In this lecture, we are going to dive deeper into Git, introducing the conepts of .gitignore, branching and forking.
We will also start looking at how to work with data using dataframes and how to perform simple operations on them to transform the data to our liking. Specifically, we will look at how to sort, select, filter, mutate and group. I will describe these operations briefly below.
In the previous lecture, we talked about some of the basic concepts of Git such as commiting, pushing, creating a repository and so on, these concepts are sufficient to use Git. However, to harness the full capabilities of Git we need to introduce a few more concepts.
As you might have already seen, when working on project, there are a lot of
unnecessary files that enter the project. For instance, if you use jupyter notebooks, a file named
.ipynb_checkpoints will be generated automatically. In most cases, tracking these types of files
and/or folders with Git is not needed. To ignore these
particular files and/or folders we can use .gitignore. The .gitignore file is an index for all the
files and folders you do not wish to track.
A simple example of a .gitignore file, in our example, would be
.ipynb_checkpoints
Sometimes, it is typical to also ignore the .gitignore file. It depends on the project. You can ignore as many file and folders you
deem unnecessary to track.
You can create this file manually, or using Github desktop. Using Github
desktop, in the menu bar, go to Repository> Repository Settings > Ignored Files
and list the files and folders you wish to ignore there. If you create the file
manually, make sure to place the .gitignore file in the root of you
repository.
You can also right click the file or folder on your Github desktop app, and select ignore file.
Forking, is a concept that is central to Git. It is the action done on a Git repository, which creates a copy of a repo and detaches from the original repo. In contrast, when you clone a repository, you make a copy, but the copy is still linked to the original repo.
When you want to make changes to a project that you do not have ownership of, or, if you want to make your own version of the project and build from the original project, you can fork the repository and work on your own continued version. Be mindfull of the rights you have to the code. If the project is open-sourc, there is no issue with forking the code. However, if this is not the case, it is theft of intellectual property.
There are many popular projects that are forks. A popular example is of linux distriubtions, which are forks of older distriubtions. For instance, Linux Mint is forked from Ubuntu.
When you want to make changes to a repository that you have ownership of and you want the changes to be reflected in the original repository, then you can clone the repository. For instance, if you new member joins the project, one can clone the project to get a hold of the code.
Cloning is something you have already done, when you set up your repository on the course organization.
Although, forking and cloning seem similar, there are small but important subtleties between them.
Forking is also very popular when making contributions to open-source projects. Read more about forking here.
During development, you will most likely encouter a situation where you would
like to test a new feature in your code without changing the original code.
Imagine you would like to change the theme of your website.
Since you are in development stages, you don't want to ruin the
original working code in case you decide the feature dosen't work as intended.
A simple solution would be to make a copy of the code and develop your new feature on the copied code.
Git, defines this procedure as branching. The idea is that you "branch" of from your main
code to develop the feature. If you decide that the feature was a bad idea, you
can just delete the branch and jump to your main branch without changes being
made to your main code. On the other hand, if you decide that the features is
good, you can merge the new feature to you main branch. Introducing the
branching feature makes it seemless to work on new features conccurently and
also makes collaboration easy. Each team or member can work on a feature
independently.
The following illustration will make this more clear.
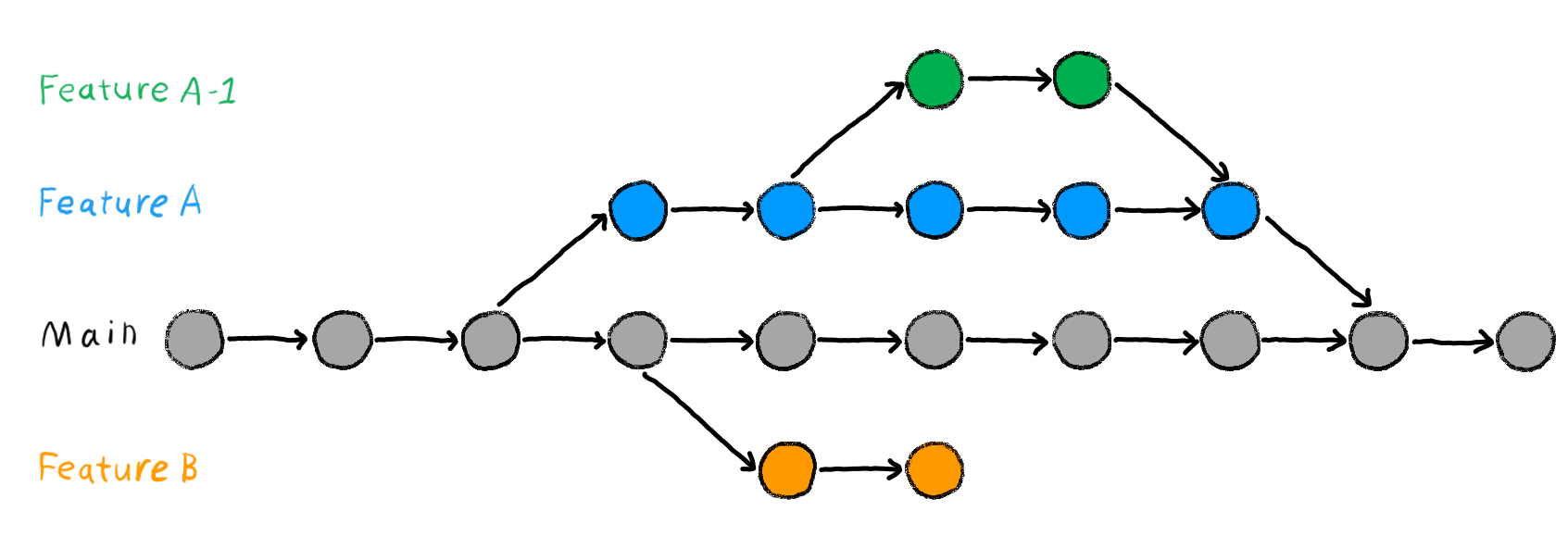
This image illustrates how the process of branching may look in a project. As you can see, two features can be developed concurrently without conflicts. This makes developing features easier as you are not restricted to a single feature at a time.
You may have noticed that you are working on the main branch of your
repository. This is the default. You can simply create a new branch whenever you
feel like you want to add a feature to your code and don't want to ruin the
original code. This is usually not an issue for very small projects. However, as
your project grows, branching becomes very important.
It is simple to create, merge or delete a branch. On Github Desktop, press
on the Current Branch tab and select new branch. Name it something that makes
sense to the feature you are going to add. Finally, choose the branch
you want base the new branch on, that is, the istance of your code that you want
to branch of from. Note that this only creates a local branch and
you need to Publish the new branch if you want it to be seen on Github.
On Github desktop, under the Current branch tab you will see all the branches of your (local)
repository. To delete a branch, simply right click on the branch and select
delete. To merge a branch, under the Current branch drop down, there is a
tab called Pull Requests, open this tab and in the bottom press the button,
Chosse a branch to merge into .... Finally, chose the branch you want to merge
and press Create merge commit. Making a pull requests is synonomous to
creating a request for merging.
If you feature branch is published on Github, you can merge on there as well. Read more about how to do that here.
Tables are the go to structure to visualize data. They consist of columns and rows labled in a particular fashion. A popular (and good) way of organizing data in tables is using columns e.g. Name, Age, Height, to represent variables and rows to represent entries.
An example of a dataset ordered in a table can look as follows.
| Name | Age | Height(cm) | Occupation | Id |
|---|---|---|---|---|
| Taariq Nazar | 26 | 187 | Phd | 0 |
| Someone Cooler | 27 | 188 | Proffesor | 1 |
| Someone Even Cooler | 28 | 189 | Emeritus | 2 |
| Someone so cool | 26 | 179 | Plumber | 3 |
A table format of the dataset is called a Dataframe. In the example above, the data
consists of three entries and 4(5) variables (one does not usually count the row
number (Id) as a variable unless it is crucial part of the dataset e.g. a
special id specific to the data). A core concept of this course is how to work
with dataframes, that is, how to perform operations on dataframes.
There are a set of operations you should be familiar with to work on
dataframes. These are sorting, selecting, filtering, mutating and grouping. Using these, you can
create complex operations by composing the operations.
Sorting is the process of ordering data by a variable or multiple variables. For instance in the above example one might want to order the entries (rows) by Height.
Selecting, is chosing variables of interest. In most datasets, there will be an excess of variables that are necessary for you analysis, then you would like select the variables that are necessary.
Just as it is named, filtering is the process of focusing on entries that
satisfy some condition. For instance, in the above example. We might only want to look at entries that are
have Age > 26.
Mutating, is to change the original dataset by creating new variables which can be functions of old variables.
Grouping, is a process of aggregating data. In most cases, it is accompanied
with a following operations, for instance, sum, count, max. In the above
example, if we group by Age, and accompany the grouping by count, this will
result in a dataframe that contains the number of people in each Age-group.
The syntax for performing these operations on dataframes looks as follows :::code-group
import pandas as pd
# The standard library for datframes in Python is Pandas
df = pd.DataFrame(data)
# Sorting
#Set ascending False if the order you want is descending.
df.sort_values(column_name, ascending=True)
# Selecting
# Selecting range can be done using :
df.loc[[row1_1, row_2,...],[column_name1, column_name2,... ]] #Choose the columns of interest.
# Filtering
#Condition of your filter, for instance condition = df["age"]>=10 && df["age"]<20.
df.loc[condition]
# Mutating
# Using lambda-functions makes it easy to mutate. You can also you regular functions.
df[new_col] = df.loc[[cols_to_mutate]].apply(lambda x: x[col_1]/x[col_2])
# Grouping accompanied with count
df.groupby(cols_to_group).count()
# Example of composing operations
df
.loc(...)
.apply(...)
.groupby(...).max()#In R, the operations is packaged into the `dplyr` library. The pipe operator %>%`, makes operating on dataframes very easy.
library(dplyr)
df = tibble(data)
# Sorting
#Change column_name -> desc(column_name) if you want it to be descending.
df %>% arrange(column_name, .by_group = FALSE)
# Selecting
# You can build complex selection methods using select(), e.g. criterion = column_name
df %>% select(criterion)
# Filtering
# Bulding complex filters is very simple, for instance criterion: age>=mean(age)
df %>% filter(crtierion)
# Mutating
# You can mutate the current df by using mutate()
df %>% mutate(new_col = f(column_variables))
# Grouping accompanied with count
df %>%
group_by(cols_to_group) %>%
tally()
# Due to pipes, one can compose operations in a simple manner.
# Example of composing
df %>%
select(column_1, column_5, column_2) %>%
arrange(column_2, .by_group=FALSE) %>%
filter(column_5 >= 10) %>%
mutate(column_99 = column_1/column_5 * column_2. .keep="all"):::
You can combine these simple operations to compose very complex ones. To find the documentation, google "package method doucmentation". E.g. "pandas loc documentation", the first link is usually the original documentation and will contain indepth information for the specific method you are searching for.