including the Protein Ontology and the Information Artifact Ontology. Since its creation, DrOn has been semi-automatically built from the RxNorm drug terminology provided by the National Library of Medicine. Because RxNorm is not available in OWL or OBO format, DrOn represents the only principled OWL and OBO format version of key RxNorm content useful for automatically classifying drugs by their various properties. This process of automatically creating large segments of DrOn from RxNorm recently underwent a major overhaul in collaboration with James Overton. The entire DrOn build from monthly RxNorm releases is now encapsulated in DrOn’s Ontology Development Kit (ODK) processes in Docker, enabling DrOn curators with less technical expertise to manage the entire monthly build and release process
+
+
+---
+
+
+## Members and Volunteers
+
+The OBO Foundry is honored to highlight two valued community members.
+
+
+### Bill Hogan
+
+
+
+ +
+
+William “Bill” Hogan, MD, MS is currently Professor and Director of the Data Science Institute at the Medical College of Wisconsin, where he holds the Mary T. and Ted D. Kellner Chair of Data Science. He has over 20 years of experience in the development, maintenance, and application of biomedical data standards, especially ontologies. He created the Drug Ontology (DrOn) to facilitate comparative effectiveness research on medications. He also leads the development of the Ontology for Modeling and Representation of Social Entities (OMRSE), which represents healthcare organizations, healthcare providers, healthcare roles, as well as numerous social determinants of health including education, language and literacy, socio-economic status, social identities and categories, and intimate partner violence. He previously held R01 funding from NIGMS to create data standards for epidemic simulators and disease transmission models, which includes the Apollo-SV ontology. Apollo-SV represents numerous aspects of the infectious disease epidemiology domain.
+
+
+### Sarah Gehrke
+
+
+###
+
+
+
+
+William “Bill” Hogan, MD, MS is currently Professor and Director of the Data Science Institute at the Medical College of Wisconsin, where he holds the Mary T. and Ted D. Kellner Chair of Data Science. He has over 20 years of experience in the development, maintenance, and application of biomedical data standards, especially ontologies. He created the Drug Ontology (DrOn) to facilitate comparative effectiveness research on medications. He also leads the development of the Ontology for Modeling and Representation of Social Entities (OMRSE), which represents healthcare organizations, healthcare providers, healthcare roles, as well as numerous social determinants of health including education, language and literacy, socio-economic status, social identities and categories, and intimate partner violence. He previously held R01 funding from NIGMS to create data standards for epidemic simulators and disease transmission models, which includes the Apollo-SV ontology. Apollo-SV represents numerous aspects of the infectious disease epidemiology domain.
+
+
+### Sarah Gehrke
+
+
+###
+
+ +
+Sarah (she/they) is a Research Program Project Manager with the Translational and Integrative Sciences Lab (TISLab) at the University of North Carolina at Chapel Hill's Department of Genetics. With a background as an analytical chemist, Sarah brings diverse experience from industry labs which focused on coal and natural gas remediation and biomass conversion technologies. She also participated in metabolomics mass spectrometry research at the University of Colorado Anschutz Medical Campus (CU-AMC) and served as Finance Manager for the Structural Biology and Biochemistry Cores at the CU-AMC School of Medicine.
+
+In 2022, Sarah transitioned to her current role as Project Manager for the TISLab. Sarah manages the Monarch Initiative, providing key support for the Mondo Disease Ontology, the Monarch Knowledge Graph, and Human Phenotype Ontology. She is a driving force behind OBO Academy, managing the [ongoing Seminar series](https://oboacademy.github.io/obook/courses/monarch-obo-training/), curating training materials and recruiting speakers. Sarah is passionate about connecting people to drive fundamental, positive changes that enhance community wellness.
+
+
+---
+
+
+## Spotlight on Research in the OBO community
+
+
+### [A Change Language for Ontologies and Knowledge Graphs](https://arxiv.org/abs/2409.13906)
+
+The research outlines the Knowledge Graph Change Language (KGCL), a new standard data model for describing changes to knowledge graphs (KGs) and ontologies. KGCL aims to standardize and simplify the process of managing changes in these complex data structures, making it easier for stakeholders to understand and contribute to the development of ontologies and KGs. The preprint details the structure of KGCL, its controlled natural language (CNL), and various tools developed to support the use of KGCL, such as the Ontobot automated agent and a BioPortal widget for requesting changes to ontologies. The paper also discusses future directions for KGCL, including its use in AI applications and integration with other frameworks.
+
+
+
+Sarah (she/they) is a Research Program Project Manager with the Translational and Integrative Sciences Lab (TISLab) at the University of North Carolina at Chapel Hill's Department of Genetics. With a background as an analytical chemist, Sarah brings diverse experience from industry labs which focused on coal and natural gas remediation and biomass conversion technologies. She also participated in metabolomics mass spectrometry research at the University of Colorado Anschutz Medical Campus (CU-AMC) and served as Finance Manager for the Structural Biology and Biochemistry Cores at the CU-AMC School of Medicine.
+
+In 2022, Sarah transitioned to her current role as Project Manager for the TISLab. Sarah manages the Monarch Initiative, providing key support for the Mondo Disease Ontology, the Monarch Knowledge Graph, and Human Phenotype Ontology. She is a driving force behind OBO Academy, managing the [ongoing Seminar series](https://oboacademy.github.io/obook/courses/monarch-obo-training/), curating training materials and recruiting speakers. Sarah is passionate about connecting people to drive fundamental, positive changes that enhance community wellness.
+
+
+---
+
+
+## Spotlight on Research in the OBO community
+
+
+### [A Change Language for Ontologies and Knowledge Graphs](https://arxiv.org/abs/2409.13906)
+
+The research outlines the Knowledge Graph Change Language (KGCL), a new standard data model for describing changes to knowledge graphs (KGs) and ontologies. KGCL aims to standardize and simplify the process of managing changes in these complex data structures, making it easier for stakeholders to understand and contribute to the development of ontologies and KGs. The preprint details the structure of KGCL, its controlled natural language (CNL), and various tools developed to support the use of KGCL, such as the Ontobot automated agent and a BioPortal widget for requesting changes to ontologies. The paper also discusses future directions for KGCL, including its use in AI applications and integration with other frameworks.
+
+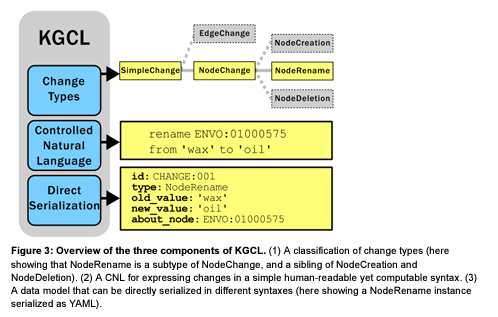 +
+
+(Figure from Harshad Hegde, Jennifer Vendetti, Damien Goutte-Gattat, J Harry Caufield, John B Graybeal, Nomi L Harris, Naouel Karam, Christian Kindermann, Nicolas Matentzoglu, James A Overton, Mark A Musen, Christopher J Mungall. A Change Language for Ontologies and Knowledge Graphs. 2024 Sep 20; [https://arxiv.org/abs/2409.13906](https://arxiv.org/abs/2409.13906))
+
+
+### [The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+This preprint describes the development of the Unified Phenotype Ontology (uPheno), a framework designed to integrate and standardize phenotype data across multiple species. uPheno addresses the challenge of integrating phenotype data from different sources by providing a consistent and logical framework for describing phenotypes. It utilizes a library of design patterns to define phenotype terms, enabling the development of a hierarchical vocabulary that groups species-specific phenotype terms under species-neutral categories. uPheno also includes mappings between species-specific ontologies, facilitating cross-species comparisons and enabling researchers to identify genes with similar phenotypic effects across different organisms. The paper highlights the potential applications of uPheno in various areas of biological research, including clinical diagnostics, variant prioritization, and disease modeling.
+
+
+
+
+(Figure from Harshad Hegde, Jennifer Vendetti, Damien Goutte-Gattat, J Harry Caufield, John B Graybeal, Nomi L Harris, Naouel Karam, Christian Kindermann, Nicolas Matentzoglu, James A Overton, Mark A Musen, Christopher J Mungall. A Change Language for Ontologies and Knowledge Graphs. 2024 Sep 20; [https://arxiv.org/abs/2409.13906](https://arxiv.org/abs/2409.13906))
+
+
+### [The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+This preprint describes the development of the Unified Phenotype Ontology (uPheno), a framework designed to integrate and standardize phenotype data across multiple species. uPheno addresses the challenge of integrating phenotype data from different sources by providing a consistent and logical framework for describing phenotypes. It utilizes a library of design patterns to define phenotype terms, enabling the development of a hierarchical vocabulary that groups species-specific phenotype terms under species-neutral categories. uPheno also includes mappings between species-specific ontologies, facilitating cross-species comparisons and enabling researchers to identify genes with similar phenotypic effects across different organisms. The paper highlights the potential applications of uPheno in various areas of biological research, including clinical diagnostics, variant prioritization, and disease modeling.
+
+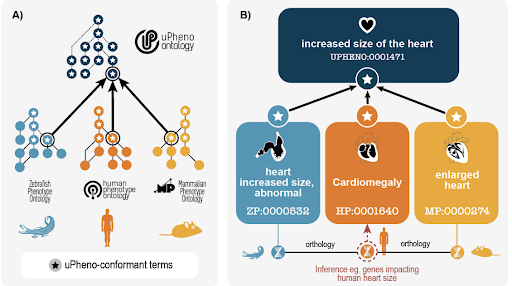 +
+
+(Figure from Nicolas Matentzoglu, Susan M Bello, Ray Stefancsik, Sarah M. Alghamdi, Anna V. Anagnostopoulos, James P. Balhoff, Meghan A. Balk, Yvonne M. Bradford, Yasemin Bridges, Tiffany J. Callahan, Harry Caufield, Alayne Cuzick, Leigh C Carmody, Anita R. Caron, Vinicius de Souza, Stacia R. Engel, Petra Fey, Malcolm Fisher, Sarah Gehrke, Christian Grove, Peter Hansen, Nomi L. Harris, Midori A. Harris, Laura Harris, Arwa Ibrahim, Julius O.B. Jacobsen, Sebastian Köhler, Julie A. McMurry, Violeta Munoz-Fuentes, Monica C. Munoz-Torres, Helen Parkinson, Zoë M Pendlington, Clare Pilgrim, Sofia MC Robb, Peter N. Robinson, James Seager, Erik Segerdell, Damian Smedley, Elliot Sollis, Sabrina Toro, Nicole Vasilevsky, Valerie Wood, Melissa A. Haendel, Christopher J. Mungall, James A. McLaughlin, and David Osumi-Sutherland. The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics. 2024 Sep 20. [https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+
+---
+
+
+## Spotlight on OBO Principles
+
+In this issue, we continue with [Principle 5 ](https://obofoundry.org/principles/fp-005-delineated-content.html)of the OBO Foundry's series of principles.
+
+
+### OBO Foundry Principle 5: Scope
+
+OBO Principle 5 addresses the importance of a clearly defined scope for ontologies. It emphasizes that each ontology should have a well-specified domain or subject matter it intends to cover, and its content should adhere strictly to that scope. This principle aims to avoid overlaps between ontologies, facilitate user searches, and enable quick selection of relevant ontologies while still allowing for the creation of new terms through cross-products. The scope of an ontology should ideally be narrow, with out-of-scope terms imported from appropriate existing ontologies when possible. If an immediate need arises for out-of-scope terms, they should be placed in a separate, shareable module. The ontology's domain should be clearly stated in a brief, jargon-free manner, and its content should remain within these defined boundaries. This principle also encourages consideration of generic and mid/upper level terms for potential inclusion in the Core Ontology for Biology and Biomedicine (COB), ensuring that the ontology serves both its specific purpose and broader community needs.
+
+
+---
+
+
+## Spotlight on Tools: Recent updates
+
+
+### Simple Standard for Sharing Ontological Mappings (SSSOM) v. 1.0.
+
+While the goal of the OBO community is to develop a set of interoperable ontologies covering the biomedical domain, many vocabularies, terminologies and ontologies are used globally which need to be linked to OBO ontologies. The [SSSOM ](https://mapping-commons.github.io/sssom/)community standard was developed to provide an easy way to specify and exchange such mappings with rich provenance. After more than 4 years of work, with 79 contributors writing more than 1200 comments the first stable release is finally out:
+
+[https://github.com/mapping-commons/sssom/releases/tag/v1.0.0](https://github.com/mapping-commons/sssom/releases/tag/v1.0.0). A short post about the release was [published on LinkedIn by the Monarch Initiative](https://www.linkedin.com/posts/the-monarch-initiative_sssom-monarchinitiative-semantic-activity-7228793130441916416-roIB/?utm_source=share&utm_medium=member_desktop).
+
+
+---
+
+
+## Spotlight on Tools: Ontology Browsers
+
+Here and in upcoming issues of this newsletter, we would like to highlight some key ontology browsers: the Ontology Lookup Service (OLS), Ontobee, and BioPortal. We spotlight BioPortal in this issue; watch for spotlights on Ontobee and OLS in future issues.
+
+
+### BioPortal
+
+[BioPortal ](https://bioportal.bioontology.org/)is a web-based repository and collaborative platform for biomedical ontologies and terminologies developed by the National Center for Biomedical Ontology (NCBO). It is a centralized resource for accessing, browsing, and integrating standardized biomedical vocabularies and ontologies. The platform utilizes semantic web technologies to enhance interoperability and data integration across biomedical domains. BioPortal's collection contains numerous ontologies covering fields such as anatomy, phenotypes, experimental conditions, and molecular functions. BioPortal also offers ontology visualization tools, mapping services, and programmatic access via web services, facilitating knowledge management and semantic annotation of biomedical data.
+
+
+---
+
+
+## Events
+
+
+
+* **COB Workshop.** Purpose: To advance the development of COB.
+
+ The 2024 COB Workshop was held September 23–24 at The La Jolla Institute for Immunology in San Diego, CA.
+
+
+ The workshop included 10 in-person and 8 remote participants. Both days were quite productive with lively discussions and consensus building. Decisions were made and documented as described below and tabled discussions were tracked for future work, with the goal of focusing discussion on topics that were most likely to result in actionable decisions. The mechanism was to go through terms included in the present COB release, and review each term to decide whether it should be part of COB base (with COB IDs, label and definitions that would replace existing terms), COB full (imported terms from existing ontologies that can be used as is, which are needed in the axiomatization of COB), and COB root (imported terms from existing ontologies that are not needed to define COB, but that are included to illustrate how existing OBO ontologies will be able to connect). Much of the discussion on Day 1 focused on macromolecules and cells, working through many of the significant material entities for the remainder of the day. Day 2 started with a focus on human-made devices and material processing terms, with the goal of using COB to align existing terms from OBI and FOODON. Specimens and their collection from anatomical entities were also briefly discussed. Finally, a proposal on how to consistently define stages, phases, and cycles was discussed; different groups have modeled them as material entities, characteristics, biological processes, or time intervals/processes. The proposal was to consistently capture these as subclasses of processes in COB and to characteristics such as PATO:duration to link these to time intervals. Overall, more than half of the COB terms were discussed and a (preliminary) classification was made. Many participants requested future focused virtual meetings to continue COB development using the same strategies, to tackle the remaining terms, and get input from a broader community.
+
+
+ If you are interested in this work, please watch and contribute to the COB repository: [https://github.com/OBOFoundry/COB](https://github.com/OBOFoundry/COB), and stay tuned for a more detailed workshop summary, which will be available in Issue 7 of the newsletter!
+
+* [OBO Academy: Monarch Seminar Series](https://oboacademy.github.io/obook/courses/monarch-obo-training/) (ongoing)
+* Schedule:[ https://oboacademy.github.io/obook/courses/monarch-obo-training/](https://oboacademy.github.io/obook/courses/monarch-obo-training)
+* You can vote for future training topics here: [https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests](https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests)
+
+
+### Past Events
+
+**Fostering Open Science Collaborative Workflows: Insights from the ICBO OBO Tutorial**
+
+By Nicole Vasilevsky and Sabrina Toro
+
+The [OBO Foundry Training at ICBO 2024](https://oboacademy.github.io/obook/courses/icbo2024/) took a close look at the social workflows surrounding ontology development. Topics such as GitHub, Pull Requests, review and principles of good collaborative work were discussed in depth. A recording is available at the above link.
+
+Every year at the International Conference on Biological and Biomedical Ontology (ICBO), members of the Open Biological and Biomedical Ontology (OBO) Foundry host an OBO Tutorial to share insights into our latest work on ontology development, share best practices, and exchange knowledge. These OBO tutorials have become part of the OBO Academy ([https://oboacademy.github.io/obook/](https://oboacademy.github.io/obook/)), with special attention to training newcomers to semantic engineering. This year’s virtual tutorial was organized by Nico Matenzoglu (Semanticly) and Nicole Vasilevsky (Critical Path Institute) and focused on “Fostering Open Science Collaborative Workflows”. More details, including a recording of the session, are available here: [https://oboacademy.github.io/obook/courses/icbo2024/](https://oboacademy.github.io/obook/courses/icbo2024/)
+
+We discussed the importance of collaboration in the development of OBO ontologies. James Overton introduced the OBO Foundry, Sabrina Toro discussed Git and GitHub, Nico Matentzoglu discussed best practices in collaborative workflows and Nicole Vasilevsky shared best practices in creating GitHub tickets, submitting pull requests (PRs), and reviewing PRs in ontology repositories.
+
+Thanks to a generous microgrant from the [International Society for Biocuration](https://www.biocuration.org/) we were able to offer small prizes for participating in a training quiz integrated into the tutorial. We awarded prizes to the top 5 participants with the most correct answers and 5 prizes to participants who answered the most questions, regardless of their answers. The winners were (in alphabetical order): Hannah Blau, Spencer Bliven, Yvonne Bradford, Lauren Chan, John Graybeal, Harshad Hegde, Bryan Laraway, Katie Mullen, Sumir Pandit, and Umasri Sankarlal.
+
+Following the session, we hosted the [OBO Community Challenge](https://github.com/OBOAcademy/obook/issues/508), which was open to all. The challenge provided an opportunity to apply the lessons learned from the OBO tutorial at ICBO. Participants were asked to choose one or several OBO ontology projects in which they were not involved yet, and complete a set of tasks (including opening an issue and creating a pull request) demonstrating the basic principles of collaborative workflows introduced at the tutorial. Ontology contributions were reviewed for the quality of the commit messages, and issue and pull request descriptions. We are very grateful to everyone who participated. The prizer was awarded to Paula Duek Roggli and Katie Mullen.
+
+This was our inaugural opportunity to award prizes to participants in the OBO tutorial and OBO challenge. We believe these prizes enhanced engagement and boosted participation within the community. We extend our heartfelt gratitude to the ISB and the OBO Foundry community for their support, and to the ISB for backing the microgrant initiative. We also encourage everyone to get involved with and support the ISB—you might be the next recipient of a similar microgrant.
+
+
+---
+
+
+## Ways to be part of the OBO Foundry community
+
+There are many ways to be part of the OBO Foundry community, beyond [using our website to find ontologies of interest](https://obofoundry.org). For example:
+
+
+
+* Join the [obo-discuss mailing list](https://groups.google.com/forum/#!forum/obo-discuss)
+* If you are interested in the technical aspects, you can also join the [obo-tools mailing list](https://groups.google.com/forum/#!members/obo-tools)
+* Join the conversation in our [Slack workspace](https://join.slack.com/t/obo-communitygroup/shared_invite/zt-1oq48ttk7-kKo0i6TwntYtAq~Jcjjg4g).
+* Use our public [issue tracker](https://github.com/OBOFoundry/OBOFoundry.github.io/issues) to report a problem you discovered on obofoundry.org or request a new feature
+ * Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
+* Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of [our code is publicly readable](https://github.com/OBOFoundry), you may be able to pinpoint where a bug fix needs to go.
+* [Request that your ontology be considered for inclusion in the Foundry](https://obofoundry.org/faq/how-do-i-register-my-ontology.html)
+* If you want to take your contributions to OBO Foundry to the next level, you can [nominate yourself to be considered for the OBO Operations Committee](https://obofoundry.org/docs/NewOBOFC.html). There are many ways in which you can contribute, including assisting with the production of this newsletter.
diff --git a/images/newsletter/hogan.png b/images/newsletter/hogan.png
new file mode 100644
index 000000000..01e34157d
Binary files /dev/null and b/images/newsletter/hogan.png differ
diff --git a/images/newsletter/kgcl.png b/images/newsletter/kgcl.png
new file mode 100644
index 000000000..3b1839726
Binary files /dev/null and b/images/newsletter/kgcl.png differ
diff --git a/images/newsletter/sgehrke.png b/images/newsletter/sgehrke.png
new file mode 100644
index 000000000..c992d7be7
Binary files /dev/null and b/images/newsletter/sgehrke.png differ
diff --git a/images/newsletter/upheno.png b/images/newsletter/upheno.png
new file mode 100644
index 000000000..d8fe58e75
Binary files /dev/null and b/images/newsletter/upheno.png differ
+
+
+(Figure from Nicolas Matentzoglu, Susan M Bello, Ray Stefancsik, Sarah M. Alghamdi, Anna V. Anagnostopoulos, James P. Balhoff, Meghan A. Balk, Yvonne M. Bradford, Yasemin Bridges, Tiffany J. Callahan, Harry Caufield, Alayne Cuzick, Leigh C Carmody, Anita R. Caron, Vinicius de Souza, Stacia R. Engel, Petra Fey, Malcolm Fisher, Sarah Gehrke, Christian Grove, Peter Hansen, Nomi L. Harris, Midori A. Harris, Laura Harris, Arwa Ibrahim, Julius O.B. Jacobsen, Sebastian Köhler, Julie A. McMurry, Violeta Munoz-Fuentes, Monica C. Munoz-Torres, Helen Parkinson, Zoë M Pendlington, Clare Pilgrim, Sofia MC Robb, Peter N. Robinson, James Seager, Erik Segerdell, Damian Smedley, Elliot Sollis, Sabrina Toro, Nicole Vasilevsky, Valerie Wood, Melissa A. Haendel, Christopher J. Mungall, James A. McLaughlin, and David Osumi-Sutherland. The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics. 2024 Sep 20. [https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+
+---
+
+
+## Spotlight on OBO Principles
+
+In this issue, we continue with [Principle 5 ](https://obofoundry.org/principles/fp-005-delineated-content.html)of the OBO Foundry's series of principles.
+
+
+### OBO Foundry Principle 5: Scope
+
+OBO Principle 5 addresses the importance of a clearly defined scope for ontologies. It emphasizes that each ontology should have a well-specified domain or subject matter it intends to cover, and its content should adhere strictly to that scope. This principle aims to avoid overlaps between ontologies, facilitate user searches, and enable quick selection of relevant ontologies while still allowing for the creation of new terms through cross-products. The scope of an ontology should ideally be narrow, with out-of-scope terms imported from appropriate existing ontologies when possible. If an immediate need arises for out-of-scope terms, they should be placed in a separate, shareable module. The ontology's domain should be clearly stated in a brief, jargon-free manner, and its content should remain within these defined boundaries. This principle also encourages consideration of generic and mid/upper level terms for potential inclusion in the Core Ontology for Biology and Biomedicine (COB), ensuring that the ontology serves both its specific purpose and broader community needs.
+
+
+---
+
+
+## Spotlight on Tools: Recent updates
+
+
+### Simple Standard for Sharing Ontological Mappings (SSSOM) v. 1.0.
+
+While the goal of the OBO community is to develop a set of interoperable ontologies covering the biomedical domain, many vocabularies, terminologies and ontologies are used globally which need to be linked to OBO ontologies. The [SSSOM ](https://mapping-commons.github.io/sssom/)community standard was developed to provide an easy way to specify and exchange such mappings with rich provenance. After more than 4 years of work, with 79 contributors writing more than 1200 comments the first stable release is finally out:
+
+[https://github.com/mapping-commons/sssom/releases/tag/v1.0.0](https://github.com/mapping-commons/sssom/releases/tag/v1.0.0). A short post about the release was [published on LinkedIn by the Monarch Initiative](https://www.linkedin.com/posts/the-monarch-initiative_sssom-monarchinitiative-semantic-activity-7228793130441916416-roIB/?utm_source=share&utm_medium=member_desktop).
+
+
+---
+
+
+## Spotlight on Tools: Ontology Browsers
+
+Here and in upcoming issues of this newsletter, we would like to highlight some key ontology browsers: the Ontology Lookup Service (OLS), Ontobee, and BioPortal. We spotlight BioPortal in this issue; watch for spotlights on Ontobee and OLS in future issues.
+
+
+### BioPortal
+
+[BioPortal ](https://bioportal.bioontology.org/)is a web-based repository and collaborative platform for biomedical ontologies and terminologies developed by the National Center for Biomedical Ontology (NCBO). It is a centralized resource for accessing, browsing, and integrating standardized biomedical vocabularies and ontologies. The platform utilizes semantic web technologies to enhance interoperability and data integration across biomedical domains. BioPortal's collection contains numerous ontologies covering fields such as anatomy, phenotypes, experimental conditions, and molecular functions. BioPortal also offers ontology visualization tools, mapping services, and programmatic access via web services, facilitating knowledge management and semantic annotation of biomedical data.
+
+
+---
+
+
+## Events
+
+
+
+* **COB Workshop.** Purpose: To advance the development of COB.
+
+ The 2024 COB Workshop was held September 23–24 at The La Jolla Institute for Immunology in San Diego, CA.
+
+
+ The workshop included 10 in-person and 8 remote participants. Both days were quite productive with lively discussions and consensus building. Decisions were made and documented as described below and tabled discussions were tracked for future work, with the goal of focusing discussion on topics that were most likely to result in actionable decisions. The mechanism was to go through terms included in the present COB release, and review each term to decide whether it should be part of COB base (with COB IDs, label and definitions that would replace existing terms), COB full (imported terms from existing ontologies that can be used as is, which are needed in the axiomatization of COB), and COB root (imported terms from existing ontologies that are not needed to define COB, but that are included to illustrate how existing OBO ontologies will be able to connect). Much of the discussion on Day 1 focused on macromolecules and cells, working through many of the significant material entities for the remainder of the day. Day 2 started with a focus on human-made devices and material processing terms, with the goal of using COB to align existing terms from OBI and FOODON. Specimens and their collection from anatomical entities were also briefly discussed. Finally, a proposal on how to consistently define stages, phases, and cycles was discussed; different groups have modeled them as material entities, characteristics, biological processes, or time intervals/processes. The proposal was to consistently capture these as subclasses of processes in COB and to characteristics such as PATO:duration to link these to time intervals. Overall, more than half of the COB terms were discussed and a (preliminary) classification was made. Many participants requested future focused virtual meetings to continue COB development using the same strategies, to tackle the remaining terms, and get input from a broader community.
+
+
+ If you are interested in this work, please watch and contribute to the COB repository: [https://github.com/OBOFoundry/COB](https://github.com/OBOFoundry/COB), and stay tuned for a more detailed workshop summary, which will be available in Issue 7 of the newsletter!
+
+* [OBO Academy: Monarch Seminar Series](https://oboacademy.github.io/obook/courses/monarch-obo-training/) (ongoing)
+* Schedule:[ https://oboacademy.github.io/obook/courses/monarch-obo-training/](https://oboacademy.github.io/obook/courses/monarch-obo-training)
+* You can vote for future training topics here: [https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests](https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests)
+
+
+### Past Events
+
+**Fostering Open Science Collaborative Workflows: Insights from the ICBO OBO Tutorial**
+
+By Nicole Vasilevsky and Sabrina Toro
+
+The [OBO Foundry Training at ICBO 2024](https://oboacademy.github.io/obook/courses/icbo2024/) took a close look at the social workflows surrounding ontology development. Topics such as GitHub, Pull Requests, review and principles of good collaborative work were discussed in depth. A recording is available at the above link.
+
+Every year at the International Conference on Biological and Biomedical Ontology (ICBO), members of the Open Biological and Biomedical Ontology (OBO) Foundry host an OBO Tutorial to share insights into our latest work on ontology development, share best practices, and exchange knowledge. These OBO tutorials have become part of the OBO Academy ([https://oboacademy.github.io/obook/](https://oboacademy.github.io/obook/)), with special attention to training newcomers to semantic engineering. This year’s virtual tutorial was organized by Nico Matenzoglu (Semanticly) and Nicole Vasilevsky (Critical Path Institute) and focused on “Fostering Open Science Collaborative Workflows”. More details, including a recording of the session, are available here: [https://oboacademy.github.io/obook/courses/icbo2024/](https://oboacademy.github.io/obook/courses/icbo2024/)
+
+We discussed the importance of collaboration in the development of OBO ontologies. James Overton introduced the OBO Foundry, Sabrina Toro discussed Git and GitHub, Nico Matentzoglu discussed best practices in collaborative workflows and Nicole Vasilevsky shared best practices in creating GitHub tickets, submitting pull requests (PRs), and reviewing PRs in ontology repositories.
+
+Thanks to a generous microgrant from the [International Society for Biocuration](https://www.biocuration.org/) we were able to offer small prizes for participating in a training quiz integrated into the tutorial. We awarded prizes to the top 5 participants with the most correct answers and 5 prizes to participants who answered the most questions, regardless of their answers. The winners were (in alphabetical order): Hannah Blau, Spencer Bliven, Yvonne Bradford, Lauren Chan, John Graybeal, Harshad Hegde, Bryan Laraway, Katie Mullen, Sumir Pandit, and Umasri Sankarlal.
+
+Following the session, we hosted the [OBO Community Challenge](https://github.com/OBOAcademy/obook/issues/508), which was open to all. The challenge provided an opportunity to apply the lessons learned from the OBO tutorial at ICBO. Participants were asked to choose one or several OBO ontology projects in which they were not involved yet, and complete a set of tasks (including opening an issue and creating a pull request) demonstrating the basic principles of collaborative workflows introduced at the tutorial. Ontology contributions were reviewed for the quality of the commit messages, and issue and pull request descriptions. We are very grateful to everyone who participated. The prizer was awarded to Paula Duek Roggli and Katie Mullen.
+
+This was our inaugural opportunity to award prizes to participants in the OBO tutorial and OBO challenge. We believe these prizes enhanced engagement and boosted participation within the community. We extend our heartfelt gratitude to the ISB and the OBO Foundry community for their support, and to the ISB for backing the microgrant initiative. We also encourage everyone to get involved with and support the ISB—you might be the next recipient of a similar microgrant.
+
+
+---
+
+
+## Ways to be part of the OBO Foundry community
+
+There are many ways to be part of the OBO Foundry community, beyond [using our website to find ontologies of interest](https://obofoundry.org). For example:
+
+
+
+* Join the [obo-discuss mailing list](https://groups.google.com/forum/#!forum/obo-discuss)
+* If you are interested in the technical aspects, you can also join the [obo-tools mailing list](https://groups.google.com/forum/#!members/obo-tools)
+* Join the conversation in our [Slack workspace](https://join.slack.com/t/obo-communitygroup/shared_invite/zt-1oq48ttk7-kKo0i6TwntYtAq~Jcjjg4g).
+* Use our public [issue tracker](https://github.com/OBOFoundry/OBOFoundry.github.io/issues) to report a problem you discovered on obofoundry.org or request a new feature
+ * Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
+* Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of [our code is publicly readable](https://github.com/OBOFoundry), you may be able to pinpoint where a bug fix needs to go.
+* [Request that your ontology be considered for inclusion in the Foundry](https://obofoundry.org/faq/how-do-i-register-my-ontology.html)
+* If you want to take your contributions to OBO Foundry to the next level, you can [nominate yourself to be considered for the OBO Operations Committee](https://obofoundry.org/docs/NewOBOFC.html). There are many ways in which you can contribute, including assisting with the production of this newsletter.
diff --git a/images/newsletter/hogan.png b/images/newsletter/hogan.png
new file mode 100644
index 000000000..01e34157d
Binary files /dev/null and b/images/newsletter/hogan.png differ
diff --git a/images/newsletter/kgcl.png b/images/newsletter/kgcl.png
new file mode 100644
index 000000000..3b1839726
Binary files /dev/null and b/images/newsletter/kgcl.png differ
diff --git a/images/newsletter/sgehrke.png b/images/newsletter/sgehrke.png
new file mode 100644
index 000000000..c992d7be7
Binary files /dev/null and b/images/newsletter/sgehrke.png differ
diff --git a/images/newsletter/upheno.png b/images/newsletter/upheno.png
new file mode 100644
index 000000000..d8fe58e75
Binary files /dev/null and b/images/newsletter/upheno.png differ
 +
+
+William “Bill” Hogan, MD, MS is currently Professor and Director of the Data Science Institute at the Medical College of Wisconsin, where he holds the Mary T. and Ted D. Kellner Chair of Data Science. He has over 20 years of experience in the development, maintenance, and application of biomedical data standards, especially ontologies. He created the Drug Ontology (DrOn) to facilitate comparative effectiveness research on medications. He also leads the development of the Ontology for Modeling and Representation of Social Entities (OMRSE), which represents healthcare organizations, healthcare providers, healthcare roles, as well as numerous social determinants of health including education, language and literacy, socio-economic status, social identities and categories, and intimate partner violence. He previously held R01 funding from NIGMS to create data standards for epidemic simulators and disease transmission models, which includes the Apollo-SV ontology. Apollo-SV represents numerous aspects of the infectious disease epidemiology domain.
+
+
+### Sarah Gehrke
+
+
+###
+
+
+
+
+William “Bill” Hogan, MD, MS is currently Professor and Director of the Data Science Institute at the Medical College of Wisconsin, where he holds the Mary T. and Ted D. Kellner Chair of Data Science. He has over 20 years of experience in the development, maintenance, and application of biomedical data standards, especially ontologies. He created the Drug Ontology (DrOn) to facilitate comparative effectiveness research on medications. He also leads the development of the Ontology for Modeling and Representation of Social Entities (OMRSE), which represents healthcare organizations, healthcare providers, healthcare roles, as well as numerous social determinants of health including education, language and literacy, socio-economic status, social identities and categories, and intimate partner violence. He previously held R01 funding from NIGMS to create data standards for epidemic simulators and disease transmission models, which includes the Apollo-SV ontology. Apollo-SV represents numerous aspects of the infectious disease epidemiology domain.
+
+
+### Sarah Gehrke
+
+
+###
+
+ +
+Sarah (she/they) is a Research Program Project Manager with the Translational and Integrative Sciences Lab (TISLab) at the University of North Carolina at Chapel Hill's Department of Genetics. With a background as an analytical chemist, Sarah brings diverse experience from industry labs which focused on coal and natural gas remediation and biomass conversion technologies. She also participated in metabolomics mass spectrometry research at the University of Colorado Anschutz Medical Campus (CU-AMC) and served as Finance Manager for the Structural Biology and Biochemistry Cores at the CU-AMC School of Medicine.
+
+In 2022, Sarah transitioned to her current role as Project Manager for the TISLab. Sarah manages the Monarch Initiative, providing key support for the Mondo Disease Ontology, the Monarch Knowledge Graph, and Human Phenotype Ontology. She is a driving force behind OBO Academy, managing the [ongoing Seminar series](https://oboacademy.github.io/obook/courses/monarch-obo-training/), curating training materials and recruiting speakers. Sarah is passionate about connecting people to drive fundamental, positive changes that enhance community wellness.
+
+
+---
+
+
+## Spotlight on Research in the OBO community
+
+
+### [A Change Language for Ontologies and Knowledge Graphs](https://arxiv.org/abs/2409.13906)
+
+The research outlines the Knowledge Graph Change Language (KGCL), a new standard data model for describing changes to knowledge graphs (KGs) and ontologies. KGCL aims to standardize and simplify the process of managing changes in these complex data structures, making it easier for stakeholders to understand and contribute to the development of ontologies and KGs. The preprint details the structure of KGCL, its controlled natural language (CNL), and various tools developed to support the use of KGCL, such as the Ontobot automated agent and a BioPortal widget for requesting changes to ontologies. The paper also discusses future directions for KGCL, including its use in AI applications and integration with other frameworks.
+
+
+
+Sarah (she/they) is a Research Program Project Manager with the Translational and Integrative Sciences Lab (TISLab) at the University of North Carolina at Chapel Hill's Department of Genetics. With a background as an analytical chemist, Sarah brings diverse experience from industry labs which focused on coal and natural gas remediation and biomass conversion technologies. She also participated in metabolomics mass spectrometry research at the University of Colorado Anschutz Medical Campus (CU-AMC) and served as Finance Manager for the Structural Biology and Biochemistry Cores at the CU-AMC School of Medicine.
+
+In 2022, Sarah transitioned to her current role as Project Manager for the TISLab. Sarah manages the Monarch Initiative, providing key support for the Mondo Disease Ontology, the Monarch Knowledge Graph, and Human Phenotype Ontology. She is a driving force behind OBO Academy, managing the [ongoing Seminar series](https://oboacademy.github.io/obook/courses/monarch-obo-training/), curating training materials and recruiting speakers. Sarah is passionate about connecting people to drive fundamental, positive changes that enhance community wellness.
+
+
+---
+
+
+## Spotlight on Research in the OBO community
+
+
+### [A Change Language for Ontologies and Knowledge Graphs](https://arxiv.org/abs/2409.13906)
+
+The research outlines the Knowledge Graph Change Language (KGCL), a new standard data model for describing changes to knowledge graphs (KGs) and ontologies. KGCL aims to standardize and simplify the process of managing changes in these complex data structures, making it easier for stakeholders to understand and contribute to the development of ontologies and KGs. The preprint details the structure of KGCL, its controlled natural language (CNL), and various tools developed to support the use of KGCL, such as the Ontobot automated agent and a BioPortal widget for requesting changes to ontologies. The paper also discusses future directions for KGCL, including its use in AI applications and integration with other frameworks.
+
+ +
+
+(Figure from Harshad Hegde, Jennifer Vendetti, Damien Goutte-Gattat, J Harry Caufield, John B Graybeal, Nomi L Harris, Naouel Karam, Christian Kindermann, Nicolas Matentzoglu, James A Overton, Mark A Musen, Christopher J Mungall. A Change Language for Ontologies and Knowledge Graphs. 2024 Sep 20; [https://arxiv.org/abs/2409.13906](https://arxiv.org/abs/2409.13906))
+
+
+### [The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+This preprint describes the development of the Unified Phenotype Ontology (uPheno), a framework designed to integrate and standardize phenotype data across multiple species. uPheno addresses the challenge of integrating phenotype data from different sources by providing a consistent and logical framework for describing phenotypes. It utilizes a library of design patterns to define phenotype terms, enabling the development of a hierarchical vocabulary that groups species-specific phenotype terms under species-neutral categories. uPheno also includes mappings between species-specific ontologies, facilitating cross-species comparisons and enabling researchers to identify genes with similar phenotypic effects across different organisms. The paper highlights the potential applications of uPheno in various areas of biological research, including clinical diagnostics, variant prioritization, and disease modeling.
+
+
+
+
+(Figure from Harshad Hegde, Jennifer Vendetti, Damien Goutte-Gattat, J Harry Caufield, John B Graybeal, Nomi L Harris, Naouel Karam, Christian Kindermann, Nicolas Matentzoglu, James A Overton, Mark A Musen, Christopher J Mungall. A Change Language for Ontologies and Knowledge Graphs. 2024 Sep 20; [https://arxiv.org/abs/2409.13906](https://arxiv.org/abs/2409.13906))
+
+
+### [The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+This preprint describes the development of the Unified Phenotype Ontology (uPheno), a framework designed to integrate and standardize phenotype data across multiple species. uPheno addresses the challenge of integrating phenotype data from different sources by providing a consistent and logical framework for describing phenotypes. It utilizes a library of design patterns to define phenotype terms, enabling the development of a hierarchical vocabulary that groups species-specific phenotype terms under species-neutral categories. uPheno also includes mappings between species-specific ontologies, facilitating cross-species comparisons and enabling researchers to identify genes with similar phenotypic effects across different organisms. The paper highlights the potential applications of uPheno in various areas of biological research, including clinical diagnostics, variant prioritization, and disease modeling.
+
+ +
+
+(Figure from Nicolas Matentzoglu, Susan M Bello, Ray Stefancsik, Sarah M. Alghamdi, Anna V. Anagnostopoulos, James P. Balhoff, Meghan A. Balk, Yvonne M. Bradford, Yasemin Bridges, Tiffany J. Callahan, Harry Caufield, Alayne Cuzick, Leigh C Carmody, Anita R. Caron, Vinicius de Souza, Stacia R. Engel, Petra Fey, Malcolm Fisher, Sarah Gehrke, Christian Grove, Peter Hansen, Nomi L. Harris, Midori A. Harris, Laura Harris, Arwa Ibrahim, Julius O.B. Jacobsen, Sebastian Köhler, Julie A. McMurry, Violeta Munoz-Fuentes, Monica C. Munoz-Torres, Helen Parkinson, Zoë M Pendlington, Clare Pilgrim, Sofia MC Robb, Peter N. Robinson, James Seager, Erik Segerdell, Damian Smedley, Elliot Sollis, Sabrina Toro, Nicole Vasilevsky, Valerie Wood, Melissa A. Haendel, Christopher J. Mungall, James A. McLaughlin, and David Osumi-Sutherland. The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics. 2024 Sep 20. [https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+
+---
+
+
+## Spotlight on OBO Principles
+
+In this issue, we continue with [Principle 5 ](https://obofoundry.org/principles/fp-005-delineated-content.html)of the OBO Foundry's series of principles.
+
+
+### OBO Foundry Principle 5: Scope
+
+OBO Principle 5 addresses the importance of a clearly defined scope for ontologies. It emphasizes that each ontology should have a well-specified domain or subject matter it intends to cover, and its content should adhere strictly to that scope. This principle aims to avoid overlaps between ontologies, facilitate user searches, and enable quick selection of relevant ontologies while still allowing for the creation of new terms through cross-products. The scope of an ontology should ideally be narrow, with out-of-scope terms imported from appropriate existing ontologies when possible. If an immediate need arises for out-of-scope terms, they should be placed in a separate, shareable module. The ontology's domain should be clearly stated in a brief, jargon-free manner, and its content should remain within these defined boundaries. This principle also encourages consideration of generic and mid/upper level terms for potential inclusion in the Core Ontology for Biology and Biomedicine (COB), ensuring that the ontology serves both its specific purpose and broader community needs.
+
+
+---
+
+
+## Spotlight on Tools: Recent updates
+
+
+### Simple Standard for Sharing Ontological Mappings (SSSOM) v. 1.0.
+
+While the goal of the OBO community is to develop a set of interoperable ontologies covering the biomedical domain, many vocabularies, terminologies and ontologies are used globally which need to be linked to OBO ontologies. The [SSSOM ](https://mapping-commons.github.io/sssom/)community standard was developed to provide an easy way to specify and exchange such mappings with rich provenance. After more than 4 years of work, with 79 contributors writing more than 1200 comments the first stable release is finally out:
+
+[https://github.com/mapping-commons/sssom/releases/tag/v1.0.0](https://github.com/mapping-commons/sssom/releases/tag/v1.0.0). A short post about the release was [published on LinkedIn by the Monarch Initiative](https://www.linkedin.com/posts/the-monarch-initiative_sssom-monarchinitiative-semantic-activity-7228793130441916416-roIB/?utm_source=share&utm_medium=member_desktop).
+
+
+---
+
+
+## Spotlight on Tools: Ontology Browsers
+
+Here and in upcoming issues of this newsletter, we would like to highlight some key ontology browsers: the Ontology Lookup Service (OLS), Ontobee, and BioPortal. We spotlight BioPortal in this issue; watch for spotlights on Ontobee and OLS in future issues.
+
+
+### BioPortal
+
+[BioPortal ](https://bioportal.bioontology.org/)is a web-based repository and collaborative platform for biomedical ontologies and terminologies developed by the National Center for Biomedical Ontology (NCBO). It is a centralized resource for accessing, browsing, and integrating standardized biomedical vocabularies and ontologies. The platform utilizes semantic web technologies to enhance interoperability and data integration across biomedical domains. BioPortal's collection contains numerous ontologies covering fields such as anatomy, phenotypes, experimental conditions, and molecular functions. BioPortal also offers ontology visualization tools, mapping services, and programmatic access via web services, facilitating knowledge management and semantic annotation of biomedical data.
+
+
+---
+
+
+## Events
+
+
+
+* **COB Workshop.** Purpose: To advance the development of COB.
+
+ The 2024 COB Workshop was held September 23–24 at The La Jolla Institute for Immunology in San Diego, CA.
+
+
+ The workshop included 10 in-person and 8 remote participants. Both days were quite productive with lively discussions and consensus building. Decisions were made and documented as described below and tabled discussions were tracked for future work, with the goal of focusing discussion on topics that were most likely to result in actionable decisions. The mechanism was to go through terms included in the present COB release, and review each term to decide whether it should be part of COB base (with COB IDs, label and definitions that would replace existing terms), COB full (imported terms from existing ontologies that can be used as is, which are needed in the axiomatization of COB), and COB root (imported terms from existing ontologies that are not needed to define COB, but that are included to illustrate how existing OBO ontologies will be able to connect). Much of the discussion on Day 1 focused on macromolecules and cells, working through many of the significant material entities for the remainder of the day. Day 2 started with a focus on human-made devices and material processing terms, with the goal of using COB to align existing terms from OBI and FOODON. Specimens and their collection from anatomical entities were also briefly discussed. Finally, a proposal on how to consistently define stages, phases, and cycles was discussed; different groups have modeled them as material entities, characteristics, biological processes, or time intervals/processes. The proposal was to consistently capture these as subclasses of processes in COB and to characteristics such as PATO:duration to link these to time intervals. Overall, more than half of the COB terms were discussed and a (preliminary) classification was made. Many participants requested future focused virtual meetings to continue COB development using the same strategies, to tackle the remaining terms, and get input from a broader community.
+
+
+ If you are interested in this work, please watch and contribute to the COB repository: [https://github.com/OBOFoundry/COB](https://github.com/OBOFoundry/COB), and stay tuned for a more detailed workshop summary, which will be available in Issue 7 of the newsletter!
+
+* [OBO Academy: Monarch Seminar Series](https://oboacademy.github.io/obook/courses/monarch-obo-training/) (ongoing)
+* Schedule:[ https://oboacademy.github.io/obook/courses/monarch-obo-training/](https://oboacademy.github.io/obook/courses/monarch-obo-training)
+* You can vote for future training topics here: [https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests](https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests)
+
+
+### Past Events
+
+**Fostering Open Science Collaborative Workflows: Insights from the ICBO OBO Tutorial**
+
+By Nicole Vasilevsky and Sabrina Toro
+
+The [OBO Foundry Training at ICBO 2024](https://oboacademy.github.io/obook/courses/icbo2024/) took a close look at the social workflows surrounding ontology development. Topics such as GitHub, Pull Requests, review and principles of good collaborative work were discussed in depth. A recording is available at the above link.
+
+Every year at the International Conference on Biological and Biomedical Ontology (ICBO), members of the Open Biological and Biomedical Ontology (OBO) Foundry host an OBO Tutorial to share insights into our latest work on ontology development, share best practices, and exchange knowledge. These OBO tutorials have become part of the OBO Academy ([https://oboacademy.github.io/obook/](https://oboacademy.github.io/obook/)), with special attention to training newcomers to semantic engineering. This year’s virtual tutorial was organized by Nico Matenzoglu (Semanticly) and Nicole Vasilevsky (Critical Path Institute) and focused on “Fostering Open Science Collaborative Workflows”. More details, including a recording of the session, are available here: [https://oboacademy.github.io/obook/courses/icbo2024/](https://oboacademy.github.io/obook/courses/icbo2024/)
+
+We discussed the importance of collaboration in the development of OBO ontologies. James Overton introduced the OBO Foundry, Sabrina Toro discussed Git and GitHub, Nico Matentzoglu discussed best practices in collaborative workflows and Nicole Vasilevsky shared best practices in creating GitHub tickets, submitting pull requests (PRs), and reviewing PRs in ontology repositories.
+
+Thanks to a generous microgrant from the [International Society for Biocuration](https://www.biocuration.org/) we were able to offer small prizes for participating in a training quiz integrated into the tutorial. We awarded prizes to the top 5 participants with the most correct answers and 5 prizes to participants who answered the most questions, regardless of their answers. The winners were (in alphabetical order): Hannah Blau, Spencer Bliven, Yvonne Bradford, Lauren Chan, John Graybeal, Harshad Hegde, Bryan Laraway, Katie Mullen, Sumir Pandit, and Umasri Sankarlal.
+
+Following the session, we hosted the [OBO Community Challenge](https://github.com/OBOAcademy/obook/issues/508), which was open to all. The challenge provided an opportunity to apply the lessons learned from the OBO tutorial at ICBO. Participants were asked to choose one or several OBO ontology projects in which they were not involved yet, and complete a set of tasks (including opening an issue and creating a pull request) demonstrating the basic principles of collaborative workflows introduced at the tutorial. Ontology contributions were reviewed for the quality of the commit messages, and issue and pull request descriptions. We are very grateful to everyone who participated. The prizer was awarded to Paula Duek Roggli and Katie Mullen.
+
+This was our inaugural opportunity to award prizes to participants in the OBO tutorial and OBO challenge. We believe these prizes enhanced engagement and boosted participation within the community. We extend our heartfelt gratitude to the ISB and the OBO Foundry community for their support, and to the ISB for backing the microgrant initiative. We also encourage everyone to get involved with and support the ISB—you might be the next recipient of a similar microgrant.
+
+
+---
+
+
+## Ways to be part of the OBO Foundry community
+
+There are many ways to be part of the OBO Foundry community, beyond [using our website to find ontologies of interest](https://obofoundry.org). For example:
+
+
+
+* Join the [obo-discuss mailing list](https://groups.google.com/forum/#!forum/obo-discuss)
+* If you are interested in the technical aspects, you can also join the [obo-tools mailing list](https://groups.google.com/forum/#!members/obo-tools)
+* Join the conversation in our [Slack workspace](https://join.slack.com/t/obo-communitygroup/shared_invite/zt-1oq48ttk7-kKo0i6TwntYtAq~Jcjjg4g).
+* Use our public [issue tracker](https://github.com/OBOFoundry/OBOFoundry.github.io/issues) to report a problem you discovered on obofoundry.org or request a new feature
+ * Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
+* Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of [our code is publicly readable](https://github.com/OBOFoundry), you may be able to pinpoint where a bug fix needs to go.
+* [Request that your ontology be considered for inclusion in the Foundry](https://obofoundry.org/faq/how-do-i-register-my-ontology.html)
+* If you want to take your contributions to OBO Foundry to the next level, you can [nominate yourself to be considered for the OBO Operations Committee](https://obofoundry.org/docs/NewOBOFC.html). There are many ways in which you can contribute, including assisting with the production of this newsletter.
diff --git a/images/newsletter/hogan.png b/images/newsletter/hogan.png
new file mode 100644
index 000000000..01e34157d
Binary files /dev/null and b/images/newsletter/hogan.png differ
diff --git a/images/newsletter/kgcl.png b/images/newsletter/kgcl.png
new file mode 100644
index 000000000..3b1839726
Binary files /dev/null and b/images/newsletter/kgcl.png differ
diff --git a/images/newsletter/sgehrke.png b/images/newsletter/sgehrke.png
new file mode 100644
index 000000000..c992d7be7
Binary files /dev/null and b/images/newsletter/sgehrke.png differ
diff --git a/images/newsletter/upheno.png b/images/newsletter/upheno.png
new file mode 100644
index 000000000..d8fe58e75
Binary files /dev/null and b/images/newsletter/upheno.png differ
+
+
+(Figure from Nicolas Matentzoglu, Susan M Bello, Ray Stefancsik, Sarah M. Alghamdi, Anna V. Anagnostopoulos, James P. Balhoff, Meghan A. Balk, Yvonne M. Bradford, Yasemin Bridges, Tiffany J. Callahan, Harry Caufield, Alayne Cuzick, Leigh C Carmody, Anita R. Caron, Vinicius de Souza, Stacia R. Engel, Petra Fey, Malcolm Fisher, Sarah Gehrke, Christian Grove, Peter Hansen, Nomi L. Harris, Midori A. Harris, Laura Harris, Arwa Ibrahim, Julius O.B. Jacobsen, Sebastian Köhler, Julie A. McMurry, Violeta Munoz-Fuentes, Monica C. Munoz-Torres, Helen Parkinson, Zoë M Pendlington, Clare Pilgrim, Sofia MC Robb, Peter N. Robinson, James Seager, Erik Segerdell, Damian Smedley, Elliot Sollis, Sabrina Toro, Nicole Vasilevsky, Valerie Wood, Melissa A. Haendel, Christopher J. Mungall, James A. McLaughlin, and David Osumi-Sutherland. The Unified Phenotype Ontology (uPheno): A framework for cross-species integrative phenomics. 2024 Sep 20. [https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1](https://www.biorxiv.org/content/10.1101/2024.09.18.613276v1)
+
+
+---
+
+
+## Spotlight on OBO Principles
+
+In this issue, we continue with [Principle 5 ](https://obofoundry.org/principles/fp-005-delineated-content.html)of the OBO Foundry's series of principles.
+
+
+### OBO Foundry Principle 5: Scope
+
+OBO Principle 5 addresses the importance of a clearly defined scope for ontologies. It emphasizes that each ontology should have a well-specified domain or subject matter it intends to cover, and its content should adhere strictly to that scope. This principle aims to avoid overlaps between ontologies, facilitate user searches, and enable quick selection of relevant ontologies while still allowing for the creation of new terms through cross-products. The scope of an ontology should ideally be narrow, with out-of-scope terms imported from appropriate existing ontologies when possible. If an immediate need arises for out-of-scope terms, they should be placed in a separate, shareable module. The ontology's domain should be clearly stated in a brief, jargon-free manner, and its content should remain within these defined boundaries. This principle also encourages consideration of generic and mid/upper level terms for potential inclusion in the Core Ontology for Biology and Biomedicine (COB), ensuring that the ontology serves both its specific purpose and broader community needs.
+
+
+---
+
+
+## Spotlight on Tools: Recent updates
+
+
+### Simple Standard for Sharing Ontological Mappings (SSSOM) v. 1.0.
+
+While the goal of the OBO community is to develop a set of interoperable ontologies covering the biomedical domain, many vocabularies, terminologies and ontologies are used globally which need to be linked to OBO ontologies. The [SSSOM ](https://mapping-commons.github.io/sssom/)community standard was developed to provide an easy way to specify and exchange such mappings with rich provenance. After more than 4 years of work, with 79 contributors writing more than 1200 comments the first stable release is finally out:
+
+[https://github.com/mapping-commons/sssom/releases/tag/v1.0.0](https://github.com/mapping-commons/sssom/releases/tag/v1.0.0). A short post about the release was [published on LinkedIn by the Monarch Initiative](https://www.linkedin.com/posts/the-monarch-initiative_sssom-monarchinitiative-semantic-activity-7228793130441916416-roIB/?utm_source=share&utm_medium=member_desktop).
+
+
+---
+
+
+## Spotlight on Tools: Ontology Browsers
+
+Here and in upcoming issues of this newsletter, we would like to highlight some key ontology browsers: the Ontology Lookup Service (OLS), Ontobee, and BioPortal. We spotlight BioPortal in this issue; watch for spotlights on Ontobee and OLS in future issues.
+
+
+### BioPortal
+
+[BioPortal ](https://bioportal.bioontology.org/)is a web-based repository and collaborative platform for biomedical ontologies and terminologies developed by the National Center for Biomedical Ontology (NCBO). It is a centralized resource for accessing, browsing, and integrating standardized biomedical vocabularies and ontologies. The platform utilizes semantic web technologies to enhance interoperability and data integration across biomedical domains. BioPortal's collection contains numerous ontologies covering fields such as anatomy, phenotypes, experimental conditions, and molecular functions. BioPortal also offers ontology visualization tools, mapping services, and programmatic access via web services, facilitating knowledge management and semantic annotation of biomedical data.
+
+
+---
+
+
+## Events
+
+
+
+* **COB Workshop.** Purpose: To advance the development of COB.
+
+ The 2024 COB Workshop was held September 23–24 at The La Jolla Institute for Immunology in San Diego, CA.
+
+
+ The workshop included 10 in-person and 8 remote participants. Both days were quite productive with lively discussions and consensus building. Decisions were made and documented as described below and tabled discussions were tracked for future work, with the goal of focusing discussion on topics that were most likely to result in actionable decisions. The mechanism was to go through terms included in the present COB release, and review each term to decide whether it should be part of COB base (with COB IDs, label and definitions that would replace existing terms), COB full (imported terms from existing ontologies that can be used as is, which are needed in the axiomatization of COB), and COB root (imported terms from existing ontologies that are not needed to define COB, but that are included to illustrate how existing OBO ontologies will be able to connect). Much of the discussion on Day 1 focused on macromolecules and cells, working through many of the significant material entities for the remainder of the day. Day 2 started with a focus on human-made devices and material processing terms, with the goal of using COB to align existing terms from OBI and FOODON. Specimens and their collection from anatomical entities were also briefly discussed. Finally, a proposal on how to consistently define stages, phases, and cycles was discussed; different groups have modeled them as material entities, characteristics, biological processes, or time intervals/processes. The proposal was to consistently capture these as subclasses of processes in COB and to characteristics such as PATO:duration to link these to time intervals. Overall, more than half of the COB terms were discussed and a (preliminary) classification was made. Many participants requested future focused virtual meetings to continue COB development using the same strategies, to tackle the remaining terms, and get input from a broader community.
+
+
+ If you are interested in this work, please watch and contribute to the COB repository: [https://github.com/OBOFoundry/COB](https://github.com/OBOFoundry/COB), and stay tuned for a more detailed workshop summary, which will be available in Issue 7 of the newsletter!
+
+* [OBO Academy: Monarch Seminar Series](https://oboacademy.github.io/obook/courses/monarch-obo-training/) (ongoing)
+* Schedule:[ https://oboacademy.github.io/obook/courses/monarch-obo-training/](https://oboacademy.github.io/obook/courses/monarch-obo-training)
+* You can vote for future training topics here: [https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests](https://github.com/OBOAcademy/obook/discussions/categories/tutorial-requests)
+
+
+### Past Events
+
+**Fostering Open Science Collaborative Workflows: Insights from the ICBO OBO Tutorial**
+
+By Nicole Vasilevsky and Sabrina Toro
+
+The [OBO Foundry Training at ICBO 2024](https://oboacademy.github.io/obook/courses/icbo2024/) took a close look at the social workflows surrounding ontology development. Topics such as GitHub, Pull Requests, review and principles of good collaborative work were discussed in depth. A recording is available at the above link.
+
+Every year at the International Conference on Biological and Biomedical Ontology (ICBO), members of the Open Biological and Biomedical Ontology (OBO) Foundry host an OBO Tutorial to share insights into our latest work on ontology development, share best practices, and exchange knowledge. These OBO tutorials have become part of the OBO Academy ([https://oboacademy.github.io/obook/](https://oboacademy.github.io/obook/)), with special attention to training newcomers to semantic engineering. This year’s virtual tutorial was organized by Nico Matenzoglu (Semanticly) and Nicole Vasilevsky (Critical Path Institute) and focused on “Fostering Open Science Collaborative Workflows”. More details, including a recording of the session, are available here: [https://oboacademy.github.io/obook/courses/icbo2024/](https://oboacademy.github.io/obook/courses/icbo2024/)
+
+We discussed the importance of collaboration in the development of OBO ontologies. James Overton introduced the OBO Foundry, Sabrina Toro discussed Git and GitHub, Nico Matentzoglu discussed best practices in collaborative workflows and Nicole Vasilevsky shared best practices in creating GitHub tickets, submitting pull requests (PRs), and reviewing PRs in ontology repositories.
+
+Thanks to a generous microgrant from the [International Society for Biocuration](https://www.biocuration.org/) we were able to offer small prizes for participating in a training quiz integrated into the tutorial. We awarded prizes to the top 5 participants with the most correct answers and 5 prizes to participants who answered the most questions, regardless of their answers. The winners were (in alphabetical order): Hannah Blau, Spencer Bliven, Yvonne Bradford, Lauren Chan, John Graybeal, Harshad Hegde, Bryan Laraway, Katie Mullen, Sumir Pandit, and Umasri Sankarlal.
+
+Following the session, we hosted the [OBO Community Challenge](https://github.com/OBOAcademy/obook/issues/508), which was open to all. The challenge provided an opportunity to apply the lessons learned from the OBO tutorial at ICBO. Participants were asked to choose one or several OBO ontology projects in which they were not involved yet, and complete a set of tasks (including opening an issue and creating a pull request) demonstrating the basic principles of collaborative workflows introduced at the tutorial. Ontology contributions were reviewed for the quality of the commit messages, and issue and pull request descriptions. We are very grateful to everyone who participated. The prizer was awarded to Paula Duek Roggli and Katie Mullen.
+
+This was our inaugural opportunity to award prizes to participants in the OBO tutorial and OBO challenge. We believe these prizes enhanced engagement and boosted participation within the community. We extend our heartfelt gratitude to the ISB and the OBO Foundry community for their support, and to the ISB for backing the microgrant initiative. We also encourage everyone to get involved with and support the ISB—you might be the next recipient of a similar microgrant.
+
+
+---
+
+
+## Ways to be part of the OBO Foundry community
+
+There are many ways to be part of the OBO Foundry community, beyond [using our website to find ontologies of interest](https://obofoundry.org). For example:
+
+
+
+* Join the [obo-discuss mailing list](https://groups.google.com/forum/#!forum/obo-discuss)
+* If you are interested in the technical aspects, you can also join the [obo-tools mailing list](https://groups.google.com/forum/#!members/obo-tools)
+* Join the conversation in our [Slack workspace](https://join.slack.com/t/obo-communitygroup/shared_invite/zt-1oq48ttk7-kKo0i6TwntYtAq~Jcjjg4g).
+* Use our public [issue tracker](https://github.com/OBOFoundry/OBOFoundry.github.io/issues) to report a problem you discovered on obofoundry.org or request a new feature
+ * Note that most issues relating to individual ontologies (e.g., a request to add a new term) should be added to the issue tracker for the specific ontology
+* Propose a fix to an issue you see on our issue tracker (this is done via a GitHub Pull Request, which will be checked and approved by someone in the Foundry). Since all of [our code is publicly readable](https://github.com/OBOFoundry), you may be able to pinpoint where a bug fix needs to go.
+* [Request that your ontology be considered for inclusion in the Foundry](https://obofoundry.org/faq/how-do-i-register-my-ontology.html)
+* If you want to take your contributions to OBO Foundry to the next level, you can [nominate yourself to be considered for the OBO Operations Committee](https://obofoundry.org/docs/NewOBOFC.html). There are many ways in which you can contribute, including assisting with the production of this newsletter.
diff --git a/images/newsletter/hogan.png b/images/newsletter/hogan.png
new file mode 100644
index 000000000..01e34157d
Binary files /dev/null and b/images/newsletter/hogan.png differ
diff --git a/images/newsletter/kgcl.png b/images/newsletter/kgcl.png
new file mode 100644
index 000000000..3b1839726
Binary files /dev/null and b/images/newsletter/kgcl.png differ
diff --git a/images/newsletter/sgehrke.png b/images/newsletter/sgehrke.png
new file mode 100644
index 000000000..c992d7be7
Binary files /dev/null and b/images/newsletter/sgehrke.png differ
diff --git a/images/newsletter/upheno.png b/images/newsletter/upheno.png
new file mode 100644
index 000000000..d8fe58e75
Binary files /dev/null and b/images/newsletter/upheno.png differ