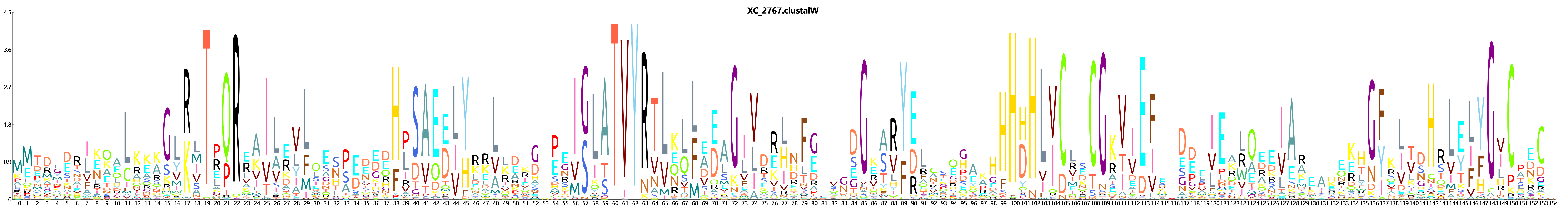
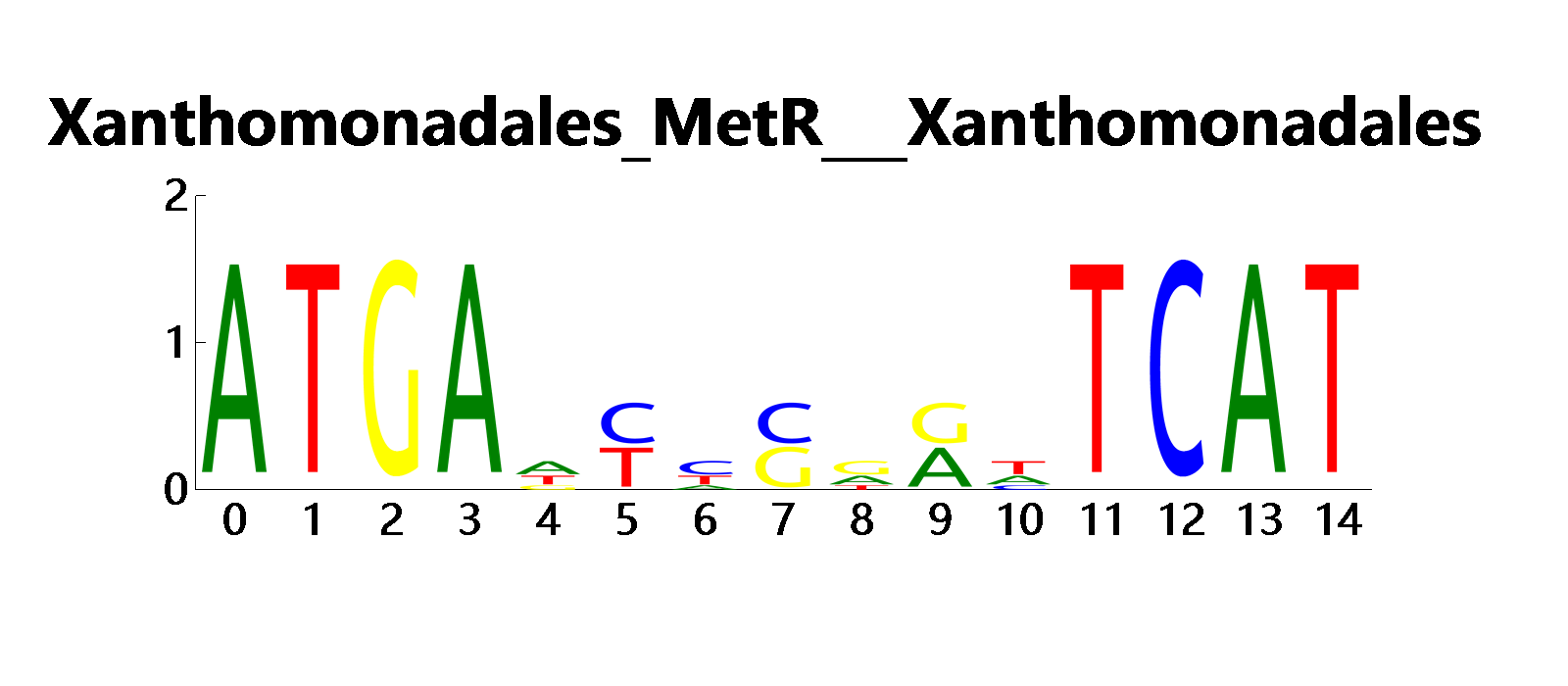Hidden Markov Model for Pfam-A
HMM data can be download from EMBL ftp website:
ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam29.0
Multiple align of the sequence, this can be applied by the clustal program
Using /logo command from the seequence CLI tools to draw the sequence logo from the multiple sequence alignment result
G:\4.15\MEME\footprints>seqtools ? /logo
Help for command '/logo':
Information:
Usage: F:\GCModeller\GCModeller-x64\seqtools.exe /logo /in <clustal.fasta> [/out <out.png>]
Example: seqtools /logo