diff --git a/docs/zh/examples/pirbn.md b/docs/zh/examples/pirbn.md
new file mode 100644
index 000000000..5ca74379d
--- /dev/null
+++ b/docs/zh/examples/pirbn.md
@@ -0,0 +1,148 @@
+# PIRBN
+
+## 1. 背景简介
+
+我们最近发现经过训练,物理信息神经网络(PINN)往往会成为局部近似函数。这一观察结果促使我们开发了一种新型的物理-信息径向基网络(PIRBN),该网络在整个训练过程中都能够维持局部近似性质。与深度神经网络不同,PIRBN 仅包含一个隐藏层和一个径向基“激活”函数。在适当的条件下,我们证明了使用梯度下降方法训练 PIRBN 可以收敛到高斯过程。此外,我们还通过神经邻近核(NTK)理论研究了 PIRBN 的训练动态。此外,我们还对 PIRBN 的初始化策略进行了全面调查。基于数值示例,我们发现 PIRBN 在解决具有高频特征和病态计算域的非线性偏微分方程方面比PINN更有效。此外,现有的 PINN 数值技术,如自适应学习、分解和不同类型的损失函数,也适用于 PIRBN。
+
+
+ 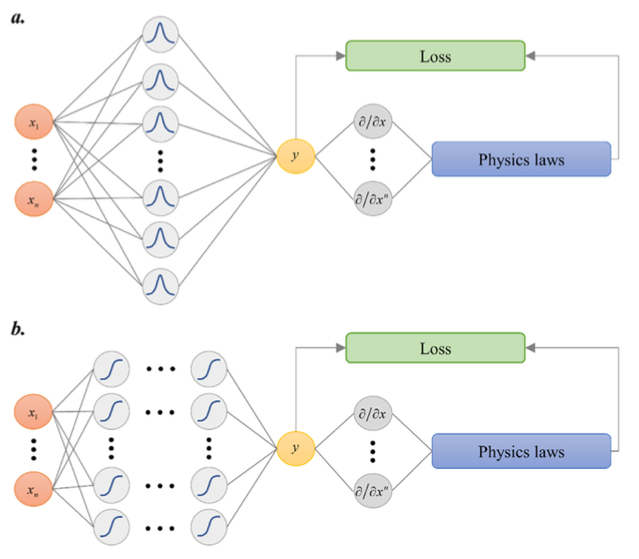{ loading=lazy }
+ 网络的结构
+
+图片左侧为常见神经网络结构的输入层,隐藏层,输出层,隐藏层包含激活层,a 中为单层隐藏层,b 中为多层隐藏层,图片右侧为 PIRBN 网络的激活函数,计算网络的损失 Loss 并反向传递。图片说明当使用 PIRBN 时,每个 RBF 神经元仅在输入接近神经元中心时被激活。直观地说,PIRBN 具有局部逼近特性。通过梯度下降算法训练一个 PIRBN 也可以通过 NTK 理论进行分析。
+
+
+ 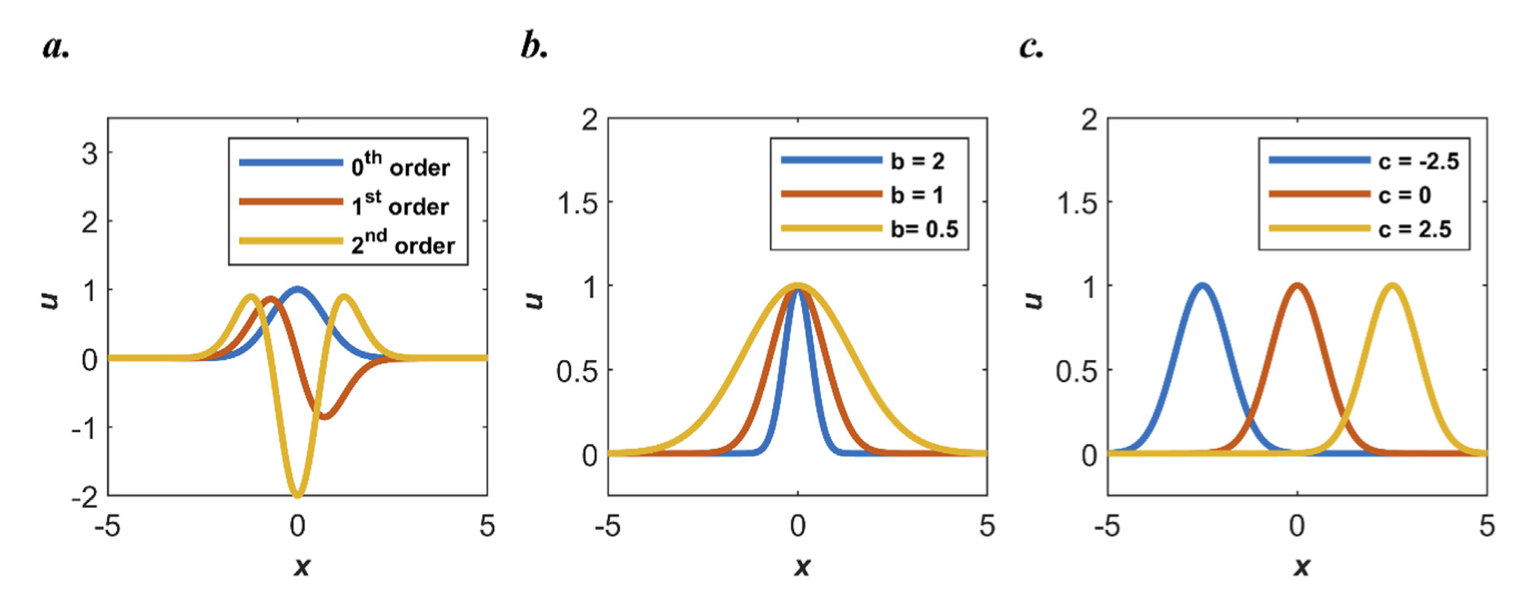{ loading=lazy }
+ 不同阶数的高斯激活函数
+
+(a) 0, 1, 2 阶高斯激活函数
+(b) 设置不同 b 值
+(c) 设置不同 c 值
+
+当使用高斯函数作为激活函数时,输入与输出之间的映射关系可以数学上表示为高斯函数的某种形式。RBF 网络是一种常用于模式识别、数据插值和函数逼近的神经网络,其关键特征是使用径向基函数作为激活函数,使得网络具有更好的全局逼近能力和灵活性。
+
+## 2. 问题定义
+
+在 NTK 和基于 NTK 的适应性训练方法的帮助下,PINN 在处理具有高频特征的问题时的性能可以得到显著提升。例如,考虑一个偏微分方程及其边界条件:
+
+$$
+\begin{aligned}
+& \frac{\mathrm{d}^2}{\mathrm{~d} x^2} u(x)-4 \mu^2 \pi^2 \sin (2 \mu \pi x)=0, \text { for } x \in[0,1] \\
+& u(0)=u(1)=0
+\end{aligned}
+$$
+
+其中μ是一个控制PDE解的频率特征的常数。
+
+## 3. 问题求解
+
+接下来开始讲解如何将问题一步一步地转化为 PaddlePaddle 代码,用深度学习的方法求解该问题。
+为了快速理解 PaddlePaddle,接下来仅对模型构建、方程构建、计算域构建等关键步骤进行阐述,而其余细节请参考 [API文档](../api/arch.md)。
+
+### 3.1 模型构建
+
+在 PIRBN 问题中,建立网络,用 PaddlePaddle 代码表示如下
+
+``` py linenums="40"
+--8<--
+jointContribution/PIRBN/main.py:40:42
+--8<--
+```
+
+### 3.2 数据构建
+
+本案例涉及读取数据构建,如下所示
+
+``` py linenums="18"
+--8<--
+jointContribution/PIRBN/main.py:18:38
+--8<--
+```
+
+### 3.3 训练和评估构建
+
+训练和评估构建,设置损失计算函数,返回字段,代码如下所示:
+
+``` py linenums="52"
+--8<--
+jointContribution/PIRBN/train.py:52:90
+--8<--
+```
+
+### 3.4 超参数设定
+
+接下来我们需要指定训练轮数,此处我们按实验经验,使用 20001 轮训练轮数。
+
+``` py linenums="43"
+--8<--
+jointContribution/PIRBN/main.py:43:43
+--8<--
+```
+
+### 3.5 优化器构建
+
+训练过程会调用优化器来更新模型参数,此处选择 `Adam` 优化器并设定 `learning_rate` 为 1e-3。
+
+``` py linenums="33"
+--8<--
+jointContribution/PIRBN/train.py:33:35
+--8<--
+```
+
+### 3.6 模型训练与评估
+
+模型训练与评估
+
+``` py linenums="92"
+--8<--
+jointContribution/PIRBN/train.py:92:99
+--8<--
+```
+
+## 4. 完整代码
+
+``` py linenums="1" title="main.py"
+--8<--
+jointContribution/PIRBN/main.py
+--8<--
+```
+
+## 5. 结果展示
+
+PINN 案例针对 epoch=20001 和 learning\_rate=1e-3 的参数配置进行了实验,结果返回Loss为 0.13567。
+
+PIRBN 案例针对 epoch=20001 和 learning\_rate=1e-3 的参数配置进行了实验,结果返回Loss为 0.59471。
+
+
+ 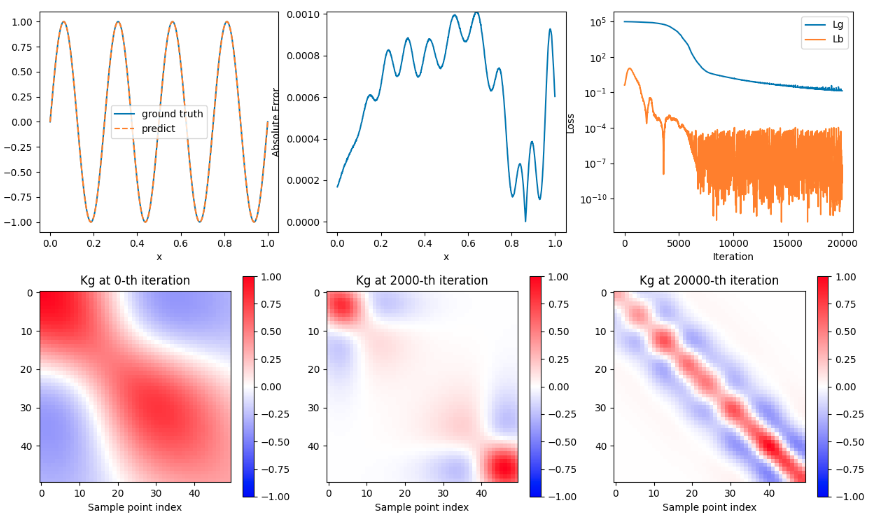{ loading=lazy }
+ PINN 结果图
+
+图为使用双曲正切函数(tanh)作为激活函数(activation function),并且使用 LuCun 初始化方法来初始化神经网络中的所有参数。
+
+- 图中子图 1 为预测值和真实值的曲线比较
+- 图中子图 2 为误差值
+- 图中子图 3 为损失值
+- 图中子图 4 为训练 1 次的 Kg 图
+- 图中子图 5 为训练 2000 次的 Kg 图
+- 图中子图 6 为训练 20000 次的 Kg 图
+
+可以看到预测值和真实值可以匹配,误差值逐渐升高然后逐渐减少,Loss 历史降低后波动,Kg 图随训练次数增加而逐渐收敛。
+
+
+ 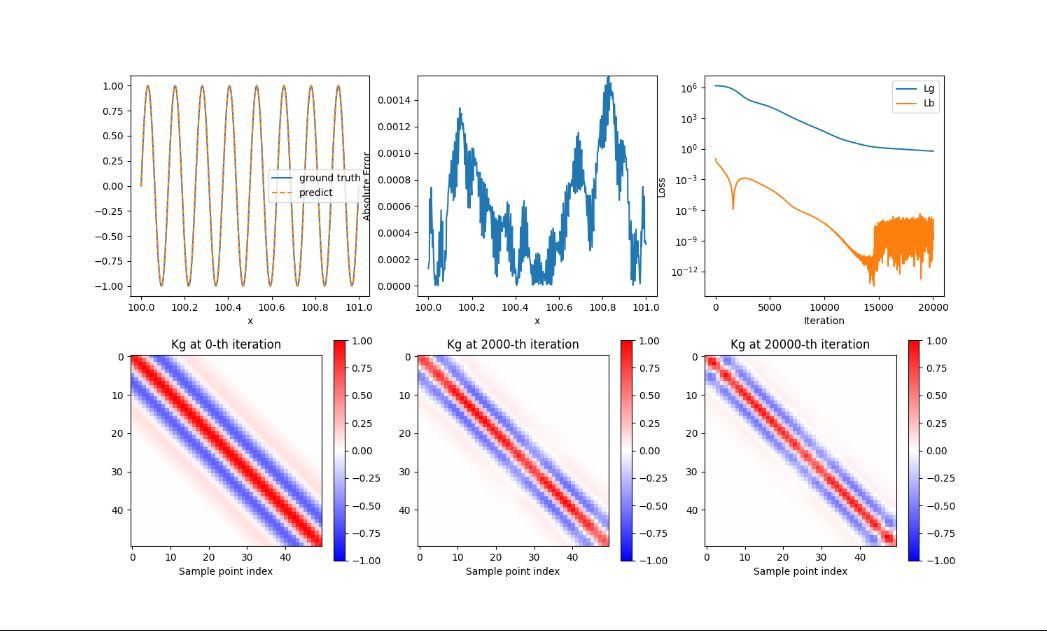{ loading=lazy }
+ PIRBN 结果图
+
+图为使用高斯函数(gaussian function)作为激活函数(activation function)生成的数据,并且使用 LuCun 初始化方法来初始化神经网络中的所有参数。
+
+- 图中子图 1 为预测值和真实值的曲线比较
+- 图中子图 2 为误差值
+- 图中子图 3 为损失值
+- 图中子图 4 为训练 1 次的 Kg 图
+- 图中子图 5 为训练 2000 次的 Kg 图
+- 图中子图 6 为训练 20000 次的 Kg 图
+
+可以看到预测值和真实值可以匹配,误差值逐渐升高然后逐渐减少再升高,Loss 历史降低后波动,Kg 图随训练次数增加而逐渐收敛。
+
+## 6. 参考资料
+
+- [Physics-informed radial basis network (PIRBN): A local approximating neural network for solving nonlinear PDEs](https://arxiv.org/abs/2304.06234)
+-
diff --git a/jointContribution/PIRBN/README.md b/jointContribution/PIRBN/README.md
new file mode 100644
index 000000000..52e017e01
--- /dev/null
+++ b/jointContribution/PIRBN/README.md
@@ -0,0 +1,45 @@
+# Physics-informed radial basis network (PIRBN)
+
+This repository provides numerical examples of the **physics-informed radial basis network** (**PIRBN**).
+
+Physics-informed neural network (PINN) has recently gained increasing interest in computational mechanics.
+
+This work starts from studying the training dynamics of PINNs via the nerual tangent kernel (NTK) theory. Based on numerical experiments, we found:
+
+- PINNs tend to be a **local approximator** during the training
+- For PINNs who fail to be a local apprixmator, the physics-informed loss can be hardly minimised through training
+
+Inspired by findings, we proposed the PIRBN, which can exhibit the local property intuitively. It has been demonstrated that the NTK theory is applicable for PIRBN. Besides, other PINN techniques can be directly migrated to PIRBNs.
+
+Numerical examples include:
+
+ - 1D sine funtion (**Eq. 13** in the manuscript)
+
+ **PDE**: $\frac{\partial^2 }{\partial x^2}u(x)-4\mu^2\pi^2 sin(2\mu\pi(x))=0, x\in[0,1]$
+
+ **BC**: $u(0)=u(1)=0.$
+
+ - 1D sine funtion (**Eq. 15** in the manuscript)
+ **PDE**: $\frac{\partial^2 }{\partial x^2}u(x-100)-4\mu^2\pi^2 sin(2\mu\pi(x-100))=0, x\in[100,101]$
+
+ **BC**: $u(100)=u(101)=0.$
+
+For more details in terms of mathematical proofs and numerical examples, please refer to our paper.
+
+## Link
+
+
+
+
+
+## Enviornmental settings
+
+``` shell
+pip install -r requirements.txt
+```
+
+## Train
+
+``` python
+python main.py
+```
diff --git a/jointContribution/PIRBN/analytical_solution.py b/jointContribution/PIRBN/analytical_solution.py
new file mode 100644
index 000000000..76304b5ef
--- /dev/null
+++ b/jointContribution/PIRBN/analytical_solution.py
@@ -0,0 +1,84 @@
+import os
+
+import matplotlib.pyplot as plt
+import numpy as np
+import paddle
+
+
+def output_fig(train_obj, mu, b, right_by, activation_function, output_Kgg):
+ plt.figure(figsize=(15, 9))
+ rbn = train_obj.pirbn.rbn
+
+ output_dir = os.path.join(os.path.dirname(__file__), "output")
+ if not os.path.exists(output_dir):
+ os.mkdir(output_dir)
+
+ # Comparisons between the network predictions and the ground truth.
+ plt.subplot(2, 3, 1)
+ ns = 1001
+ dx = 1 / (ns - 1)
+ xy = np.zeros((ns, 1)).astype(np.float32)
+ for i in range(0, ns):
+ xy[i, 0] = i * dx + right_by
+ y = rbn(paddle.to_tensor(xy))
+ y = y.numpy()
+ y_true = np.sin(2 * mu * np.pi * xy)
+ plt.plot(xy, y_true)
+ plt.plot(xy, y, linestyle="--")
+ plt.legend(["ground truth", "predict"])
+ plt.xlabel("x")
+
+ # Point-wise absolute error plot.
+ plt.subplot(2, 3, 2)
+ xy_y = np.abs(y_true - y)
+ plt.plot(xy, xy_y)
+ plt.ylim(top=np.max(xy_y))
+ plt.ylabel("Absolute Error")
+ plt.xlabel("x")
+
+ # Loss history of the network during the training process.
+ plt.subplot(2, 3, 3)
+ loss_g = train_obj.loss_g
+ x = range(len(loss_g))
+ plt.yscale("log")
+ plt.plot(x, loss_g)
+ plt.plot(x, train_obj.loss_b)
+ plt.legend(["Lg", "Lb"])
+ plt.ylabel("Loss")
+ plt.xlabel("Iteration")
+
+ # Visualise NTK after initialisation, The normalised Kg at 0th iteration.
+ plt.subplot(2, 3, 4)
+ index = str(output_Kgg[0])
+ K = train_obj.ntk_list[index].numpy()
+ plt.imshow(K / (np.max(abs(K))), cmap="bwr", vmax=1, vmin=-1)
+ plt.colorbar()
+ plt.title(f"Kg at {index}-th iteration")
+ plt.xlabel("Sample point index")
+
+ # Visualise NTK after training, The normalised Kg at 2000th iteration.
+ plt.subplot(2, 3, 5)
+ index = str(output_Kgg[1])
+ K = train_obj.ntk_list[index].numpy()
+ plt.imshow(K / (np.max(abs(K))), cmap="bwr", vmax=1, vmin=-1)
+ plt.colorbar()
+ plt.title(f"Kg at {index}-th iteration")
+ plt.xlabel("Sample point index")
+
+ # The normalised Kg at 20000th iteration.
+ plt.subplot(2, 3, 6)
+ index = str(output_Kgg[2])
+ K = train_obj.ntk_list[index].numpy()
+ plt.imshow(K / (np.max(abs(K))), cmap="bwr", vmax=1, vmin=-1)
+ plt.colorbar()
+ plt.title(f"Kg at {index}-th iteration")
+ plt.xlabel("Sample point index")
+
+ plt.savefig(
+ os.path.join(
+ output_dir, f"sine_function_{mu}_{b}_{right_by}_{activation_function}.png"
+ )
+ )
+
+ # Save data
+ # scipy.io.savemat(os.path.join(output_dir, "out.mat"), {"NTK": a, "x": xy, "y": y})
diff --git a/jointContribution/PIRBN/jacobian_function.py b/jointContribution/PIRBN/jacobian_function.py
new file mode 100644
index 000000000..526c27f53
--- /dev/null
+++ b/jointContribution/PIRBN/jacobian_function.py
@@ -0,0 +1,36 @@
+import paddle
+
+
+def flat(x, start_axis=0, stop_axis=None):
+ # TODO Error if use paddle.flatten -> The Op flatten_grad doesn't have any gradop
+ stop_axis = None if stop_axis is None else stop_axis + 1
+ shape = x.shape
+
+ # [3, 1] --flat--> [3]
+ # [2, 2] --flat--> [4]
+ temp = shape[start_axis:stop_axis]
+ temp = [0 if x == 1 else x for x in temp] # kill invalid axis
+ flat_sum = sum(temp)
+ head = shape[0:start_axis]
+ body = [flat_sum]
+ tail = [] if stop_axis is None else shape[stop_axis:]
+ new_shape = head + body + tail
+ x_flat = x.reshape(new_shape)
+ return x_flat
+
+
+def jacobian(y, x):
+ J_shape = y.shape + x.shape
+ J = paddle.zeros(J_shape)
+ y_flat = flat(y)
+ J_flat = flat(
+ J, start_axis=0, stop_axis=len(y.shape) - 1
+ ) # partialy flatten as y_flat
+ for i, y_i in enumerate(y_flat):
+ grad = paddle.grad(y_i, x, allow_unused=True)[
+ 0
+ ] # grad[i] == sum by j (dy[j] / dx[i])
+ if grad is None:
+ grad = paddle.zeros_like(x)
+ J_flat[i] = grad
+ return J_flat.reshape(J_shape)
diff --git a/jointContribution/PIRBN/main.py b/jointContribution/PIRBN/main.py
new file mode 100644
index 000000000..149dd829e
--- /dev/null
+++ b/jointContribution/PIRBN/main.py
@@ -0,0 +1,68 @@
+import analytical_solution
+import numpy as np
+import pirbn
+import rbn_net
+import train
+
+import ppsci
+
+# set random seed for reproducibility
+SEED = 2023
+ppsci.utils.misc.set_random_seed(SEED)
+
+# mu, Fig.1, Page5
+# right_by, Formula (15) Page5
+def sine_function_main(
+ mu, adaptive_weights=True, right_by=0, activation_function="gaussian"
+):
+ # Define the number of sample points
+ ns = 50
+
+ # Define the sample points' interval
+ dx = 1.0 / (ns - 1)
+
+ # Initialise sample points' coordinates
+ x_eq = np.linspace(0.0, 1.0, ns)[:, None]
+
+ for i in range(0, ns):
+ x_eq[i, 0] = i * dx + right_by
+ x_bc = np.array([[right_by + 0.0], [right_by + 1.0]])
+ x = [x_eq, x_bc]
+ y = -4 * mu**2 * np.pi**2 * np.sin(2 * mu * np.pi * x_eq)
+
+ # Set up radial basis network
+ n_in = 1
+ n_out = 1
+ n_neu = 61
+ b = 10.0
+ c = [right_by - 0.1, right_by + 1.1]
+
+ # Set up PIRBN
+ rbn = rbn_net.RBN_Net(n_in, n_out, n_neu, b, c, activation_function)
+ rbn_loss = pirbn.PIRBN(rbn, activation_function)
+ maxiter = 20001
+ output_Kgg = [0, int(0.1 * maxiter), maxiter - 1]
+ train_obj = train.Trainer(
+ rbn_loss,
+ x,
+ y,
+ learning_rate=0.001,
+ maxiter=maxiter,
+ adaptive_weights=adaptive_weights,
+ )
+ train_obj.fit(output_Kgg)
+
+ # Visualise results
+ analytical_solution.output_fig(
+ train_obj, mu, b, right_by, activation_function, output_Kgg
+ )
+
+
+# Fig.1
+sine_function_main(mu=4, right_by=0, activation_function="tanh")
+# Fig.2
+sine_function_main(mu=8, right_by=0, activation_function="tanh")
+# Fig.3
+sine_function_main(mu=4, right_by=100, activation_function="tanh")
+# Fig.6
+sine_function_main(mu=8, right_by=100, activation_function="gaussian")
diff --git a/jointContribution/PIRBN/pirbn.py b/jointContribution/PIRBN/pirbn.py
new file mode 100644
index 000000000..f6179e15e
--- /dev/null
+++ b/jointContribution/PIRBN/pirbn.py
@@ -0,0 +1,106 @@
+import paddle
+from jacobian_function import jacobian
+
+
+class PIRBN(paddle.nn.Layer):
+ def __init__(self, rbn, activation_function="gaussian"):
+ super().__init__()
+ self.rbn = rbn
+ self.activation_function = activation_function
+
+ def forward(self, input_data):
+ xy, xy_b = input_data
+ # initialize the differential operators
+ u_b = self.rbn(xy_b)
+ # obtain partial derivatives of u with respect to x
+ xy.stop_gradient = False
+ # Obtain the output from the RBN
+ u = self.rbn(xy)
+ # Obtain the first-order derivative of the output with respect to the input
+ u_x = paddle.grad(u, xy, retain_graph=True, create_graph=True)[0]
+ # Obtain the second-order derivative of the output with respect to the input
+ u_xx = paddle.grad(u_x, xy, retain_graph=True, create_graph=True)[0]
+ return u_xx, u_b, u
+
+ def cal_K(self, x):
+ u_xx, _, _ = self.forward(x)
+ w, b = [], []
+
+ if self.activation_function == "gaussian":
+ b.append(self.rbn.activation.b)
+ w.append(self.rbn.last_fc_layer.weight)
+ elif self.activation_function == "tanh":
+ w.append(self.rbn.hidden_layer.weight)
+ b.append(self.rbn.hidden_layer.bias)
+ w.append(self.rbn.last_fc_layer.weight)
+
+ J_list = []
+
+ for w_i in w:
+ J_w = jacobian(u_xx, w_i).squeeze()
+ J_list.append(J_w)
+
+ for b_i in b:
+ J_b = jacobian(u_xx, b_i).squeeze()
+ J_list.append(J_b)
+
+ n_input = x[0].shape[0] # ns in main.py
+ K = paddle.zeros((n_input, n_input))
+
+ for J in J_list:
+ K += J @ J.T
+
+ return K
+
+ def cal_ntk(self, x):
+ # Formula (4), Page3, \lambda variable
+ # Lambda represents the eigenvalues of the matrix(Kg)
+ lambda_g = 0.0
+ lambda_b = 0.0
+ n_neu = self.rbn.n_neu
+
+ # in-domain
+ n1 = x[0].shape[0]
+ for i in range(n1):
+ temp_x = [x[0][i, ...].unsqueeze(0), paddle.to_tensor([[0.0]])]
+ y = self.forward(temp_x)
+ l1t = paddle.grad(y[0], self.parameters(), allow_unused=True)
+ for j, grad in enumerate(l1t):
+ if grad is None:
+ grad = paddle.to_tensor([0.0]).broadcast_to(
+ self.parameters()[j].shape
+ )
+ l1t[j] = grad
+ lambda_g = lambda_g + paddle.sum(grad**2) / n1
+
+ # When use tanh activation function, the value may be None
+ if self.activation_function == "tanh":
+ temp = paddle.concat(
+ # (l1t[0], l1t[1], l1t[2].reshape((1, n_neu))), axis=1
+ # Not select last_fc_bias
+ (l1t[1], l1t[2], l1t[3].reshape((1, n_neu))),
+ axis=1,
+ )
+ else:
+ temp = paddle.concat((l1t[0], l1t[1].reshape((1, n_neu))), axis=1)
+ if i == 0:
+ # Fig.1, Page5, Kg variable
+ Kg = temp
+ else:
+ Kg = paddle.concat((Kg, temp), axis=0)
+
+ # bound
+ n2 = x[1].shape[0]
+ for i in range(n2):
+ temp_x = [paddle.to_tensor([[0.0]]), x[1][i, ...].unsqueeze(0)]
+ y = self.forward(temp_x)
+ l1t = paddle.grad(y[1], self.rbn.parameters(), allow_unused=True)
+ for j in l1t:
+ lambda_b = lambda_b + paddle.sum(j**2) / n2
+
+ # calculate adapt factors
+ temp = lambda_g + lambda_b
+ lambda_g = temp / lambda_g
+ lambda_b = temp / lambda_b
+
+ return lambda_g, lambda_b, Kg
diff --git a/jointContribution/PIRBN/rbn_net.py b/jointContribution/PIRBN/rbn_net.py
new file mode 100644
index 000000000..a79898a66
--- /dev/null
+++ b/jointContribution/PIRBN/rbn_net.py
@@ -0,0 +1,172 @@
+import math
+
+import numpy as np
+import paddle
+
+
+class RBN_Net(paddle.nn.Layer):
+ """This class is to build a radial basis network (RBN).
+
+ Args:
+ n_in (int): Number of input of the RBN.
+ n_out (int): Number of output of the RBN.
+ n_neu (int): Number of neurons in the hidden layer.
+ b (Union[List[float], float]): Initial value for hyperparameter b.
+ c (List[float]): Initial value for hyperparameter c.
+ activation_function (str, optional): The activation function, tanh or gaussian. Defaults to "gaussian".
+ """
+
+ def __init__(self, n_in, n_out, n_neu, b, c, activation_function="gaussian"):
+ super().__init__()
+ self.n_in = n_in
+ self.n_out = n_out
+ self.n_neu = n_neu
+ self.b = paddle.to_tensor(b)
+ self.c = paddle.to_tensor(c)
+ self.activation_function = activation_function
+ self.activation = Activation(self.n_neu, self.c, n_in, activation_function)
+
+ # LeCun normal initialization
+ std = math.sqrt(1 / self.n_neu)
+ ini = paddle.ParamAttr(
+ initializer=paddle.nn.initializer.Normal(mean=0.0, std=std)
+ )
+
+ if self.activation_function == "gaussian":
+ # gaussian activation_function need to set self.b
+ self.init_ab()
+ self.last_fc_layer = paddle.nn.Linear(
+ self.n_neu,
+ self.n_out,
+ weight_attr=ini,
+ bias_attr=False,
+ )
+
+ elif self.activation_function == "tanh":
+ w, b = self.initialize_NN([self.n_in, self.n_neu, self.n_out])
+ w_0 = paddle.ParamAttr(initializer=paddle.nn.initializer.Assign(w[0]))
+ b_0 = paddle.ParamAttr(initializer=paddle.nn.initializer.Assign(b[0]))
+ w_1 = paddle.ParamAttr(initializer=paddle.nn.initializer.Assign(w[1]))
+
+ self.hidden_layer = paddle.nn.Linear(
+ self.n_in, self.n_neu, weight_attr=w_0, bias_attr=b_0
+ )
+
+ self.last_fc_layer = paddle.nn.Linear(
+ self.n_neu,
+ self.n_out,
+ weight_attr=w_1,
+ bias_attr=False,
+ )
+
+ self.last_fc_bias = self.create_parameter(
+ shape=b[1].shape,
+ default_initializer=paddle.nn.initializer.Assign(b[1]),
+ dtype=paddle.get_default_dtype(),
+ )
+ else:
+ raise ("Not implemented yet")
+
+ def NTK_init(self, size):
+ in_dim = size[0]
+ out_dim = size[1]
+ std = 1.0 / np.sqrt(in_dim)
+ return self.create_parameter(
+ shape=[in_dim, out_dim],
+ default_initializer=paddle.nn.initializer.Assign(
+ paddle.normal(shape=[in_dim, out_dim]) * std
+ ),
+ dtype=paddle.get_default_dtype(),
+ )
+
+ # Initialize network weights and biases using Xavier initialization
+ def initialize_NN(self, layers):
+ weights = []
+ biases = []
+ num_layers = len(layers)
+ for l in range(0, num_layers - 1):
+ W = self.NTK_init(size=[layers[l], layers[l + 1]])
+ b = self.create_parameter(
+ shape=[1, layers[l + 1]],
+ default_initializer=paddle.nn.initializer.Assign(
+ paddle.normal(shape=[1, layers[l + 1]])
+ ),
+ dtype=paddle.get_default_dtype(),
+ )
+ weights.append(W)
+ biases.append(b)
+ return weights, biases
+
+ def forward(self, x):
+ if self.activation_function == "gaussian":
+ y = self.activation(x)
+ y = self.last_fc_layer(y)
+ elif self.activation_function == "tanh":
+ # input : n x 1
+ # hidden layer : 1 x 61
+ # last fc layer : 61 x 1
+ y = self.hidden_layer(x)
+ y = self.activation(y)
+ y = self.last_fc_layer(y)
+ y = paddle.add(y, self.last_fc_bias)
+ else:
+ raise ("Not implemented yet")
+ return y
+
+ # gaussian activation_function need to set self.b
+ def init_ab(self):
+ b = np.ones((1, self.n_neu)) * self.b
+ self.activation.b = self.activation.create_parameter(
+ (1, self.n_neu), default_initializer=paddle.nn.initializer.Assign(b)
+ )
+
+
+class Activation(paddle.nn.Layer):
+ """This class is to create the hidden layer of a radial basis network.
+
+ Args:
+ n_neu (int): Number of neurons in the hidden layer.
+ c (List[float32]): Initial value for hyperparameter b.
+ n_in (int): Last item of input shape.
+ """
+
+ def __init__(self, n_neu, c, n_in, activation_function="gaussian"):
+ super(Activation, self).__init__()
+ self.activation_function = activation_function
+ # PINN y = w2 * (tanh(w1 * x + b1)) + b2 w,b are trainable parameters, b is bais
+ # PIRBN y = w * exp(b^2 * |x-c|^2) w,b are trainable parameters, c is constant, b is not bias
+
+ if self.activation_function == "gaussian":
+ self.n_neu = n_neu
+ self.c = c
+ self.b = self.create_parameter(
+ shape=(n_in, self.n_neu),
+ dtype=paddle.get_default_dtype(),
+ # Convert from tensorflow tf.random_normal_initializer(), default value mean=0.0, std=0.05
+ default_initializer=paddle.nn.initializer.Normal(mean=0.0, std=0.05),
+ )
+
+ def forward(self, inputs):
+ if self.activation_function == "gaussian":
+ return self.gaussian_function(inputs)
+ elif self.activation_function == "tanh":
+ return self.tanh_function(inputs)
+
+ # Gaussian function,Formula (19), Page7
+ def gaussian_function(self, inputs):
+ temp_x = paddle.matmul(inputs, paddle.ones((1, self.n_neu)))
+ x0 = (
+ paddle.reshape(
+ paddle.arange(self.n_neu, dtype=paddle.get_default_dtype()),
+ (1, self.n_neu),
+ )
+ * (self.c[1] - self.c[0])
+ / (self.n_neu - 1)
+ + self.c[0]
+ )
+ x_new = temp_x - x0
+ s = self.b * self.b
+ return paddle.exp(-(x_new * x_new) * s)
+
+ def tanh_function(self, inputs):
+ return paddle.tanh(inputs)
diff --git a/jointContribution/PIRBN/train.py b/jointContribution/PIRBN/train.py
new file mode 100644
index 000000000..cce269044
--- /dev/null
+++ b/jointContribution/PIRBN/train.py
@@ -0,0 +1,99 @@
+import paddle
+
+paddle.framework.core.set_prim_eager_enabled(True)
+
+
+class Trainer:
+ def __init__(
+ self,
+ pirbn,
+ x_train,
+ y_train,
+ learning_rate=0.001,
+ maxiter=10000,
+ adaptive_weights=True,
+ ):
+ # set attributes
+ self.pirbn = pirbn
+
+ self.learning_rate = learning_rate
+ self.x_train = [
+ paddle.to_tensor(x, dtype=paddle.get_default_dtype()) for x in x_train
+ ]
+ self.y_train = paddle.to_tensor(y_train, dtype=paddle.get_default_dtype())
+
+ self.maxiter = maxiter
+ self.loss_g = [] # eq loss
+ self.loss_b = [] # boundary loss
+ self.iter = 0
+ self.a_g = paddle.to_tensor(1.0)
+ self.a_b = paddle.to_tensor(1.0)
+ self.his_a_g = []
+ self.his_a_b = []
+ self.optimizer = paddle.optimizer.Adam(
+ learning_rate=0.001, parameters=self.pirbn.parameters()
+ )
+ self.ntk_list = {}
+ # Update loss by calculate ntk
+ self.adaptive_weights = (
+ adaptive_weights # Adaptive weights for physics-informed neural networks
+ )
+
+ def Loss(self, x, y, a_g, a_b):
+ u_xx, u_b, _ = self.pirbn(x)
+ loss_g = 0.5 * paddle.mean(paddle.square(u_xx - y))
+ loss_b = 0.5 * paddle.mean(paddle.square(u_b))
+ if self.adaptive_weights:
+ loss = loss_g * a_g + loss_b * a_b
+ else:
+ loss = loss_g + 100 * loss_b
+ return loss, loss_g, loss_b
+
+ def evaluate(self):
+ # compute loss
+ loss, loss_g, loss_b = self.Loss(self.x_train, self.y_train, self.a_g, self.a_b)
+ loss_g_numpy = float(loss_g)
+ loss_b_numpy = float(loss_b)
+ # eq loss
+ self.loss_g.append(loss_g_numpy)
+ # boundary loss
+ self.loss_b.append(loss_b_numpy)
+ if self.iter % 100 == 0:
+ if self.adaptive_weights:
+ self.a_g, self.a_b, _ = self.pirbn.cal_ntk(self.x_train)
+ print(
+ "Iter : ",
+ self.iter,
+ "\tloss : ",
+ float(loss),
+ "\tboundary loss : ",
+ float(loss_b),
+ "\teq loss : ",
+ float(loss_g),
+ )
+ print("\ta_g =", float(self.a_g), "\ta_b =", float(self.a_b))
+ else:
+ print(
+ "Iter : ",
+ self.iter,
+ "\tloss : ",
+ float(loss),
+ "\tboundary loss : ",
+ float(loss_b),
+ "\teq loss : ",
+ float(loss_g),
+ )
+ self.his_a_g.append(self.a_g)
+ self.his_a_b.append(self.a_b)
+
+ self.iter = self.iter + 1
+ return loss
+
+ def fit(self, output_Kgg):
+ for i in range(0, self.maxiter):
+ loss = self.evaluate()
+ loss.backward()
+ if i in output_Kgg:
+ self.ntk_list[f"{i}"] = self.pirbn.cal_K(self.x_train)
+ self.optimizer.step()
+ self.optimizer.clear_grad()
diff --git a/mkdocs.yml b/mkdocs.yml
index 4cf50e9d4..690f89c09 100644
--- a/mkdocs.yml
+++ b/mkdocs.yml
@@ -39,6 +39,7 @@ nav:
- 方程求解:
- Volterra_IDE: zh/examples/volterra_ide.md
- Laplace2D: zh/examples/laplace2d.md
+ - PIRBN: zh/examples/pirbn.md
- 振动:
- ViV: zh/examples/viv.md
- 流体: