diff --git a/.gitignore b/.gitignore
index 7daa4363..7f89125e 100644
--- a/.gitignore
+++ b/.gitignore
@@ -3,3 +3,4 @@ __pycache__/
*.pth
*.tif
examples/data/*
+*.out
diff --git a/README.md b/README.md
index aa1c6f56..be7b6331 100644
--- a/README.md
+++ b/README.md
@@ -16,31 +16,26 @@ We implement napari applications for:
 -**Beta version**
-
-This is an advanced beta version. While many features are still under development, we aim to keep the user interface and python library stable.
-Any feedback is welcome, but please be aware that the functionality is under active development and that some features may not be thoroughly tested yet.
-We will soon provide a stand-alone application for running the `micro_sam` annotation tools, and plan to also release it as [napari plugin](https://napari.org/stable/plugins/index.html) in the future.
-
-If you run into any problems or have questions please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
+If you run into any problems or have questions regarding our tool please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
## Installation and Usage
You can install `micro_sam` via conda:
```
-conda install -c conda-forge micro_sam
+conda install -c conda-forge micro_sam napari pyqt
```
You can then start the `micro_sam` tools by running `$ micro_sam.annotator` in the command line.
+For an introduction in how to use the napari based annotation tools check out [the video tutorials](https://www.youtube.com/watch?v=ket7bDUP9tI&list=PLwYZXQJ3f36GQPpKCrSbHjGiH39X4XjSO&pp=gAQBiAQB).
Please check out [the documentation](https://computational-cell-analytics.github.io/micro-sam/) for more details on the installation and usage of `micro_sam`.
## Citation
If you are using this repository in your research please cite
-- [SegmentAnything](https://arxiv.org/abs/2304.02643)
-- and our repository on [zenodo](https://doi.org/10.5281/zenodo.7919746) (we are working on a publication)
+- Our [preprint](https://doi.org/10.1101/2023.08.21.554208)
+- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643)
## Related Projects
@@ -56,6 +51,17 @@ Compared to these we support more applications (2d, 3d and tracking), and provid
## Release Overview
+**New in version 0.2.1 and 0.2.2**
+
+- Several bugfixes for the newly introduced functionality in 0.2.0.
+
+**New in version 0.2.0**
+
+- Functionality for training / finetuning and evaluation of Segment Anything Models
+- Full support for our finetuned segment anything models
+- Improvements of the automated instance segmentation functionality in the 2d annotator
+- And several other small improvements
+
**New in version 0.1.1**
- Fine-tuned segment anything models for microscopy (experimental)
diff --git a/deployment/construct.yaml b/deployment/construct.yaml
index 5231b738..d9db54df 100644
--- a/deployment/construct.yaml
+++ b/deployment/construct.yaml
@@ -8,8 +8,6 @@ header_image: ../doc/images/micro-sam-logo.png
icon_image: ../doc/images/micro-sam-logo.png
channels:
- conda-forge
-welcome_text: Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod
- tempor incididunt ut labore et dolore magna aliqua.
-conclusion_text: Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris
- nisi ut aliquip ex ea commodo consequat.
-initialize_by_default: false
\ No newline at end of file
+welcome_text: Install Segment Anything for Microscopy.
+conclusion_text: Segment Anything for Microscopy has been installed.
+initialize_by_default: false
diff --git a/doc/annotation_tools.md b/doc/annotation_tools.md
index 921e1112..7d6a4027 100644
--- a/doc/annotation_tools.md
+++ b/doc/annotation_tools.md
@@ -13,9 +13,22 @@ The annotation tools can be started from the `micro_sam` GUI, the command line o
$ micro_sam.annotator
```
-They are built with [napari](https://napari.org/stable/) to implement the viewer and user interaction.
+They are built using [napari](https://napari.org/stable/) and [magicgui](https://pyapp-kit.github.io/magicgui/) to provide the viewer and user interface.
If you are not familiar with napari yet, [start here](https://napari.org/stable/tutorials/fundamentals/quick_start.html).
-The `micro_sam` applications are mainly based on [the point layer](https://napari.org/stable/howtos/layers/points.html), [the shape layer](https://napari.org/stable/howtos/layers/shapes.html) and [the label layer](https://napari.org/stable/howtos/layers/labels.html).
+The `micro_sam` tools use [the point layer](https://napari.org/stable/howtos/layers/points.html), [shape layer](https://napari.org/stable/howtos/layers/shapes.html) and [label layer](https://napari.org/stable/howtos/layers/labels.html).
+
+The annotation tools are explained in detail below. In addition to the documentation here we also provide [video tutorials](https://www.youtube.com/watch?v=ket7bDUP9tI&list=PLwYZXQJ3f36GQPpKCrSbHjGiH39X4XjSO).
+
+
+## Starting via GUI
+
+The annotation toools can be started from a central GUI, which can be started with the command `$ micro_sam.annotator` or using the executable [from an installer](#from-installer).
+
+In the GUI you can select with of the four annotation tools you want to use:
+
-**Beta version**
-
-This is an advanced beta version. While many features are still under development, we aim to keep the user interface and python library stable.
-Any feedback is welcome, but please be aware that the functionality is under active development and that some features may not be thoroughly tested yet.
-We will soon provide a stand-alone application for running the `micro_sam` annotation tools, and plan to also release it as [napari plugin](https://napari.org/stable/plugins/index.html) in the future.
-
-If you run into any problems or have questions please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
+If you run into any problems or have questions regarding our tool please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
## Installation and Usage
You can install `micro_sam` via conda:
```
-conda install -c conda-forge micro_sam
+conda install -c conda-forge micro_sam napari pyqt
```
You can then start the `micro_sam` tools by running `$ micro_sam.annotator` in the command line.
+For an introduction in how to use the napari based annotation tools check out [the video tutorials](https://www.youtube.com/watch?v=ket7bDUP9tI&list=PLwYZXQJ3f36GQPpKCrSbHjGiH39X4XjSO&pp=gAQBiAQB).
Please check out [the documentation](https://computational-cell-analytics.github.io/micro-sam/) for more details on the installation and usage of `micro_sam`.
## Citation
If you are using this repository in your research please cite
-- [SegmentAnything](https://arxiv.org/abs/2304.02643)
-- and our repository on [zenodo](https://doi.org/10.5281/zenodo.7919746) (we are working on a publication)
+- Our [preprint](https://doi.org/10.1101/2023.08.21.554208)
+- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643)
## Related Projects
@@ -56,6 +51,17 @@ Compared to these we support more applications (2d, 3d and tracking), and provid
## Release Overview
+**New in version 0.2.1 and 0.2.2**
+
+- Several bugfixes for the newly introduced functionality in 0.2.0.
+
+**New in version 0.2.0**
+
+- Functionality for training / finetuning and evaluation of Segment Anything Models
+- Full support for our finetuned segment anything models
+- Improvements of the automated instance segmentation functionality in the 2d annotator
+- And several other small improvements
+
**New in version 0.1.1**
- Fine-tuned segment anything models for microscopy (experimental)
diff --git a/deployment/construct.yaml b/deployment/construct.yaml
index 5231b738..d9db54df 100644
--- a/deployment/construct.yaml
+++ b/deployment/construct.yaml
@@ -8,8 +8,6 @@ header_image: ../doc/images/micro-sam-logo.png
icon_image: ../doc/images/micro-sam-logo.png
channels:
- conda-forge
-welcome_text: Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod
- tempor incididunt ut labore et dolore magna aliqua.
-conclusion_text: Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris
- nisi ut aliquip ex ea commodo consequat.
-initialize_by_default: false
\ No newline at end of file
+welcome_text: Install Segment Anything for Microscopy.
+conclusion_text: Segment Anything for Microscopy has been installed.
+initialize_by_default: false
diff --git a/doc/annotation_tools.md b/doc/annotation_tools.md
index 921e1112..7d6a4027 100644
--- a/doc/annotation_tools.md
+++ b/doc/annotation_tools.md
@@ -13,9 +13,22 @@ The annotation tools can be started from the `micro_sam` GUI, the command line o
$ micro_sam.annotator
```
-They are built with [napari](https://napari.org/stable/) to implement the viewer and user interaction.
+They are built using [napari](https://napari.org/stable/) and [magicgui](https://pyapp-kit.github.io/magicgui/) to provide the viewer and user interface.
If you are not familiar with napari yet, [start here](https://napari.org/stable/tutorials/fundamentals/quick_start.html).
-The `micro_sam` applications are mainly based on [the point layer](https://napari.org/stable/howtos/layers/points.html), [the shape layer](https://napari.org/stable/howtos/layers/shapes.html) and [the label layer](https://napari.org/stable/howtos/layers/labels.html).
+The `micro_sam` tools use [the point layer](https://napari.org/stable/howtos/layers/points.html), [shape layer](https://napari.org/stable/howtos/layers/shapes.html) and [label layer](https://napari.org/stable/howtos/layers/labels.html).
+
+The annotation tools are explained in detail below. In addition to the documentation here we also provide [video tutorials](https://www.youtube.com/watch?v=ket7bDUP9tI&list=PLwYZXQJ3f36GQPpKCrSbHjGiH39X4XjSO).
+
+
+## Starting via GUI
+
+The annotation toools can be started from a central GUI, which can be started with the command `$ micro_sam.annotator` or using the executable [from an installer](#from-installer).
+
+In the GUI you can select with of the four annotation tools you want to use:
+ +
+And after selecting them a new window will open where you can select the input file path and other optional parameter. Then click the top button to start the tool. **Note: If you are not starting the annotation tool with a path to pre-computed embeddings then it can take several minutes to open napari after pressing the button because the embeddings are being computed.**
+
## Annotator 2D
@@ -44,7 +57,7 @@ It contains the following elements:
Note that point prompts and box prompts can be combined. When you're using point prompts you can only segment one object at a time. With box prompts you can segment several objects at once.
-Check out [this video](https://youtu.be/DfWE_XRcqN8) for an example of how to use the interactive 2d annotator.
+Check out [this video](https://youtu.be/ket7bDUP9tI) for a tutorial for the 2d annotation tool.
We also provide the `image series annotator`, which can be used for running the 2d annotator for several images in a folder. You can start by clicking `Image series annotator` in the GUI, running `micro_sam.image_series_annotator` in the command line or from a [python script](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/image_series_annotator.py).
@@ -69,7 +82,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that you can only segment one object at a time with the 3d annotator.
-Check out [this video](https://youtu.be/5Jo_CtIefTM) for an overview of the interactive 3d segmentation functionality.
+Check out [this video](https://youtu.be/PEy9-rTCdS4) for a tutorial for the 3d annotation tool.
## Annotator Tracking
@@ -93,7 +106,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that the tracking annotator only supports 2d image data, volumetric data is not supported.
-Check out [this video](https://youtu.be/PBPW0rDOn9w) for an overview of the interactive tracking functionality.
+Check out [this video](https://youtu.be/Xi5pRWMO6_w) for a tutorial for how to use the tracking annotation tool.
## Tips & Tricks
@@ -105,7 +118,7 @@ You can activate tiling by passing the parameters `tile_shape`, which determines
- If you're using the command line functions you can pass them via the options `--tile_shape 1024 1024 --halo 128 128`
- Note that prediction with tiling only works when the embeddings are cached to file, so you must specify an `embedding_path` (`-e` in the CLI).
- You should choose the `halo` such that it is larger than half of the maximal radius of the objects your segmenting.
-- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_embeddings` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
+- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_state` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
- Most other processing steps are very fast even on a CPU, so interactive annotation is possible. An exception is the automatic segmentation step (2d segmentation), which takes several minutes without a GPU (depending on the image size). For large volumes and timeseries segmenting an object in 3d / tracking across time can take a couple settings with a CPU (it is very fast with a GPU).
- You can also try using a smaller version of the SegmentAnything model to speed up the computations. For this you can pass the `model_type` argument and either set it to `vit_b` or to `vit_l` (default is `vit_h`). However, this may lead to worse results.
- You can save and load the results from the `committed_objects` / `committed_tracks` layer to correct segmentations you obtained from another tool (e.g. CellPose) or to save intermediate annotation results. The results can be saved via `File -> Save Selected Layer(s) ...` in the napari menu (see the tutorial videos for details). They can be loaded again by specifying the corresponding location via the `segmentation_result` (2d and 3d segmentation) or `tracking_result` (tracking) argument.
diff --git a/doc/finetuned_models.md b/doc/finetuned_models.md
new file mode 100644
index 00000000..4fc9fb13
--- /dev/null
+++ b/doc/finetuned_models.md
@@ -0,0 +1,36 @@
+# Finetuned models
+
+We provide models that were finetuned on microscopy data using `micro_sam.training`. They are hosted on zenodo. We currently offer the following models:
+- `vit_h`: Default Segment Anything model with vit-h backbone.
+- `vit_l`: Default Segment Anything model with vit-l backbone.
+- `vit_b`: Default Segment Anything model with vit-b backbone.
+- `vit_h_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-h backbone.
+- `vit_b_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-b backbone.
+- `vit_h_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-h backbone.
+- `vit_b_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-b backbone.
+
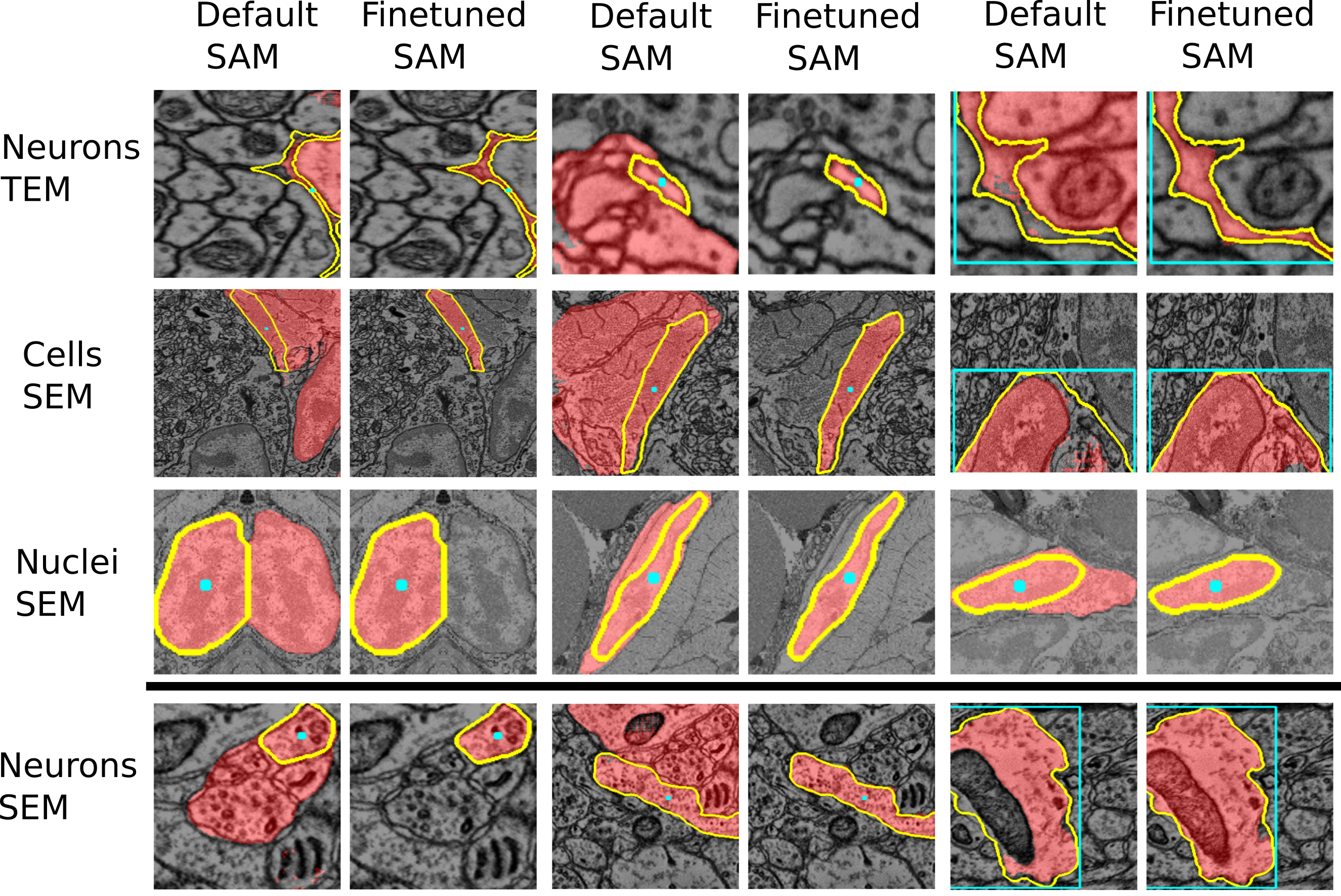
+See the two figures below of the improvements through the finetuned model for LM and EM data.
+
+
+
+And after selecting them a new window will open where you can select the input file path and other optional parameter. Then click the top button to start the tool. **Note: If you are not starting the annotation tool with a path to pre-computed embeddings then it can take several minutes to open napari after pressing the button because the embeddings are being computed.**
+
## Annotator 2D
@@ -44,7 +57,7 @@ It contains the following elements:
Note that point prompts and box prompts can be combined. When you're using point prompts you can only segment one object at a time. With box prompts you can segment several objects at once.
-Check out [this video](https://youtu.be/DfWE_XRcqN8) for an example of how to use the interactive 2d annotator.
+Check out [this video](https://youtu.be/ket7bDUP9tI) for a tutorial for the 2d annotation tool.
We also provide the `image series annotator`, which can be used for running the 2d annotator for several images in a folder. You can start by clicking `Image series annotator` in the GUI, running `micro_sam.image_series_annotator` in the command line or from a [python script](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/image_series_annotator.py).
@@ -69,7 +82,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that you can only segment one object at a time with the 3d annotator.
-Check out [this video](https://youtu.be/5Jo_CtIefTM) for an overview of the interactive 3d segmentation functionality.
+Check out [this video](https://youtu.be/PEy9-rTCdS4) for a tutorial for the 3d annotation tool.
## Annotator Tracking
@@ -93,7 +106,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that the tracking annotator only supports 2d image data, volumetric data is not supported.
-Check out [this video](https://youtu.be/PBPW0rDOn9w) for an overview of the interactive tracking functionality.
+Check out [this video](https://youtu.be/Xi5pRWMO6_w) for a tutorial for how to use the tracking annotation tool.
## Tips & Tricks
@@ -105,7 +118,7 @@ You can activate tiling by passing the parameters `tile_shape`, which determines
- If you're using the command line functions you can pass them via the options `--tile_shape 1024 1024 --halo 128 128`
- Note that prediction with tiling only works when the embeddings are cached to file, so you must specify an `embedding_path` (`-e` in the CLI).
- You should choose the `halo` such that it is larger than half of the maximal radius of the objects your segmenting.
-- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_embeddings` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
+- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_state` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
- Most other processing steps are very fast even on a CPU, so interactive annotation is possible. An exception is the automatic segmentation step (2d segmentation), which takes several minutes without a GPU (depending on the image size). For large volumes and timeseries segmenting an object in 3d / tracking across time can take a couple settings with a CPU (it is very fast with a GPU).
- You can also try using a smaller version of the SegmentAnything model to speed up the computations. For this you can pass the `model_type` argument and either set it to `vit_b` or to `vit_l` (default is `vit_h`). However, this may lead to worse results.
- You can save and load the results from the `committed_objects` / `committed_tracks` layer to correct segmentations you obtained from another tool (e.g. CellPose) or to save intermediate annotation results. The results can be saved via `File -> Save Selected Layer(s) ...` in the napari menu (see the tutorial videos for details). They can be loaded again by specifying the corresponding location via the `segmentation_result` (2d and 3d segmentation) or `tracking_result` (tracking) argument.
diff --git a/doc/finetuned_models.md b/doc/finetuned_models.md
new file mode 100644
index 00000000..4fc9fb13
--- /dev/null
+++ b/doc/finetuned_models.md
@@ -0,0 +1,36 @@
+# Finetuned models
+
+We provide models that were finetuned on microscopy data using `micro_sam.training`. They are hosted on zenodo. We currently offer the following models:
+- `vit_h`: Default Segment Anything model with vit-h backbone.
+- `vit_l`: Default Segment Anything model with vit-l backbone.
+- `vit_b`: Default Segment Anything model with vit-b backbone.
+- `vit_h_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-h backbone.
+- `vit_b_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-b backbone.
+- `vit_h_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-h backbone.
+- `vit_b_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-b backbone.
+
+See the two figures below of the improvements through the finetuned model for LM and EM data.
+
+ +
+
+
+ +
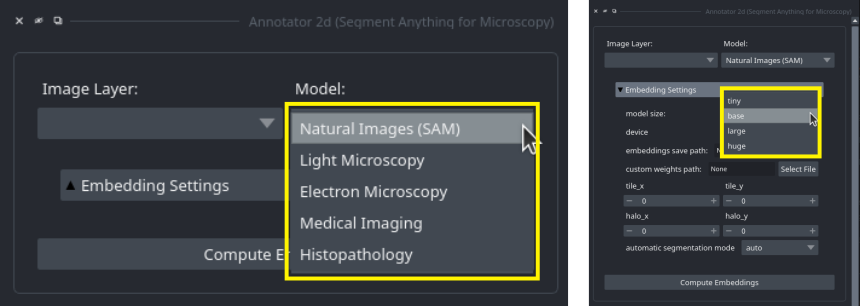
+You can select which of the models is used in the annotation tools by selecting the corresponding name from the `Model Type` menu:
+
+
+
+You can select which of the models is used in the annotation tools by selecting the corresponding name from the `Model Type` menu:
+
+ +
+To use a specific model in the python library you need to pass the corresponding name as value to the `model_type` parameter exposed by all relevant functions.
+See for example the [2d annotator example](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/annotator_2d.py#L62) where `use_finetuned_model` can be set to `True` to use the `vit_h_lm` model.
+
+## Which model should I choose?
+
+As a rule of thumb:
+- Use the `_lm` models for segmenting cells or nuclei in light microscopy.
+- Use the `_em` models for segmenting ceells or neurites in electron microscopy.
+ - Note that this model does not work well for segmenting mitochondria or other organelles becuase it is biased towards segmenting the full cell / cellular compartment.
+- For other cases use the default models.
+
+See also the figures above for examples where the finetuned models work better than the vanilla models.
+Currently the model `vit_h` is used by default.
+
+We are working on releasing more fine-tuned models, in particular for mitochondria and other organelles in EM.
diff --git a/doc/images/em_comparison.png b/doc/images/em_comparison.png
new file mode 100644
index 00000000..86b2d66c
Binary files /dev/null and b/doc/images/em_comparison.png differ
diff --git a/doc/images/lm_comparison.png b/doc/images/lm_comparison.png
new file mode 100644
index 00000000..4e115d61
Binary files /dev/null and b/doc/images/lm_comparison.png differ
diff --git a/doc/images/micro-sam-gui.png b/doc/images/micro-sam-gui.png
new file mode 100644
index 00000000..26254490
Binary files /dev/null and b/doc/images/micro-sam-gui.png differ
diff --git a/doc/images/model-type-selector.png b/doc/images/model-type-selector.png
new file mode 100644
index 00000000..cab3b077
Binary files /dev/null and b/doc/images/model-type-selector.png differ
diff --git a/doc/images/vanilla-v-finetuned.png b/doc/images/vanilla-v-finetuned.png
deleted file mode 100644
index a72446a4..00000000
Binary files a/doc/images/vanilla-v-finetuned.png and /dev/null differ
diff --git a/doc/installation.md b/doc/installation.md
index 12bda71e..0b49edc5 100644
--- a/doc/installation.md
+++ b/doc/installation.md
@@ -1,16 +1,38 @@
# Installation
-`micro_sam` requires the following dependencies:
+We provide three different ways of installing `micro_sam`:
+- [From conda](#from-conda) is the recommended way if you want to use all functionality.
+- [From source](#from-source) for setting up a development environment to change and potentially contribute to our software.
+- [From installer](#from-installer) to install without having to use conda. This mode of installation is still experimental! It only provides the annotation tools and does not enable model finetuning.
+
+Our software requires the following dependencies:
- [PyTorch](https://pytorch.org/get-started/locally/)
- [SegmentAnything](https://github.com/facebookresearch/segment-anything#installation)
-- [napari](https://napari.org/stable/)
- [elf](https://github.com/constantinpape/elf)
+- [napari](https://napari.org/stable/) (for the interactive annotation tools)
+- [torch_em](https://github.com/constantinpape/torch-em) (for the training functionality)
+
+## From conda
-It is available as a conda package and can be installed via
+`micro_sam` is available as a conda package and can be installed via
```
$ conda install -c conda-forge micro_sam
```
+This command will not install the required dependencies for the annotation tools and for training / finetuning.
+To use the annotation functionality you also need to install `napari`:
+```
+$ conda install -c conda-forge napari pyqt
+```
+And to use the training functionality `torch_em`:
+```
+$ conda install -c conda-forge torch_em
+```
+
+In case the installation via conda takes too long consider using [mamba](https://mamba.readthedocs.io/en/latest/).
+Once you have it installed you can simply replace the `conda` commands with `mamba`.
+
+
## From source
To install `micro_sam` from source, we recommend to first set up a conda environment with the necessary requirements:
@@ -54,3 +76,48 @@ $ pip install -e .
- Install `micro_sam` by running `pip install -e .` in this folder.
- **Note:** we have seen many issues with the pytorch installation on MAC. If a wrong pytorch version is installed for you (which will cause pytorch errors once you run the application) please try again with a clean `mambaforge` installation. Please install the `OS X, arm64` version from [here](https://github.com/conda-forge/miniforge#mambaforge).
- Some MACs require a specific installation order of packages. If the steps layed out above don't work for you please check out the procedure described [in this github issue](https://github.com/computational-cell-analytics/micro-sam/issues/77).
+
+
+## From installer
+
+We also provide installers for Linuxand Windows:
+- [Linux](https://owncloud.gwdg.de/index.php/s/Cw9RmA3BlyqKJeU)
+- [Windows](https://owncloud.gwdg.de/index.php/s/1iD1eIcMZvEyE6d)
+
+
+**The installers are stil experimental and not fully tested.** Mac is not supported yet, but we are working on also providing an installer for it.
+
+If you encounter problems with them then please consider installing `micro_sam` via [conda](#from-conda) instead.
+
+**Linux Installer:**
+
+To use the installer:
+- Unpack the zip file you have downloaded.
+- Make the installer executable: `$ chmod +x micro_sam-0.2.0post1-Linux-x86_64.sh`
+- Run the installer: `$./micro_sam-0.2.0post1-Linux-x86_64.sh$`
+ - You can select where to install `micro_sam` during the installation. By default it will be installed in `$HOME/micro_sam`.
+ - The installer will unpack all `micro_sam` files to the installation directory.
+- After the installation you can start the annotator with the command `.../micro_sam/bin/micro_sam.annotator`.
+ - To make it easier to run the annotation tool you can add `.../micro_sam/bin` to your `PATH` or set a softlink to `.../micro_sam/bin/micro_sam.annotator`.
+
+
+
+**Windows Installer:**
+
+- Unpack the zip file you have downloaded.
+- Run the installer by double clicking on it.
+- Choose installation type: `Just Me(recommended)` or `All Users(requires admin privileges)`.
+- Choose installation path. By default it will be installed in `C:\Users\\micro_sam` for `Just Me` installation or in `C:\ProgramData\micro_sam` for `All Users`.
+ - The installer will unpack all micro_sam files to the installation directory.
+- After the installation you can start the annotator by double clicking on `.\micro_sam\Scripts\micro_sam.annotator.exe` or with the command `.\micro_sam\Scripts\micro_sam.annotator.exe` from the Command Prompt.
diff --git a/doc/python_library.md b/doc/python_library.md
index a34b1966..dbab4b0a 100644
--- a/doc/python_library.md
+++ b/doc/python_library.md
@@ -5,24 +5,25 @@ The python library can be imported via
import micro_sam
```
-It implements functionality for running Segment Anything for 2d and 3d data, provides more instance segmentation functionality and several other helpful functions for using Segment Anything.
-This functionality is used to implement the `micro_sam` annotation tools, but you can also use it as a standalone python library. Check out the documentation under `Submodules` for more details on the python library.
+The library
+- implements function to apply Segment Anything to 2d and 3d data more conviently in `micro_sam.prompt_based_segmentation`.
+- provides more and imporoved automatic instance segmentation functionality in `micro_sam.instance_segmentation`.
+- implements training functionality that can be used for finetuning on your own data in `micro_sam.training`.
+- provides functionality for quantitative and qualitative evaluation of Segment Anything models in `micro_sam.evaluation`.
-## Finetuned models
+This functionality is used to implement the interactive annotation tools and can also be used as a standalone python library.
+Some preliminary examples for how to use the python library can be found [here](https://github.com/computational-cell-analytics/micro-sam/tree/master/examples/use_as_library). Check out the `Submodules` documentation for more details.
-We provide finetuned Segment Anything models for microscopy data. They are still in an experimental stage and we will upload more and better models soon, as well as the code for fine-tuning.
-For using the preliminary models, check out the [2d annotator example](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/annotator_2d.py#L62) and set `use_finetuned_model` to `True`.
+## Training your own model
-We currently provide support for the following models:
-- `vit_h`: The default Segment Anything model with vit-h backbone.
-- `vit_l`: The default Segment Anything model with vit-l backbone.
-- `vit_b`: The default Segment Anything model with vit-b backbone.
-- `vit_h_lm`: The preliminary finetuned Segment Anything model for light microscopy data with vit-h backbone.
-- `vit_b_lm`: The preliminary finetuned Segment Anything model for light microscopy data with vit-b backbone.
+We reimplement the training logic described in the [Segment Anything publication](https://arxiv.org/abs/2304.02643) to enable finetuning on custom data.
+We use this functionality to provide the [finetuned microscopy models](#finetuned-models) and it can also be used to finetune models on your own data.
+In fact the best results can be expected when finetuning on your own data, and we found that it does not require much annotated training data to get siginficant improvements in model performance.
+So a good strategy is to annotate a few images with one of the provided models using one of the interactive annotation tools and, if the annotation is not working as good as expected yet, finetune on the annotated data.
+
-These are also the valid names for the `model_type` parameter in `micro_sam`. The library will automatically choose and if necessary download the corresponding model.
-
-See the difference between the normal and finetuned Segment Anything ViT-h model on an image from [LiveCELL](https://sartorius-research.github.io/LIVECell/):
-
-
+
+To use a specific model in the python library you need to pass the corresponding name as value to the `model_type` parameter exposed by all relevant functions.
+See for example the [2d annotator example](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/annotator_2d.py#L62) where `use_finetuned_model` can be set to `True` to use the `vit_h_lm` model.
+
+## Which model should I choose?
+
+As a rule of thumb:
+- Use the `_lm` models for segmenting cells or nuclei in light microscopy.
+- Use the `_em` models for segmenting ceells or neurites in electron microscopy.
+ - Note that this model does not work well for segmenting mitochondria or other organelles becuase it is biased towards segmenting the full cell / cellular compartment.
+- For other cases use the default models.
+
+See also the figures above for examples where the finetuned models work better than the vanilla models.
+Currently the model `vit_h` is used by default.
+
+We are working on releasing more fine-tuned models, in particular for mitochondria and other organelles in EM.
diff --git a/doc/images/em_comparison.png b/doc/images/em_comparison.png
new file mode 100644
index 00000000..86b2d66c
Binary files /dev/null and b/doc/images/em_comparison.png differ
diff --git a/doc/images/lm_comparison.png b/doc/images/lm_comparison.png
new file mode 100644
index 00000000..4e115d61
Binary files /dev/null and b/doc/images/lm_comparison.png differ
diff --git a/doc/images/micro-sam-gui.png b/doc/images/micro-sam-gui.png
new file mode 100644
index 00000000..26254490
Binary files /dev/null and b/doc/images/micro-sam-gui.png differ
diff --git a/doc/images/model-type-selector.png b/doc/images/model-type-selector.png
new file mode 100644
index 00000000..cab3b077
Binary files /dev/null and b/doc/images/model-type-selector.png differ
diff --git a/doc/images/vanilla-v-finetuned.png b/doc/images/vanilla-v-finetuned.png
deleted file mode 100644
index a72446a4..00000000
Binary files a/doc/images/vanilla-v-finetuned.png and /dev/null differ
diff --git a/doc/installation.md b/doc/installation.md
index 12bda71e..0b49edc5 100644
--- a/doc/installation.md
+++ b/doc/installation.md
@@ -1,16 +1,38 @@
# Installation
-`micro_sam` requires the following dependencies:
+We provide three different ways of installing `micro_sam`:
+- [From conda](#from-conda) is the recommended way if you want to use all functionality.
+- [From source](#from-source) for setting up a development environment to change and potentially contribute to our software.
+- [From installer](#from-installer) to install without having to use conda. This mode of installation is still experimental! It only provides the annotation tools and does not enable model finetuning.
+
+Our software requires the following dependencies:
- [PyTorch](https://pytorch.org/get-started/locally/)
- [SegmentAnything](https://github.com/facebookresearch/segment-anything#installation)
-- [napari](https://napari.org/stable/)
- [elf](https://github.com/constantinpape/elf)
+- [napari](https://napari.org/stable/) (for the interactive annotation tools)
+- [torch_em](https://github.com/constantinpape/torch-em) (for the training functionality)
+
+## From conda
-It is available as a conda package and can be installed via
+`micro_sam` is available as a conda package and can be installed via
```
$ conda install -c conda-forge micro_sam
```
+This command will not install the required dependencies for the annotation tools and for training / finetuning.
+To use the annotation functionality you also need to install `napari`:
+```
+$ conda install -c conda-forge napari pyqt
+```
+And to use the training functionality `torch_em`:
+```
+$ conda install -c conda-forge torch_em
+```
+
+In case the installation via conda takes too long consider using [mamba](https://mamba.readthedocs.io/en/latest/).
+Once you have it installed you can simply replace the `conda` commands with `mamba`.
+
+
## From source
To install `micro_sam` from source, we recommend to first set up a conda environment with the necessary requirements:
@@ -54,3 +76,48 @@ $ pip install -e .
- Install `micro_sam` by running `pip install -e .` in this folder.
- **Note:** we have seen many issues with the pytorch installation on MAC. If a wrong pytorch version is installed for you (which will cause pytorch errors once you run the application) please try again with a clean `mambaforge` installation. Please install the `OS X, arm64` version from [here](https://github.com/conda-forge/miniforge#mambaforge).
- Some MACs require a specific installation order of packages. If the steps layed out above don't work for you please check out the procedure described [in this github issue](https://github.com/computational-cell-analytics/micro-sam/issues/77).
+
+
+## From installer
+
+We also provide installers for Linuxand Windows:
+- [Linux](https://owncloud.gwdg.de/index.php/s/Cw9RmA3BlyqKJeU)
+- [Windows](https://owncloud.gwdg.de/index.php/s/1iD1eIcMZvEyE6d)
+
+
+**The installers are stil experimental and not fully tested.** Mac is not supported yet, but we are working on also providing an installer for it.
+
+If you encounter problems with them then please consider installing `micro_sam` via [conda](#from-conda) instead.
+
+**Linux Installer:**
+
+To use the installer:
+- Unpack the zip file you have downloaded.
+- Make the installer executable: `$ chmod +x micro_sam-0.2.0post1-Linux-x86_64.sh`
+- Run the installer: `$./micro_sam-0.2.0post1-Linux-x86_64.sh$`
+ - You can select where to install `micro_sam` during the installation. By default it will be installed in `$HOME/micro_sam`.
+ - The installer will unpack all `micro_sam` files to the installation directory.
+- After the installation you can start the annotator with the command `.../micro_sam/bin/micro_sam.annotator`.
+ - To make it easier to run the annotation tool you can add `.../micro_sam/bin` to your `PATH` or set a softlink to `.../micro_sam/bin/micro_sam.annotator`.
+
+
+
+**Windows Installer:**
+
+- Unpack the zip file you have downloaded.
+- Run the installer by double clicking on it.
+- Choose installation type: `Just Me(recommended)` or `All Users(requires admin privileges)`.
+- Choose installation path. By default it will be installed in `C:\Users\\micro_sam` for `Just Me` installation or in `C:\ProgramData\micro_sam` for `All Users`.
+ - The installer will unpack all micro_sam files to the installation directory.
+- After the installation you can start the annotator by double clicking on `.\micro_sam\Scripts\micro_sam.annotator.exe` or with the command `.\micro_sam\Scripts\micro_sam.annotator.exe` from the Command Prompt.
diff --git a/doc/python_library.md b/doc/python_library.md
index a34b1966..dbab4b0a 100644
--- a/doc/python_library.md
+++ b/doc/python_library.md
@@ -5,24 +5,25 @@ The python library can be imported via
import micro_sam
```
-It implements functionality for running Segment Anything for 2d and 3d data, provides more instance segmentation functionality and several other helpful functions for using Segment Anything.
-This functionality is used to implement the `micro_sam` annotation tools, but you can also use it as a standalone python library. Check out the documentation under `Submodules` for more details on the python library.
+The library
+- implements function to apply Segment Anything to 2d and 3d data more conviently in `micro_sam.prompt_based_segmentation`.
+- provides more and imporoved automatic instance segmentation functionality in `micro_sam.instance_segmentation`.
+- implements training functionality that can be used for finetuning on your own data in `micro_sam.training`.
+- provides functionality for quantitative and qualitative evaluation of Segment Anything models in `micro_sam.evaluation`.
-## Finetuned models
+This functionality is used to implement the interactive annotation tools and can also be used as a standalone python library.
+Some preliminary examples for how to use the python library can be found [here](https://github.com/computational-cell-analytics/micro-sam/tree/master/examples/use_as_library). Check out the `Submodules` documentation for more details.
-We provide finetuned Segment Anything models for microscopy data. They are still in an experimental stage and we will upload more and better models soon, as well as the code for fine-tuning.
-For using the preliminary models, check out the [2d annotator example](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/annotator_2d.py#L62) and set `use_finetuned_model` to `True`.
+## Training your own model
-We currently provide support for the following models:
-- `vit_h`: The default Segment Anything model with vit-h backbone.
-- `vit_l`: The default Segment Anything model with vit-l backbone.
-- `vit_b`: The default Segment Anything model with vit-b backbone.
-- `vit_h_lm`: The preliminary finetuned Segment Anything model for light microscopy data with vit-h backbone.
-- `vit_b_lm`: The preliminary finetuned Segment Anything model for light microscopy data with vit-b backbone.
+We reimplement the training logic described in the [Segment Anything publication](https://arxiv.org/abs/2304.02643) to enable finetuning on custom data.
+We use this functionality to provide the [finetuned microscopy models](#finetuned-models) and it can also be used to finetune models on your own data.
+In fact the best results can be expected when finetuning on your own data, and we found that it does not require much annotated training data to get siginficant improvements in model performance.
+So a good strategy is to annotate a few images with one of the provided models using one of the interactive annotation tools and, if the annotation is not working as good as expected yet, finetune on the annotated data.
+
-These are also the valid names for the `model_type` parameter in `micro_sam`. The library will automatically choose and if necessary download the corresponding model.
-
-See the difference between the normal and finetuned Segment Anything ViT-h model on an image from [LiveCELL](https://sartorius-research.github.io/LIVECell/):
-
- +The training logic is implemented in `micro_sam.training` and is based on [torch-em](https://github.com/constantinpape/torch-em). Please check out [examples/finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/examples/finetuning) to see how you can finetune on your own data with it. The script `finetune_hela.py` contains an example for finetuning on a small custom microscopy dataset and `use_finetuned_model.py` shows how this model can then be used in the interactive annotatin tools.
+More advanced examples, including quantitative and qualitative evaluation, of finetuned models can be found in [finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/finetuning), which contains the code for training and evaluating our microscopy models.
diff --git a/doc/start_page.md b/doc/start_page.md
index b5fab2a9..840a650b 100644
--- a/doc/start_page.md
+++ b/doc/start_page.md
@@ -2,7 +2,7 @@
Segment Anything for Microscopy implements automatic and interactive annotation for microscopy data. It is built on top of [Segment Anything](https://segment-anything.com/) by Meta AI and specializes it for microscopy and other bio-imaging data.
Its core components are:
-- The `micro_sam` annotator tools for interactive data annotation with [napari](https://napari.org/stable/).
+- The `micro_sam` tools for interactive data annotation with [napari](https://napari.org/stable/).
- The `micro_sam` library to apply Segment Anything to 2d and 3d data or fine-tune it on your data.
- The `micro_sam` models that are fine-tuned on publicly available microscopy data.
@@ -19,20 +19,18 @@ On our roadmap for more functionality are:
If you run into any problems or have questions please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
-
## Quickstart
You can install `micro_sam` via conda:
```
-$ conda install -c conda-forge micro_sam
+$ conda install -c conda-forge micro_sam napari pyqt
```
-For more installation options check out [Installation](#installation)
+We also provide experimental installers for all operating systems.
+For more details on the available installation options check out [the installation section](#installation).
After installing `micro_sam` you can run the annotation tool via `$ micro_sam.annotator`, which opens a menu for selecting the annotation tool and its inputs.
-See [Annotation Tools](#annotation-tools) for an overview and explanation of the annotation functionality.
+See [the annotation tool section](#annotation-tools) for an overview and explanation of the annotation functionality.
The `micro_sam` python library can be used via
```python
@@ -47,7 +45,5 @@ For now, check out [the example script for the 2d annotator](https://github.com/
## Citation
If you are using `micro_sam` in your research please cite
-- [SegmentAnything](https://arxiv.org/abs/2304.02643)
-- and our repository on [zenodo](https://doi.org/10.5281/zenodo.7919746)
-
-We will release a preprint soon that describes this work and can be cited instead of zenodo.
+- Our [preprint](https://doi.org/10.1101/2023.08.21.554208)
+- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643)
diff --git a/environment_cpu.yaml b/environment_cpu.yaml
index 4d5f8ea7..ad91dab9 100644
--- a/environment_cpu.yaml
+++ b/environment_cpu.yaml
@@ -11,6 +11,7 @@ dependencies:
- pytorch
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/environment_gpu.yaml b/environment_gpu.yaml
index 21d5df53..57700759 100644
--- a/environment_gpu.yaml
+++ b/environment_gpu.yaml
@@ -12,6 +12,7 @@ dependencies:
- pytorch-cuda>=11.7 # you may need to update the cuda version to match your system
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/examples/README.md b/examples/README.md
index 87b3a963..4844167f 100644
--- a/examples/README.md
+++ b/examples/README.md
@@ -6,5 +6,8 @@ Examples for using the micro_sam annotation tools:
- `annotator_tracking.py`: run the interactive tracking annotation tool
- `image_series_annotator.py`: run the annotation tool for a series of images
+The folder `finetuning` contains example scripts that show how a Segment Anything model can be fine-tuned
+on custom data with the `micro_sam.train` library, and how the finetuned models can then be used within the annotatin tools.
+
The folder `use_as_library` contains example scripts that show how `micro_sam` can be used as a python
-library to apply Segment Anything on mult-dimensional data.
+library to apply Segment Anything to mult-dimensional data.
diff --git a/examples/annotator_2d.py b/examples/annotator_2d.py
index f0d6d44b..8f4930f2 100644
--- a/examples/annotator_2d.py
+++ b/examples/annotator_2d.py
@@ -34,7 +34,7 @@ def hela_2d_annotator(use_finetuned_model):
embedding_path = "./embeddings/embeddings-hela2d.zarr"
model_type = "vit_h"
- annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type)
+ annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type, precompute_amg_state=True)
def wholeslide_annotator(use_finetuned_model):
diff --git a/examples/annotator_with_custom_model.py b/examples/annotator_with_custom_model.py
new file mode 100644
index 00000000..ceb8b2cb
--- /dev/null
+++ b/examples/annotator_with_custom_model.py
@@ -0,0 +1,23 @@
+import h5py
+import micro_sam.sam_annotator as annotator
+from micro_sam.util import get_sam_model
+
+# TODO add an example for the 2d annotator with a custom model
+
+
+def annotator_3d_with_custom_model():
+ with h5py.File("./data/gut1_block_1.h5") as f:
+ raw = f["raw"][:]
+
+ custom_model = "/home/pape/Work/data/models/sam/user-study/vit_h_nuclei_em_finetuned.pt"
+ embedding_path = "./embeddings/nuclei3d-custom-vit-h.zarr"
+ predictor = get_sam_model(checkpoint_path=custom_model, model_type="vit_h")
+ annotator.annotator_3d(raw, embedding_path, predictor=predictor)
+
+
+def main():
+ annotator_3d_with_custom_model()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/finetuning/.gitignore b/examples/finetuning/.gitignore
new file mode 100644
index 00000000..ec28c5ea
--- /dev/null
+++ b/examples/finetuning/.gitignore
@@ -0,0 +1,2 @@
+checkpoints/
+logs/
diff --git a/examples/finetuning/finetune_hela.py b/examples/finetuning/finetune_hela.py
new file mode 100644
index 00000000..f83bd8cb
--- /dev/null
+++ b/examples/finetuning/finetune_hela.py
@@ -0,0 +1,140 @@
+import os
+
+import numpy as np
+import torch
+import torch_em
+
+import micro_sam.training as sam_training
+from micro_sam.sample_data import fetch_tracking_example_data, fetch_tracking_segmentation_data
+from micro_sam.util import export_custom_sam_model
+
+DATA_FOLDER = "data"
+
+
+def get_dataloader(split, patch_shape, batch_size):
+ """Return train or val data loader for finetuning SAM.
+
+ The data loader must be a torch data loader that retuns `x, y` tensors,
+ where `x` is the image data and `y` are the labels.
+ The labels have to be in a label mask instance segmentation format.
+ I.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
+ Important: the ID 0 is reseved for background, and the IDs must be consecutive
+
+ Here, we use `torch_em.default_segmentation_loader` for creating a suitable data loader from
+ the example hela data. You can either adapt this for your own data (see comments below)
+ or write a suitable torch dataloader yourself.
+ """
+ assert split in ("train", "val")
+ os.makedirs(DATA_FOLDER, exist_ok=True)
+
+ # This will download the image and segmentation data for training.
+ image_dir = fetch_tracking_example_data(DATA_FOLDER)
+ segmentation_dir = fetch_tracking_segmentation_data(DATA_FOLDER)
+
+ # torch_em.default_segmentation_loader is a convenience function to build a torch dataloader
+ # from image data and labels for training segmentation models.
+ # It supports image data in various formats. Here, we load image data and labels from the two
+ # folders with tif images that were downloaded by the example data functionality, by specifying
+ # `raw_key` and `label_key` as `*.tif`. This means all images in the respective folders that end with
+ # .tif will be loadded.
+ # The function supports many other file formats. For example, if you have tif stacks with multiple slices
+ # instead of multiple tif images in a foldder, then you can pass raw_key=label_key=None.
+
+ # Load images from multiple files in folder via pattern (here: all tif files)
+ raw_key, label_key = "*.tif", "*.tif"
+ # Alternative: if you have tif stacks you can just set raw_key and label_key to None
+ # raw_key, label_key= None, None
+
+ # The 'roi' argument can be used to subselect parts of the data.
+ # Here, we use it to select the first 70 frames fro the test split and the other frames for the val split.
+ if split == "train":
+ roi = np.s_[:70, :, :]
+ else:
+ roi = np.s_[70:, :, :]
+
+ loader = torch_em.default_segmentation_loader(
+ raw_paths=image_dir, raw_key=raw_key,
+ label_paths=segmentation_dir, label_key=label_key,
+ patch_shape=patch_shape, batch_size=batch_size,
+ ndim=2, is_seg_dataset=True, rois=roi,
+ label_transform=torch_em.transform.label.connected_components,
+ )
+ return loader
+
+
+def run_training(checkpoint_name, model_type):
+ """Run the actual model training."""
+
+ # All hyperparameters for training.
+ batch_size = 1 # the training batch size
+ patch_shape = (1, 512, 512) # the size of patches for training
+ n_objects_per_batch = 25 # the number of objects per batch that will be sampled
+ device = torch.device("cuda") # the device/GPU used for training
+ n_iterations = 10000 # how long we train (in iterations)
+
+ # Get the dataloaders.
+ train_loader = get_dataloader("train", patch_shape, batch_size)
+ val_loader = get_dataloader("val", patch_shape, batch_size)
+
+ # Get the segment anything model, the optimizer and the LR scheduler
+ model = sam_training.get_trainable_sam_model(model_type=model_type, device=device)
+ optimizer = torch.optim.Adam(model.parameters(), lr=1e-5)
+ scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode="min", factor=0.9, patience=10, verbose=True)
+
+ # This class creates all the training data for a batch (inputs, prompts and labels).
+ convert_inputs = sam_training.ConvertToSamInputs()
+
+ # the trainer which performs training and validation (implemented using "torch_em")
+ trainer = sam_training.SamTrainer(
+ name=checkpoint_name,
+ train_loader=train_loader,
+ val_loader=val_loader,
+ model=model,
+ optimizer=optimizer,
+ # currently we compute loss batch-wise, else we pass channelwise True
+ loss=torch_em.loss.DiceLoss(channelwise=False),
+ metric=torch_em.loss.DiceLoss(),
+ device=device,
+ lr_scheduler=scheduler,
+ logger=sam_training.SamLogger,
+ log_image_interval=10,
+ mixed_precision=True,
+ convert_inputs=convert_inputs,
+ n_objects_per_batch=n_objects_per_batch,
+ n_sub_iteration=8,
+ compile_model=False
+ )
+ trainer.fit(n_iterations)
+
+
+def export_model(checkpoint_name, model_type):
+ """Export the trained model."""
+ # export the model after training so that it can be used by the rest of the micro_sam library
+ export_path = "./finetuned_hela_model.pth"
+ checkpoint_path = os.path.join("checkpoints", checkpoint_name, "best.pt")
+ export_custom_sam_model(
+ checkpoint_path=checkpoint_path,
+ model_type=model_type,
+ save_path=export_path,
+ )
+
+
+def main():
+ """Finetune a Segment Anything model.
+
+ This example uses image data and segmentations from the cell tracking challenge,
+ but can easily be adapted for other data (including data you have annoated with micro_sam beforehand).
+ """
+ # The model_type determines which base model is used to initialize the weights that are finetuned.
+ # We use vit_b here because it can be trained faster. Note that vit_h usually yields higher quality results.
+ model_type = "vit_b"
+
+ # The name of the checkpoint. The checkpoints will be stored in './checkpoints/'
+ checkpoint_name = "sam_hela"
+
+ run_training(checkpoint_name, model_type)
+ export_model(checkpoint_name, model_type)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/finetuning/use_finetuned_model.py b/examples/finetuning/use_finetuned_model.py
new file mode 100644
index 00000000..19600241
--- /dev/null
+++ b/examples/finetuning/use_finetuned_model.py
@@ -0,0 +1,33 @@
+import imageio.v3 as imageio
+
+import micro_sam.util as util
+from micro_sam.sam_annotator import annotator_2d
+
+
+def run_annotator_with_custom_model():
+ """Run the 2d anntator with a custom (finetuned) model.
+
+ Here, we use the model that is produced by `finetuned_hela.py` and apply it
+ for an image from the validation set.
+ """
+ # take the last frame, which is part of the val set, so the model was not directly trained on it
+ im = imageio.imread("./data/DIC-C2DH-HeLa.zip.unzip/DIC-C2DH-HeLa/01/t083.tif")
+
+ # set the checkpoint and the path for caching the embeddings
+ checkpoint = "./finetuned_hela_model.pth"
+ embedding_path = "./embeddings/embeddings-finetuned.zarr"
+
+ model_type = "vit_b" # We finetune a vit_b in the example script.
+ # Adapt this if you finetune a different model type, e.g. vit_h.
+
+ # Load the custom model.
+ predictor = util.get_sam_model(model_type=model_type, checkpoint_path=checkpoint)
+
+ # Run the 2d annotator with the custom model.
+ annotator_2d(
+ im, embedding_path=embedding_path, predictor=predictor, precompute_amg_state=True,
+ )
+
+
+if __name__ == "__main__":
+ run_annotator_with_custom_model()
diff --git a/examples/image_series_annotator.py b/examples/image_series_annotator.py
index 2a1b65e7..1632d793 100644
--- a/examples/image_series_annotator.py
+++ b/examples/image_series_annotator.py
@@ -16,7 +16,8 @@ def series_annotation(use_finetuned_model):
example_data = fetch_image_series_example_data("./data")
image_folder_annotator(
example_data, "./data/series-segmentation-result", embedding_path=embedding_path,
- pattern="*.tif", model_type=model_type
+ pattern="*.tif", model_type=model_type,
+ precompute_amg_state=True,
)
diff --git a/examples/use_as_library/instance_segmentation.py b/examples/use_as_library/instance_segmentation.py
index 89b95900..be9300d3 100644
--- a/examples/use_as_library/instance_segmentation.py
+++ b/examples/use_as_library/instance_segmentation.py
@@ -50,7 +50,8 @@ def cell_segmentation():
# Generate the instance segmentation. You can call this again for different values of 'pred_iou_thresh'
# without having to call initialize again.
- # NOTE: the main advantage of this method is that it's considerably faster than the original implementation.
+ # NOTE: the main advantage of this method is that it's faster than the original implementation,
+ # however the quality is not as high as the original instance segmentation quality yet.
instances_mws = amg_mws.generate(pred_iou_thresh=0.88)
instances_mws = instance_segmentation.mask_data_to_segmentation(
instances_mws, shape=image.shape, with_background=True
@@ -64,7 +65,7 @@ def cell_segmentation():
napari.run()
-def segmentation_with_tiling():
+def cell_segmentation_with_tiling():
"""Run the instance segmentation functionality from micro_sam for segmentation of
cells in a large image. You need to run examples/annotator_2d.py:wholeslide_annotator once before
running this script so that all required data is downloaded and pre-computed.
@@ -111,20 +112,21 @@ def segmentation_with_tiling():
# Generate the instance segmentation. You can call this again for different values of 'pred_iou_thresh'
# without having to call initialize again.
- # NOTE: the main advantage of this method is that it's considerably faster than the original implementation.
+ # NOTE: the main advantage of this method is that it's faster than the original implementation.
+ # however the quality is not as high as the original instance segmentation quality yet.
instances_mws = amg_mws.generate(pred_iou_thresh=0.88)
# Show the results.
v = napari.Viewer()
v.add_image(image)
- # v.add_labels(instances_amg)
+ v.add_labels(instances_amg)
v.add_labels(instances_mws)
napari.run()
def main():
cell_segmentation()
- # segmentation_with_tiling()
+ # cell_segmentation_with_tiling()
if __name__ == "__main__":
diff --git a/finetuning/.gitignore b/finetuning/.gitignore
new file mode 100644
index 00000000..60fd41c2
--- /dev/null
+++ b/finetuning/.gitignore
@@ -0,0 +1,4 @@
+checkpoints/
+logs/
+sam_embeddings/
+results/
diff --git a/finetuning/README.md b/finetuning/README.md
new file mode 100644
index 00000000..6164837d
--- /dev/null
+++ b/finetuning/README.md
@@ -0,0 +1,58 @@
+# Segment Anything Finetuning
+
+Code for finetuning segment anything data on microscopy data and evaluating the finetuned models.
+
+## Example: LiveCELL
+
+**Finetuning**
+
+Run the script `livecell_finetuning.py` for fine-tuning a model on LiveCELL.
+
+**Inference**
+
+The script `livecell_inference.py` can be used to run inference on the test set. It supports different arguments for inference with different configurations.
+For example run
+```
+python livecell_inference.py -c checkpoints/livecell_sam/best.pt -m vit_b -e experiment -i /scratch/projects/nim00007/data/LiveCELL --points --positive 1 --negative 0
+```
+for inference with 1 positive point prompt and no negative point prompt (the prompts are derived from ground-truth).
+
+The arguments `-c`, `-e` and `-i` specify where the checkpoint for the model is, where the predictions from the model and other experiment data will be saved, and where the input dataset (LiveCELL) is stored.
+
+To run the default set of experiments from our publication use the command
+```
+python livecell_inference.py -c checkpoints/livecell_sam/best.pt -m vit_b -e experiment -i /scratch/projects/nim00007/data/LiveCELL -d --prompt_folder /scratch/projects/nim00007/sam/experiments/prompts/livecell
+```
+
+Here `-d` automatically runs the evaluation for these settings:
+- `--points --positive 1 --negative 0` (using point prompts with a single positive point)
+- `--points --positive 2 --negative 4` (using point prompts with two positive points and four negative points)
+- `--points --positive 4 --negative 8` (using point prompts with four positive points and eight negative points)
+- `--box` (using box prompts)
+
+In addition `--prompt_folder` specifies a folder with precomputed prompts. Using pre-computed prompts significantly speeds up the experiments and enables running them in a reproducible manner. (Without it the prompts will be recalculated each time.)
+
+You can also evaluate the automatic instance segmentation functionality, by running
+```
+python livecell_inference.py -c checkpoints/livecell_sam/best.pt -m vit_b -e experiment -i /scratch/projects/nim00007/data/LiveCELL -a
+```
+
+This will first perform a grid-search for the best parameters on a subset of the validation set and then run inference on the test set. This can take up to a day.
+
+**Evaluation**
+
+The script `livecell_evaluation.py` can then be used to evaluate the results from the inference runs.
+E.g. run the script like below to evaluate the previous predictions.
+```
+python livecell_evaluation.py -i /scratch/projects/nim00007/data/LiveCELL -e experiment
+```
+This will create a folder `experiment/results` with csv tables with the results per cell type and averaged over all images.
+
+
+## Finetuning and evaluation code
+
+The subfolders contain the code for different finetuning and evaluation experiments for microscopy data:
+- `livecell`: TODO
+- `generalist`: TODO
+
+Note: we still need to clean up most of this code and will add it later.
diff --git a/finetuning/generalists/cellpose_baseline.py b/finetuning/generalists/cellpose_baseline.py
new file mode 100644
index 00000000..f1a294b0
--- /dev/null
+++ b/finetuning/generalists/cellpose_baseline.py
@@ -0,0 +1,124 @@
+import argparse
+import os
+from glob import glob
+from subprocess import run
+
+import imageio.v3 as imageio
+
+from tqdm import tqdm
+
+DATA_ROOT = "/scratch/projects/nim00007/sam/datasets"
+EXP_ROOT = "/scratch/projects/nim00007/sam/experiments/cellpose"
+
+DATASETS = (
+ "covid-if",
+ "deepbacs",
+ "hpa",
+ "livecell",
+ "lizard",
+ "mouse-embryo",
+ "plantseg-ovules",
+ "plantseg-root",
+ "tissuenet",
+)
+
+
+def load_cellpose_model():
+ from cellpose import models
+
+ device, gpu = models.assign_device(True, True)
+ model = models.Cellpose(gpu=gpu, model_type="cyto", device=device)
+ return model
+
+
+def run_cellpose_segmentation(datasets, job_id):
+ dataset = datasets[job_id]
+ experiment_folder = os.path.join(EXP_ROOT, dataset)
+
+ prediction_folder = os.path.join(experiment_folder, "predictions")
+ os.makedirs(prediction_folder, exist_ok=True)
+
+ image_paths = sorted(glob(os.path.join(DATA_ROOT, dataset, "test", "image*.tif")))
+ model = load_cellpose_model()
+
+ for path in tqdm(image_paths, desc=f"Segmenting {dataset} with cellpose"):
+ fname = os.path.basename(path)
+ out_path = os.path.join(prediction_folder, fname)
+ if os.path.exists(out_path):
+ continue
+ image = imageio.imread(path)
+ if image.ndim == 3:
+ assert image.shape[-1] == 3
+ image = image.mean(axis=-1)
+ assert image.ndim == 2
+ seg = model.eval(image, diameter=None, flow_threshold=None, channels=[0, 0])[0]
+ assert seg.shape == image.shape
+ imageio.imwrite(out_path, seg, compression=5)
+
+
+def submit_array_job(datasets):
+ n_datasets = len(datasets)
+ cmd = ["sbatch", "-a", f"0-{n_datasets-1}", "cellpose_baseline.sbatch"]
+ run(cmd)
+
+
+def evaluate_dataset(dataset):
+ from micro_sam.evaluation.evaluation import run_evaluation
+
+ gt_paths = sorted(glob(os.path.join(DATA_ROOT, dataset, "test", "label*.tif")))
+ experiment_folder = os.path.join(EXP_ROOT, dataset)
+ pred_paths = sorted(glob(os.path.join(experiment_folder, "predictions", "*.tif")))
+ assert len(gt_paths) == len(pred_paths), f"{len(gt_paths)}, {len(pred_paths)}"
+ result_path = os.path.join(experiment_folder, "cellpose.csv")
+ run_evaluation(gt_paths, pred_paths, result_path)
+
+
+def evaluate_segmentations(datasets):
+ for dataset in datasets:
+ # we skip livecell, which has already been processed by cellpose
+ if dataset == "livecell":
+ continue
+ evaluate_dataset(dataset)
+
+
+def check_results(datasets):
+ for ds in datasets:
+ # we skip livecell, which has already been processed by cellpose
+ if ds == "livecell":
+ continue
+ result_path = os.path.join(EXP_ROOT, ds, "cellpose.csv")
+ if not os.path.exists(result_path):
+ print("Cellpose results missing for", ds)
+ print("All checks passed")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("--segment", "-s", action="store_true")
+ parser.add_argument("--evaluate", "-e", action="store_true")
+ parser.add_argument("--check", "-c", action="store_true")
+ parser.add_argument("--datasets", nargs="+")
+ args = parser.parse_args()
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+
+ if args.datasets is None:
+ datasets = DATASETS
+ else:
+ datasets = args.datasets
+ assert all(ds in DATASETS for ds in datasets)
+
+ if job_id is not None:
+ run_cellpose_segmentation(datasets, int(job_id))
+ elif args.segment:
+ submit_array_job(datasets)
+ elif args.evaluate:
+ evaluate_segmentations(datasets)
+ elif args.check:
+ check_results(datasets)
+ else:
+ raise ValueError("Doing nothing")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/cellpose_baseline.sbatch b/finetuning/generalists/cellpose_baseline.sbatch
new file mode 100755
index 00000000..c839abab
--- /dev/null
+++ b/finetuning/generalists/cellpose_baseline.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 300
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate cellpose
+python cellpose_baseline.py $@
diff --git a/finetuning/generalists/compile_results.py b/finetuning/generalists/compile_results.py
new file mode 100644
index 00000000..06165a75
--- /dev/null
+++ b/finetuning/generalists/compile_results.py
@@ -0,0 +1,104 @@
+import os
+from glob import glob
+
+import pandas as pd
+
+from evaluate_generalist import EXPERIMENT_ROOT
+from util import EM_DATASETS, LM_DATASETS
+
+
+def get_results(model, ds):
+ res_folder = os.path.join(EXPERIMENT_ROOT, model, ds, "results")
+ res_paths = sorted(glob(os.path.join(res_folder, "box", "*.csv"))) +\
+ sorted(glob(os.path.join(res_folder, "points", "*.csv")))
+
+ amg_res = os.path.join(res_folder, "amg.csv")
+ if os.path.exists(amg_res):
+ res_paths.append(amg_res)
+
+ results = []
+ for path in res_paths:
+ prompt_res = pd.read_csv(path)
+ prompt_name = os.path.splitext(os.path.relpath(path, res_folder))[0]
+ prompt_res.insert(0, "prompt", [prompt_name])
+ results.append(prompt_res)
+ results = pd.concat(results)
+ results.insert(0, "dataset", results.shape[0] * [ds])
+
+ return results
+
+
+def compile_results(models, datasets, out_path, load_results=False):
+ results = []
+
+ for model in models:
+ model_results = []
+
+ for ds in datasets:
+ ds_results = get_results(model, ds)
+ model_results.append(ds_results)
+
+ model_results = pd.concat(model_results)
+ model_results.insert(0, "model", [model] * model_results.shape[0])
+ results.append(model_results)
+
+ results = pd.concat(results)
+ if load_results:
+ assert os.path.exists(out_path)
+ all_results = pd.read_csv(out_path)
+ results = pd.concat([all_results, results])
+
+ results.to_csv(out_path, index=False)
+
+
+def compile_em():
+ compile_results(
+ ["vit_h", "vit_h_em", "vit_b", "vit_b_em"],
+ EM_DATASETS,
+ os.path.join(EXPERIMENT_ROOT, "evaluation-em.csv")
+ )
+
+
+def add_cellpose_results(datasets, out_path):
+ cp_root = "/scratch/projects/nim00007/sam/experiments/cellpose"
+
+ results = []
+ for dataset in datasets:
+ if dataset == "livecell":
+ continue
+ res_path = os.path.join(cp_root, dataset, "cellpose.csv")
+ ds_res = pd.read_csv(res_path)
+ ds_res.insert(0, "prompt", ["cellpose"] * ds_res.shape[0])
+ ds_res.insert(0, "dataset", [dataset] * ds_res.shape[0])
+ results.append(ds_res)
+
+ results = pd.concat(results)
+ results.insert(0, "model", ["cellpose"] * results.shape[0])
+
+ all_results = pd.read_csv(out_path)
+ results = pd.concat([all_results, results])
+ results.to_csv(out_path, index=False)
+
+
+def compile_lm():
+ res_path = os.path.join(EXPERIMENT_ROOT, "evaluation-lm.csv")
+ compile_results(
+ ["vit_h", "vit_h_lm", "vit_b", "vit_b_lm"], LM_DATASETS, res_path
+ )
+
+ # add the deepbacs and tissuenet specialist results
+ assert os.path.exists(res_path)
+ compile_results(["vit_h_tissuenet", "vit_b_tissuenet"], ["tissuenet"], res_path, True)
+ compile_results(["vit_h_deepbacs", "vit_b_deepbacs"], ["deepbacs"], res_path, True)
+
+ # add the cellpose results
+ add_cellpose_results(LM_DATASETS, res_path)
+
+
+def main():
+ # compile_em()
+ compile_lm()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/create_tissuenet_data.py b/finetuning/generalists/create_tissuenet_data.py
new file mode 100644
index 00000000..18114973
--- /dev/null
+++ b/finetuning/generalists/create_tissuenet_data.py
@@ -0,0 +1,63 @@
+
+import os
+from tqdm import tqdm
+import imageio.v2 as imageio
+import numpy as np
+
+from torch_em.data import MinInstanceSampler
+from torch_em.transform.label import label_consecutive
+from torch_em.data.datasets import get_tissuenet_loader
+from torch_em.transform.raw import standardize, normalize_percentile
+
+
+def rgb_to_gray_transform(raw):
+ raw = normalize_percentile(raw, axis=(1, 2))
+ raw = np.mean(raw, axis=0)
+ raw = standardize(raw)
+ return raw
+
+
+def get_tissuenet_loaders(input_path):
+ sampler = MinInstanceSampler()
+ label_transform = label_consecutive
+ raw_transform = rgb_to_gray_transform
+ val_loader = get_tissuenet_loader(path=input_path, split="val", raw_channel="rgb", label_channel="cell",
+ batch_size=1, patch_shape=(256, 256), num_workers=0,
+ sampler=sampler, label_transform=label_transform, raw_transform=raw_transform)
+ test_loader = get_tissuenet_loader(path=input_path, split="test", raw_channel="rgb", label_channel="cell",
+ batch_size=1, patch_shape=(256, 256), num_workers=0,
+ sampler=sampler, label_transform=label_transform, raw_transform=raw_transform)
+ return val_loader, test_loader
+
+
+def extract_images(loader, out_folder):
+ os.makedirs(out_folder, exist_ok=True)
+ for i, (x, y) in tqdm(enumerate(loader), total=len(loader)):
+ img_path = os.path.join(out_folder, "image_{:04d}.tif".format(i))
+ gt_path = os.path.join(out_folder, "label_{:04d}.tif".format(i))
+
+ img = x.squeeze().detach().cpu().numpy()
+ gt = y.squeeze().detach().cpu().numpy()
+
+ imageio.imwrite(img_path, img)
+ imageio.imwrite(gt_path, gt)
+
+
+def main():
+ val_loader, test_loader = get_tissuenet_loaders("/scratch-grete/projects/nim00007/data/tissuenet")
+ print("Length of val loader is:", len(val_loader))
+ print("Length of test loader is:", len(test_loader))
+
+ root_save_dir = "/scratch/projects/nim00007/sam/datasets/tissuenet"
+
+ # we use the val set for test because there are some issues with the test set
+ # out_folder = os.path.join(root_save_dir, "test")
+ # extract_images(val_loader, out_folder)
+
+ # we use the test folder for val and just use as many images as we can sample
+ out_folder = os.path.join(root_save_dir, "val")
+ extract_images(test_loader, out_folder)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/evaluate_generalist.py b/finetuning/generalists/evaluate_generalist.py
new file mode 100644
index 00000000..af217c0e
--- /dev/null
+++ b/finetuning/generalists/evaluate_generalist.py
@@ -0,0 +1,109 @@
+import argparse
+import os
+from subprocess import run
+
+from util import evaluate_checkpoint_for_dataset, ALL_DATASETS, EM_DATASETS, LM_DATASETS
+from micro_sam.evaluation import default_experiment_settings, get_experiment_setting_name
+
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/generalists"
+CHECKPOINTS = {
+ # Vanilla models
+ "vit_b": "/home/nimcpape/.sam_models/sam_vit_b_01ec64.pth",
+ "vit_h": "/home/nimcpape/.sam_models/sam_vit_h_4b8939.pth",
+ # Generalist LM models
+ "vit_b_lm": "/scratch/projects/nim00007/sam/models/LM/generalist/v2/vit_b/best.pt",
+ "vit_h_lm": "/scratch/projects/nim00007/sam/models/LM/generalist/v2/vit_h/best.pt",
+ # Generalist EM models
+ "vit_b_em": "/scratch/projects/nim00007/sam/models/EM/generalist/v2/vit_b/best.pt",
+ "vit_h_em": "/scratch/projects/nim00007/sam/models/EM/generalist/v2/vit_h/best.pt",
+ # Specialist Models (we don't add livecell, because these results are all computed already)
+ "vit_b_tissuenet": "/scratch/projects/nim00007/sam/models/LM/TissueNet/vit_b/best.pt",
+ "vit_h_tissuenet": "/scratch/projects/nim00007/sam/models/LM/TissueNet/vit_h/best.pt",
+ "vit_b_deepbacs": "/scratch/projects/nim00007/sam/models/LM/DeepBacs/vit_b/best.pt",
+ "vit_h_deepbacs": "/scratch/projects/nim00007/sam/models/LM/DeepBacs/vit_h/best.pt",
+}
+
+
+def submit_array_job(model_name, datasets):
+ n_datasets = len(datasets)
+ cmd = ["sbatch", "-a", f"0-{n_datasets-1}", "evaluate_generalist.sbatch", model_name, "--datasets"]
+ cmd.extend(datasets)
+ run(cmd)
+
+
+def evaluate_dataset_slurm(model_name, dataset):
+ if dataset in EM_DATASETS:
+ run_amg = False
+ max_num_val_images = None
+ else:
+ run_amg = True
+ max_num_val_images = 64
+
+ is_custom_model = model_name not in ("vit_h", "vit_b")
+ checkpoint = CHECKPOINTS[model_name]
+ model_type = model_name[:5]
+
+ experiment_folder = os.path.join(EXPERIMENT_ROOT, model_name, dataset)
+ evaluate_checkpoint_for_dataset(
+ checkpoint, model_type, dataset, experiment_folder,
+ run_default_evaluation=True, run_amg=run_amg,
+ is_custom_model=is_custom_model,
+ max_num_val_images=max_num_val_images,
+ )
+
+
+def _get_datasets(lm, em):
+ assert lm or em
+ datasets = []
+ if lm:
+ datasets.extend(LM_DATASETS)
+ if em:
+ datasets.extend(EM_DATASETS)
+ return datasets
+
+
+def check_computation(model_name, datasets):
+ prompt_settings = default_experiment_settings()
+ for ds in datasets:
+ experiment_folder = os.path.join(EXPERIMENT_ROOT, model_name, ds)
+ for setting in prompt_settings:
+ setting_name = get_experiment_setting_name(setting)
+ expected_path = os.path.join(experiment_folder, "results", f"{setting_name}.csv")
+ if not os.path.exists(expected_path):
+ print("Missing results for:", expected_path)
+ if ds in LM_DATASETS:
+ expected_path = os.path.join(experiment_folder, "results", "amg.csv")
+ if not os.path.exists(expected_path):
+ print("Missing results for:", expected_path)
+ print("All checks_run")
+
+
+# evaluation on slurm
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("model_name")
+ parser.add_argument("--check", "-c", action="store_true")
+ parser.add_argument("--lm", action="store_true")
+ parser.add_argument("--em", action="store_true")
+ parser.add_argument("--datasets", nargs="+")
+ args = parser.parse_args()
+
+ datasets = args.datasets
+ if datasets is None or len(datasets) == 0:
+ datasets = _get_datasets(args.lm, args.em)
+ assert all(ds in ALL_DATASETS for ds in datasets)
+
+ if args.check:
+ check_computation(args.model_name, datasets)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(args.model_name, datasets)
+ else: # we're in a slurm job
+ job_id = int(job_id)
+ evaluate_dataset_slurm(args.model_name, datasets[job_id])
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/evaluate_generalist.sbatch b/finetuning/generalists/evaluate_generalist.sbatch
new file mode 100755
index 00000000..121ba55f
--- /dev/null
+++ b/finetuning/generalists/evaluate_generalist.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 2800
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python evaluate_generalist.py $@
diff --git a/finetuning/generalists/evaluate_training_evolution.py b/finetuning/generalists/evaluate_training_evolution.py
new file mode 100644
index 00000000..c1435593
--- /dev/null
+++ b/finetuning/generalists/evaluate_training_evolution.py
@@ -0,0 +1,78 @@
+import argparse
+import os
+
+from glob import glob
+from subprocess import run
+
+import pandas as pd
+from util import evaluate_checkpoint_for_datasets, get_generalist_predictor
+
+CHECKPOINT_ROOT = "/scratch/projects/nim00007/sam/models/LM/generalist/v2"
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/training-evolution"
+# We evaluate these three datasets for the training evolution.
+# These are chosen based on observations from preliminary experiments.
+# - covid-if: out-of-domain dataset that shows the expected improvement (over vanilla).
+# - deepbacs: in domain dataset where we see the biggest gap to the specialist.
+# - lizard: out-of-domain that is furthest from the training data.
+EVAL_DATASETS = ("covid-if", "deepbacs", "lizard")
+
+
+def evaluate_checkpoint_slurm(model_type, job_id, checkpoints):
+ checkpoint = checkpoints[job_id]
+
+ predictor, state = get_generalist_predictor(
+ checkpoint, model_type, is_custom_model=True, return_state=True
+ )
+ epoch = state["epoch"] + 1
+
+ print("Run evaluation for", model_type, "epoch", epoch)
+ experiment_root = os.path.join(EXPERIMENT_ROOT, f"{model_type}-epoch-{epoch}")
+ result = evaluate_checkpoint_for_datasets(
+ None, None, experiment_root, EVAL_DATASETS,
+ run_default_evaluation=True, run_amg=False,
+ is_custom_model=True, predictor=predictor,
+ )
+
+ result.insert(0, "epoch", [epoch] * result.shape[0])
+ return result
+
+
+def evaluate_training_evolution(model_type, checkpoints):
+ results = []
+ for i in range(len(checkpoints)):
+ result = evaluate_checkpoint_slurm(model_type, i, checkpoints)
+ results.append(result)
+ results = pd.concat(results)
+ save_path = os.path.join(EXPERIMENT_ROOT, f"{model_type}.csv")
+ results.to_csv(save_path, index=False)
+
+
+def submit_array_job(model_type, checkpoints):
+ n_checkpoints = len(checkpoints)
+ cmd = ["sbatch", "-a", f"0-{n_checkpoints-1}", "evaluate_training_evolution.sbatch", model_type]
+ run(cmd)
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("model_type")
+ parser.add_argument("-e", "--evaluate", action="store_true")
+ args = parser.parse_args()
+
+ checkpoints = sorted(glob(os.path.join(CHECKPOINT_ROOT, args.model_type, "epoch-*.pt")))
+ assert len(checkpoints) > 0
+
+ if args.evaluate:
+ evaluate_training_evolution(args.model_type, checkpoints)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(args.model_type, checkpoints)
+ else: # we're in a slurm job
+ job_id = int(job_id)
+ evaluate_checkpoint_slurm(args.model_type, job_id, checkpoints)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/evaluate_training_evolution.sbatch b/finetuning/generalists/evaluate_training_evolution.sbatch
new file mode 100755
index 00000000..e628f44f
--- /dev/null
+++ b/finetuning/generalists/evaluate_training_evolution.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 96G
+#SBATCH -t 240
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python evaluate_training_evolution.py $@
diff --git a/finetuning/generalists/export_generalist_model.py b/finetuning/generalists/export_generalist_model.py
new file mode 100644
index 00000000..11073364
--- /dev/null
+++ b/finetuning/generalists/export_generalist_model.py
@@ -0,0 +1,24 @@
+import os
+from micro_sam.util import export_custom_sam_model
+from evaluate_generalist import CHECKPOINTS, EXPERIMENT_ROOT
+
+OUT_ROOT = os.path.join(EXPERIMENT_ROOT, "exported")
+os.makedirs(OUT_ROOT, exist_ok=True)
+
+
+def export_generalist(model):
+ checkpoint_path = CHECKPOINTS[model]
+ model_type = model[:5]
+ save_path = os.path.join(OUT_ROOT, f"{model}.pth")
+ export_custom_sam_model(checkpoint_path, model_type, save_path)
+
+
+def main():
+ export_generalist("vit_b_em")
+ export_generalist("vit_h_em")
+ export_generalist("vit_b_lm")
+ export_generalist("vit_h_lm")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/generate_model_comparison.py b/finetuning/generalists/generate_model_comparison.py
new file mode 100644
index 00000000..f0ed3a9a
--- /dev/null
+++ b/finetuning/generalists/generate_model_comparison.py
@@ -0,0 +1,82 @@
+import os
+
+import imageio.v3 as imageio
+import micro_sam.evaluation.model_comparison as comparison
+import torch_em
+
+from util import get_data_paths, EM_DATASETS, LM_DATASETS
+
+OUTPUT_ROOT = "/scratch-grete/projects/nim00007/sam/experiments/model_comparison"
+
+
+def _get_patch_shape(path):
+ im_shape = imageio.imread(path).shape[:2]
+ patch_shape = tuple(min(sh, 512) for sh in im_shape)
+ return patch_shape
+
+
+def raw_trafo(raw):
+ raw = raw.transpose((2, 0, 1))
+ print(raw.shape)
+ return raw
+
+
+def get_loader(dataset):
+ image_paths, gt_paths = get_data_paths(dataset, split="test")
+ image_paths, gt_paths = image_paths[:100], gt_paths[:100]
+
+ with_channels = dataset in ("hpa", "lizard")
+
+ label_transform = torch_em.transform.label.connected_components
+ loader = torch_em.default_segmentation_loader(
+ image_paths, None, gt_paths, None,
+ batch_size=1, patch_shape=_get_patch_shape(image_paths[0]),
+ shuffle=True, n_samples=25, label_transform=label_transform,
+ with_channels=with_channels, is_seg_dataset=not with_channels
+ )
+ return loader
+
+
+def generate_comparison_for_dataset(dataset, model1, model2):
+ output_folder = os.path.join(OUTPUT_ROOT, dataset)
+ if os.path.exists(output_folder):
+ return output_folder
+ print("Generate model comparison data for", dataset)
+ loader = get_loader(dataset)
+ comparison.generate_data_for_model_comparison(loader, output_folder, model1, model2, n_samples=25)
+ return output_folder
+
+
+def create_comparison_images(output_folder, dataset):
+ plot_folder = os.path.join(OUTPUT_ROOT, "images", dataset)
+ if os.path.exists(plot_folder):
+ return
+ comparison.model_comparison(
+ output_folder, n_images_per_sample=25, min_size=100,
+ plot_folder=plot_folder, outline_dilation=1
+ )
+
+
+def generate_comparison_em():
+ model1 = "vit_h"
+ model2 = "vit_h_em"
+ for dataset in EM_DATASETS:
+ folder = generate_comparison_for_dataset(dataset, model1, model2)
+ create_comparison_images(folder, dataset)
+
+
+def generate_comparison_lm():
+ model1 = "vit_h"
+ model2 = "vit_h_lm"
+ for dataset in LM_DATASETS:
+ folder = generate_comparison_for_dataset(dataset, model1, model2)
+ create_comparison_images(folder, dataset)
+
+
+def main():
+ generate_comparison_lm()
+ # generate_comparison_em()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/precompute_prompts.py b/finetuning/generalists/precompute_prompts.py
new file mode 100644
index 00000000..73d5e4ed
--- /dev/null
+++ b/finetuning/generalists/precompute_prompts.py
@@ -0,0 +1,102 @@
+import argparse
+import os
+import pickle
+
+from subprocess import run
+
+import micro_sam.evaluation as evaluation
+from util import get_data_paths, ALL_DATASETS, LM_DATASETS
+from tqdm import tqdm
+
+PROMPT_ROOT = "/scratch/projects/nim00007/sam/experiments/prompts"
+
+
+def precompute_prompts(dataset):
+ # everything for livecell has been computed already
+ if dataset == "livecell":
+ return
+
+ prompt_folder = os.path.join(PROMPT_ROOT, dataset)
+ _, gt_paths = get_data_paths(dataset, "test")
+
+ settings = evaluation.default_experiment_settings()
+ evaluation.precompute_all_prompts(gt_paths, prompt_folder, settings)
+
+
+def precompute_prompts_slurm(job_id):
+ dataset = ALL_DATASETS[job_id]
+ precompute_prompts(dataset)
+
+
+def submit_array_job():
+ n_datasets = len(ALL_DATASETS)
+ cmd = ["sbatch", "-a", f"0-{n_datasets-1}", "precompute_prompts.sbatch"]
+ run(cmd)
+
+
+def _check_prompts(dataset, settings, expected_len):
+ prompt_folder = os.path.join(PROMPT_ROOT, dataset)
+
+ def check_prompt_file(prompt_file):
+ assert os.path.exists(prompt_file), prompt_file
+ with open(prompt_file, "rb") as f:
+ prompts = pickle.load(f)
+ assert len(prompts) == expected_len, f"{len(prompts)}, {expected_len}"
+
+ for setting in settings:
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ prompt_file = os.path.join(prompt_folder, f"points-p{pos}-n{neg}.pkl")
+ if pos == 0 and neg == 0:
+ prompt_file = os.path.join(prompt_folder, "boxes.pkl")
+ check_prompt_file(prompt_file)
+
+
+def check_prompts_and_datasets():
+
+ def check_dataset(dataset):
+ try:
+ images, _ = get_data_paths(dataset, "test")
+ except AssertionError as e:
+ print("Checking test split failed for datasset", dataset, "due to", e)
+
+ if dataset not in LM_DATASETS:
+ return len(images)
+
+ try:
+ get_data_paths(dataset, "val")
+ except AssertionError as e:
+ print("Checking val split failed for datasset", dataset, "due to", e)
+
+ return len(images)
+
+ settings = evaluation.default_experiment_settings()
+ for ds in tqdm(ALL_DATASETS, desc="Checking datasets"):
+ n_images = check_dataset(ds)
+ _check_prompts(ds, settings, n_images)
+ print("All checks done!")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-d", "--dataset")
+ parser.add_argument("--check", "-c", action="store_true")
+ args = parser.parse_args()
+
+ if args.check:
+ check_prompts_and_datasets()
+ return
+
+ if args.dataset is not None:
+ precompute_prompts(args.dataset)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job()
+ else: # we're in a slurm job and precompute a setting
+ job_id = int(job_id)
+ precompute_prompts_slurm(job_id)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/precompute_prompts.sbatch b/finetuning/generalists/precompute_prompts.sbatch
new file mode 100755
index 00000000..2b04cc96
--- /dev/null
+++ b/finetuning/generalists/precompute_prompts.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 2000
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python precompute_prompts.py $@
diff --git a/finetuning/generalists/util.py b/finetuning/generalists/util.py
new file mode 100644
index 00000000..d354d882
--- /dev/null
+++ b/finetuning/generalists/util.py
@@ -0,0 +1,195 @@
+import json
+import os
+import warnings
+
+from glob import glob
+from pathlib import Path
+
+import pandas as pd
+from micro_sam.evaluation import (
+ automatic_mask_generation, inference, evaluation,
+ default_experiment_settings, get_experiment_setting_name
+)
+from micro_sam.evaluation.livecell import _get_livecell_paths
+
+DATA_ROOT = "/scratch/projects/nim00007/sam/datasets"
+LIVECELL_ROOT = "/scratch/projects/nim00007/data/LiveCELL"
+PROMPT_ROOT = "/scratch-grete/projects/nim00007/sam/experiments/prompts"
+
+LM_DATASETS = (
+ "covid-if",
+ "deepbacs",
+ "hpa",
+ "livecell",
+ "lizard",
+ "mouse-embryo",
+ "plantseg-ovules",
+ "plantseg-root",
+ "tissuenet",
+)
+
+EM_DATASETS = (
+ "cremi",
+ "lucchi",
+ "mitoem",
+ "nuc_mm/mouse",
+ "nuc_mm/zebrafish",
+ "platy-cell",
+ "platy-cuticle",
+ "platy-nuclei",
+ "snemi",
+ "sponge-em",
+ "vnc",
+)
+ALL_DATASETS = EM_DATASETS + LM_DATASETS
+
+
+###
+# Dataset functionality
+###
+
+
+def get_data_paths(dataset, split, max_num_images=None):
+ if dataset == "livecell":
+ n_val_per_cell_type = None if max_num_images is None else int(max_num_images / 8)
+ return _get_livecell_paths(LIVECELL_ROOT, split=split, n_val_per_cell_type=n_val_per_cell_type)
+
+ image_pattern = os.path.join(DATA_ROOT, dataset, split, "image*.tif")
+ image_paths = sorted(glob(image_pattern))
+ gt_paths = sorted(glob(os.path.join(DATA_ROOT, dataset, split, "label*.tif")))
+ assert len(image_paths) == len(gt_paths)
+ assert len(image_paths) > 0, image_pattern
+ if max_num_images is not None:
+ image_paths, gt_paths = image_paths[:max_num_images], gt_paths[:max_num_images]
+ return image_paths, gt_paths
+
+
+###
+# Evaluation functionality
+###
+
+
+def get_generalist_predictor(checkpoint, model_type, is_custom_model, return_state=False):
+ with warnings.catch_warnings():
+ warnings.simplefilter("ignore")
+ return inference.get_predictor(
+ checkpoint, model_type=model_type,
+ return_state=return_state, is_custom_model=is_custom_model
+ )
+
+
+def evaluate_checkpoint_for_dataset(
+ checkpoint, model_type, dataset, experiment_folder,
+ run_default_evaluation, run_amg, is_custom_model,
+ predictor=None, max_num_val_images=None,
+):
+ """Evaluate a generalist checkpoint for a given dataset.
+ """
+ assert run_default_evaluation or run_amg
+
+ prompt_dir = os.path.join(PROMPT_ROOT, dataset)
+
+ if predictor is None:
+ predictor = get_generalist_predictor(checkpoint, model_type, is_custom_model)
+ test_image_paths, test_gt_paths = get_data_paths(dataset, "test")
+
+ embedding_dir = os.path.join(experiment_folder, "test", "embeddings")
+ os.makedirs(embedding_dir, exist_ok=True)
+ result_dir = os.path.join(experiment_folder, "results")
+
+ results = []
+ if run_default_evaluation:
+ prompt_settings = default_experiment_settings()
+ for setting in prompt_settings:
+
+ setting_name = get_experiment_setting_name(setting)
+ prediction_dir = os.path.join(experiment_folder, "test", setting_name)
+ os.makedirs(prediction_dir, exist_ok=True)
+
+ inference.run_inference_with_prompts(
+ predictor, test_image_paths, test_gt_paths,
+ embedding_dir, prediction_dir,
+ use_points=setting["use_points"], use_boxes=setting["use_boxes"],
+ n_positives=setting["n_positives"], n_negatives=setting["n_negatives"],
+ prompt_save_dir=prompt_dir,
+ )
+
+ if dataset == "livecell":
+ pred_paths = [
+ os.path.join(prediction_dir, os.path.basename(gt_path)) for gt_path in test_gt_paths
+ ]
+ assert all(os.path.exists(pred_path) for pred_path in pred_paths)
+ else:
+ pred_paths = sorted(glob(os.path.join(prediction_dir, "*.tif")))
+ result_path = os.path.join(result_dir, f"{setting_name}.csv")
+ os.makedirs(Path(result_path).parent, exist_ok=True)
+
+ result = evaluation.run_evaluation(test_gt_paths, pred_paths, result_path)
+ result.insert(0, "setting", [setting_name])
+ results.append(result)
+
+ if run_amg:
+ val_embedding_dir = os.path.join(experiment_folder, "val", "embeddings")
+ val_result_dir = os.path.join(experiment_folder, "val", "results")
+ os.makedirs(val_embedding_dir, exist_ok=True)
+
+ val_image_paths, val_gt_paths = get_data_paths(dataset, "val", max_num_images=max_num_val_images)
+ automatic_mask_generation.run_amg_grid_search(
+ predictor, val_image_paths, val_gt_paths, val_embedding_dir,
+ val_result_dir, verbose_gs=True,
+ )
+
+ best_iou_thresh, best_stability_thresh, _ = automatic_mask_generation.evaluate_amg_grid_search(val_result_dir)
+ best_settings = {"pred_iou_thresh": best_iou_thresh, "stability_score_thresh": best_stability_thresh}
+ gs_result_path = os.path.join(experiment_folder, "best_gs_params.json")
+ with open(gs_result_path, "w") as f:
+ json.dump(best_settings, f)
+
+ prediction_dir = os.path.join(experiment_folder, "test", "amg")
+ os.makedirs(prediction_dir, exist_ok=True)
+ automatic_mask_generation.run_amg_inference(
+ predictor, test_image_paths, embedding_dir, prediction_dir,
+ amg_generate_kwargs=best_settings,
+ )
+
+ if dataset == "livecell":
+ pred_paths = [
+ os.path.join(prediction_dir, os.path.basename(gt_path)) for gt_path in test_gt_paths
+ ]
+ assert all(os.path.exists(pred_path) for pred_path in pred_paths)
+ else:
+ pred_paths = sorted(glob(os.path.join(prediction_dir, "*.tif")))
+
+ result_path = os.path.join(result_dir, "amg.csv")
+ os.makedirs(Path(result_path).parent, exist_ok=True)
+
+ result = evaluation.run_evaluation(test_gt_paths, pred_paths, result_path)
+ result.insert(0, "setting", ["amg"])
+ results.append(result)
+
+ results = pd.concat(results)
+ results.insert(0, "dataset", [dataset] * results.shape[0])
+ return results
+
+
+def evaluate_checkpoint_for_datasets(
+ checkpoint, model_type, experiment_root, datasets,
+ run_default_evaluation, run_amg, is_custom_model,
+ predictor=None, max_num_val_images=None,
+):
+ if predictor is None:
+ predictor = get_generalist_predictor(checkpoint, model_type, is_custom_model)
+
+ results = []
+ for dataset in datasets:
+ experiment_folder = os.path.join(experiment_root, dataset)
+ os.makedirs(experiment_folder, exist_ok=True)
+ result = evaluate_checkpoint_for_dataset(
+ None, None, dataset, experiment_folder,
+ run_default_evaluation=run_default_evaluation,
+ run_amg=run_amg, is_custom_model=is_custom_model,
+ predictor=predictor, max_num_val_images=max_num_val_images,
+ )
+ results.append(result)
+
+ return pd.concat(results)
diff --git a/finetuning/livecell/README.md b/finetuning/livecell/README.md
new file mode 100644
index 00000000..fceedcb7
--- /dev/null
+++ b/finetuning/livecell/README.md
@@ -0,0 +1,27 @@
+# Finetuning Segment Anything for LiveCELL
+
+TODO: explain the set-up
+
+These experiments are implemented for a slurm cluster with access to GPUs (and you ideally need access to A100s or H100s with 80GB of memory, if you only use ViT-b then using a GPU with 32 GB or 40 GB should suffice.)
+
+## Training
+
+TODO: add training code and explain how to run it
+
+## Evaluation
+
+To run the evaluation experiments for the Segment Anything Models on LiveCELL follow these steps:
+
+- Preparation:
+ - Go to the `evaluation` directory.
+ - Adapt the path to the models, the data folder and the experiment folder in `util.py`
+ - Make sure you have the LiveCELL data downloaded in the data folder. If not you can run `python util.py` and it will automatically downloaded.
+ - Adapt the settings in `util/precompute_embeddings.sh` and `util/precompute_prompts.sh` to your slurm set-up.
+- Precompute the embeddings by running `sbatch precompute_embeddings.sbatch ` for all the models you want to evaluate.
+ - This will submit a slurm job that needs access to a GPU.
+- Precompute the prompts by running `python precompute_prompts.py -f`.
+ - This will submit a slurm array job that precomputes the prompts for all images. In total 31 jobs will be started. Note: these jobs do not need access to a GPU, the computation is purely CPU based.
+- Run inference via `python inference.py -n `.
+ - This will submit a slurm array job that runs the prediction for all prompt settings for this model. In total 61 jobs will be started, but each should only take 10-20 minutes (on a A100 and depending on the model type).
+- Run the evaluation of inference results via `sbatch evaluation.sbatch -n `.
+ - This will submit a single slurm job that does not need a GPU.
diff --git a/finetuning/livecell/amg/grid_search_and_inference.py b/finetuning/livecell/amg/grid_search_and_inference.py
new file mode 100644
index 00000000..1a04d763
--- /dev/null
+++ b/finetuning/livecell/amg/grid_search_and_inference.py
@@ -0,0 +1,40 @@
+import argparse
+from micro_sam.evaluation.livecell import run_livecell_amg
+from util import DATA_ROOT, get_checkpoint, get_experiment_folder, check_model
+
+
+def run_job(model_name, use_mws):
+ checkpoint, model_type = get_checkpoint(model_name)
+ experiment_folder = get_experiment_folder(model_name)
+ input_folder = DATA_ROOT
+
+ run_livecell_amg(
+ checkpoint, model_type, input_folder, experiment_folder,
+ n_val_per_cell_type=25, use_mws=use_mws,
+ )
+
+
+# TODO
+def check_amg(model_name, use_mws):
+ pass
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ parser.add_argument("--mws", action="store_true")
+ parser.add_argument("-c", "--check", action="store_true")
+ args = parser.parse_args()
+
+ model_name = args.name
+ use_mws = args.mws
+ check_model(model_name)
+
+ if args.check:
+ check_amg(model_name, use_mws)
+ else:
+ run_job(model_name, use_mws)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/amg/grid_search_and_inference.sbatch b/finetuning/livecell/amg/grid_search_and_inference.sbatch
new file mode 100755
index 00000000..a3692182
--- /dev/null
+++ b/finetuning/livecell/amg/grid_search_and_inference.sbatch
@@ -0,0 +1,9 @@
+#! /bin/bash
+#SBATCH -c 8
+#SBATCH --mem 96G
+#SBATCH -t 2880
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python grid_search_and_inference.py $@
diff --git a/finetuning/livecell/amg/util.py b/finetuning/livecell/amg/util.py
new file mode 100644
index 00000000..eff50eac
--- /dev/null
+++ b/finetuning/livecell/amg/util.py
@@ -0,0 +1,30 @@
+import os
+
+DATA_ROOT = "/scratch/projects/nim00007/data/LiveCELL"
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/livecell"
+MODELS = {
+ "vit_b": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_b_01ec64.pth",
+ "vit_h": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_h_4b8939.pth",
+ "vit_b_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_b/best.pt",
+ "vit_h_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_h/best.pt",
+ "vit_b_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_b/best.pt",
+ "vit_h_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_h/best.pt",
+}
+
+
+def get_checkpoint(name):
+ assert name in MODELS, name
+ ckpt = MODELS[name]
+ assert os.path.exists(ckpt), ckpt
+ model_type = name[:5]
+ assert model_type in ("vit_b", "vit_h"), model_type
+ return ckpt, model_type
+
+
+def get_experiment_folder(name):
+ return os.path.join(EXPERIMENT_ROOT, name)
+
+
+def check_model(name):
+ if name not in MODELS:
+ raise ValueError(f"Invalid model {name}, expect one of {MODELS.keys()}")
diff --git a/finetuning/livecell/evaluation/evaluation.py b/finetuning/livecell/evaluation/evaluation.py
new file mode 100644
index 00000000..18e93eb5
--- /dev/null
+++ b/finetuning/livecell/evaluation/evaluation.py
@@ -0,0 +1,58 @@
+import argparse
+import os
+from glob import glob
+
+import pandas as pd
+from tqdm import tqdm
+
+from micro_sam.evaluation.livecell import evaluate_livecell_predictions
+from util import get_experiment_folder, DATA_ROOT
+
+
+def run_eval(gt_dir, experiment_folder, prompt_prefix):
+ result_dir = os.path.join(experiment_folder, "results", prompt_prefix)
+ os.makedirs(result_dir, exist_ok=True)
+
+ pred_dirs = sorted(glob(os.path.join(experiment_folder, prompt_prefix, "*")))
+
+ results = []
+ for pred_dir in tqdm(pred_dirs, desc=f"Run livecell evaluation for all {prompt_prefix}-prompt settings"):
+ setting_name = os.path.basename(pred_dir)
+ save_path = os.path.join(result_dir, f"{setting_name}.csv")
+ if os.path.exists(save_path):
+ result = pd.read_csv(save_path)
+ else:
+ result = evaluate_livecell_predictions(gt_dir, pred_dir, verbose=False)
+ result.to_csv(save_path, index=False)
+ result.insert(0, "setting", [setting_name] * result.shape[0])
+ results.append(result)
+
+ results = pd.concat(results)
+ return results
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ args = parser.parse_args()
+
+ name = args.name
+
+ gt_dir = os.path.join(DATA_ROOT, "annotations", "livecell_test_images")
+ assert os.path.exists(gt_dir), "The LiveCELL Dataset is incomplete"
+
+ experiment_folder = get_experiment_folder(name)
+
+ result_box = run_eval(gt_dir, experiment_folder, "box")
+ result_box.insert(0, "prompt", ["box"] * result_box.shape[0])
+
+ result_point = run_eval(gt_dir, experiment_folder, "points")
+ result_point.insert(0, "prompt", ["points"] * result_point.shape[0])
+
+ result = pd.concat([result_box, result_point])
+ save_path = os.path.join(experiment_folder, f"result_{name}.csv")
+ result.to_csv(save_path, index=False)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/evaluation.sbatch b/finetuning/livecell/evaluation/evaluation.sbatch
new file mode 100755
index 00000000..8306a1df
--- /dev/null
+++ b/finetuning/livecell/evaluation/evaluation.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -N 1
+#SBATCH -c 4
+#SBATCH --mem 32G
+#SBATCH -t 720
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python evaluation.py $@
diff --git a/finetuning/livecell/evaluation/inference.py b/finetuning/livecell/evaluation/inference.py
new file mode 100644
index 00000000..00e1b894
--- /dev/null
+++ b/finetuning/livecell/evaluation/inference.py
@@ -0,0 +1,79 @@
+import argparse
+import os
+from glob import glob
+from subprocess import run
+
+import micro_sam.evaluation as evaluation
+from micro_sam.evaluation.livecell import livecell_inference
+from util import check_model, get_checkpoint, get_experiment_folder, DATA_ROOT, PROMPT_FOLDER
+
+
+def inference_job(prompt_settings, model_name):
+ experiment_folder = get_experiment_folder(model_name)
+ checkpoint, model_type = get_checkpoint(model_name)
+ livecell_inference(
+ checkpoint,
+ input_folder=DATA_ROOT,
+ model_type=model_type,
+ experiment_folder=experiment_folder,
+ use_points=prompt_settings["use_points"],
+ use_boxes=prompt_settings["use_boxes"],
+ n_positives=prompt_settings["n_positives"],
+ n_negatives=prompt_settings["n_negatives"],
+ prompt_folder=PROMPT_FOLDER,
+ )
+
+
+def submit_array_job(prompt_settings, model_name, test_run):
+ n_settings = len(prompt_settings)
+ cmd = ["sbatch", "-a", f"0-{n_settings-1}", "inference.sbatch", "-n", model_name]
+ if test_run:
+ cmd.append("-t")
+ run(cmd)
+
+
+def check_inference(settings, model_name):
+ experiment_folder = get_experiment_folder(model_name)
+ for setting in settings:
+ prefix = "box" if setting["use_boxes"] else "points"
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ pred_folder = os.path.join(experiment_folder, prefix, f"p{pos}-n{neg}")
+ assert os.path.exists(pred_folder), pred_folder
+ n_files = len(glob(os.path.join(pred_folder, "*.tif")))
+ assert n_files == 1512, str(n_files)
+
+ print("Inference checks successful!")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ parser.add_argument("-t", "--test_run", action="store_true")
+ parser.add_argument("-c", "--check", action="store_true")
+ args = parser.parse_args()
+
+ if args.test_run: # test run with only the three default experiment settings
+ settings = evaluation.default_experiment_settings()
+ else: # all experiment settings
+ settings = evaluation.full_experiment_settings()
+ settings.extend(evaluation.full_experiment_settings(use_boxes=True))
+
+ model_name = args.name
+ check_model(model_name)
+
+ if args.check:
+ check_inference(settings, model_name)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(settings, model_name, args.test_run)
+ else: # we're in a slurm job and run inference for a setting
+ job_id = int(job_id)
+ this_settings = settings[job_id]
+ inference_job(this_settings, model_name)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/inference.sbatch b/finetuning/livecell/evaluation/inference.sbatch
new file mode 100755
index 00000000..62d926eb
--- /dev/null
+++ b/finetuning/livecell/evaluation/inference.sbatch
@@ -0,0 +1,9 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 60
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python inference.py $@
diff --git a/finetuning/livecell/evaluation/iterative_prompting.py b/finetuning/livecell/evaluation/iterative_prompting.py
new file mode 100644
index 00000000..0278ff38
--- /dev/null
+++ b/finetuning/livecell/evaluation/iterative_prompting.py
@@ -0,0 +1,52 @@
+import os
+from glob import glob
+
+from micro_sam.evaluation.inference import run_inference_with_iterative_prompting
+from micro_sam.evaluation.evaluation import run_evaluation
+
+from util import get_checkpoint, get_paths
+
+LIVECELL_GT_ROOT = "/scratch-grete/projects/nim00007/data/LiveCELL/annotations_corrected/livecell_test_images"
+# TODO update to make fit other models
+PREDICTION_ROOT = "./pred_interactive_prompting"
+
+
+def run_interactive_prompting():
+ prediction_root = PREDICTION_ROOT
+
+ checkpoint, model_type = get_checkpoint("vit_b")
+ image_paths, gt_paths = get_paths()
+
+ run_inference_with_iterative_prompting(
+ checkpoint, model_type, image_paths, gt_paths,
+ prediction_root, use_boxes=False, batch_size=16,
+ )
+
+
+def get_pg_paths(pred_folder):
+ pred_paths = sorted(glob(os.path.join(pred_folder, "*.tif")))
+ names = [os.path.split(path)[1] for path in pred_paths]
+ gt_paths = [
+ os.path.join(LIVECELL_GT_ROOT, name.split("_")[0], name) for name in names
+ ]
+ assert all(os.path.exists(pp) for pp in gt_paths)
+ return pred_paths, gt_paths
+
+
+def evaluate_interactive_prompting():
+ prediction_root = PREDICTION_ROOT
+ prediction_folders = sorted(glob(os.path.join(prediction_root, "iteration*")))
+ for pred_folder in prediction_folders:
+ print("Evaluating", pred_folder)
+ pred_paths, gt_paths = get_pg_paths(pred_folder)
+ res = run_evaluation(gt_paths, pred_paths, save_path=None)
+ print(res)
+
+
+def main():
+ # run_interactive_prompting()
+ evaluate_interactive_prompting()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/precompute_embeddings.py b/finetuning/livecell/evaluation/precompute_embeddings.py
new file mode 100644
index 00000000..3d53ed83
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_embeddings.py
@@ -0,0 +1,24 @@
+import argparse
+import os
+
+from micro_sam.evaluation import precompute_all_embeddings
+from util import get_paths, get_model, get_experiment_folder
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ args = parser.parse_args()
+
+ name = args.name
+
+ image_paths, _ = get_paths()
+ predictor = get_model(name)
+ exp_folder = get_experiment_folder(name)
+ embedding_dir = os.path.join(exp_folder, "embeddings")
+ os.makedirs(embedding_dir, exist_ok=True)
+ precompute_all_embeddings(predictor, image_paths, embedding_dir)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/precompute_embeddings.sbatch b/finetuning/livecell/evaluation/precompute_embeddings.sbatch
new file mode 100755
index 00000000..70faec19
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_embeddings.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -N 1
+#SBATCH -c 4
+#SBATCH --mem 64G
+#SBATCH -t 120
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python precompute_embeddings.py -n $1
diff --git a/finetuning/livecell/evaluation/precompute_prompts.py b/finetuning/livecell/evaluation/precompute_prompts.py
new file mode 100644
index 00000000..f22ccdc6
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_prompts.py
@@ -0,0 +1,68 @@
+import argparse
+import os
+import pickle
+from subprocess import run
+
+import micro_sam.evaluation as evaluation
+from tqdm import tqdm
+from util import get_paths, PROMPT_FOLDER
+
+
+def precompute_setting(prompt_settings):
+ _, gt_paths = get_paths()
+ evaluation.precompute_all_prompts(gt_paths, PROMPT_FOLDER, prompt_settings)
+
+
+def submit_array_job(prompt_settings, full_settings):
+ n_settings = len(prompt_settings)
+ cmd = ["sbatch", "-a", f"0-{n_settings-1}", "precompute_prompts.sbatch"]
+ if full_settings:
+ cmd.append("-f")
+ run(cmd)
+
+
+def check_settings(settings):
+
+ def check_prompt_file(prompt_file):
+ assert os.path.exists(prompt_file), prompt_file
+ with open(prompt_file, "rb") as f:
+ prompts = pickle.load(f)
+ assert len(prompts) == 1512, f"{len(prompts)}"
+
+ for setting in tqdm(settings, desc="Check prompt files"):
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ prompt_file = os.path.join(PROMPT_FOLDER, f"points-p{pos}-n{neg}.pkl")
+ if pos == 0 and neg == 0:
+ prompt_file = os.path.join(PROMPT_FOLDER, "boxes.pkl")
+ check_prompt_file(prompt_file)
+
+ print("All files checked!")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-f", "--full_settings", action="store_true")
+ parser.add_argument("-c", "--check", action="store_true")
+ args = parser.parse_args()
+
+ if args.full_settings:
+ settings = evaluation.full_experiment_settings(use_boxes=True)
+ else:
+ settings = evaluation.default_experiment_settings()
+
+ if args.check:
+ check_settings(settings)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(settings, args.full_settings)
+ else: # we're in a slurm job and precompute a setting
+ job_id = int(job_id)
+ this_settings = [settings[job_id]]
+ precompute_setting(this_settings)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/precompute_prompts.sbatch b/finetuning/livecell/evaluation/precompute_prompts.sbatch
new file mode 100755
index 00000000..2a9e2df0
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_prompts.sbatch
@@ -0,0 +1,8 @@
+#! /bin/bash
+#SBATCH -p standard96
+#SBATCH -c 4
+#SBATCH --mem 64G
+#SBATCH -t 720
+
+source activate sam
+python precompute_prompts.py $@
diff --git a/finetuning/livecell/evaluation/util.py b/finetuning/livecell/evaluation/util.py
new file mode 100644
index 00000000..48767a38
--- /dev/null
+++ b/finetuning/livecell/evaluation/util.py
@@ -0,0 +1,55 @@
+import os
+
+from micro_sam.evaluation import get_predictor
+from micro_sam.evaluation.livecell import _get_livecell_paths
+
+DATA_ROOT = "/scratch/projects/nim00007/data/LiveCELL"
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/livecell"
+PROMPT_FOLDER = "/scratch-grete/projects/nim00007/sam/experiments/prompts/livecell"
+MODELS = {
+ "vit_b": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_b_01ec64.pth",
+ "vit_h": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_h_4b8939.pth",
+ "vit_b_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_b/best.pt",
+ "vit_h_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_h/best.pt",
+ "vit_b_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_b/best.pt",
+ "vit_h_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_h/best.pt",
+}
+
+
+def get_paths():
+ return _get_livecell_paths(DATA_ROOT)
+
+
+def get_checkpoint(name):
+ assert name in MODELS, name
+ ckpt = MODELS[name]
+ assert os.path.exists(ckpt), ckpt
+ model_type = name[:5]
+ assert model_type in ("vit_b", "vit_h"), model_type
+ return ckpt, model_type
+
+
+def get_model(name):
+ ckpt, model_type = get_checkpoint(name)
+ predictor = get_predictor(ckpt, model_type)
+ return predictor
+
+
+def get_experiment_folder(name):
+ return os.path.join(EXPERIMENT_ROOT, name)
+
+
+def check_model(name):
+ if name not in MODELS:
+ raise ValueError(f"Invalid model {name}, expect one of {MODELS.keys()}")
+
+
+def download_livecell():
+ from torch_em.data.datasets import get_livecell_loader
+ get_livecell_loader(DATA_ROOT, "train", (512, 512), 1, download=True)
+ get_livecell_loader(DATA_ROOT, "val", (512, 512), 1, download=True)
+ get_livecell_loader(DATA_ROOT, "test", (512, 512), 1, download=True)
+
+
+if __name__ == "__main__":
+ download_livecell()
diff --git a/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.py b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.py
new file mode 100644
index 00000000..031984dd
--- /dev/null
+++ b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.py
@@ -0,0 +1,119 @@
+import argparse
+import os
+import warnings
+from subprocess import run
+
+import pandas as pd
+
+from micro_sam.evaluation import (
+ inference,
+ evaluation,
+ default_experiment_settings,
+ get_experiment_setting_name
+)
+from micro_sam.evaluation.livecell import _get_livecell_paths
+
+DATA_ROOT = "/scratch-grete/projects/nim00007/data/LiveCELL"
+EXPERIMENT_ROOT = "/scratch-grete/projects/nim00007/sam/experiments/livecell/partial-finetuning"
+PROMPT_DIR = "/scratch-grete/projects/nim00007/sam/experiments/prompts/livecell"
+MODELS = {
+ "freeze-image_encoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-image_encoder",
+ "freeze-image_encoder-mask_decoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-image_encoder-mask_decoder",
+ "freeze-image_encoder-prompt_encoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-image_encoder-prompt_encoder",
+ "freeze-mask_decoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-mask_decoder",
+ "freeze-None": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-None",
+ "freeze-prompt_encoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-prompt_encoder",
+ "freeze-prompt_encoder-mask_decoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-prompt_encoder-mask_decoder",
+ "vanilla": "/home/nimcpape/.sam_models/sam_vit_b_01ec64.pth",
+}
+
+
+def evaluate_model(model_id):
+ model_name = list(MODELS.keys())[model_id]
+ print("Evaluating", model_name)
+
+ try:
+ checkpoint = os.path.join(MODELS[model_name], "best.pt")
+ assert os.path.exists(checkpoint)
+ except AssertionError:
+ checkpoint = MODELS[model_name]
+
+ print("Evalute", model_name, "from", checkpoint)
+ model_type = "vit_b"
+ with warnings.catch_warnings():
+ warnings.simplefilter("ignore")
+ predictor = inference.get_predictor(checkpoint, model_type=model_type)
+
+ experiment_dir = os.path.join(EXPERIMENT_ROOT, model_name)
+
+ embedding_dir = os.path.join(experiment_dir, "embeddings")
+ os.makedirs(embedding_dir, exist_ok=True)
+
+ result_dir = os.path.join(experiment_dir, "results")
+ os.makedirs(result_dir, exist_ok=True)
+
+ image_paths, gt_paths = _get_livecell_paths(DATA_ROOT)
+ experiment_settings = default_experiment_settings()
+
+ results = []
+ for setting in experiment_settings:
+ setting_name = get_experiment_setting_name(setting)
+ prediction_dir = os.path.join(experiment_dir, setting_name)
+
+ os.makedirs(prediction_dir, exist_ok=True)
+ inference.run_inference_with_prompts(
+ predictor, image_paths, gt_paths,
+ embedding_dir, prediction_dir,
+ prompt_save_dir=PROMPT_DIR, **setting
+ )
+
+ pred_paths = [os.path.join(prediction_dir, os.path.basename(gt_path)) for gt_path in gt_paths]
+ assert len(pred_paths) == len(gt_paths)
+ result_path = os.path.join(result_dir, f"{setting_name}.csv")
+
+ if os.path.exists(result_path):
+ result = pd.read_csv(result_path)
+ else:
+ result = evaluation.run_evaluation(gt_paths, pred_paths, result_path)
+ result.insert(0, "setting", [setting_name])
+ results.append(result)
+
+ results = pd.concat(results)
+ return results
+
+
+def combine_results():
+ results = []
+ for model_id, model_name in enumerate(MODELS):
+ res = evaluate_model(model_id)
+ res.insert(0, "frozen", res.shape[0] * [model_name.lstrip("freeze-")])
+ results.append(res)
+ results = pd.concat(results)
+ res_path = os.path.join(EXPERIMENT_ROOT, "partial_finetuning_results.csv")
+ results.to_csv(res_path, index=False)
+
+
+def submit_array_job():
+ n_models = len(MODELS)
+ cmd = ["sbatch", "-a", f"0-{n_models-1}", "evaluate_partially_finetuned.sbatch"]
+ run(cmd)
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-e", "--evaluate", action="store_true")
+ args = parser.parse_args()
+
+ if args.evaluate:
+ combine_results()
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None:
+ submit_array_job()
+ else:
+ evaluate_model(int(job_id))
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.sbatch b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.sbatch
new file mode 100755
index 00000000..68fee4fa
--- /dev/null
+++ b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 96G
+#SBATCH -t 2880
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python evaluate_partially_finetuned.py $@
diff --git a/finetuning/livecell_evaluation.py b/finetuning/livecell_evaluation.py
new file mode 100644
index 00000000..3f545304
--- /dev/null
+++ b/finetuning/livecell_evaluation.py
@@ -0,0 +1,3 @@
+from micro_sam.evaluation.livecell import run_livecell_evaluation
+
+run_livecell_evaluation()
diff --git a/finetuning/livecell_finetuning.py b/finetuning/livecell_finetuning.py
new file mode 100644
index 00000000..3521db4b
--- /dev/null
+++ b/finetuning/livecell_finetuning.py
@@ -0,0 +1,115 @@
+import argparse
+import os
+
+import micro_sam.training as sam_training
+import torch
+import torch_em
+
+from torch_em.data.datasets import get_livecell_loader
+from micro_sam.util import export_custom_sam_model
+
+
+def get_dataloaders(patch_shape, data_path, cell_type=None):
+ """This returns the livecell data loaders implemented in torch_em:
+ https://github.com/constantinpape/torch-em/blob/main/torch_em/data/datasets/livecell.py
+ It will automatically download the livecell data.
+
+ Note: to replace this with another data loader you need to return a torch data loader
+ that retuns `x, y` tensors, where `x` is the image data and `y` are the labels.
+ The labels have to be in a label mask instance segmentation format.
+ I.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
+ Important: the ID 0 is reseved for background, and the IDs must be consecutive
+ """
+ label_transform = torch_em.transform.label.label_consecutive # to ensure consecutive IDs
+ train_loader = get_livecell_loader(path=data_path, patch_shape=patch_shape, split="train", batch_size=2,
+ num_workers=8, cell_types=cell_type, download=True,
+ label_transform=label_transform)
+ val_loader = get_livecell_loader(path=data_path, patch_shape=patch_shape, split="val", batch_size=1,
+ num_workers=8, cell_types=cell_type, download=True,
+ label_transform=label_transform)
+ return train_loader, val_loader
+
+
+def finetune_livecell(args):
+ """Example code for finetuning SAM on LiveCELL"""
+
+ # training settings:
+ model_type = args.model_type
+ checkpoint_path = None # override this to start training from a custom checkpoint
+ device = "cuda" # override this if you have some more complex set-up and need to specify the exact gpu
+ patch_shape = (520, 740) # the patch shape for training
+ n_objects_per_batch = 25 # this is the number of objects per batch that will be sampled
+
+ # get the trainable segment anything model
+ model = sam_training.get_trainable_sam_model(model_type, checkpoint_path, device=device)
+
+ # all the stuff we need for training
+ optimizer = torch.optim.Adam(model.parameters(), lr=1e-5)
+ scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode="min", factor=0.9, patience=10, verbose=True)
+ train_loader, val_loader = get_dataloaders(patch_shape=patch_shape, data_path=args.input_path)
+
+ # this class creates all the training data for a batch (inputs, prompts and labels)
+ convert_inputs = sam_training.ConvertToSamInputs()
+
+ checkpoint_name = "livecell_sam"
+ # the trainer which performs training and validation (implemented using "torch_em")
+ trainer = sam_training.SamTrainer(
+ name=checkpoint_name,
+ save_root=args.save_root,
+ train_loader=train_loader,
+ val_loader=val_loader,
+ model=model,
+ optimizer=optimizer,
+ # currently we compute loss batch-wise, else we pass channelwise True
+ loss=torch_em.loss.DiceLoss(channelwise=False),
+ metric=torch_em.loss.DiceLoss(),
+ device=device,
+ lr_scheduler=scheduler,
+ logger=sam_training.SamLogger,
+ log_image_interval=10,
+ mixed_precision=True,
+ convert_inputs=convert_inputs,
+ n_objects_per_batch=n_objects_per_batch,
+ n_sub_iteration=8,
+ compile_model=False
+ )
+ trainer.fit(args.iterations)
+ if args.export_path is not None:
+ checkpoint_path = os.path.join(
+ "" if args.save_root is None else args.save_root, "checkpoints", checkpoint_name, "best.pt"
+ )
+ export_custom_sam_model(
+ checkpoint_path=checkpoint_path,
+ model_type=model_type,
+ save_path=args.export_path,
+ )
+
+
+def main():
+ parser = argparse.ArgumentParser(description="Finetune Segment Anything for the LiveCELL dataset.")
+ parser.add_argument(
+ "--input_path", "-i", default="",
+ help="The filepath to the LiveCELL data. If the data does not exist yet it will be downloaded."
+ )
+ parser.add_argument(
+ "--model_type", "-m", default="vit_b",
+ help="The model type to use for fine-tuning. Either vit_h, vit_b or vit_l."
+ )
+ parser.add_argument(
+ "--save_root", "-s",
+ help="Where to save the checkpoint and logs. By default they will be saved where this script is run."
+ )
+ parser.add_argument(
+ "--iterations", type=int, default=int(1e5),
+ help="For how many iterations should the model be trained? By default 100k."
+ )
+ parser.add_argument(
+ "--export_path", "-e",
+ help="Where to export the finetuned model to. The exported model can be use din the annotation tools."
+ )
+ args = parser.parse_args()
+ finetune_livecell(args)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell_inference.py b/finetuning/livecell_inference.py
new file mode 100644
index 00000000..3ec2f6ce
--- /dev/null
+++ b/finetuning/livecell_inference.py
@@ -0,0 +1,3 @@
+from micro_sam.evaluation.livecell import run_livecell_inference
+
+run_livecell_inference()
diff --git a/micro_sam/__init__.py b/micro_sam/__init__.py
index 5e90fe5b..9a135385 100644
--- a/micro_sam/__init__.py
+++ b/micro_sam/__init__.py
@@ -3,6 +3,7 @@
.. include:: ../doc/installation.md
.. include:: ../doc/annotation_tools.md
.. include:: ../doc/python_library.md
+.. include:: ../doc/finetuned_models.md
"""
from .__version__ import __version__
diff --git a/micro_sam/__version__.py b/micro_sam/__version__.py
index 02e905d7..b5fdc753 100644
--- a/micro_sam/__version__.py
+++ b/micro_sam/__version__.py
@@ -1 +1 @@
-__version__ = "0.1.2.post1"
+__version__ = "0.2.2"
diff --git a/micro_sam/evaluation/__init__.py b/micro_sam/evaluation/__init__.py
new file mode 100644
index 00000000..b16a45c1
--- /dev/null
+++ b/micro_sam/evaluation/__init__.py
@@ -0,0 +1,19 @@
+"""Functionality for evaluating Segment Anything models on microscopy data.
+"""
+
+from .automatic_mask_generation import (
+ run_amg_inference,
+ run_amg_grid_search,
+ run_amg_grid_search_and_inference,
+)
+from .inference import (
+ get_predictor,
+ run_inference_with_prompts,
+ precompute_all_embeddings,
+ precompute_all_prompts,
+)
+from .experiments import (
+ default_experiment_settings,
+ full_experiment_settings,
+ get_experiment_setting_name,
+)
diff --git a/micro_sam/evaluation/automatic_mask_generation.py b/micro_sam/evaluation/automatic_mask_generation.py
new file mode 100644
index 00000000..46b12ef5
--- /dev/null
+++ b/micro_sam/evaluation/automatic_mask_generation.py
@@ -0,0 +1,272 @@
+"""Inference and evaluation for the automatic instance segmentation functionality.
+"""
+
+import os
+from glob import glob
+from pathlib import Path
+from typing import Any, Dict, List, Optional, Tuple, Union
+
+import imageio.v3 as imageio
+import numpy as np
+import pandas as pd
+
+from elf.evaluation import mean_segmentation_accuracy
+from segment_anything import SamPredictor
+from tqdm import tqdm
+
+from .. import instance_segmentation
+from .. import util
+
+
+def _get_range_of_search_values(input_vals, step):
+ if isinstance(input_vals, list):
+ search_range = np.arange(input_vals[0], input_vals[1] + step, step)
+ search_range = [round(e, 2) for e in search_range]
+ else:
+ search_range = [input_vals]
+ return search_range
+
+
+def _grid_search(
+ amg, gt, image_name, iou_thresh_values, stability_score_values, result_path, amg_generate_kwargs, verbose,
+):
+ net_list = []
+ gs_combinations = [(r1, r2) for r1 in iou_thresh_values for r2 in stability_score_values]
+
+ for iou_thresh, stability_thresh in tqdm(gs_combinations, disable=not verbose):
+ masks = amg.generate(
+ pred_iou_thresh=iou_thresh, stability_score_thresh=stability_thresh, **amg_generate_kwargs
+ )
+ instance_labels = instance_segmentation.mask_data_to_segmentation(
+ masks, gt.shape, with_background=True,
+ min_object_size=amg_generate_kwargs.get("min_mask_region_area", 0),
+ )
+ m_sas, sas = mean_segmentation_accuracy(instance_labels, gt, return_accuracies=True) # type: ignore
+
+ result_dict = {
+ "image_name": image_name,
+ "pred_iou_thresh": iou_thresh,
+ "stability_score_thresh": stability_thresh,
+ "mSA": m_sas,
+ "SA50": sas[0],
+ "SA75": sas[5]
+ }
+ tmp_df = pd.DataFrame([result_dict])
+ net_list.append(tmp_df)
+
+ img_gs_df = pd.concat(net_list)
+ img_gs_df.to_csv(result_path, index=False)
+
+
+# ideally we would generalize the parameters that GS runs over
+def run_amg_grid_search(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ gt_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ result_dir: Union[str, os.PathLike],
+ iou_thresh_values: Optional[List[float]] = None,
+ stability_score_values: Optional[List[float]] = None,
+ amg_kwargs: Optional[Dict[str, Any]] = None,
+ amg_generate_kwargs: Optional[Dict[str, Any]] = None,
+ AMG: instance_segmentation.AMGBase = instance_segmentation.AutomaticMaskGenerator,
+ verbose_gs: bool = False,
+) -> None:
+ """Run grid search for automatic mask generation.
+
+ The grid search goes over the two most important parameters:
+ - `pred_iou_thresh`, the threshold for keeping objects according to the IoU predicted by the model
+ - `stability_score_thresh`, the theshold for keepong objects according to their stability
+
+ Args:
+ predictor: The segment anything predictor.
+ image_paths: The input images for the grid search.
+ gt_paths: The ground-truth segmentation for the grid search.
+ embedding_dir: Folder to cache the image embeddings.
+ result_dir: Folder to cache the evaluation results per image.
+ iou_thresh_values: The values for `pred_iou_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ stability_score_values: The values for `stability_score_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ amg_kwargs: The keyword arguments for the automatic mask generator class.
+ amg_generate_kwargs: The keyword arguments for the `generate` method of the mask generator.
+ This must not contain `pred_iou_thresh` or `stability_score_thresh`.
+ AMG: The automatic mask generator. By default `micro_sam.instance_segmentation.AutomaticMaskGenerator`.
+ verbose_gs: Whether to run the gridsearch for individual images in a verbose mode.
+ """
+ assert len(image_paths) == len(gt_paths)
+ amg_kwargs = {} if amg_kwargs is None else amg_kwargs
+ amg_generate_kwargs = {} if amg_generate_kwargs is None else amg_generate_kwargs
+ if "pred_iou_thresh" in amg_generate_kwargs or "stability_score_thresh" in amg_generate_kwargs:
+ raise ValueError("The threshold parameters are optimized in the grid-search. You must not pass them as kwargs.")
+
+ if iou_thresh_values is None:
+ iou_thresh_values = _get_range_of_search_values([0.6, 0.9], step=0.025)
+ if stability_score_values is None:
+ stability_score_values = _get_range_of_search_values([0.6, 0.95], step=0.025)
+
+ os.makedirs(result_dir, exist_ok=True)
+ amg = AMG(predictor, **amg_kwargs)
+
+ for image_path, gt_path in tqdm(
+ zip(image_paths, gt_paths), desc="Run grid search for AMG", total=len(image_paths)
+ ):
+ image_name = Path(image_path).stem
+ result_path = os.path.join(result_dir, f"{image_name}.csv")
+
+ # We skip images for which the grid search was done already.
+ if os.path.exists(result_path):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ assert os.path.exists(gt_path), gt_path
+
+ image = imageio.imread(image_path)
+ gt = imageio.imread(gt_path)
+
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ image_embeddings = util.precompute_image_embeddings(predictor, image, embedding_path, ndim=2)
+ amg.initialize(image, image_embeddings)
+
+ _grid_search(
+ amg, gt, image_name,
+ iou_thresh_values, stability_score_values,
+ result_path, amg_generate_kwargs, verbose=verbose_gs,
+ )
+
+
+def run_amg_inference(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ prediction_dir: Union[str, os.PathLike],
+ amg_kwargs: Optional[Dict[str, Any]] = None,
+ amg_generate_kwargs: Optional[Dict[str, Any]] = None,
+ AMG: instance_segmentation.AMGBase = instance_segmentation.AutomaticMaskGenerator,
+) -> None:
+ """Run inference for automatic mask generation.
+
+ Args:
+ predictor: The segment anything predictor.
+ image_paths: The input images.
+ embedding_dir: Folder to cache the image embeddings.
+ prediction_dir: Folder to save the predictions.
+ amg_kwargs: The keyword arguments for the automatic mask generator class.
+ amg_generate_kwargs: The keyword arguments for the `generate` method of the mask generator.
+ This must not contain `pred_iou_thresh` or `stability_score_thresh`.
+ AMG: The automatic mask generator. By default `micro_sam.instance_segmentation.AutomaticMaskGenerator`.
+ """
+ amg_kwargs = {} if amg_kwargs is None else amg_kwargs
+ amg_generate_kwargs = {} if amg_generate_kwargs is None else amg_generate_kwargs
+
+ amg = AMG(predictor, **amg_kwargs)
+
+ for image_path in tqdm(image_paths, desc="Run inference for automatic mask generation"):
+ image_name = os.path.basename(image_path)
+
+ # We skip the images that already have been segmented.
+ prediction_path = os.path.join(prediction_dir, image_name)
+ if os.path.exists(prediction_path):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ image = imageio.imread(image_path)
+
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ image_embeddings = util.precompute_image_embeddings(predictor, image, embedding_path, ndim=2)
+
+ amg.initialize(image, image_embeddings)
+ masks = amg.generate(**amg_generate_kwargs)
+ instances = instance_segmentation.mask_data_to_segmentation(
+ masks, image.shape, with_background=True, min_object_size=amg_generate_kwargs.get("min_mask_region_area", 0)
+ )
+
+ # It's important to compress here, otherwise the predictions would take up a lot of space.
+ imageio.imwrite(prediction_path, instances, compression=5)
+
+
+def evaluate_amg_grid_search(result_dir: Union[str, os.PathLike], criterion: str = "mSA") -> Tuple[float, float, float]:
+ """Evaluate gridsearch results.
+
+ Args:
+ result_dir: The folder with the gridsearch results.
+ criterion: The metric to use for determining the best parameters.
+
+ Returns:
+ - The best value for `pred_iou_thresh`.
+ - The best value for `stability_score_thresh`.
+ - The evaluation score for the best setting.
+ """
+
+ # load all the grid search results
+ gs_files = glob(os.path.join(result_dir, "*.csv"))
+ gs_result = pd.concat([pd.read_csv(gs_file) for gs_file in gs_files])
+
+ # contain only the relevant columns and group by the gridsearch columns
+ gs_col1 = "pred_iou_thresh"
+ gs_col2 = "stability_score_thresh"
+ gs_result = gs_result[[gs_col1, gs_col2, criterion]]
+
+ # compute the mean over the grouped columns
+ grouped_result = gs_result.groupby([gs_col1, gs_col2]).mean()
+
+ # find the best grouped result and return the corresponding thresholds
+ best_score = grouped_result.max().values[0]
+ best_result = grouped_result.idxmax()
+ best_iou_thresh, best_stability_score = best_result.values[0]
+ return best_iou_thresh, best_stability_score, best_score
+
+
+def run_amg_grid_search_and_inference(
+ predictor: SamPredictor,
+ val_image_paths: List[Union[str, os.PathLike]],
+ val_gt_paths: List[Union[str, os.PathLike]],
+ test_image_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ prediction_dir: Union[str, os.PathLike],
+ result_dir: Union[str, os.PathLike],
+ iou_thresh_values: Optional[List[float]] = None,
+ stability_score_values: Optional[List[float]] = None,
+ amg_kwargs: Optional[Dict[str, Any]] = None,
+ amg_generate_kwargs: Optional[Dict[str, Any]] = None,
+ AMG: instance_segmentation.AMGBase = instance_segmentation.AutomaticMaskGenerator,
+ verbose_gs: bool = True,
+) -> None:
+ """Run grid search and inference for automatic mask generation.
+
+ Args:
+ predictor: The segment anything predictor.
+ val_image_paths: The input images for the grid search.
+ val_gt_paths: The ground-truth segmentation for the grid search.
+ test_image_paths: The input images for inference.
+ embedding_dir: Folder to cache the image embeddings.
+ prediction_dir: Folder to save the predictions.
+ result_dir: Folder to cache the evaluation results per image.
+ iou_thresh_values: The values for `pred_iou_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ stability_score_values: The values for `stability_score_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ amg_kwargs: The keyword arguments for the automatic mask generator class.
+ amg_generate_kwargs: The keyword arguments for the `generate` method of the mask generator.
+ This must not contain `pred_iou_thresh` or `stability_score_thresh`.
+ AMG: The automatic mask generator. By default `micro_sam.instance_segmentation.AutomaticMaskGenerator`.
+ verbose_gs: Whether to run the gridsearch for individual images in a verbose mode.
+ """
+ run_amg_grid_search(
+ predictor, val_image_paths, val_gt_paths, embedding_dir, result_dir,
+ iou_thresh_values=iou_thresh_values, stability_score_values=stability_score_values,
+ amg_kwargs=amg_kwargs, amg_generate_kwargs=amg_generate_kwargs, AMG=AMG, verbose_gs=verbose_gs,
+ )
+
+ amg_generate_kwargs = {} if amg_generate_kwargs is None else amg_generate_kwargs
+ best_iou_thresh, best_stability_score, best_msa = evaluate_amg_grid_search(result_dir)
+ print(
+ "Best grid-search result:", best_msa,
+ f"@ iou_thresh = {best_iou_thresh}, stability_score = {best_stability_score}"
+ )
+ amg_generate_kwargs["pred_iou_thresh"] = best_iou_thresh
+ amg_generate_kwargs["stability_score_thresh"] = best_stability_score
+
+ run_amg_inference(
+ predictor, test_image_paths, embedding_dir, prediction_dir, amg_kwargs, amg_generate_kwargs, AMG
+ )
diff --git a/micro_sam/evaluation/evaluation.py b/micro_sam/evaluation/evaluation.py
new file mode 100644
index 00000000..854804ae
--- /dev/null
+++ b/micro_sam/evaluation/evaluation.py
@@ -0,0 +1,78 @@
+"""Evaluation functionality for segmentation predictions from `micro_sam.evaluation.automatic_mask_generation`
+and `micro_sam.evaluation.inference`.
+"""
+
+import os
+from pathlib import Path
+from typing import List, Optional, Union
+
+import imageio.v3 as imageio
+import numpy as np
+import pandas as pd
+
+from elf.evaluation import mean_segmentation_accuracy
+from skimage.measure import label
+from tqdm import tqdm
+
+
+def _run_evaluation(gt_paths, prediction_paths, verbose=True):
+ assert len(gt_paths) == len(prediction_paths)
+ msas, sa50s, sa75s = [], [], []
+
+ for gt_path, pred_path in tqdm(
+ zip(gt_paths, prediction_paths), desc="Evaluate predictions", total=len(gt_paths), disable=not verbose
+ ):
+ assert os.path.exists(gt_path), gt_path
+ assert os.path.exists(pred_path), pred_path
+
+ gt = imageio.imread(gt_path)
+ gt = label(gt)
+ pred = imageio.imread(pred_path)
+
+ msa, scores = mean_segmentation_accuracy(pred, gt, return_accuracies=True)
+ sa50, sa75 = scores[0], scores[5]
+ msas.append(msa), sa50s.append(sa50), sa75s.append(sa75)
+
+ return msas, sa50s, sa75s
+
+
+def run_evaluation(
+ gt_paths: List[Union[os.PathLike, str]],
+ prediction_paths: List[Union[os.PathLike, str]],
+ save_path: Optional[Union[os.PathLike, str]] = None,
+ verbose: bool = True,
+) -> pd.DataFrame:
+ """Run evaluation for instance segmentation predictions.
+
+ Args:
+ gt_folder: The folder with ground-truth images.
+ prediction_folder: The folder with the instance segmentations to evaluate.
+ save_path: Optional path for saving the results.
+ pattern: Optional pattern for selecting the images to evaluate via glob.
+ By default all images with ending .tif will be evaluated.
+ verbose: Whether to print the progress.
+
+ Returns:
+ A DataFrame that contains the evaluation results.
+ """
+ assert len(gt_paths) == len(prediction_paths)
+ # if a save_path is given and it already exists then just load it instead of running the eval
+ if save_path is not None and os.path.exists(save_path):
+ return pd.read_csv(save_path)
+
+ msas, sa50s, sa75s = _run_evaluation(gt_paths, prediction_paths, verbose=verbose)
+
+ results = pd.DataFrame.from_dict({
+ "msa": [np.mean(msas)],
+ "sa50": [np.mean(sa50s)],
+ "sa75": [np.mean(sa75s)],
+ })
+
+ if save_path is not None:
+ os.makedirs(Path(save_path).parent, exist_ok=True)
+ results.to_csv(save_path, index=False)
+
+ return results
+
+
+# TODO function to evaluate full experiment and resave in one table
diff --git a/micro_sam/evaluation/experiments.py b/micro_sam/evaluation/experiments.py
new file mode 100644
index 00000000..4646af52
--- /dev/null
+++ b/micro_sam/evaluation/experiments.py
@@ -0,0 +1,83 @@
+"""Predefined experiment settings for experiments with different prompt strategies.
+"""
+
+from typing import Dict, List, Optional
+
+# TODO fully define the dict type
+ExperimentSetting = Dict
+ExperimentSettings = List[ExperimentSetting]
+"""@private"""
+
+
+def full_experiment_settings(
+ use_boxes: bool = False,
+ positive_range: Optional[List[int]] = None,
+ negative_range: Optional[List[int]] = None,
+) -> ExperimentSettings:
+ """The full experiment settings.
+
+ Args:
+ use_boxes: Whether to run the experiments with or without boxes.
+ positive_range: The different number of positive points that will be used.
+ By defaul the values are set to [1, 2, 4, 8, 16].
+ negative_range: The different number of negative points that will be used.
+ By defaul the values are set to [0, 1, 2, 4, 8, 16].
+
+ Returns:
+ The list of experiment settings.
+ """
+ experiment_settings = []
+ if use_boxes:
+ experiment_settings.append(
+ {"use_points": False, "use_boxes": True, "n_positives": 0, "n_negatives": 0}
+ )
+
+ # set default values for the ranges if none were passed
+ if positive_range is None:
+ positive_range = [1, 2, 4, 8, 16]
+ if negative_range is None:
+ negative_range = [0, 1, 2, 4, 8, 16]
+
+ for n_positives in positive_range:
+ for n_negatives in negative_range:
+ if n_positives == 0 and n_negatives == 0:
+ continue
+ experiment_settings.append(
+ {"use_points": True, "use_boxes": use_boxes, "n_positives": n_positives, "n_negatives": n_negatives}
+ )
+
+ return experiment_settings
+
+
+def default_experiment_settings() -> ExperimentSettings:
+ """The three default experiment settings.
+
+ For the default experiments we use a single positive prompt,
+ two positive and four negative prompts and box prompts.
+
+ Returns:
+ The list of experiment settings.
+ """
+ experiment_settings = [
+ {"use_points": True, "use_boxes": False, "n_positives": 1, "n_negatives": 0}, # p1-n0
+ {"use_points": True, "use_boxes": False, "n_positives": 2, "n_negatives": 4}, # p2-n4
+ {"use_points": True, "use_boxes": False, "n_positives": 4, "n_negatives": 8}, # p4-n8
+ {"use_points": False, "use_boxes": True, "n_positives": 0, "n_negatives": 0}, # only box prompts
+ ]
+ return experiment_settings
+
+
+def get_experiment_setting_name(setting: ExperimentSetting) -> str:
+ """Get the name for the given experiment setting.
+
+ Args:
+ setting: The experiment setting.
+ Returns:
+ The name for this experiment setting.
+ """
+ use_points, use_boxes = setting["use_points"], setting["use_boxes"]
+ assert use_points or use_boxes
+ prefix = "points" if use_points else "box"
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ name = f"p{pos}-n{neg}" if use_points else "p0-n0"
+ return f"{prefix}/{name}"
diff --git a/micro_sam/evaluation/inference.py b/micro_sam/evaluation/inference.py
new file mode 100644
index 00000000..e490b6c6
--- /dev/null
+++ b/micro_sam/evaluation/inference.py
@@ -0,0 +1,553 @@
+"""Inference with Segment Anything models and different prompt strategies.
+"""
+
+import os
+import pickle
+import warnings
+
+from copy import deepcopy
+from typing import Any, Dict, List, Optional, Union
+
+import imageio.v3 as imageio
+import numpy as np
+import torch
+
+from skimage.segmentation import relabel_sequential
+from tqdm import tqdm
+
+from segment_anything import SamPredictor
+from segment_anything.utils.transforms import ResizeLongestSide
+
+from .. import util as util
+from ..instance_segmentation import mask_data_to_segmentation
+from ..prompt_generators import PointAndBoxPromptGenerator, IterativePromptGenerator
+from ..training import get_trainable_sam_model, ConvertToSamInputs
+
+
+def _load_prompts(
+ cached_point_prompts, save_point_prompts,
+ cached_box_prompts, save_box_prompts,
+ image_name
+):
+
+ def load_prompt_type(cached_prompts, save_prompts):
+ # Check if we have saved prompts.
+ if cached_prompts is None or save_prompts: # we don't have cached prompts
+ return cached_prompts, None
+
+ # we have cached prompts, but they have not been loaded yet
+ if isinstance(cached_prompts, str):
+ with open(cached_prompts, "rb") as f:
+ cached_prompts = pickle.load(f)
+
+ prompts = cached_prompts[image_name]
+ return cached_prompts, prompts
+
+ cached_point_prompts, point_prompts = load_prompt_type(cached_point_prompts, save_point_prompts)
+ cached_box_prompts, box_prompts = load_prompt_type(cached_box_prompts, save_box_prompts)
+
+ # we don't have anything cached
+ if point_prompts is None and box_prompts is None:
+ return None, cached_point_prompts, cached_box_prompts
+
+ if point_prompts is None:
+ input_point, input_label = [], []
+ else:
+ input_point, input_label = point_prompts
+
+ if box_prompts is None:
+ input_box = []
+ else:
+ input_box = box_prompts
+
+ prompts = (input_point, input_label, input_box)
+ return prompts, cached_point_prompts, cached_box_prompts
+
+
+def _get_batched_prompts(
+ gt,
+ gt_ids,
+ use_points,
+ use_boxes,
+ n_positives,
+ n_negatives,
+ dilation,
+ transform_function,
+):
+ input_point, input_label, input_box = [], [], []
+
+ # Initialize the prompt generator.
+ center_coordinates, bbox_coordinates = util.get_centers_and_bounding_boxes(gt)
+ prompt_generator = PointAndBoxPromptGenerator(
+ n_positive_points=n_positives, n_negative_points=n_negatives,
+ dilation_strength=dilation, get_point_prompts=use_points,
+ get_box_prompts=use_boxes
+ )
+
+ # Iterate over the gt ids, generate the corresponding prompts and combine them to batched input.
+ for gt_id in gt_ids:
+ centers, bboxes = center_coordinates.get(gt_id), bbox_coordinates.get(gt_id)
+ input_point_list, input_label_list, input_box_list, objm = prompt_generator(gt, gt_id, bboxes, centers)
+
+ if use_boxes:
+ # indexes hard-coded to adapt with SAM's bbox format
+ # default format: [a, b, c, d] -> SAM's format: [b, a, d, c]
+ _ib = [input_box_list[0][1], input_box_list[0][0],
+ input_box_list[0][3], input_box_list[0][2]]
+ # transform boxes to the expected format - see predictor.predict function for details
+ _ib = transform_function.apply_boxes(np.array(_ib), gt.shape)
+ input_box.append(_ib)
+
+ if use_points:
+ assert len(input_point_list) == (n_positives + n_negatives)
+ _ip = [ip[::-1] for ip in input_point_list] # to match the coordinate system used by SAM
+
+ # transform coords to the expected format - see predictor.predict function for details
+ _ip = transform_function.apply_coords(np.array(_ip), gt.shape)
+ input_point.append(_ip)
+ input_label.append(input_label_list)
+
+ return input_point, input_label, input_box
+
+
+def _run_inference_with_prompts_for_image(
+ predictor,
+ gt,
+ use_points,
+ use_boxes,
+ n_positives,
+ n_negatives,
+ dilation,
+ batch_size,
+ cached_prompts,
+):
+ # We need the resize transformation for the expected model input size.
+ transform_function = ResizeLongestSide(1024)
+ gt_ids = np.unique(gt)[1:]
+
+ if cached_prompts is None:
+ input_point, input_label, input_box = _get_batched_prompts(
+ gt, gt_ids, use_points, use_boxes, n_positives, n_negatives, dilation, transform_function,
+ )
+ else:
+ input_point, input_label, input_box = cached_prompts
+
+ # Make a copy of the point prompts to return them at the end.
+ prompts = deepcopy((input_point, input_label, input_box))
+
+ # Transform the prompts into batches
+ device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
+ input_point = torch.tensor(np.array(input_point)).to(device) if len(input_point) > 0 else None
+ input_label = torch.tensor(np.array(input_label)).to(device) if len(input_label) > 0 else None
+ input_box = torch.tensor(np.array(input_box)).to(device) if len(input_box) > 0 else None
+
+ # Use multi-masking only if we have a single positive point without box
+ multimasking = False
+ if not use_boxes and (n_positives == 1 and n_negatives == 0):
+ multimasking = True
+
+ # Run the batched inference.
+ n_samples = input_box.shape[0] if input_point is None else input_point.shape[0]
+ n_batches = int(np.ceil(float(n_samples) / batch_size))
+ masks, ious = [], []
+ with torch.no_grad():
+ for batch_idx in range(n_batches):
+ batch_start = batch_idx * batch_size
+ batch_stop = min((batch_idx + 1) * batch_size, n_samples)
+
+ batch_points = None if input_point is None else input_point[batch_start:batch_stop]
+ batch_labels = None if input_label is None else input_label[batch_start:batch_stop]
+ batch_boxes = None if input_box is None else input_box[batch_start:batch_stop]
+
+ batch_masks, batch_ious, _ = predictor.predict_torch(
+ point_coords=batch_points, point_labels=batch_labels,
+ boxes=batch_boxes, multimask_output=multimasking
+ )
+ masks.append(batch_masks)
+ ious.append(batch_ious)
+ masks = torch.cat(masks)
+ ious = torch.cat(ious)
+ assert len(masks) == len(ious) == n_samples
+
+ # TODO we should actually use non-max suppression here
+ # I will implement it somewhere to have it refactored
+ instance_labels = np.zeros_like(gt, dtype=int)
+ for m, iou, gt_idx in zip(masks, ious, gt_ids):
+ best_idx = torch.argmax(iou)
+ best_mask = m[best_idx]
+ instance_labels[best_mask.detach().cpu().numpy()] = gt_idx
+
+ return instance_labels, prompts
+
+
+def get_predictor(
+ checkpoint_path: Union[str, os.PathLike],
+ model_type: str,
+ return_state: bool = False,
+ is_custom_model: Optional[bool] = None,
+) -> SamPredictor:
+ """Get the segment anything predictor from an exported or custom checkpoint.
+
+ Args:
+ checkpoint_path: The checkpoint filepath.
+ model_type: The type of the model, either vit_h, vit_b or vit_l.
+ return_state: Whether to return the complete state of the checkpoint in addtion to the predictor.
+ is_custom_model: Whether this is a custom model or not.
+ Returns:
+ The segment anything predictor.
+ """
+ # By default we check if the model follows the torch_em checkpint naming scheme to check whether it is a
+ # custom model or not. This can be over-ridden by passing True or False for is_custom_model.
+ is_custom_model = checkpoint_path.split("/")[-1] == "best.pt" if is_custom_model is None else is_custom_model
+
+ if is_custom_model: # Finetuned SAM model
+ predictor = util.get_custom_sam_model(
+ checkpoint_path=checkpoint_path, model_type=model_type, return_state=return_state
+ )
+ else: # Vanilla SAM model
+ assert not return_state
+ predictor = util.get_sam_model(model_type=model_type, checkpoint_path=checkpoint_path) # type: ignore
+ return predictor
+
+
+def precompute_all_embeddings(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+) -> None:
+ """Precompute all image embeddings.
+
+ To enable running different inference tasks in parallel afterwards.
+
+ Args:
+ predictor: The SegmentAnything predictor.
+ image_paths: The image file paths.
+ embedding_dir: The directory where the embeddings will be saved.
+ """
+ for image_path in tqdm(image_paths, desc="Precompute embeddings"):
+ image_name = os.path.basename(image_path)
+ im = imageio.imread(image_path)
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ util.precompute_image_embeddings(predictor, im, embedding_path, ndim=2)
+
+
+def _precompute_prompts(gt_path, use_points, use_boxes, n_positives, n_negatives, dilation, transform_function):
+ name = os.path.basename(gt_path)
+
+ gt = imageio.imread(gt_path).astype("uint32")
+ gt = relabel_sequential(gt)[0]
+ gt_ids = np.unique(gt)[1:]
+
+ input_point, input_label, input_box = _get_batched_prompts(
+ gt, gt_ids, use_points, use_boxes, n_positives, n_negatives, dilation, transform_function
+ )
+
+ if use_boxes and not use_points:
+ return name, input_box
+ return name, (input_point, input_label)
+
+
+def precompute_all_prompts(
+ gt_paths: List[Union[str, os.PathLike]],
+ prompt_save_dir: Union[str, os.PathLike],
+ prompt_settings: List[Dict[str, Any]],
+) -> None:
+ """Precompute all point prompts.
+
+ To enable running different inference tasks in parallel afterwards.
+
+ Args:
+ gt_paths: The file paths to the ground-truth segmentations.
+ prompt_save_dir: The directory where the prompt files will be saved.
+ prompt_settings: The settings for which the prompts will be computed.
+ """
+ os.makedirs(prompt_save_dir, exist_ok=True)
+ transform_function = ResizeLongestSide(1024)
+
+ for settings in tqdm(prompt_settings, desc="Precompute prompts"):
+
+ use_points, use_boxes = settings["use_points"], settings["use_boxes"]
+ n_positives, n_negatives = settings["n_positives"], settings["n_negatives"]
+ dilation = settings.get("dilation", 5)
+
+ # check if the prompts were already computed
+ if use_boxes and not use_points:
+ prompt_save_path = os.path.join(prompt_save_dir, "boxes.pkl")
+ else:
+ prompt_save_path = os.path.join(prompt_save_dir, f"points-p{n_positives}-n{n_negatives}.pkl")
+ if os.path.exists(prompt_save_path):
+ continue
+
+ results = []
+ for gt_path in tqdm(gt_paths, desc=f"Precompute prompts for p{n_positives}-n{n_negatives}"):
+ prompts = _precompute_prompts(
+ gt_path,
+ use_points=use_points,
+ use_boxes=use_boxes,
+ n_positives=n_positives,
+ n_negatives=n_negatives,
+ dilation=dilation,
+ transform_function=transform_function,
+ )
+ results.append(prompts)
+
+ saved_prompts = {res[0]: res[1] for res in results}
+ with open(prompt_save_path, "wb") as f:
+ pickle.dump(saved_prompts, f)
+
+
+def _get_prompt_caching(prompt_save_dir, use_points, use_boxes, n_positives, n_negatives):
+
+ def get_prompt_type_caching(use_type, save_name):
+ if not use_type:
+ return None, False, None
+
+ prompt_save_path = os.path.join(prompt_save_dir, save_name)
+ if os.path.exists(prompt_save_path):
+ print("Using precomputed prompts from", prompt_save_path)
+ # We delay loading the prompts, so we only have to load them once they're needed the first time.
+ # This avoids loading the prompts (which are in a big pickle file) if all predictions are done already.
+ cached_prompts = prompt_save_path
+ save_prompts = False
+ else:
+ print("Saving prompts in", prompt_save_path)
+ cached_prompts = {}
+ save_prompts = True
+ return cached_prompts, save_prompts, prompt_save_path
+
+ # Check if prompt serialization is enabled.
+ # If it is then load the prompts if they are already cached and otherwise store them.
+ if prompt_save_dir is None:
+ print("Prompts are not cached.")
+ cached_point_prompts, cached_box_prompts = None, None
+ save_point_prompts, save_box_prompts = False, False
+ point_prompt_save_path, box_prompt_save_path = None, None
+ else:
+ cached_point_prompts, save_point_prompts, point_prompt_save_path = get_prompt_type_caching(
+ use_points, f"points-p{n_positives}-n{n_negatives}.pkl"
+ )
+ cached_box_prompts, save_box_prompts, box_prompt_save_path = get_prompt_type_caching(
+ use_boxes, "boxes.pkl"
+ )
+
+ return (cached_point_prompts, save_point_prompts, point_prompt_save_path,
+ cached_box_prompts, save_box_prompts, box_prompt_save_path)
+
+
+def run_inference_with_prompts(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ gt_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ prediction_dir: Union[str, os.PathLike],
+ use_points: bool,
+ use_boxes: bool,
+ n_positives: int,
+ n_negatives: int,
+ dilation: int = 5,
+ prompt_save_dir: Optional[Union[str, os.PathLike]] = None,
+ batch_size: int = 512,
+) -> None:
+ """Run segment anything inference for multiple images using prompts derived form groundtruth.
+
+ Args:
+ predictor: The SegmentAnything predictor.
+ image_paths: The image file paths.
+ gt_paths: The ground-truth segmentation file paths.
+ embedding_dir: The directory where the image embddings will be saved or are already saved.
+ use_points: Whether to use point prompts.
+ use_boxes: Whetehr to use box prompts
+ n_positives: The number of positive point prompts that will be sampled.
+ n_negativess: The number of negative point prompts that will be sampled.
+ dilation: The dilation factor for the radius around the ground-truth object
+ around which points will not be sampled.
+ prompt_save_dir: The directory where point prompts will be saved or are already saved.
+ This enables running multiple experiments in a reproducible manner.
+ batch_size: The batch size used for batched prediction.
+ """
+ if not (use_points or use_boxes):
+ raise ValueError("You need to use at least one of point or box prompts.")
+
+ if len(image_paths) != len(gt_paths):
+ raise ValueError(f"Expect same number of images and gt images, got {len(image_paths)}, {len(gt_paths)}")
+
+ (cached_point_prompts, save_point_prompts, point_prompt_save_path,
+ cached_box_prompts, save_box_prompts, box_prompt_save_path) = _get_prompt_caching(
+ prompt_save_dir, use_points, use_boxes, n_positives, n_negatives
+ )
+
+ for image_path, gt_path in tqdm(
+ zip(image_paths, gt_paths), total=len(image_paths), desc="Run inference with prompts"
+ ):
+ image_name = os.path.basename(image_path)
+ label_name = os.path.basename(gt_path)
+
+ # We skip the images that already have been segmented.
+ prediction_path = os.path.join(prediction_dir, image_name)
+ if os.path.exists(prediction_path):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ assert os.path.exists(gt_path), gt_path
+
+ im = imageio.imread(image_path)
+ gt = imageio.imread(gt_path).astype("uint32")
+ gt = relabel_sequential(gt)[0]
+
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ image_embeddings = util.precompute_image_embeddings(predictor, im, embedding_path, ndim=2)
+ util.set_precomputed(predictor, image_embeddings)
+
+ this_prompts, cached_point_prompts, cached_box_prompts = _load_prompts(
+ cached_point_prompts, save_point_prompts,
+ cached_box_prompts, save_box_prompts,
+ label_name
+ )
+ instances, this_prompts = _run_inference_with_prompts_for_image(
+ predictor, gt, n_positives=n_positives, n_negatives=n_negatives,
+ dilation=dilation, use_points=use_points, use_boxes=use_boxes,
+ batch_size=batch_size, cached_prompts=this_prompts
+ )
+
+ if save_point_prompts:
+ cached_point_prompts[label_name] = this_prompts[:2]
+ if save_box_prompts:
+ cached_box_prompts[label_name] = this_prompts[-1]
+
+ # It's important to compress here, otherwise the predictions would take up a lot of space.
+ imageio.imwrite(prediction_path, instances, compression=5)
+
+ # Save the prompts if we run experiments with prompt caching and have computed them
+ # for the first time.
+ if save_point_prompts:
+ with open(point_prompt_save_path, "wb") as f:
+ pickle.dump(cached_point_prompts, f)
+ if save_box_prompts:
+ with open(box_prompt_save_path, "wb") as f:
+ pickle.dump(cached_box_prompts, f)
+
+
+def _save_segmentation(masks, prediction_path):
+ # masks to segmentation
+ masks = masks.cpu().numpy().squeeze().astype("bool")
+ shape = masks.shape[-2:]
+ masks = [{"segmentation": mask, "area": mask.sum()} for mask in masks]
+ segmentation = mask_data_to_segmentation(masks, shape, with_background=True)
+ imageio.imwrite(prediction_path, segmentation)
+
+
+def _run_inference_with_iterative_prompting_for_image(
+ model,
+ image,
+ gt,
+ n_iterations,
+ device,
+ use_boxes,
+ prediction_paths,
+ batch_size,
+):
+ assert len(prediction_paths) == n_iterations, f"{len(prediction_paths)}, {n_iterations}"
+ to_sam_inputs = ConvertToSamInputs()
+
+ image = torch.from_numpy(
+ image[None, None] if image.ndim == 2 else image[None]
+ )
+ gt = torch.from_numpy(gt[None].astype("int32"))
+
+ n_pos = 0 if use_boxes else 1
+ batched_inputs, sampled_ids = to_sam_inputs(image, gt, n_pos=n_pos, n_neg=0, get_boxes=use_boxes)
+
+ input_images = torch.stack([model.preprocess(x=x["image"].to(device)) for x in batched_inputs], dim=0)
+ image_embeddings = model.image_embeddings_oft(input_images)
+
+ multimasking = n_pos == 1
+ prompt_generator = IterativePromptGenerator()
+
+ n_samples = len(sampled_ids[0])
+ n_batches = int(np.ceil(float(n_samples) / batch_size))
+
+ for iteration in range(n_iterations):
+ final_masks = []
+ for batch_idx in range(n_batches):
+ batch_start = batch_idx * batch_size
+ batch_stop = min((batch_idx + 1) * batch_size, n_samples)
+
+ this_batched_inputs = [{
+ k: v[batch_start:batch_stop] if k in ("point_coords", "point_labels") else v
+ for k, v in batched_inputs[0].items()
+ }]
+
+ sampled_binary_y = torch.stack([
+ torch.stack([_gt == idx for idx in sampled[batch_start:batch_stop]])[:, None]
+ for _gt, sampled in zip(gt, sampled_ids)
+ ]).to(torch.float32)
+
+ batched_outputs = model(
+ this_batched_inputs,
+ multimask_output=multimasking if iteration == 0 else False,
+ image_embeddings=image_embeddings
+ )
+
+ masks, logits_masks = [], []
+ for m in batched_outputs:
+ mask, l_mask = [], []
+ for _m, _l, _iou in zip(m["masks"], m["low_res_masks"], m["iou_predictions"]):
+ best_iou_idx = torch.argmax(_iou)
+ mask.append(torch.sigmoid(_m[best_iou_idx][None]))
+ l_mask.append(_l[best_iou_idx][None])
+ mask, l_mask = torch.stack(mask), torch.stack(l_mask)
+ masks.append(mask)
+ logits_masks.append(l_mask)
+
+ masks, logits_masks = torch.stack(masks), torch.stack(logits_masks)
+ masks = (masks > 0.5).to(torch.float32)
+ final_masks.append(masks)
+
+ for _pred, _gt, _inp, logits in zip(masks, sampled_binary_y, this_batched_inputs, logits_masks):
+ next_coords, next_labels = prompt_generator(_gt, _pred, _inp["point_coords"], _inp["point_labels"])
+ _inp["point_coords"], _inp["point_labels"], _inp["mask_inputs"] = next_coords, next_labels, logits
+
+ final_masks = torch.cat(final_masks, dim=1)
+ _save_segmentation(final_masks, prediction_paths[iteration])
+
+
+def run_inference_with_iterative_prompting(
+ checkpoint_path: Union[str, os.PathLike],
+ model_type: str,
+ image_paths: List[Union[str, os.PathLike]],
+ gt_paths: List[Union[str, os.PathLike]],
+ prediction_root: Union[str, os.PathLike],
+ use_boxes: bool,
+ n_iterations: int = 8,
+ batch_size: int = 32,
+) -> None:
+ """@private"""
+ warnings.warn("The iterative prompting functionality is not working correctly yet.")
+
+ device = torch.device("cuda")
+ model = get_trainable_sam_model(model_type, checkpoint_path)
+
+ # create all prediction folders
+ for i in range(n_iterations):
+ os.makedirs(os.path.join(prediction_root, f"iteration{i:02}"), exist_ok=True)
+
+ for image_path, gt_path in tqdm(
+ zip(image_paths, gt_paths), total=len(image_paths), desc="Run inference with prompts"
+ ):
+ image_name = os.path.basename(image_path)
+
+ prediction_paths = [os.path.join(prediction_root, f"iteration{i:02}", image_name) for i in range(n_iterations)]
+ if all(os.path.exists(prediction_path) for prediction_path in prediction_paths):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ assert os.path.exists(gt_path), gt_path
+
+ image = imageio.imread(image_path)
+ gt = imageio.imread(gt_path).astype("uint32")
+ gt = relabel_sequential(gt)[0]
+
+ with torch.no_grad():
+ _run_inference_with_iterative_prompting_for_image(
+ model, image, gt, n_iterations, device, use_boxes, prediction_paths, batch_size,
+ )
diff --git a/micro_sam/evaluation/livecell.py b/micro_sam/evaluation/livecell.py
new file mode 100644
index 00000000..752a0359
--- /dev/null
+++ b/micro_sam/evaluation/livecell.py
@@ -0,0 +1,362 @@
+"""Inference and evaluation for the [LiveCELL dataset](https://www.nature.com/articles/s41592-021-01249-6) and
+the different cell lines contained in it.
+"""
+
+import argparse
+import json
+import os
+
+from glob import glob
+from typing import List, Optional, Union
+
+import numpy as np
+import pandas as pd
+
+from segment_anything import SamPredictor
+from tqdm import tqdm
+
+from ..instance_segmentation import AutomaticMaskGenerator, EmbeddingMaskGenerator
+from . import automatic_mask_generation, inference, evaluation
+from .experiments import default_experiment_settings, full_experiment_settings
+
+CELL_TYPES = ["A172", "BT474", "BV2", "Huh7", "MCF7", "SHSY5Y", "SkBr3", "SKOV3"]
+
+
+#
+# Inference
+#
+
+
+def _get_livecell_paths(input_folder, split="test", n_val_per_cell_type=None):
+ assert split in ["val", "test"]
+ assert os.path.exists(input_folder), "Please download the LIVECell Dataset"
+
+ if split == "test":
+
+ img_dir = os.path.join(input_folder, "images", "livecell_test_images")
+ assert os.path.exists(img_dir), "The LIVECell Dataset is incomplete"
+ gt_dir = os.path.join(input_folder, "annotations", "livecell_test_images")
+ assert os.path.exists(gt_dir), "The LIVECell Dataset is incomplete"
+ image_paths, gt_paths = [], []
+ for ctype in CELL_TYPES:
+ for img_path in glob(os.path.join(img_dir, f"{ctype}*")):
+ image_paths.append(img_path)
+ img_name = os.path.basename(img_path)
+ gt_path = os.path.join(gt_dir, ctype, img_name)
+ assert os.path.exists(gt_path), gt_path
+ gt_paths.append(gt_path)
+ else:
+
+ with open(os.path.join(input_folder, "val.json")) as f:
+ data = json.load(f)
+ livecell_val_ids = [i["file_name"] for i in data["images"]]
+
+ img_dir = os.path.join(input_folder, "images", "livecell_train_val_images")
+ assert os.path.exists(img_dir), "The LIVECell Dataset is incomplete"
+ gt_dir = os.path.join(input_folder, "annotations", "livecell_train_val_images")
+ assert os.path.exists(gt_dir), "The LIVECell Dataset is incomplete"
+
+ image_paths, gt_paths = [], []
+ count_per_cell_type = {ct: 0 for ct in CELL_TYPES}
+
+ for img_name in livecell_val_ids:
+ cell_type = img_name.split("_")[0]
+ if n_val_per_cell_type is not None and count_per_cell_type[cell_type] >= n_val_per_cell_type:
+ continue
+
+ image_paths.append(os.path.join(img_dir, img_name))
+ gt_paths.append(os.path.join(gt_dir, cell_type, img_name))
+ count_per_cell_type[cell_type] += 1
+
+ return image_paths, gt_paths
+
+
+def livecell_inference(
+ checkpoint: Union[str, os.PathLike],
+ input_folder: Union[str, os.PathLike],
+ model_type: str,
+ experiment_folder: Union[str, os.PathLike],
+ use_points: bool,
+ use_boxes: bool,
+ n_positives: Optional[int] = None,
+ n_negatives: Optional[int] = None,
+ prompt_folder: Optional[Union[str, os.PathLike]] = None,
+ predictor: Optional[SamPredictor] = None,
+) -> None:
+ """Run inference for livecell with a fixed prompt setting.
+
+ Args:
+ checkpoint: The segment anything model checkpoint.
+ input_folder: The folder with the livecell data.
+ model_type: The type of the segment anything model.
+ experiment_folder: The folder where to save all data associated with the experiment.
+ use_points: Whether to use point prompts.
+ use_boxes: Whether to use box prompts.
+ n_positives: The number of positive point prompts.
+ n_negatives: The number of negative point prompts.
+ prompt_folder: The folder where the prompts should be saved.
+ predictor: The segment anything predictor.
+ """
+ image_paths, gt_paths = _get_livecell_paths(input_folder)
+ if predictor is None:
+ predictor = inference.get_predictor(checkpoint, model_type)
+
+ if use_boxes and use_points:
+ assert (n_positives is not None) and (n_negatives is not None)
+ setting_name = f"box/p{n_positives}-n{n_negatives}"
+ elif use_boxes:
+ setting_name = "box/p0-n0"
+ elif use_points:
+ assert (n_positives is not None) and (n_negatives is not None)
+ setting_name = f"points/p{n_positives}-n{n_negatives}"
+ else:
+ raise ValueError("You need to use at least one of point or box prompts.")
+
+ # we organize all folders with data from this experiment beneath 'experiment_folder'
+ prediction_folder = os.path.join(experiment_folder, setting_name) # where the predicted segmentations are saved
+ os.makedirs(prediction_folder, exist_ok=True)
+ embedding_folder = os.path.join(experiment_folder, "embeddings") # where the precomputed embeddings are saved
+ os.makedirs(embedding_folder, exist_ok=True)
+
+ # NOTE: we can pass an external prompt folder, to make re-use prompts from another experiment
+ # for reproducibility / fair comparison of results
+ if prompt_folder is None:
+ prompt_folder = os.path.join(experiment_folder, "prompts")
+ os.makedirs(prompt_folder, exist_ok=True)
+
+ inference.run_inference_with_prompts(
+ predictor,
+ image_paths,
+ gt_paths,
+ embedding_dir=embedding_folder,
+ prediction_dir=prediction_folder,
+ prompt_save_dir=prompt_folder,
+ use_points=use_points,
+ use_boxes=use_boxes,
+ n_positives=n_positives,
+ n_negatives=n_negatives,
+ )
+
+
+def run_livecell_amg(
+ checkpoint: Union[str, os.PathLike],
+ input_folder: Union[str, os.PathLike],
+ model_type: str,
+ experiment_folder: Union[str, os.PathLike],
+ iou_thresh_values: Optional[List[float]] = None,
+ stability_score_values: Optional[List[float]] = None,
+ verbose_gs: bool = False,
+ n_val_per_cell_type: int = 25,
+ use_mws: bool = False,
+) -> None:
+ """Run automatic mask generation grid-search and inference for livecell.
+
+ Args:
+ checkpoint: The segment anything model checkpoint.
+ input_folder: The folder with the livecell data.
+ model_type: The type of the segmenta anything model.
+ experiment_folder: The folder where to save all data associated with the experiment.
+ iou_thresh_values: The values for `pred_iou_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ stability_score_values: The values for `stability_score_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ verbose_gs: Whether to run the gridsearch for individual images in a verbose mode.
+ n_val_per_cell_type: The number of validation images per cell type.
+ use_mws: Whether to use the mutex watershed based automatic mask generator approach.
+ """
+ embedding_folder = os.path.join(experiment_folder, "embeddings") # where the precomputed embeddings are saved
+ os.makedirs(embedding_folder, exist_ok=True)
+
+ if use_mws:
+ amg_prefix = "amg_mws"
+ AMG = EmbeddingMaskGenerator
+ else:
+ amg_prefix = "amg"
+ AMG = AutomaticMaskGenerator
+
+ # where the predictions are saved
+ prediction_folder = os.path.join(experiment_folder, amg_prefix, "inference")
+ os.makedirs(prediction_folder, exist_ok=True)
+
+ # where the grid-search results are saved
+ gs_result_folder = os.path.join(experiment_folder, amg_prefix, "grid_search")
+ os.makedirs(gs_result_folder, exist_ok=True)
+
+ val_image_paths, val_gt_paths = _get_livecell_paths(input_folder, "val", n_val_per_cell_type=n_val_per_cell_type)
+ test_image_paths, _ = _get_livecell_paths(input_folder, "test")
+
+ predictor = inference.get_predictor(checkpoint, model_type)
+ automatic_mask_generation.run_amg_grid_search_and_inference(
+ predictor, val_image_paths, val_gt_paths, test_image_paths,
+ embedding_folder, prediction_folder, gs_result_folder,
+ iou_thresh_values=iou_thresh_values, stability_score_values=stability_score_values,
+ AMG=AMG, verbose_gs=verbose_gs,
+ )
+
+
+def _run_multiple_prompt_settings(args, prompt_settings):
+ predictor = inference.get_predictor(args.ckpt, args.model)
+ for settings in prompt_settings:
+ livecell_inference(
+ args.ckpt,
+ args.input,
+ args.model,
+ args.experiment_folder,
+ use_points=settings["use_points"],
+ use_boxes=settings["use_boxes"],
+ n_positives=settings["n_positives"],
+ n_negatives=settings["n_negatives"],
+ prompt_folder=args.prompt_folder,
+ predictor=predictor
+ )
+
+
+def run_livecell_inference() -> None:
+ """Run LiveCELL inference with command line tool."""
+ parser = argparse.ArgumentParser()
+
+ # the checkpoint, input and experiment folder
+ parser.add_argument("-c", "--ckpt", type=str, required=True,
+ help="Provide model checkpoints (vanilla / finetuned).")
+ parser.add_argument("-i", "--input", type=str, required=True,
+ help="Provide the data directory for LIVECell Dataset.")
+ parser.add_argument("-e", "--experiment_folder", type=str, required=True,
+ help="Provide the path where all data for the inference run will be stored.")
+ parser.add_argument("-m", "--model", type=str, required=True,
+ help="Pass the checkpoint-specific model name being used for inference.")
+
+ # the experiment type:
+ # - default settings (p1-n0, p2-n4, box)
+ # - full experiment (ranges: p:1-16, n:0-16)
+ # - automatic mask generation (auto)
+ # if none of the two are active then the prompt setting arguments will be parsed
+ # and used to run inference for a single prompt setting
+ parser.add_argument("-f", "--full_experiment", action="store_true")
+ parser.add_argument("-d", "--default_experiment", action="store_true")
+ parser.add_argument("-a", "--auto_mask_generation", action="store_true")
+
+ # the prompt settings for an individual inference run
+ parser.add_argument("--box", action="store_true", help="Activate box-prompted based inference")
+ parser.add_argument("--points", action="store_true", help="Activate point-prompt based inference")
+ parser.add_argument("-p", "--positive", type=int, default=1, help="No. of positive prompts")
+ parser.add_argument("-n", "--negative", type=int, default=0, help="No. of negative prompts")
+
+ # optional external prompt folder
+ parser.add_argument("--prompt_folder", help="")
+
+ args = parser.parse_args()
+ if sum([args.full_experiment, args.default_experiment, args.auto_mask_generation]) > 2:
+ raise ValueError("Can only run one of 'full_experiment', 'default_experiment' or 'auto_mask_generation'.")
+
+ if args.full_experiment:
+ prompt_settings = full_experiment_settings(args.box)
+ _run_multiple_prompt_settings(args, prompt_settings)
+ elif args.default_experiment:
+ prompt_settings = default_experiment_settings()
+ _run_multiple_prompt_settings(args, prompt_settings)
+ elif args.auto_mask_generation:
+ run_livecell_amg(args.ckpt, args.input, args.model, args.experiment_folder)
+ else:
+ livecell_inference(
+ args.ckpt, args.input, args.model, args.experiment_folder,
+ args.points, args.box, args.positive, args.negative, args.prompt_folder,
+ )
+
+
+#
+# Evaluation
+#
+
+
+def evaluate_livecell_predictions(
+ gt_dir: Union[os.PathLike, str],
+ pred_dir: Union[os.PathLike, str],
+ verbose: bool,
+) -> None:
+ """Evaluate LiveCELL predictions.
+
+ Args:
+ gt_dir: The folder with the groundtruth segmentations.
+ pred_dir: The folder with the segmentation predictions.
+ verbose: Whether to run the evaluation in verbose mode.
+ """
+ assert os.path.exists(gt_dir), gt_dir
+ assert os.path.exists(pred_dir), pred_dir
+
+ msas, sa50s, sa75s = [], [], []
+ msas_ct, sa50s_ct, sa75s_ct = [], [], []
+
+ for ct in tqdm(CELL_TYPES, desc="Evaluate livecell predictions", disable=not verbose):
+
+ gt_pattern = os.path.join(gt_dir, f"{ct}/*.tif")
+ gt_paths = glob(gt_pattern)
+ assert len(gt_paths) > 0, "gt_pattern"
+
+ pred_paths = [
+ os.path.join(pred_dir, os.path.basename(path)) for path in gt_paths
+ ]
+
+ this_msas, this_sa50s, this_sa75s = evaluation._run_evaluation(
+ gt_paths, pred_paths, False
+ )
+
+ msas.extend(this_msas), sa50s.extend(this_sa50s), sa75s.extend(this_sa75s)
+ msas_ct.append(np.mean(this_msas))
+ sa50s_ct.append(np.mean(this_sa50s))
+ sa75s_ct.append(np.mean(this_sa75s))
+
+ result_dict = {
+ "cell_type": CELL_TYPES + ["Total"],
+ "msa": msas_ct + [np.mean(msas)],
+ "sa50": sa50s_ct + [np.mean(sa50s_ct)],
+ "sa75": sa75s_ct + [np.mean(sa75s_ct)],
+ }
+ df = pd.DataFrame.from_dict(result_dict)
+ df = df.round(decimals=4)
+ return df
+
+
+def run_livecell_evaluation() -> None:
+ """Run LiveCELL evaluation with command line tool."""
+ parser = argparse.ArgumentParser()
+ parser.add_argument(
+ "-i", "--input", required=True, help="Provide the data directory for LIVECell Dataset"
+ )
+ parser.add_argument(
+ "-e", "--experiment_folder", required=True,
+ help="Provide the path where the inference data is stored."
+ )
+ parser.add_argument(
+ "-f", "--force", action="store_true",
+ help="Force recomputation of already cached eval results."
+ )
+ args = parser.parse_args()
+
+ gt_dir = os.path.join(args.input, "annotations", "livecell_test_images")
+ assert os.path.exists(gt_dir), "The LiveCELL Dataset is incomplete"
+
+ experiment_folder = args.experiment_folder
+ save_root = os.path.join(experiment_folder, "results")
+
+ inference_root_names = ["points", "box", "amg/inference"]
+ for inf_root in inference_root_names:
+
+ pred_root = os.path.join(experiment_folder, inf_root)
+ if inf_root.startswith("amg"):
+ pred_folders = [pred_root]
+ else:
+ pred_folders = sorted(glob(os.path.join(pred_root, "*")))
+
+ if inf_root == "amg/inference":
+ save_folder = os.path.join(save_root, "amg")
+ else:
+ save_folder = os.path.join(save_root, inf_root)
+ os.makedirs(save_folder, exist_ok=True)
+
+ for pred_folder in tqdm(pred_folders, desc=f"Evaluate predictions for {inf_root} prompt settings"):
+ exp_name = os.path.basename(pred_folder)
+ save_path = os.path.join(save_folder, f"{exp_name}.csv")
+ if os.path.exists(save_path) and not args.force:
+ continue
+ results = evaluate_livecell_predictions(gt_dir, pred_folder, verbose=False)
+ results.to_csv(save_path, index=False)
diff --git a/micro_sam/evaluation/model_comparison.py b/micro_sam/evaluation/model_comparison.py
new file mode 100644
index 00000000..a65437b7
--- /dev/null
+++ b/micro_sam/evaluation/model_comparison.py
@@ -0,0 +1,429 @@
+"""Functionality for qualitative comparison of Segment Anything models on microscopy data.
+"""
+
+import os
+from functools import partial
+from glob import glob
+from pathlib import Path
+
+import h5py
+import matplotlib.pyplot as plt
+import numpy as np
+import pandas as pd
+import torch
+
+import skimage.draw as draw
+from scipy.ndimage import binary_dilation
+from skimage import exposure
+from skimage.segmentation import relabel_sequential, find_boundaries
+
+from tqdm import tqdm
+from typing import Optional, Union
+
+from .. import util
+from ..prompt_generators import PointAndBoxPromptGenerator
+from ..prompt_based_segmentation import segment_from_box, segment_from_points
+
+
+#
+# Compute all required data for the model comparison
+#
+
+
+def _predict_models_with_loader(loader, n_samples, prompt_generator, predictor1, predictor2, output_folder):
+ i = 0
+ os.makedirs(output_folder, exist_ok=True)
+
+ for x, y in tqdm(loader, total=n_samples):
+ out_path = os.path.join(output_folder, f"sample_{i}.h5")
+
+ im = x.numpy().squeeze()
+ if im.ndim == 3 and im.shape[0] == 3:
+ im = im.transpose((1, 2, 0))
+
+ gt = y.numpy().squeeze().astype("uint32")
+ gt = relabel_sequential(gt)[0]
+
+ emb1 = util.precompute_image_embeddings(predictor1, im, ndim=2)
+ util.set_precomputed(predictor1, emb1)
+
+ emb2 = util.precompute_image_embeddings(predictor2, im, ndim=2)
+ util.set_precomputed(predictor2, emb2)
+
+ centers, boxes = util.get_centers_and_bounding_boxes(gt)
+
+ with h5py.File(out_path, "a") as f:
+ f.create_dataset("image", data=im, compression="gzip")
+
+ gt_ids = np.unique(gt)[1:]
+ for gt_id in tqdm(gt_ids):
+
+ gt_mask = (gt == gt_id).astype("uint8")
+ point_coords, point_labels, box, _ = prompt_generator(gt, gt_id, boxes[gt_id], centers[gt_id])
+
+ box = np.array(box[0])
+ mask1_box = segment_from_box(predictor1, box)
+ mask2_box = segment_from_box(predictor2, box)
+ mask1_box, mask2_box = mask1_box.squeeze(), mask2_box.squeeze()
+
+ point_coords, point_labels = np.array(point_coords), np.array(point_labels)
+ mask1_points = segment_from_points(predictor1, point_coords, point_labels)
+ mask2_points = segment_from_points(predictor2, point_coords, point_labels)
+ mask1_points, mask2_points = mask1_points.squeeze(), mask2_points.squeeze()
+
+ with h5py.File(out_path, "a") as f:
+ g = f.create_group(str(gt_id))
+ g.attrs["point_coords"] = point_coords
+ g.attrs["point_labels"] = point_labels
+ g.attrs["box"] = box
+
+ g.create_dataset("gt_mask", data=gt_mask, compression="gzip")
+ g.create_dataset("box/mask1", data=mask1_box.astype("uint8"), compression="gzip")
+ g.create_dataset("box/mask2", data=mask2_box.astype("uint8"), compression="gzip")
+ g.create_dataset("points/mask1", data=mask1_points.astype("uint8"), compression="gzip")
+ g.create_dataset("points/mask2", data=mask2_points.astype("uint8"), compression="gzip")
+
+ i += 1
+ if i >= n_samples:
+ return
+
+
+def generate_data_for_model_comparison(
+ loader: torch.utils.data.DataLoader,
+ output_folder: Union[str, os.PathLike],
+ model_type1: str,
+ model_type2: str,
+ n_samples: int,
+) -> None:
+ """Generate samples for qualitative model comparison.
+
+ This precomputes the input for `model_comparison` and `model_comparison_with_napari`.
+
+ Args:
+ loader: The torch dataloader from which samples are drawn.
+ output_folder: The folder where the samples will be saved.
+ model_type1: The first model to use for comparison.
+ The value needs to be a valid model_type for `micro_sam.util.get_sam_model`.
+ model_type1: The second model to use for comparison.
+ The value needs to be a valid model_type for `micro_sam.util.get_sam_model`.
+ n_samples: The number of samples to draw from the dataloader.
+ """
+ prompt_generator = PointAndBoxPromptGenerator(
+ n_positive_points=1,
+ n_negative_points=0,
+ dilation_strength=3,
+ get_point_prompts=True,
+ get_box_prompts=True,
+ )
+ predictor1 = util.get_sam_model(model_type=model_type1)
+ predictor2 = util.get_sam_model(model_type=model_type2)
+ _predict_models_with_loader(loader, n_samples, prompt_generator, predictor1, predictor2, output_folder)
+
+
+#
+# Visual evaluation accroding to metrics
+#
+
+
+def _evaluate_samples(f, prefix, min_size):
+ eval_result = {
+ "gt_id": [],
+ "score1": [],
+ "score2": [],
+ }
+ for name, group in f.items():
+ if name == "image":
+ continue
+
+ gt_mask = group["gt_mask"][:]
+
+ size = gt_mask.sum()
+ if size < min_size:
+ continue
+
+ m1 = group[f"{prefix}/mask1"][:]
+ m2 = group[f"{prefix}/mask2"][:]
+
+ score1 = util.compute_iou(gt_mask, m1)
+ score2 = util.compute_iou(gt_mask, m2)
+
+ eval_result["gt_id"].append(name)
+ eval_result["score1"].append(score1)
+ eval_result["score2"].append(score2)
+
+ eval_result = pd.DataFrame.from_dict(eval_result)
+ eval_result["advantage1"] = eval_result["score1"] - eval_result["score2"]
+ eval_result["advantage2"] = eval_result["score2"] - eval_result["score1"]
+ return eval_result
+
+
+def _overlay_mask(image, mask):
+ assert image.ndim in (2, 3)
+ # overlay the mask
+ if image.ndim == 2:
+ overlay = np.stack([image, image, image]).transpose((1, 2, 0))
+ else:
+ overlay = image
+ assert overlay.shape[-1] == 3
+ mask_overlay = np.zeros_like(overlay)
+ mask_overlay[mask == 1] = [255, 0, 0]
+ alpha = 0.6
+ overlay = alpha * overlay + (1.0 - alpha) * mask_overlay
+ return overlay.astype("uint8")
+
+
+def _enhance_image(im):
+ # apply CLAHE to improve the image quality
+ im -= im.min(axis=(0, 1), keepdims=True)
+ im /= (im.max(axis=(0, 1), keepdims=True) + 1e-6)
+ im = exposure.equalize_adapthist(im)
+ im *= 255
+ return im
+
+
+def _overlay_outline(im, mask, outline_dilation):
+ outline = find_boundaries(mask)
+ if outline_dilation > 0:
+ outline = binary_dilation(outline, iterations=outline_dilation)
+ overlay = im.copy()
+ overlay[outline] = [255, 255, 0]
+ return overlay
+
+
+def _overlay_box(im, prompt, outline_dilation):
+ start, end = prompt
+ rr, cc = draw.rectangle_perimeter(start, end=end, shape=im.shape[:2])
+
+ box_outline = np.zeros(im.shape[:2], dtype="bool")
+ box_outline[rr, cc] = 1
+ if outline_dilation > 0:
+ box_outline = binary_dilation(box_outline, iterations=outline_dilation)
+
+ overlay = im.copy()
+ overlay[box_outline] = [0, 255, 255]
+
+ return overlay
+
+
+# NOTE: we currently only support a single point
+def _overlay_points(im, prompt, radius):
+ coords, labels = prompt
+ # make sure we have a single positive prompt, other options are
+ # currently not supported
+ assert coords.shape[0] == labels.shape[0] == 1
+ assert labels[0] == 1
+
+ rr, cc = draw.disk(coords[0], radius, shape=im.shape[:2])
+ overlay = im.copy()
+ draw.set_color(overlay, (rr, cc), [0, 255, 255], alpha=1.0)
+
+ return overlay
+
+
+def _compare_eval(
+ f, eval_result, advantage_column,
+ n_images_per_sample, prefix,
+ sample_name, plot_folder,
+ point_radius, outline_dilation,
+):
+ result = eval_result.sort_values(advantage_column, ascending=False).iloc[:n_images_per_sample]
+ n_rows = result.shape[0]
+
+ image = f["image"][:]
+ is_box_prompt = prefix == "box"
+ overlay_prompts = partial(_overlay_box, outline_dilation=outline_dilation) if is_box_prompt else\
+ partial(_overlay_points, radius=point_radius)
+
+ def make_square(bb, shape):
+ box_shape = [b.stop - b.start for b in bb]
+ longest_side = max(box_shape)
+ padding = [(longest_side - sh) // 2 for sh in box_shape]
+ bb = tuple(
+ slice(max(b.start - pad, 0), min(b.stop + pad, sh)) for b, pad, sh in zip(bb, padding, shape)
+ )
+ return bb
+
+ def plot_ax(axis, i, row):
+ g = f[row.gt_id]
+
+ gt = g["gt_mask"][:]
+ mask1 = g[f"{prefix}/mask1"][:]
+ mask2 = g[f"{prefix}/mask2"][:]
+
+ fg_mask = (gt + mask1 + mask2) > 0
+ # if this is a box prompt we dilate the mask so that the bounding box
+ # can be seen
+ if is_box_prompt:
+ fg_mask = binary_dilation(fg_mask, iterations=5)
+ bb = np.where(fg_mask)
+ bb = tuple(
+ slice(int(b.min()), int(b.max() + 1)) for b in bb
+ )
+ bb = make_square(bb, fg_mask.shape)
+
+ offset = np.array([b.start for b in bb])
+ if is_box_prompt:
+ prompt = g.attrs["box"]
+ prompt = np.array(
+ [prompt[:2], prompt[2:]]
+ ) - offset
+ else:
+ prompt = (g.attrs["point_coords"] - offset, g.attrs["point_labels"])
+
+ im = _enhance_image(image[bb])
+ gt, mask1, mask2 = gt[bb], mask1[bb], mask2[bb]
+
+ im1 = _overlay_mask(im, mask1)
+ im1 = _overlay_outline(im1, gt, outline_dilation)
+ im1 = overlay_prompts(im1, prompt)
+ ax = axis[0] if i is None else axis[i, 0]
+ ax.axis("off")
+ ax.imshow(im1)
+
+ im2 = _overlay_mask(im, mask2)
+ im2 = _overlay_outline(im2, gt, outline_dilation)
+ im2 = overlay_prompts(im2, prompt)
+ ax = axis[1] if i is None else axis[i, 1]
+ ax.axis("off")
+ ax.imshow(im2)
+
+ if plot_folder is None:
+ fig, axis = plt.subplots(n_rows, 2)
+ for i, (_, row) in enumerate(result.iterrows()):
+ plot_ax(axis, i, row)
+ plt.show()
+ else:
+ for i, (_, row) in enumerate(result.iterrows()):
+ fig, axis = plt.subplots(1, 2)
+ plot_ax(axis, None, row)
+ plt.subplots_adjust(wspace=0.05, hspace=0)
+ plt.savefig(os.path.join(plot_folder, f"{sample_name}_{i}.png"), bbox_inches="tight")
+ plt.close()
+
+
+def _compare_prompts(
+ f, prefix, n_images_per_sample, min_size, sample_name, plot_folder,
+ point_radius, outline_dilation
+):
+ box_eval = _evaluate_samples(f, prefix, min_size)
+ if plot_folder is None:
+ plot_folder1, plot_folder2 = None, None
+ else:
+ plot_folder1 = os.path.join(plot_folder, "advantage1")
+ plot_folder2 = os.path.join(plot_folder, "advantage2")
+ os.makedirs(plot_folder1, exist_ok=True)
+ os.makedirs(plot_folder2, exist_ok=True)
+ _compare_eval(
+ f, box_eval, "advantage1", n_images_per_sample, prefix, sample_name, plot_folder1,
+ point_radius, outline_dilation
+ )
+ _compare_eval(
+ f, box_eval, "advantage2", n_images_per_sample, prefix, sample_name, plot_folder2,
+ point_radius, outline_dilation
+ )
+
+
+def _compare_models(
+ path, n_images_per_sample, min_size, plot_folder, point_radius, outline_dilation
+):
+ sample_name = Path(path).stem
+ with h5py.File(path, "r") as f:
+ if plot_folder is None:
+ plot_folder_points, plot_folder_box = None, None
+ else:
+ plot_folder_points = os.path.join(plot_folder, "points")
+ plot_folder_box = os.path.join(plot_folder, "box")
+ _compare_prompts(
+ f, "points", n_images_per_sample, min_size, sample_name, plot_folder_points,
+ point_radius, outline_dilation
+ )
+ _compare_prompts(
+ f, "box", n_images_per_sample, min_size, sample_name, plot_folder_box,
+ point_radius, outline_dilation
+ )
+
+
+def model_comparison(
+ output_folder: Union[str, os.PathLike],
+ n_images_per_sample: int,
+ min_size: int,
+ plot_folder: Optional[Union[str, os.PathLike]] = None,
+ point_radius: int = 4,
+ outline_dilation: int = 0,
+) -> None:
+ """Create images for a qualitative model comparision.
+
+ Args:
+ output_folder: The folder with the data precomputed by `generate_data_for_model_comparison`.
+ n_images_per_sample: The number of images to generate per precomputed sample.
+ min_size: The min size of ground-truth objects to take into account.
+ plot_folder: The folder where to save the plots. If not given the plots will be displayed.
+ point_radius: The radius of the point overlay.
+ outline_dilation: The dilation factor of the outline overlay.
+ """
+ files = glob(os.path.join(output_folder, "*.h5"))
+ for path in tqdm(files):
+ _compare_models(
+ path, n_images_per_sample, min_size, plot_folder, point_radius, outline_dilation
+ )
+
+
+#
+# Quick visual evaluation with napari
+#
+
+
+def _check_group(g, show_points):
+ import napari
+
+ image = g["image"][:]
+ gt = g["gt_mask"][:]
+ if show_points:
+ m1 = g["points/mask1"][:]
+ m2 = g["points/mask2"][:]
+ points = g.attrs["point_coords"]
+ else:
+ m1 = g["box/mask1"][:]
+ m2 = g["box/mask2"][:]
+ box = g.attrs["box"]
+ box = np.array([
+ [box[0], box[1]], [box[2], box[3]]
+ ])
+
+ v = napari.Viewer()
+ v.add_image(image)
+ v.add_labels(gt)
+ v.add_labels(m1)
+ v.add_labels(m2)
+ if show_points:
+ # TODO use point labels for coloring
+ v.add_points(
+ points,
+ edge_color="#00FF00",
+ symbol="o",
+ face_color="transparent",
+ edge_width=0.5,
+ size=12,
+ )
+ else:
+ v.add_shapes(
+ box, face_color="transparent", edge_color="green", edge_width=4,
+ )
+ napari.run()
+
+
+def model_comparison_with_napari(output_folder: Union[str, os.PathLike], show_points: bool = True) -> None:
+ """Use napari to display the qualtiative comparison results for two models.
+
+ Args:
+ output_folder: The folder with the data precomputed by `generate_data_for_model_comparison`.
+ show_points: Whether to show the results for point or for box prompts.
+ """
+ files = glob(os.path.join(output_folder, "*.h5"))
+ for path in files:
+ print("Comparing models in", path)
+ with h5py.File(path, "r") as f:
+ for name, g in f.items():
+ if name == "image":
+ continue
+ _check_group(g, show_points=show_points)
diff --git a/micro_sam/instance_segmentation.py b/micro_sam/instance_segmentation.py
index f21ad9fa..f1c5e644 100644
--- a/micro_sam/instance_segmentation.py
+++ b/micro_sam/instance_segmentation.py
@@ -54,6 +54,7 @@ def mask_data_to_segmentation(
masks: List[Dict[str, Any]],
shape: tuple[int, ...],
with_background: bool,
+ min_object_size: int = 0,
) -> np.ndarray:
"""Convert the output of the automatic mask generation to an instance segmentation.
@@ -63,6 +64,7 @@ def mask_data_to_segmentation(
shape: The image shape.
with_background: Whether the segmentation has background. If yes this function assures that the largest
object in the output will be mapped to zero (the background value).
+ min_object_size: The minimal size of an object in pixels.
Returns:
The instance segmentation.
"""
@@ -70,8 +72,12 @@ def mask_data_to_segmentation(
masks = sorted(masks, key=(lambda x: x["area"]), reverse=True)
segmentation = np.zeros(shape[:2], dtype="uint32")
- for seg_id, mask in enumerate(masks, 1):
+ seg_id = 1
+ for mask in masks:
+ if mask["area"] < min_object_size:
+ continue
segmentation[mask["segmentation"]] = seg_id
+ seg_id += 1
if with_background:
seg_ids, sizes = np.unique(segmentation, return_counts=True)
@@ -129,7 +135,6 @@ def _postprocess_batch(
original_size,
pred_iou_thresh,
stability_score_thresh,
- stability_score_offset,
box_nms_thresh,
):
orig_h, orig_w = original_size
@@ -139,28 +144,16 @@ def _postprocess_batch(
keep_mask = data["iou_preds"] > pred_iou_thresh
data.filter(keep_mask)
- # calculate stability score
- data["stability_score"] = amg_utils.calculate_stability_score(
- data["masks"], self._predictor.model.mask_threshold, stability_score_offset
- )
+ # filter by stability score
if stability_score_thresh > 0.0:
keep_mask = data["stability_score"] >= stability_score_thresh
data.filter(keep_mask)
- # threshold masks and calculate boxes
- data["masks"] = data["masks"] > self._predictor.model.mask_threshold
- data["boxes"] = amg_utils.batched_mask_to_box(data["masks"])
-
# filter boxes that touch crop boundaries
keep_mask = ~amg_utils.is_box_near_crop_edge(data["boxes"], crop_box, [0, 0, orig_w, orig_h])
if not torch.all(keep_mask):
data.filter(keep_mask)
- # compress to RLE
- data["masks"] = amg_utils.uncrop_masks(data["masks"], crop_box, orig_h, orig_w)
- data["rles"] = amg_utils.mask_to_rle_pytorch(data["masks"])
- del data["masks"]
-
# remove duplicates within this crop.
keep_by_nms = batched_nms(
data["boxes"].float(),
@@ -261,6 +254,32 @@ def _postprocess_masks(self, mask_data, min_mask_region_area, box_nms_thresh, cr
return curr_anns
+ def _to_mask_data(self, masks, iou_preds, crop_box, original_size, points=None):
+ orig_h, orig_w = original_size
+
+ # serialize predictions and store in MaskData
+ data = amg_utils.MaskData(masks=masks.flatten(0, 1), iou_preds=iou_preds.flatten(0, 1))
+ if points is not None:
+ data["points"] = torch.as_tensor(points.repeat(masks.shape[1], axis=0))
+
+ del masks
+
+ # calculate the stability scores
+ data["stability_score"] = amg_utils.calculate_stability_score(
+ data["masks"], self._predictor.model.mask_threshold, self._stability_score_offset
+ )
+
+ # threshold masks and calculate boxes
+ data["masks"] = data["masks"] > self._predictor.model.mask_threshold
+ data["boxes"] = amg_utils.batched_mask_to_box(data["masks"])
+
+ # compress to RLE
+ data["masks"] = amg_utils.uncrop_masks(data["masks"], crop_box, orig_h, orig_w)
+ data["rles"] = amg_utils.mask_to_rle_pytorch(data["masks"])
+ del data["masks"]
+
+ return data
+
def get_state(self) -> Dict[str, Any]:
"""Get the initialized state of the mask generator.
@@ -269,6 +288,7 @@ def get_state(self) -> Dict[str, Any]:
"""
if not self.is_initialized:
raise RuntimeError("The state has not been computed yet. Call initialize first.")
+
return {"crop_list": self.crop_list, "crop_boxes": self.crop_boxes, "original_size": self.original_size}
def set_state(self, state: Dict[str, Any]) -> None:
@@ -309,6 +329,7 @@ class AutomaticMaskGenerator(AMGBase):
crop_n_points_downscale_factor: How the number of points is downsampled when predicting with crops.
point_grids: A lisst over explicit grids of points used for sampling masks.
Normalized to [0, 1] with respect to the image coordinate system.
+ stability_score_offset: The amount to shift the cutoff when calculating the stability score.
"""
def __init__(
self,
@@ -319,6 +340,7 @@ def __init__(
crop_overlap_ratio: float = 512 / 1500,
crop_n_points_downscale_factor: int = 1,
point_grids: Optional[List[np.ndarray]] = None,
+ stability_score_offset: float = 1.0,
):
super().__init__()
@@ -339,8 +361,9 @@ def __init__(
self._crop_n_layers = crop_n_layers
self._crop_overlap_ratio = crop_overlap_ratio
self._crop_n_points_downscale_factor = crop_n_points_downscale_factor
+ self._stability_score_offset = stability_score_offset
- def _process_batch(self, points, im_size):
+ def _process_batch(self, points, im_size, crop_box, original_size):
# run model on this batch
transformed_points = self._predictor.transform.apply_coords(points, im_size)
in_points = torch.as_tensor(transformed_points, device=self._predictor.device)
@@ -351,24 +374,14 @@ def _process_batch(self, points, im_size):
multimask_output=True,
return_logits=True,
)
-
- # serialize predictions and store in MaskData
- data = amg_utils.MaskData(
- masks=masks.flatten(0, 1),
- iou_preds=iou_preds.flatten(0, 1),
- points=torch.as_tensor(points.repeat(masks.shape[1], axis=0)),
- )
+ data = self._to_mask_data(masks, iou_preds, crop_box, original_size, points=points)
del masks
-
return data
def _process_crop(self, image, crop_box, crop_layer_idx, verbose, precomputed_embeddings):
# crop the image and calculate embeddings
- if crop_box is None:
- cropped_im = image
- else:
- x0, y0, x1, y1 = crop_box
- cropped_im = image[y0:y1, x0:x1, :]
+ x0, y0, x1, y1 = crop_box
+ cropped_im = image[y0:y1, x0:x1, :]
cropped_im_size = cropped_im.shape[:2]
if not precomputed_embeddings:
@@ -387,7 +400,7 @@ def _process_crop(self, image, crop_box, crop_layer_idx, verbose, precomputed_em
disable=not verbose, total=n_batches,
desc="Predict masks for point grid prompts",
):
- batch_data = self._process_batch(points, cropped_im_size)
+ batch_data = self._process_batch(points, cropped_im_size, crop_box, self.original_size)
data.cat(batch_data)
del batch_data
@@ -415,6 +428,8 @@ def initialize(
verbose: Whether to print computation progress.
"""
original_size = image.shape[:2]
+ self._original_size = original_size
+
crop_boxes, layer_idxs = amg_utils.generate_crop_boxes(
original_size, self._crop_n_layers, self._crop_overlap_ratio
)
@@ -443,14 +458,12 @@ def initialize(
self._is_initialized = True
self._crop_list = crop_list
self._crop_boxes = crop_boxes
- self._original_size = original_size
@torch.no_grad()
def generate(
self,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- stability_score_offset: float = 1.0,
box_nms_thresh: float = 0.7,
crop_nms_thresh: float = 0.7,
min_mask_region_area: int = 0,
@@ -462,7 +475,6 @@ def generate(
pred_iou_thresh: Filter threshold in [0, 1], using the mask quality predicted by the model.
stability_score_thresh: Filter threshold in [0, 1], using the stability of the mask
under changes to the cutoff used to binarize the model prediction.
- stability_score_offset: The amount to shift the cutoff when calculating the stability score.
box_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks.
crop_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks between crops.
min_mask_region_area: Minimal size for the predicted masks.
@@ -481,7 +493,6 @@ def generate(
crop_box=crop_box, original_size=self.original_size,
pred_iou_thresh=pred_iou_thresh,
stability_score_thresh=stability_score_thresh,
- stability_score_offset=stability_score_offset,
box_nms_thresh=box_nms_thresh
)
data.cat(crop_data)
@@ -529,6 +540,7 @@ class EmbeddingMaskGenerator(AMGBase):
use_mask: Whether to use the initial segments as prompts.
use_points: Whether to use points derived from the initial segments as prompts.
box_extension: Factor for extending the bounding box prompts, given in the relative box size.
+ stability_score_offset: The amount to shift the cutoff when calculating the stability score.
"""
default_offsets = [[-1, 0], [0, -1], [-3, 0], [0, -3], [-9, 0], [0, -9]]
@@ -543,6 +555,7 @@ def __init__(
use_mask: bool = True,
use_points: bool = False,
box_extension: float = 0.05,
+ stability_score_offset: float = 1.0,
):
super().__init__()
@@ -555,6 +568,7 @@ def __init__(
self._use_mask = use_mask
self._use_points = use_points
self._box_extension = box_extension
+ self._stability_score_offset = stability_score_offset
# additional state that is set 'initialize'
self._initial_segmentation = None
@@ -581,7 +595,7 @@ def _compute_initial_segmentation(self):
return initial_segmentation
- def _compute_mask_data(self, initial_segmentation, original_size, verbose):
+ def _compute_mask_data(self, initial_segmentation, crop_box, original_size, verbose):
seg_ids = np.unique(initial_segmentation)
if seg_ids[0] == 0:
seg_ids = seg_ids[1:]
@@ -592,15 +606,13 @@ def _compute_mask_data(self, initial_segmentation, original_size, verbose):
mask = initial_segmentation == seg_id
masks, iou_preds, _ = segment_from_mask(
self._predictor, mask, original_size=original_size,
- multimask_output=True, return_logits=True, return_all=True,
+ multimask_output=False, return_logits=True, return_all=True,
use_box=self._use_box, use_mask=self._use_mask, use_points=self._use_points,
box_extension=self._box_extension,
)
- data = amg_utils.MaskData(
- masks=torch.from_numpy(masks),
- iou_preds=torch.from_numpy(iou_preds),
- seg_id=torch.from_numpy(np.full(len(masks), seg_id, dtype="int64")),
- )
+ # bring masks and iou_preds to a format compatible with _to_mask_data
+ masks, iou_preds = torch.from_numpy(masks[None]), torch.from_numpy(iou_preds[None])
+ data = self._to_mask_data(masks, iou_preds, crop_box, original_size)
del masks
mask_data.cat(data)
@@ -625,6 +637,11 @@ def initialize(
verbose: Whether to print computation progress.
"""
original_size = image.shape[:2]
+ self._original_size = original_size
+
+ # the crop box is always the full image
+ crop_box = [0, 0, original_size[1], original_size[0]]
+ self._crop_boxes = [crop_box]
if image_embeddings is None:
image_embeddings = util.precompute_image_embeddings(self._predictor, image,)
@@ -633,7 +650,7 @@ def initialize(
# compute the initial segmentation via embedding based MWS and then refine the masks
# with the segment anything model
initial_segmentation = self._compute_initial_segmentation()
- mask_data = self._compute_mask_data(initial_segmentation, original_size, verbose)
+ mask_data = self._compute_mask_data(initial_segmentation, crop_box, original_size, verbose)
# to be compatible with the file format of the super class we have to wrap the mask data in a list
crop_list = [mask_data]
@@ -641,18 +658,12 @@ def initialize(
self._is_initialized = True
self._initial_segmentation = initial_segmentation
self._crop_list = crop_list
- # the crop box is always the full image
- self._crop_boxes = [
- [0, 0, original_size[1], original_size[0]]
- ]
- self._original_size = original_size
@torch.no_grad()
def generate(
self,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- stability_score_offset: float = 1.0,
box_nms_thresh: float = 0.7,
min_mask_region_area: int = 0,
output_mode: str = "binary_mask",
@@ -663,7 +674,6 @@ def generate(
pred_iou_thresh: Filter threshold in [0, 1], using the mask quality predicted by the model.
stability_score_thresh: Filter threshold in [0, 1], using the stability of the mask
under changes to the cutoff used to binarize the model prediction.
- stability_score_offset: The amount to shift the cutoff when calculating the stability score.
box_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks.
min_mask_region_area: Minimal size for the predicted masks.
output_mode: The form masks are returned in.
@@ -679,7 +689,6 @@ def generate(
original_size=self.original_size,
pred_iou_thresh=pred_iou_thresh,
stability_score_thresh=stability_score_thresh,
- stability_score_offset=stability_score_offset,
box_nms_thresh=box_nms_thresh
)
@@ -771,6 +780,7 @@ class TiledAutomaticMaskGenerator(AutomaticMaskGenerator):
Higher numbers may be faster but use more GPU memory.
point_grids: A lisst over explicit grids of points used for sampling masks.
Normalized to [0, 1] with respect to the image coordinate system.
+ stability_score_offset: The amount to shift the cutoff when calculating the stability score.
"""
# We only expose the arguments that make sense for the tiled mask generator.
@@ -782,12 +792,14 @@ def __init__(
points_per_side: Optional[int] = 32,
points_per_batch: int = 64,
point_grids: Optional[List[np.ndarray]] = None,
+ stability_score_offset: float = 1.0,
) -> None:
super().__init__(
predictor=predictor,
points_per_side=points_per_side,
points_per_batch=points_per_batch,
point_grids=point_grids,
+ stability_score_offset=stability_score_offset,
)
@torch.no_grad()
@@ -815,6 +827,8 @@ def initialize(
embedding_save_path: Where to save the image embeddings.
"""
original_size = image.shape[:2]
+ self._original_size = original_size
+
image_embeddings, tile_shape, halo = _compute_tiled_embeddings(
self._predictor, image, image_embeddings, embedding_save_path, tile_shape, halo
)
@@ -822,13 +836,15 @@ def initialize(
tiling = blocking([0, 0], original_size, tile_shape)
n_tiles = tiling.numberOfBlocks
+ # the crop box is always the full local tile
+ tiles = [tiling.getBlockWithHalo(tile_id, list(halo)).outerBlock for tile_id in range(n_tiles)]
+ crop_boxes = [[tile.begin[1], tile.begin[0], tile.end[1], tile.end[0]] for tile in tiles]
+
+ # we need to cast to the image representation that is compatible with SAM
+ image = util._to_image(image)
+
mask_data = []
for tile_id in tqdm(range(n_tiles), total=n_tiles, desc="Compute masks for tile", disable=not verbose):
- # get the bounding box for this tile and crop the image data
- tile = tiling.getBlockWithHalo(tile_id, list(halo)).outerBlock
- tile_bb = tuple(slice(beg, end) for beg, end in zip(tile.begin, tile.end))
- tile_data = image[tile_bb]
-
# set the pre-computed embeddings for this tile
features = image_embeddings["features"][tile_id]
tile_embeddings = {
@@ -840,18 +856,14 @@ def initialize(
# compute the mask data for this tile and append it
this_mask_data = self._process_crop(
- tile_data, crop_box=None, crop_layer_idx=0, verbose=verbose, precomputed_embeddings=True
+ image, crop_box=crop_boxes[tile_id], crop_layer_idx=0, verbose=verbose, precomputed_embeddings=True
)
mask_data.append(this_mask_data)
# set the initialized data
self._is_initialized = True
self._crop_list = mask_data
- self._original_size = original_size
-
- # the crop box is always the full local tile
- tiles = [tiling.getBlockWithHalo(tile_id, list(halo)).outerBlock for tile_id in range(n_tiles)]
- self._crop_boxes = [[tile.begin[1], tile.begin[0], tile.end[1], tile.end[0]] for tile in tiles]
+ self._crop_boxes = crop_boxes
class TiledEmbeddingMaskGenerator(EmbeddingMaskGenerator):
@@ -918,7 +930,10 @@ def _compute_mask_data_tiled(self, image_embeddings, i, initial_segmentations, n
"original_size": this_tile_shape
}
util.set_precomputed(self._predictor, tile_image_embeddings, i)
- tile_data = self._compute_mask_data(initial_segmentations[tile_id], this_tile_shape, verbose=False)
+ this_crop_box = [0, 0, this_tile_shape[1], this_tile_shape[0]]
+ tile_data = self._compute_mask_data(
+ initial_segmentations[tile_id], this_crop_box, this_tile_shape, verbose=False
+ )
mask_data.append(tile_data)
return mask_data
@@ -976,7 +991,6 @@ def generate(
self,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- stability_score_offset: float = 1.0,
box_nms_thresh: float = 0.7,
min_mask_region_area: int = 0,
verbose: bool = False
@@ -987,7 +1001,6 @@ def generate(
pred_iou_thresh: Filter threshold in [0, 1], using the mask quality predicted by the model.
stability_score_thresh: Filter threshold in [0, 1], using the stability of the mask
under changes to the cutoff used to binarize the model prediction.
- stability_score_offset: The amount to shift the cutoff when calculating the stability score.
box_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks.
min_mask_region_area: Minimal size for the predicted masks.
verbose: Whether to print progress of the computation.
@@ -1008,7 +1021,6 @@ def segment_tile(_, tile_id):
data=mask_data, crop_box=crop_box, original_size=this_tile_shape,
pred_iou_thresh=pred_iou_thresh,
stability_score_thresh=stability_score_thresh,
- stability_score_offset=stability_score_offset,
box_nms_thresh=box_nms_thresh,
)
mask_data.to_numpy()
@@ -1080,6 +1092,35 @@ def set_state(self, state: Dict[str, Any]) -> None:
super().set_state(state)
+def get_amg(
+ predictor: SamPredictor,
+ is_tiled: bool,
+ embedding_based_amg: bool = False,
+ **kwargs,
+) -> AMGBase:
+ """Get the automatic mask generator class.
+
+ Args:
+ predictor: The segment anything predictor.
+ is_tiled: Whether tiled embeddings are used.
+ embedding_based_amg: Whether to use the embedding based instance segmentation functionality.
+ This functionality is still experimental.
+ kwargs: The keyword arguments for the amg class.
+
+ Returns:
+ The automatic mask generator.
+ """
+ if embedding_based_amg:
+ warnings.warn("The embedding based instance segmentation functionality is experimental.")
+ if is_tiled:
+ amg = TiledEmbeddingMaskGenerator(predictor, **kwargs) if embedding_based_amg else\
+ TiledAutomaticMaskGenerator(predictor, **kwargs)
+ else:
+ amg = EmbeddingMaskGenerator(predictor, **kwargs) if embedding_based_amg else\
+ AutomaticMaskGenerator(predictor, **kwargs)
+ return amg
+
+
#
# Experimental functionality
#
diff --git a/micro_sam/napari.yaml b/micro_sam/napari.yaml
index 6107c87e..f6cdc25b 100644
--- a/micro_sam/napari.yaml
+++ b/micro_sam/napari.yaml
@@ -2,6 +2,9 @@ name: micro-sam
display_name: SegmentAnything for Microscopy
contributions:
commands:
+ - id: micro-sam.sample_data_image_series
+ python_name: micro_sam.sample_data:sample_data_image_series
+ title: Load image series sample data from micro-sam plugin
- id: micro-sam.sample_data_wholeslide
python_name: micro_sam.sample_data:sample_data_wholeslide
title: Load WholeSlide sample data from micro-sam plugin
@@ -17,7 +20,13 @@ contributions:
- id: micro-sam.sample_data_tracking
python_name: micro_sam.sample_data:sample_data_tracking
title: Load tracking sample data from micro-sam plugin
+ - id: micro-sam.sample_data_segmentation
+ python_name: micro_sam.sample_data:sample_data_segmentation
+ title: Load segmentation sample data from micro-sam plugin
sample_data:
+ - command: micro-sam.sample_data_image_series
+ display_name: Image series example data
+ key: micro-sam-image-series
- command: micro-sam.sample_data_wholeslide
display_name: WholeSlide example data
key: micro-sam-wholeslide
@@ -33,3 +42,6 @@ contributions:
- command: micro-sam.sample_data_tracking
display_name: Tracking sample dataset
key: micro-sam-tracking
+ - command: micro-sam.sample_data_segmentation
+ display_name: Segmentation sample dataset
+ key: micro-sam-segmentation
diff --git a/micro_sam/precompute_state.py b/micro_sam/precompute_state.py
new file mode 100644
index 00000000..6844c0f9
--- /dev/null
+++ b/micro_sam/precompute_state.py
@@ -0,0 +1,173 @@
+"""Precompute image embeddings and automatic mask generator state for image data.
+"""
+
+import os
+import pickle
+
+from glob import glob
+from pathlib import Path
+from typing import Optional, Tuple, Union
+
+import numpy as np
+import torch
+from segment_anything.predictor import SamPredictor
+from tqdm import tqdm
+
+from . import instance_segmentation, util
+
+
+def cache_amg_state(
+ predictor: SamPredictor,
+ raw: np.ndarray,
+ image_embeddings: util.ImageEmbeddings,
+ save_path: Union[str, os.PathLike],
+ verbose: bool = True,
+ **kwargs,
+) -> instance_segmentation.AMGBase:
+ """Compute and cache or load the state for the automatic mask generator.
+
+ Args:
+ predictor: The segment anything predictor.
+ raw: The image data.
+ image_embeddings: The image embeddings.
+ save_path: The embedding save path. The AMG state will be stored in 'save_path/amg_state.pickle'.
+ verbose: Whether to run the computation verbose.
+ kwargs: The keyword arguments for the amg class.
+
+ Returns:
+ The automatic mask generator class with the cached state.
+ """
+ is_tiled = image_embeddings["input_size"] is None
+ amg = instance_segmentation.get_amg(predictor, is_tiled, **kwargs)
+
+ save_path_amg = os.path.join(save_path, "amg_state.pickle")
+ if os.path.exists(save_path_amg):
+ if verbose:
+ print("Load the AMG state from", save_path_amg)
+ with open(save_path_amg, "rb") as f:
+ amg_state = pickle.load(f)
+ amg.set_state(amg_state)
+ return amg
+
+ if verbose:
+ print("Precomputing the state for instance segmentation.")
+
+ amg.initialize(raw, image_embeddings=image_embeddings, verbose=verbose)
+ amg_state = amg.get_state()
+
+ # put all state onto the cpu so that the state can be deserialized without a gpu
+ new_crop_list = []
+ for mask_data in amg_state["crop_list"]:
+ for k, v in mask_data.items():
+ if torch.is_tensor(v):
+ mask_data[k] = v.cpu()
+ new_crop_list.append(mask_data)
+ amg_state["crop_list"] = new_crop_list
+
+ with open(save_path_amg, "wb") as f:
+ pickle.dump(amg_state, f)
+
+ return amg
+
+
+def _precompute_state_for_file(
+ predictor, input_path, output_path, key, ndim, tile_shape, halo, precompute_amg_state,
+):
+ image_data = util.load_image_data(input_path, key)
+ output_path = Path(output_path).with_suffix(".zarr")
+ embeddings = util.precompute_image_embeddings(
+ predictor, image_data, output_path, ndim=ndim, tile_shape=tile_shape, halo=halo,
+ )
+ if precompute_amg_state:
+ cache_amg_state(predictor, image_data, embeddings, output_path, verbose=True)
+
+
+def _precompute_state_for_files(
+ predictor, input_files, output_path, ndim, tile_shape, halo, precompute_amg_state,
+):
+ os.makedirs(output_path, exist_ok=True)
+ for file_path in tqdm(input_files, desc="Precompute state for files."):
+ out_path = os.path.join(output_path, os.path.basename(file_path))
+ _precompute_state_for_file(
+ predictor, file_path, out_path,
+ key=None, ndim=ndim, tile_shape=tile_shape, halo=halo,
+ precompute_amg_state=precompute_amg_state,
+ )
+
+
+def precompute_state(
+ input_path: Union[os.PathLike, str],
+ output_path: Union[os.PathLike, str],
+ model_type: str = util._DEFAULT_MODEL,
+ checkpoint_path: Optional[Union[os.PathLike, str]] = None,
+ key: Optional[str] = None,
+ ndim: Union[int] = None,
+ tile_shape: Optional[Tuple[int, int]] = None,
+ halo: Optional[Tuple[int, int]] = None,
+ precompute_amg_state: bool = False,
+) -> None:
+ """Precompute the image embeddings and other optional state for the input image(s).
+
+ Args:
+ input_path: The input image file(s). Can either be a single image file (e.g. tif or png),
+ a container file (e.g. hdf5 or zarr) or a folder with images files.
+ In case of a container file the argument `key` must be given. In case of a folder
+ it can be given to provide a glob pattern to subselect files from the folder.
+ output_path: The output path were the embeddings and other state will be saved.
+ model_type: The SegmentAnything model to use. Will use the standard vit_h model by default.
+ checkpoint_path: Path to a checkpoint for a custom model.
+ key: The key to the input file. This is needed for contaner files (e.g. hdf5 or zarr)
+ and can be used to provide a glob pattern if the input is a folder with image files.
+ ndim: The dimensionality of the data.
+ tile_shape: Shape of tiles for tiled prediction. By default prediction is run without tiling.
+ halo: Overlap of the tiles for tiled prediction.
+ precompute_amg_state: Whether to precompute the state for automatic instance segmentation
+ in addition to the image embeddings.
+ """
+ predictor = util.get_sam_model(model_type=model_type, checkpoint_path=checkpoint_path)
+ # check if we precompute the state for a single file or for a folder with image files
+ if os.path.isdir(input_path) and Path(input_path).suffix not in (".n5", ".zarr"):
+ pattern = "*" if key is None else key
+ input_files = glob(os.path.join(input_path, pattern))
+ _precompute_state_for_files(
+ predictor, input_files, output_path,
+ ndim=ndim, tile_shape=tile_shape, halo=halo,
+ precompute_amg_state=precompute_amg_state,
+ )
+ else:
+ _precompute_state_for_file(
+ predictor, input_path, output_path, key,
+ ndim=ndim, tile_shape=tile_shape, halo=halo,
+ precompute_amg_state=precompute_amg_state,
+ )
+
+
+def main():
+ """@private"""
+ import argparse
+
+ parser = argparse.ArgumentParser(description="Compute the embeddings for an image.")
+ parser.add_argument("-i", "--input_path", required=True)
+ parser.add_argument("-o", "--output_path", required=True)
+ parser.add_argument("-m", "--model_type", default="vit_h")
+ parser.add_argument("-c", "--checkpoint_path", default=None)
+ parser.add_argument("-k", "--key")
+ parser.add_argument(
+ "--tile_shape", nargs="+", type=int, help="The tile shape for using tiled prediction", default=None
+ )
+ parser.add_argument(
+ "--halo", nargs="+", type=int, help="The halo for using tiled prediction", default=None
+ )
+ parser.add_argument("-n", "--ndim")
+ parser.add_argument("-p", "--precompute_amg_state")
+
+ args = parser.parse_args()
+ precompute_state(
+ args.input_path, args.output_path, args.model_type, args.checkpoint_path,
+ key=args.key, tile_shape=args.tile_shape, halo=args.halo, ndim=args.ndim,
+ precompute_amg_state=args.precompute_amg_state,
+ )
+
+
+if __name__ == "__main__":
+ main()
diff --git a/micro_sam/prompt_generators.py b/micro_sam/prompt_generators.py
index 18c6e6fe..51849e3e 100644
--- a/micro_sam/prompt_generators.py
+++ b/micro_sam/prompt_generators.py
@@ -4,11 +4,14 @@
"""
from collections.abc import Mapping
-from typing import Optional
+from typing import Optional, Tuple
import numpy as np
from scipy.ndimage import binary_dilation
+import torch
+from kornia.morphology import dilation
+
class PointAndBoxPromptGenerator:
"""Generate point and/or box prompts from an instance segmentation.
@@ -52,34 +55,7 @@ def __init__(
if self.get_point_prompts is False and self.get_box_prompts is False:
raise ValueError("You need to request box prompts, point prompts or both.")
- def __call__(
- self,
- segmentation: np.ndarray,
- segmentation_id: int,
- bbox_coordinates: Mapping[int, tuple],
- center_coordinates: Optional[Mapping[int, np.ndarray]] = None
- ) -> tuple[
- Optional[list[tuple]], Optional[list[int]], Optional[list[tuple]], np.ndarray
- ]:
- """Generate the prompts for one object in the segmentation.
-
- Args:
- segmentation: The instance segmentation.
- segmentation_id: The ID of the instance.
- bbox_coordinates: The precomputed bounding boxes of all objects in the segmentation.
- center_coordinates: The precomputed center coordinates of all objects in the segmentation.
- If passed, these coordinates will be used as the first positive point prompt.
- If not passed a random point from within the object mask will be used.
-
- Returns:
- List of point coordinates. Returns None, if get_point_prompts is false.
- List of point labels. Returns None, if get_point_prompts is false.
- List containing the object bounding box. Returns None, if get_box_prompts is false.
- Object mask.
- """
- coord_list = []
- label_list = []
-
+ def _sample_positive_points(self, object_mask, center_coordinates, coord_list, label_list):
if center_coordinates is not None:
# getting the center coordinate as the first positive point (OPTIONAL)
coord_list.append(tuple(map(int, center_coordinates))) # to get int coords instead of float
@@ -93,11 +69,6 @@ def __call__(
# need to sample "self.n_positive_points" number of points
n_positive_remaining = self.n_positive_points
- if self.get_box_prompts:
- bbox_list = [bbox_coordinates]
-
- object_mask = segmentation == segmentation_id
-
if n_positive_remaining > 0:
# all coordinates of our current object
object_coordinates = np.where(object_mask)
@@ -117,11 +88,14 @@ def __call__(
coord_list.append(positive_coordinates)
label_list.append(1)
+ return coord_list, label_list
+
+ def _sample_negative_points(self, object_mask, bbox_coordinates, coord_list, label_list):
# getting the negative points
# for this we do the opposite and we set the mask to the bounding box - the object mask
# we need to dilate the object mask before doing this: we use scipy.ndimage.binary_dilation for this
dilated_object = binary_dilation(object_mask, iterations=self.dilation_strength)
- background_mask = np.zeros(segmentation.shape)
+ background_mask = np.zeros(object_mask.shape)
background_mask[bbox_coordinates[0]:bbox_coordinates[2], bbox_coordinates[1]:bbox_coordinates[3]] = 1
background_mask = binary_dilation(background_mask, iterations=self.dilation_strength)
background_mask = abs(
@@ -148,12 +122,175 @@ def __call__(
coord_list.append(negative_coordinates)
label_list.append(0)
- # returns object-level masks per instance for cross-verification (TODO: fix it later)
- if self.get_point_prompts is True and self.get_box_prompts is True: # we want points and box
- return coord_list, label_list, bbox_list, object_mask
+ return coord_list, label_list
+
+ def _ensure_num_points(self, object_mask, coord_list, label_list):
+ num_points = self.n_positive_points + self.n_negative_points
+
+ # fill up to the necessary number of points if we did not sample enough of them
+ if len(coord_list) != num_points:
+ # to stay consistent, we add random points in the background of an object
+ # if there's no neg region around the object - usually happens with small rois
+ needed_points = num_points - len(coord_list)
+ more_neg_points = np.where(object_mask == 0)
+ chosen_idx = np.random.choice(len(more_neg_points[0]), size=needed_points)
+
+ coord_list.extend([
+ (more_neg_points[0][idx], more_neg_points[1][idx]) for idx in chosen_idx
+ ])
+ label_list.extend([0] * needed_points)
+
+ assert len(coord_list) == len(label_list) == num_points
+ return coord_list, label_list
+
+ def _sample_points(self, object_mask, bbox_coordinates, center_coordinates):
+ coord_list, label_list = [], []
+
+ coord_list, label_list = self._sample_positive_points(object_mask, center_coordinates, coord_list, label_list)
+ coord_list, label_list = self._sample_negative_points(object_mask, bbox_coordinates, coord_list, label_list)
+ coord_list, label_list = self._ensure_num_points(object_mask, coord_list, label_list)
+
+ return coord_list, label_list
+
+ def __call__(
+ self,
+ segmentation: np.ndarray,
+ segmentation_id: int,
+ bbox_coordinates: Mapping[int, tuple],
+ center_coordinates: Optional[Mapping[int, np.ndarray]] = None
+ ) -> tuple[
+ Optional[list[tuple]], Optional[list[int]], Optional[list[tuple]], np.ndarray
+ ]:
+ """Generate the prompts for one object in the segmentation.
+
+ Args:
+ segmentation: The instance segmentation.
+ segmentation_id: The ID of the instance.
+ bbox_coordinates: The precomputed bounding boxes of all objects in the segmentation.
+ center_coordinates: The precomputed center coordinates of all objects in the segmentation.
+ If passed, these coordinates will be used as the first positive point prompt.
+ If not passed a random point from within the object mask will be used.
+
+ Returns:
+ List of point coordinates. Returns None, if get_point_prompts is false.
+ List of point labels. Returns None, if get_point_prompts is false.
+ List containing the object bounding box. Returns None, if get_box_prompts is false.
+ Object mask.
+ """
+ object_mask = segmentation == segmentation_id
+
+ if self.get_point_prompts:
+ coord_list, label_list = self._sample_points(object_mask, bbox_coordinates, center_coordinates)
+ else:
+ coord_list, label_list = None, None
+
+ if self.get_box_prompts:
+ bbox_list = [bbox_coordinates]
+ else:
+ bbox_list = None
+
+ return coord_list, label_list, bbox_list, object_mask
+
+
+class IterativePromptGenerator:
+ """Generate point prompts from an instance segmentation iteratively.
+ """
+ def _get_positive_points(self, pos_region, overlap_region):
+ positive_locations = [torch.where(pos_reg) for pos_reg in pos_region]
+ # we may have objects withput a positive region (= missing true foreground)
+ # in this case we just sample a point where the model was already correct
+ positive_locations = [
+ torch.where(ovlp_reg) if len(pos_loc[0]) == 0 else pos_loc
+ for pos_loc, ovlp_reg in zip(positive_locations, overlap_region)
+ ]
+ # we sample one location for each object in the batch
+ sampled_indices = [np.random.choice(len(pos_loc[0])) for pos_loc in positive_locations]
+ # get the corresponding coordinates (Note that we flip the axis order here due to the expected order of SAM)
+ pos_coordinates = [
+ [pos_loc[-1][idx], pos_loc[-2][idx]] for pos_loc, idx in zip(positive_locations, sampled_indices)
+ ]
+
+ # make sure that we still have the correct batch size
+ assert len(pos_coordinates) == pos_region.shape[0]
+ pos_labels = [1] * len(pos_coordinates)
+
+ return pos_coordinates, pos_labels
+
+ # TODO get rid of this looped implementation and use proper batched computation instead
+ def _get_negative_points(self, negative_region_batched, true_object_batched, gt_batched):
+ device = negative_region_batched.device
+
+ negative_coordinates, negative_labels = [], []
+ for neg_region, true_object, gt in zip(negative_region_batched, true_object_batched, gt_batched):
+
+ tmp_neg_loc = torch.where(neg_region)
+ if torch.stack(tmp_neg_loc).shape[-1] == 0:
+ tmp_true_loc = torch.where(true_object)
+ x_coords, y_coords = tmp_true_loc[1], tmp_true_loc[2]
+ bbox = torch.stack([torch.min(x_coords), torch.min(y_coords),
+ torch.max(x_coords) + 1, torch.max(y_coords) + 1])
+ bbox_mask = torch.zeros_like(true_object).squeeze(0)
+ bbox_mask[bbox[0]:bbox[2], bbox[1]:bbox[3]] = 1
+ bbox_mask = bbox_mask[None].to(device)
+
+ # NOTE: FIX: here we add dilation to the bbox because in some case we couldn't find objects at all
+ # TODO: just expand the pixels of bbox
+ dilated_bbox_mask = dilation(bbox_mask[None], torch.ones(3, 3).to(device)).squeeze(0)
+ background_mask = abs(dilated_bbox_mask - true_object)
+ tmp_neg_loc = torch.where(background_mask)
+
+ # there is a chance that the object is small to not return a decent-sized bounding box
+ # hence we might not find points sometimes there as well, hence we sample points from true background
+ if torch.stack(tmp_neg_loc).shape[-1] == 0:
+ tmp_neg_loc = torch.where(gt == 0)
+
+ neg_index = np.random.choice(len(tmp_neg_loc[1]))
+ neg_coordinates = [tmp_neg_loc[1][neg_index], tmp_neg_loc[2][neg_index]]
+ neg_coordinates = neg_coordinates[::-1]
+ neg_labels = 0
+
+ negative_coordinates.append(neg_coordinates)
+ negative_labels.append(neg_labels)
+
+ return negative_coordinates, negative_labels
+
+ def __call__(
+ self,
+ gt: torch.Tensor,
+ object_mask: torch.Tensor,
+ current_points: torch.Tensor,
+ current_labels: torch.Tensor
+ ) -> Tuple[torch.Tensor, torch.Tensor]:
+ """Generate the prompts for each object iteratively in the segmentation.
+
+ Args:
+ The groundtruth segmentation.
+ The predicted objects.
+ The current points.
+ Thr current labels.
+
+ Returns:
+ The updated point prompt coordinates.
+ The updated point prompt labels.
+ """
+ assert gt.shape == object_mask.shape
+ device = object_mask.device
+
+ true_object = gt.to(device)
+ expected_diff = (object_mask - true_object)
+ neg_region = (expected_diff == 1).to(torch.float)
+ pos_region = (expected_diff == -1)
+ overlap_region = torch.logical_and(object_mask == 1, true_object == 1).to(torch.float32)
+
+ pos_coordinates, pos_labels = self._get_positive_points(pos_region, overlap_region)
+ neg_coordinates, neg_labels = self._get_negative_points(neg_region, true_object, gt)
+ assert len(pos_coordinates) == len(pos_labels) == len(neg_coordinates) == len(neg_labels)
+
+ pos_coordinates = torch.tensor(pos_coordinates)[:, None]
+ neg_coordinates = torch.tensor(neg_coordinates)[:, None]
+ pos_labels, neg_labels = torch.tensor(pos_labels)[:, None], torch.tensor(neg_labels)[:, None]
- elif self.get_point_prompts is True and self.get_box_prompts is False: # we want only points
- return coord_list, label_list, None, object_mask
+ net_coords = torch.cat([current_points, pos_coordinates, neg_coordinates], dim=1)
+ net_labels = torch.cat([current_labels, pos_labels, neg_labels], dim=1)
- elif self.get_point_prompts is False and self.get_box_prompts is True: # we want only boxes
- return None, None, bbox_list, object_mask
+ return net_coords, net_labels
diff --git a/micro_sam/sam_annotator/annotator.py b/micro_sam/sam_annotator/annotator.py
index 0c8fae27..9be755f5 100644
--- a/micro_sam/sam_annotator/annotator.py
+++ b/micro_sam/sam_annotator/annotator.py
@@ -9,7 +9,7 @@
from magicgui.application import use_app
from PyQt5.QtWidgets import QFileDialog, QMessageBox
-from ..util import load_image_data, get_model_names
+from ..util import load_image_data, get_model_names, _DEFAULT_MODEL
from .annotator_2d import annotator_2d
from .annotator_3d import annotator_3d
from .image_series_annotator import image_folder_annotator
@@ -64,7 +64,7 @@ def _on_2d():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select image", labels=False)
def on_select_image():
@@ -156,7 +156,7 @@ def _on_3d():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select images", labels=False)
def on_select_image():
@@ -271,7 +271,7 @@ def _on_series():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select input directory", labels=False)
def on_select_input_dir():
@@ -364,7 +364,7 @@ def _on_tracking():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select images", labels=False)
def on_select_image():
diff --git a/micro_sam/sam_annotator/annotator_2d.py b/micro_sam/sam_annotator/annotator_2d.py
index 73fd5159..eceb19db 100644
--- a/micro_sam/sam_annotator/annotator_2d.py
+++ b/micro_sam/sam_annotator/annotator_2d.py
@@ -8,8 +8,8 @@
from napari import Viewer
from segment_anything import SamPredictor
-from .. import util
-from .. import instance_segmentation
+from .. import instance_segmentation, util
+from ..precompute_state import cache_amg_state
from ..visualization import project_embeddings_for_visualization
from . import util as vutil
from .gui_utils import show_wrong_file_warning
@@ -38,18 +38,6 @@ def _segment_widget(v: Viewer) -> None:
v.layers["current_object"].refresh()
-def _get_amg(is_tiled, with_background, min_initial_size, box_extension=0.05):
- if is_tiled:
- amg = instance_segmentation.TiledEmbeddingMaskGenerator(
- PREDICTOR, with_background=with_background, min_initial_size=min_initial_size, box_extension=box_extension,
- )
- else:
- amg = instance_segmentation.EmbeddingMaskGenerator(
- PREDICTOR, min_initial_size=min_initial_size, box_extension=box_extension,
- )
- return amg
-
-
def _changed_param(amg, **params):
if amg is None:
return None
@@ -62,29 +50,25 @@ def _changed_param(amg, **params):
@magicgui(call_button="Automatic Segmentation")
def _autosegment_widget(
v: Viewer,
- with_background: bool = True,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- min_initial_size: int = 10,
- box_extension: float = 0.05,
+ min_object_size: int = 100,
+ with_background: bool = True,
) -> None:
global AMG
is_tiled = IMAGE_EMBEDDINGS["input_size"] is None
- param_changed = _changed_param(
- AMG, with_background=with_background, min_initial_size=min_initial_size, box_extension=box_extension
- )
- if AMG is None or param_changed:
- if param_changed:
- print(f"The parameter {param_changed} was changed, so the full instance segmentation has to be recomputed.")
- AMG = _get_amg(is_tiled, with_background, min_initial_size, box_extension)
+ if AMG is None:
+ AMG = instance_segmentation.get_amg(PREDICTOR, is_tiled)
if not AMG.is_initialized:
AMG.initialize(v.layers["raw"].data, image_embeddings=IMAGE_EMBEDDINGS, verbose=True)
seg = AMG.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
- if not is_tiled:
- shape = v.layers["raw"].data.shape[:2]
- seg = instance_segmentation.mask_data_to_segmentation(seg, shape, with_background)
+
+ shape = v.layers["raw"].data.shape[:2]
+ seg = instance_segmentation.mask_data_to_segmentation(
+ seg, shape, with_background=True, min_object_size=min_object_size
+ )
assert isinstance(seg, np.ndarray)
v.layers["auto_segmentation"].data = seg
@@ -197,12 +181,13 @@ def annotator_2d(
embedding_path: Optional[str] = None,
show_embeddings: bool = False,
segmentation_result: Optional[np.ndarray] = None,
- model_type: str = "vit_h",
+ model_type: str = util._DEFAULT_MODEL,
tile_shape: Optional[Tuple[int, int]] = None,
halo: Optional[Tuple[int, int]] = None,
return_viewer: bool = False,
v: Optional[Viewer] = None,
predictor: Optional[SamPredictor] = None,
+ precompute_amg_state: bool = False,
) -> Optional[Viewer]:
"""The 2d annotation tool.
@@ -225,6 +210,9 @@ def annotator_2d(
This enables using a pre-initialized viewer, for example in `sam_annotator.image_series_annotator`.
predictor: The Segment Anything model. Passing this enables using fully custom models.
If you pass `predictor` then `model_type` will be ignored.
+ precompute_amg_state: Whether to precompute the state for automatic mask generation.
+ This will take more time when precomputing embeddings, but will then make
+ automatic mask generation much faster.
Returns:
The napari viewer, only returned if `return_viewer=True`.
@@ -237,10 +225,13 @@ def annotator_2d(
PREDICTOR = util.get_sam_model(model_type=model_type)
else:
PREDICTOR = predictor
+
IMAGE_EMBEDDINGS = util.precompute_image_embeddings(
PREDICTOR, raw, save_path=embedding_path, ndim=2, tile_shape=tile_shape, halo=halo,
wrong_file_callback=show_wrong_file_warning
)
+ if precompute_amg_state and (embedding_path is not None):
+ AMG = cache_amg_state(PREDICTOR, raw, IMAGE_EMBEDDINGS, embedding_path)
# we set the pre-computed image embeddings if we don't use tiling
# (if we use tiling we cannot directly set it because the tile will be chosen dynamically)
@@ -268,6 +259,7 @@ def annotator_2d(
def main():
"""@private"""
parser = vutil._initialize_parser(description="Run interactive segmentation for an image.")
+ parser.add_argument("--precompute_amg_state", action="store_true")
args = parser.parse_args()
raw = util.load_image_data(args.input, key=args.key)
@@ -283,4 +275,5 @@ def main():
raw, embedding_path=args.embedding_path,
show_embeddings=args.show_embeddings, segmentation_result=segmentation_result,
model_type=args.model_type, tile_shape=args.tile_shape, halo=args.halo,
+ precompute_amg_state=args.precompute_amg_state,
)
diff --git a/micro_sam/sam_annotator/annotator_3d.py b/micro_sam/sam_annotator/annotator_3d.py
index 6518336b..ba1740fd 100644
--- a/micro_sam/sam_annotator/annotator_3d.py
+++ b/micro_sam/sam_annotator/annotator_3d.py
@@ -154,7 +154,7 @@ def _segment_slice_wigdet(v: Viewer) -> None:
@magicgui(call_button="Segment Volume [V]", projection={"choices": ["default", "bounding_box", "mask", "points"]})
def _segment_volume_widget(
- v: Viewer, iou_threshold: float = 0.8, projection: str = "default", box_extension: float = 0.1
+ v: Viewer, iou_threshold: float = 0.8, projection: str = "default", box_extension: float = 0.05
) -> None:
# step 1: segment all slices with prompts
shape = v.layers["raw"].data.shape
@@ -192,7 +192,7 @@ def annotator_3d(
embedding_path: Optional[str] = None,
show_embeddings: bool = False,
segmentation_result: Optional[np.ndarray] = None,
- model_type: str = "vit_h",
+ model_type: str = util._DEFAULT_MODEL,
tile_shape: Optional[Tuple[int, int]] = None,
halo: Optional[Tuple[int, int]] = None,
return_viewer: bool = False,
diff --git a/micro_sam/sam_annotator/annotator_tracking.py b/micro_sam/sam_annotator/annotator_tracking.py
index e22a9172..691cd6ae 100644
--- a/micro_sam/sam_annotator/annotator_tracking.py
+++ b/micro_sam/sam_annotator/annotator_tracking.py
@@ -1,4 +1,6 @@
+import json
import warnings
+from pathlib import Path
from typing import Optional, Tuple
import napari
@@ -24,6 +26,9 @@
STATE_COLOR_CYCLE = ["#00FFFF", "#FF00FF", ]
"""@private"""
+COMMITTED_LINEAGES = []
+"""@private"""
+
#
# util functionality
@@ -336,6 +341,8 @@ def _reset_tracking_state():
@magicgui(call_button="Commit [C]", layer={"choices": ["current_track"]})
def _commit_tracking_widget(v: Viewer, layer: str = "current_track") -> None:
+ global COMMITTED_LINEAGES
+
seg = v.layers[layer].data
id_offset = int(v.layers["committed_tracks"].data.max())
@@ -348,6 +355,11 @@ def _commit_tracking_widget(v: Viewer, layer: str = "current_track") -> None:
v.layers[layer].data = np.zeros(shape, dtype="uint32")
v.layers[layer].refresh()
+ updated_lineage = {
+ parent + id_offset: [child + id_offset for child in children] for parent, children in LINEAGE.items()
+ }
+ COMMITTED_LINEAGES.append(updated_lineage)
+
_reset_tracking_state()
vutil.clear_annotations(v, clear_segmentations=False)
@@ -358,12 +370,19 @@ def _clear_widget_tracking(v: Viewer) -> None:
vutil.clear_annotations(v)
+@magicgui(call_button="Save Lineage")
+def _save_lineage_widget(v: Viewer, path: Path) -> None:
+ path = path.with_suffix(".json")
+ with open(path, "w") as f:
+ json.dump(COMMITTED_LINEAGES, f)
+
+
def annotator_tracking(
raw: np.ndarray,
embedding_path: Optional[str] = None,
show_embeddings: bool = False,
tracking_result: Optional[str] = None,
- model_type: str = "vit_h",
+ model_type: str = util._DEFAULT_MODEL,
tile_shape: Optional[Tuple[int, int]] = None,
halo: Optional[Tuple[int, int]] = None,
return_viewer: bool = False,
@@ -486,6 +505,7 @@ def annotator_tracking(
v.window.add_dock_widget(_segment_frame_wigdet)
v.window.add_dock_widget(_track_objet_widget)
v.window.add_dock_widget(_commit_tracking_widget)
+ v.window.add_dock_widget(_save_lineage_widget)
v.window.add_dock_widget(_clear_widget_tracking)
#
diff --git a/micro_sam/sam_annotator/gui_utils.py b/micro_sam/sam_annotator/gui_utils.py
index 8965daf6..fb412429 100644
--- a/micro_sam/sam_annotator/gui_utils.py
+++ b/micro_sam/sam_annotator/gui_utils.py
@@ -47,36 +47,26 @@ def _overwrite():
@magicgui.magicgui(call_button="Create new file", labels=False)
def _create():
- msg_box.close()
# unfortunately there exists no dialog to create a directory so we have
# to use "create new file" dialog with some adjustments.
dialog = QtWidgets.QFileDialog(None)
dialog.setFileMode(QtWidgets.QFileDialog.AnyFile)
dialog.setOption(QtWidgets.QFileDialog.ShowDirsOnly)
dialog.setNameFilter("Archives (*.zarr)")
- try_cnt = 0
- while os.path.splitext(new_path["value"])[1] != ".zarr":
- if try_cnt > 3:
- new_path["value"] = file_path
- return
- dialog.exec_()
- res = dialog.selectedFiles()
- new_path["value"] = res[0] if len(res) > 0 else ""
- try_cnt += 1
+ dialog.exec_()
+ res = dialog.selectedFiles()
+ new_path["value"] = res[0] if len(res) > 0 else ""
+ if os.path.splitext(new_path["value"])[1] != ".zarr":
+ new_path["value"] += ".zarr"
os.makedirs(new_path["value"])
+ msg_box.close()
@magicgui.magicgui(call_button="Select different file", labels=False)
def _select():
+ new_path["value"] = QtWidgets.QFileDialog.getExistingDirectory(
+ None, "Open a folder", os.path.split(file_path)[0], QtWidgets.QFileDialog.ShowDirsOnly
+ )
msg_box.close()
- try_cnt = 0
- while not os.path.exists(new_path["value"]):
- if try_cnt > 3:
- new_path["value"] = file_path
- return
- new_path["value"] = QtWidgets.QFileDialog.getExistingDirectory(
- None, "Open a folder", os.path.split(file_path)[0], QtWidgets.QFileDialog.ShowDirsOnly
- )
- try_cnt += 1
msg_box = Container(widgets=[_select, _ignore, _overwrite, _create], layout='horizontal', labels=False)
msg_box.root_native_widget.setWindowTitle("The input data does not match the embeddings file")
diff --git a/micro_sam/sam_annotator/image_series_annotator.py b/micro_sam/sam_annotator/image_series_annotator.py
index 80fa7602..d561b7d5 100644
--- a/micro_sam/sam_annotator/image_series_annotator.py
+++ b/micro_sam/sam_annotator/image_series_annotator.py
@@ -1,33 +1,19 @@
import os
import warnings
+
from glob import glob
+from pathlib import Path
from typing import List, Optional, Union
import imageio.v3 as imageio
import napari
from magicgui import magicgui
-from napari.utils import progress as tqdm
from segment_anything import SamPredictor
-from .annotator_2d import annotator_2d
from .. import util
-
-
-def _precompute_embeddings_for_image_series(predictor, image_files, embedding_root, tile_shape, halo):
- os.makedirs(embedding_root, exist_ok=True)
- embedding_paths = []
- for image_file in tqdm(image_files, desc="Precompute embeddings"):
- fname = os.path.basename(image_file)
- fname = os.path.splitext(fname)[0] + ".zarr"
- embedding_path = os.path.join(embedding_root, fname)
- image = imageio.imread(image_file)
- util.precompute_image_embeddings(
- predictor, image, save_path=embedding_path, ndim=2,
- tile_shape=tile_shape, halo=halo
- )
- embedding_paths.append(embedding_path)
- return embedding_paths
+from ..precompute_state import _precompute_state_for_files
+from .annotator_2d import annotator_2d
def image_series_annotator(
@@ -47,6 +33,7 @@ def image_series_annotator(
embedding_path: Filepath where to save the embeddings.
predictor: The Segment Anything model. Passing this enables using fully custom models.
If you pass `predictor` then `model_type` will be ignored.
+ kwargs: The keywored arguments for `micro_sam.sam_annotator.annotator_2d`.
"""
# make sure we don't set incompatible kwargs
assert kwargs.get("show_embeddings", False) is False
@@ -58,13 +45,20 @@ def image_series_annotator(
next_image_id = 0
if predictor is None:
- predictor = util.get_sam_model(model_type=kwargs.get("model_type", "vit_h"))
+ predictor = util.get_sam_model(model_type=kwargs.get("model_type", util._DEFAULT_MODEL))
if embedding_path is None:
embedding_paths = None
else:
- embedding_paths = _precompute_embeddings_for_image_series(
- predictor, image_files, embedding_path, kwargs.get("tile_shape", None), kwargs.get("halo", None)
+ _precompute_state_for_files(
+ predictor, image_files, embedding_path, ndim=2,
+ tile_shape=kwargs.get("tile_shape", None),
+ halo=kwargs.get("halo", None),
+ precompute_amg_state=kwargs.get("precompute_amg_state", False),
)
+ embedding_paths = [
+ os.path.join(embedding_path, f"{Path(path).stem}.zarr") for path in image_files
+ ]
+ assert all(os.path.exists(emb_path) for emb_path in embedding_paths)
def _save_segmentation(image_path, segmentation):
fname = os.path.basename(image_path)
@@ -127,6 +121,7 @@ def image_folder_annotator(
embedding_path: Filepath where to save the embeddings.
predictor: The Segment Anything model. Passing this enables using fully custom models.
If you pass `predictor` then `model_type` will be ignored.
+ kwargs: The keywored arguments for `micro_sam.sam_annotator.annotator_2d`.
"""
image_files = sorted(glob(os.path.join(input_folder, pattern)))
image_series_annotator(image_files, output_folder, embedding_path, predictor, **kwargs)
@@ -136,6 +131,9 @@ def main():
"""@private"""
import argparse
+ available_models = list(util.get_model_names())
+ available_models = ", ".join(available_models)
+
parser = argparse.ArgumentParser(description="Annotate a series of images from a folder.")
parser.add_argument(
"-i", "--input_folder", required=True,
@@ -157,7 +155,8 @@ def main():
"otherwise they will be recomputed every time (which can take a long time)."
)
parser.add_argument(
- "--model_type", default="vit_h", help="The segment anything model that will be used, one of vit_h,l,b."
+ "--model_type", default=util._DEFAULT_MODEL,
+ help=f"The segment anything model that will be used, one of {available_models}."
)
parser.add_argument(
"--tile_shape", nargs="+", type=int, help="The tile shape for using tiled prediction", default=None
@@ -165,6 +164,7 @@ def main():
parser.add_argument(
"--halo", nargs="+", type=int, help="The halo for using tiled prediction", default=None
)
+ parser.add_argument("--precompute_amg_state", action="store_true")
args = parser.parse_args()
@@ -175,4 +175,5 @@ def main():
args.input_folder, args.output_folder, args.pattern,
embedding_path=args.embedding_path, model_type=args.model_type,
tile_shape=args.tile_shape, halo=args.halo,
+ precompute_amg_state=args.precompute_amg_state,
)
diff --git a/micro_sam/sam_annotator/util.py b/micro_sam/sam_annotator/util.py
index e4dbf20d..48a02622 100644
--- a/micro_sam/sam_annotator/util.py
+++ b/micro_sam/sam_annotator/util.py
@@ -1,4 +1,6 @@
import argparse
+import os
+import pickle
from typing import Optional, Tuple
import napari
@@ -7,6 +9,7 @@
from magicgui import magicgui
from magicgui.widgets import ComboBox, Container
+from .. import instance_segmentation, util
from ..prompt_based_segmentation import segment_from_box, segment_from_box_and_points, segment_from_points
# Green and Red
@@ -362,6 +365,10 @@ def toggle_label(prompts):
def _initialize_parser(description, with_segmentation_result=True, with_show_embeddings=True):
+
+ available_models = list(util.get_model_names())
+ available_models = ", ".join(available_models)
+
parser = argparse.ArgumentParser(description=description)
parser.add_argument(
@@ -401,7 +408,8 @@ def _initialize_parser(description, with_segmentation_result=True, with_show_emb
help="Visualize the embeddings computed by SegmentAnything. This can be helpful for debugging."
)
parser.add_argument(
- "--model_type", default="vit_h", help="The segment anything model that will be used, one of vit_h,l,b."
+ "--model_type", default=util._DEFAULT_MODEL,
+ help=f"The segment anything model that will be used, one of {available_models}."
)
parser.add_argument(
"--tile_shape", nargs="+", type=int, help="The tile shape for using tiled prediction", default=None
diff --git a/micro_sam/sample_data.py b/micro_sam/sample_data.py
index 533d97ae..8c9523c5 100644
--- a/micro_sam/sample_data.py
+++ b/micro_sam/sample_data.py
@@ -11,7 +11,57 @@
import pooch
-def fetch_wholeslide_example_data(save_directory: Union[str, os.PathLike]) -> Union[str, os.PathLike]:
+def fetch_image_series_example_data(save_directory: Union[str, os.PathLike]) -> str:
+ """Download the sample images for the image series annotator.
+
+ Args:
+ save_directory: Root folder to save the downloaded data.
+ Returns:
+ The folder that contains the downloaded data.
+ """
+ # This sample dataset is currently not provided to napari by the micro-sam
+ # plugin, because images are not all the same shape and cannot be combined
+ # into a single layer
+ save_directory = Path(save_directory)
+ os.makedirs(save_directory, exist_ok=True)
+ print("Example data directory is:", save_directory.resolve())
+ fname = "image-series.zip"
+ unpack_filenames = [os.path.join("series", f"im{i}.tif") for i in range(3)]
+ unpack = pooch.Unzip(members=unpack_filenames)
+ pooch.retrieve(
+ url="https://owncloud.gwdg.de/index.php/s/M1zGnfkulWoAhUG/download",
+ known_hash="92346ca9770bcaf55248efee590718d54c7135b6ebca15d669f3b77b6afc8706",
+ fname=fname,
+ path=save_directory,
+ progressbar=True,
+ processor=unpack,
+ )
+ data_folder = os.path.join(save_directory, f"{fname}.unzip", "series")
+ assert os.path.exists(data_folder)
+ return data_folder
+
+
+def sample_data_image_series():
+ """Provides image series example image to napari.
+
+ Opens as three separate image layers in napari (one per image in series).
+ The third image in the series has a different size and modality.
+ """
+ # Return list of tuples
+ # [(data1, add_image_kwargs1), (data2, add_image_kwargs2)]
+ # Check the documentation for more information about the
+ # add_image_kwargs
+ # https://napari.org/stable/api/napari.Viewer.html#napari.Viewer.add_image
+ default_base_data_dir = pooch.os_cache('micro-sam')
+ data_directory = fetch_image_series_example_data(default_base_data_dir)
+ fnames = os.listdir(data_directory)
+ full_filenames = [os.path.join(data_directory, f) for f in fnames]
+ full_filenames.sort()
+ data_and_image_kwargs = [(imageio.imread(f), {"name": f"img-{i}"}) for i, f in enumerate(full_filenames)]
+ return data_and_image_kwargs
+
+
+def fetch_wholeslide_example_data(save_directory: Union[str, os.PathLike]) -> str:
"""Download the sample data for the 2d annotator.
This downloads part of a whole-slide image from the NeurIPS Cell Segmentation Challenge.
@@ -220,3 +270,49 @@ def sample_data_tracking():
data = np.stack([imageio.imread(f) for f in full_filenames], axis=0)
add_image_kwargs = {"name": "tracking"}
return [(data, add_image_kwargs)]
+
+
+def fetch_tracking_segmentation_data(save_directory: Union[str, os.PathLike]) -> str:
+ """Download groundtruth segmentation for the tracking example data.
+
+ This downloads the groundtruth segmentation for the image data from `fetch_tracking_example_data`.
+
+ Args:
+ save_directory: Root folder to save the downloaded data.
+ Returns:
+ The folder that contains the downloaded data.
+ """
+ save_directory = Path(save_directory)
+ os.makedirs(save_directory, exist_ok=True)
+ print("Example data directory is:", save_directory.resolve())
+ unpack_filenames = [os.path.join("masks", f"mask_{str(i).zfill(4)}.tif") for i in range(84)]
+ unpack = pooch.Unzip(members=unpack_filenames)
+ fname = "hela-ctc-01-gt.zip"
+ pooch.retrieve(
+ url="https://owncloud.gwdg.de/index.php/s/AWxQMblxwR99OjC/download",
+ known_hash="c0644d8ebe1390fb60125560ba15aa2342caf44f50ff0667a0318ea0ac6c958b",
+ fname=fname,
+ path=save_directory,
+ progressbar=True,
+ processor=unpack,
+ )
+ cell_tracking_dir = save_directory.joinpath(f"{fname}.unzip", "masks")
+ assert os.path.exists(cell_tracking_dir)
+ return str(cell_tracking_dir)
+
+
+def sample_data_segmentation():
+ """Provides segmentation example dataset to napari."""
+ # Return list of tuples
+ # [(data1, add_image_kwargs1), (data2, add_image_kwargs2)]
+ # Check the documentation for more information about the
+ # add_image_kwargs
+ # https://napari.org/stable/api/napari.Viewer.html#napari.Viewer.add_image
+ default_base_data_dir = pooch.os_cache("micro-sam")
+ data_directory = fetch_tracking_segmentation_data(default_base_data_dir)
+ fnames = os.listdir(data_directory)
+ full_filenames = [os.path.join(data_directory, f) for f in fnames]
+ full_filenames.sort()
+ data = np.stack([imageio.imread(f) for f in full_filenames], axis=0)
+ add_image_kwargs = {"name": "segmentation"}
+ return [(data, add_image_kwargs)]
diff --git a/micro_sam/training/__init__.py b/micro_sam/training/__init__.py
new file mode 100644
index 00000000..225b6568
--- /dev/null
+++ b/micro_sam/training/__init__.py
@@ -0,0 +1,5 @@
+"""Functionality for training Segment Anything.
+"""
+
+from .sam_trainer import SamTrainer, SamLogger
+from .util import ConvertToSamInputs, get_trainable_sam_model
diff --git a/micro_sam/training/sam_trainer.py b/micro_sam/training/sam_trainer.py
new file mode 100644
index 00000000..9a30554b
--- /dev/null
+++ b/micro_sam/training/sam_trainer.py
@@ -0,0 +1,458 @@
+import os
+import time
+from typing import Optional
+
+import numpy as np
+import torch
+import torch_em
+
+from kornia.morphology import dilation
+from torchvision.utils import make_grid
+from torch_em.trainer.logger_base import TorchEmLogger
+
+
+class SamTrainer(torch_em.trainer.DefaultTrainer):
+ """Trainer class for training the Segment Anything model.
+
+ This class is derived from `torch_em.trainer.DefaultTrainer`.
+ Check out https://github.com/constantinpape/torch-em/blob/main/torch_em/trainer/default_trainer.py
+ for details on its usage and implementation.
+
+ Args:
+ convert_inputs: Class that converts the output of the dataloader to the expected input format of SAM.
+ The class `micro_sam.training.util.ConvertToSamInputs` can be used here.
+ n_sub_iteration: The number of iteration steps for which the masks predicted for one object are updated.
+ In each sub-iteration new point prompts are sampled where the model was wrong.
+ n_objects_per_batch: If not given, we compute the loss for all objects in a sample.
+ Otherwise the loss computation is limited to n_objects_per_batch, and the objects are randomly sampled.
+ mse_loss: The regression loss to compare the IoU predicted by the model with the true IoU.
+ sigmoid: The activation function for normalizing the model output.
+ **kwargs: The keyword arguments of the DefaultTrainer super class.
+ """
+
+ def __init__(
+ self,
+ convert_inputs,
+ n_sub_iteration: int,
+ n_objects_per_batch: Optional[int] = None,
+ mse_loss: torch.nn.Module = torch.nn.MSELoss(),
+ _sigmoid: torch.nn.Module = torch.nn.Sigmoid(),
+ **kwargs
+ ):
+ super().__init__(**kwargs)
+ self.convert_inputs = convert_inputs
+ self.mse_loss = mse_loss
+ self._sigmoid = _sigmoid
+ self.n_objects_per_batch = n_objects_per_batch
+ self.n_sub_iteration = n_sub_iteration
+ self._kwargs = kwargs
+
+ def _get_prompt_and_multimasking_choices(self, current_iteration):
+ """Choose the type of prompts we sample for training, and then we call
+ 'convert_inputs' with the correct prompting from here.
+ """
+ if current_iteration % 2 == 0: # sample only a single point per object
+ n_pos, n_neg = 1, 0
+ get_boxes = False
+ multimask_output = True
+
+ else: # sample only a single box per object
+ n_pos, n_neg = 0, 0
+ get_boxes = True
+ multimask_output = False
+
+ return n_pos, n_neg, get_boxes, multimask_output
+
+ def _get_prompt_and_multimasking_choices_for_val(self, current_iteration):
+ """Choose the type of prompts we sample for validation, and then we call
+ 'convert_inputs' with the correct prompting from here.
+ """
+ if current_iteration % 4 == 0: # sample only a single point per object
+ n_pos, n_neg = 1, 0
+ get_boxes = False
+ multimask_output = True
+
+ elif current_iteration % 4 == 1: # sample only a single box per object
+ n_pos, n_neg = 0, 0
+ get_boxes = True
+ multimask_output = False
+
+ elif current_iteration % 4 == 2: # sample a random no. of points
+ pos_range, neg_range = 4, 4
+
+ n_pos = np.random.randint(1, pos_range + 1)
+ if n_pos == 1: # to avoid (1, 0) combination for redundancy but still have (n_pos, 0)
+ n_neg = np.random.randint(1, neg_range + 1)
+ else:
+ n_neg = np.random.randint(0, neg_range + 1)
+ get_boxes = False
+ multimask_output = False
+
+ else: # sample boxes AND random no. of points
+ # here we can have (1, 0) because we also have box
+ pos_range, neg_range = 4, 4
+
+ n_pos = np.random.randint(1, pos_range + 1)
+ n_neg = np.random.randint(0, neg_range + 1)
+ get_boxes = True
+ multimask_output = False
+
+ return n_pos, n_neg, get_boxes, multimask_output
+
+ def _get_dice(self, input_, target):
+ """Using the default "DiceLoss" called by the trainer from "torch_em"
+ """
+ dice_loss = self.loss(input_, target)
+ return dice_loss
+
+ def _get_iou(self, pred, true, eps=1e-7):
+ """Getting the IoU score for the predicted and true labels
+ """
+ pred_mask = pred > 0.5 # binarizing the output predictions
+ overlap = pred_mask.logical_and(true).sum()
+ union = pred_mask.logical_or(true).sum()
+ iou = overlap / (union + eps)
+ return iou
+
+ def _get_net_loss(self, batched_outputs, y, sampled_ids):
+ """What do we do here? two **separate** things
+ 1. compute the mask loss: loss between the predicted and ground-truth masks
+ for this we just use the dice of the prediction vs. the gt (binary) mask
+ 2. compute the mask for the "IOU Regression Head": so we want the iou output from the decoder to
+ match the actual IOU between predicted and (binary) ground-truth mask. And we use L2Loss / MSE for this.
+ """
+ masks = [m["masks"] for m in batched_outputs]
+ predicted_iou_values = [m["iou_predictions"] for m in batched_outputs]
+ with torch.no_grad():
+ mean_model_iou = torch.mean(torch.stack([p.mean() for p in predicted_iou_values]))
+
+ mask_loss = 0.0 # this is the loss term for 1.
+ iou_regression_loss = 0.0 # this is the loss term for 2.
+
+ # outer loop is over the batch (different image/patch predictions)
+ for m_, y_, ids_, predicted_iou_ in zip(masks, y, sampled_ids, predicted_iou_values):
+ per_object_dice_scores = []
+ per_object_iou_scores = []
+
+ # inner loop is over the channels, this corresponds to the different predicted objects
+ for i, (predicted_obj, predicted_iou) in enumerate(zip(m_, predicted_iou_)):
+ predicted_obj = self._sigmoid(predicted_obj).to(self.device)
+ true_obj = (y_ == ids_[i]).to(self.device)
+
+ # this is computing the LOSS for 1.)
+ _dice_score = min([self._get_dice(p[None], true_obj) for p in predicted_obj])
+ per_object_dice_scores.append(_dice_score)
+
+ # now we need to compute the loss for 2.)
+ with torch.no_grad():
+ true_iou = torch.stack([self._get_iou(p[None], true_obj) for p in predicted_obj])
+ _iou_score = self.mse_loss(true_iou, predicted_iou)
+ per_object_iou_scores.append(_iou_score)
+
+ mask_loss = mask_loss + torch.mean(torch.stack(per_object_dice_scores))
+ iou_regression_loss = iou_regression_loss + torch.mean(torch.stack(per_object_iou_scores))
+
+ loss = mask_loss + iou_regression_loss
+
+ return loss, mask_loss, iou_regression_loss, mean_model_iou
+
+ def _postprocess_outputs(self, masks):
+ """ masks look like -> (B, 1, X, Y)
+ where, B is the number of objects, (X, Y) is the input image shape
+ """
+ instance_labels = []
+ for m in masks:
+ instance_list = [self._sigmoid(_val) for _val in m.squeeze(1)]
+ instance_label = torch.stack(instance_list, dim=0).sum(dim=0).clip(0, 1)
+ instance_labels.append(instance_label)
+ instance_labels = torch.stack(instance_labels).unsqueeze(1)
+ return instance_labels
+
+ def _get_val_metric(self, batched_outputs, sampled_binary_y):
+ """ Tracking the validation metric based on the DiceLoss
+ """
+ masks = [m["masks"] for m in batched_outputs]
+ pred_labels = self._postprocess_outputs(masks)
+
+ # we do the condition below to adapt w.r.t. the multimask output
+ # to select the "objectively" best response
+ if pred_labels.dim() == 5:
+ metric = min([self.metric(pred_labels[:, :, i, :, :], sampled_binary_y.to(self.device))
+ for i in range(pred_labels.shape[2])])
+ else:
+ metric = self.metric(pred_labels, sampled_binary_y.to(self.device))
+
+ return metric
+
+ #
+ # Update Masks Iteratively while Training
+ #
+ def _update_masks(self, batched_inputs, y, sampled_binary_y, sampled_ids, num_subiter, multimask_output):
+ # estimating the image inputs to make the computations faster for the decoder
+ input_images = torch.stack([self.model.preprocess(x=x["image"].to(self.device)) for x in batched_inputs], dim=0)
+ image_embeddings = self.model.image_embeddings_oft(input_images)
+
+ loss = 0.0
+ mask_loss = 0.0
+ iou_regression_loss = 0.0
+ mean_model_iou = 0.0
+
+ # this loop takes care of the idea of sub-iterations, i.e. the number of times we iterate over each batch
+ for i in range(0, num_subiter):
+ # we do multimasking only in the first sub-iteration as we then pass single prompt
+ # after the first sub-iteration, we don't do multimasking because we get multiple prompts
+ batched_outputs = self.model(batched_inputs,
+ multimask_output=multimask_output if i == 0 else False,
+ image_embeddings=image_embeddings)
+
+ # we want to average the loss and then backprop over the net sub-iterations
+ net_loss, net_mask_loss, net_iou_regression_loss, net_mean_model_iou = self._get_net_loss(batched_outputs,
+ y, sampled_ids)
+ loss += net_loss
+ mask_loss += net_mask_loss
+ iou_regression_loss += net_iou_regression_loss
+ mean_model_iou += net_mean_model_iou
+
+ masks, logits_masks = [], []
+ # the loop below gets us the masks and logits from the batch-level outputs
+ for m in batched_outputs:
+ mask, l_mask = [], []
+ for _m, _l, _iou in zip(m["masks"], m["low_res_masks"], m["iou_predictions"]):
+ best_iou_idx = torch.argmax(_iou)
+
+ best_mask, best_logits = _m[best_iou_idx], _l[best_iou_idx]
+ best_mask, best_logits = best_mask[None], best_logits[None]
+ mask.append(self._sigmoid(best_mask))
+ l_mask.append(best_logits)
+
+ mask, l_mask = torch.stack(mask), torch.stack(l_mask)
+ masks.append(mask)
+ logits_masks.append(l_mask)
+
+ masks, logits_masks = torch.stack(masks), torch.stack(logits_masks)
+ masks = (masks > 0.5).to(torch.float32)
+
+ self._get_updated_points_per_mask_per_subiter(masks, sampled_binary_y, batched_inputs, logits_masks)
+
+ loss = loss / num_subiter
+ mask_loss = mask_loss / num_subiter
+ iou_regression_loss = iou_regression_loss / num_subiter
+ mean_model_iou = mean_model_iou / num_subiter
+
+ return loss, mask_loss, iou_regression_loss, mean_model_iou
+
+ def _get_updated_points_per_mask_per_subiter(self, masks, sampled_binary_y, batched_inputs, logits_masks):
+ # here, we get the pair-per-batch of predicted and true elements (and also the "batched_inputs")
+ for x1, x2, _inp, logits in zip(masks, sampled_binary_y, batched_inputs, logits_masks):
+ net_coords, net_labels = [], []
+
+ # here, we get each object in the pairs and do the point choices per-object
+ for pred_obj, true_obj in zip(x1, x2):
+ true_obj = true_obj.to(self.device)
+
+ expected_diff = (pred_obj - true_obj)
+
+ neg_region = (expected_diff == 1).to(torch.float32)
+ pos_region = (expected_diff == -1)
+ overlap_region = torch.logical_and(pred_obj == 1, true_obj == 1).to(torch.float32)
+
+ # POSITIVE POINTS
+ tmp_pos_loc = torch.where(pos_region)
+ if torch.stack(tmp_pos_loc).shape[-1] == 0:
+ tmp_pos_loc = torch.where(overlap_region)
+
+ pos_index = np.random.choice(len(tmp_pos_loc[1]))
+ pos_coordinates = int(tmp_pos_loc[1][pos_index]), int(tmp_pos_loc[2][pos_index])
+ pos_coordinates = pos_coordinates[::-1]
+ pos_labels = 1
+
+ # NEGATIVE POINTS
+ tmp_neg_loc = torch.where(neg_region)
+ if torch.stack(tmp_neg_loc).shape[-1] == 0:
+ tmp_true_loc = torch.where(true_obj)
+ x_coords, y_coords = tmp_true_loc[1], tmp_true_loc[2]
+ bbox = torch.stack([torch.min(x_coords), torch.min(y_coords),
+ torch.max(x_coords) + 1, torch.max(y_coords) + 1])
+ bbox_mask = torch.zeros_like(true_obj).squeeze(0)
+ bbox_mask[bbox[0]:bbox[2], bbox[1]:bbox[3]] = 1
+ bbox_mask = bbox_mask[None].to(self.device)
+
+ dilated_bbox_mask = dilation(bbox_mask[None], torch.ones(3, 3).to(self.device)).squeeze(0)
+ background_mask = abs(dilated_bbox_mask - true_obj)
+ tmp_neg_loc = torch.where(background_mask)
+
+ neg_index = np.random.choice(len(tmp_neg_loc[1]))
+ neg_coordinates = int(tmp_neg_loc[1][neg_index]), int(tmp_neg_loc[2][neg_index])
+ neg_coordinates = neg_coordinates[::-1]
+ neg_labels = 0
+
+ net_coords.append([pos_coordinates, neg_coordinates])
+ net_labels.append([pos_labels, neg_labels])
+
+ if "point_labels" in _inp.keys():
+ updated_point_coords = torch.cat([_inp["point_coords"], torch.tensor(net_coords)], dim=1)
+ updated_point_labels = torch.cat([_inp["point_labels"], torch.tensor(net_labels)], dim=1)
+ else:
+ updated_point_coords = torch.tensor(net_coords)
+ updated_point_labels = torch.tensor(net_labels)
+
+ _inp["point_coords"] = updated_point_coords
+ _inp["point_labels"] = updated_point_labels
+ _inp["mask_inputs"] = logits
+
+ #
+ # Training Loop
+ #
+
+ def _update_samples_for_gt_instances(self, y, n_samples):
+ num_instances_gt = [len(torch.unique(_y)) for _y in y]
+ if n_samples > min(num_instances_gt):
+ n_samples = min(num_instances_gt) - 1
+ return n_samples
+
+ def _train_epoch_impl(self, progress, forward_context, backprop):
+ self.model.train()
+
+ n_iter = 0
+ t_per_iter = time.time()
+ for x, y in self.train_loader:
+
+ self.optimizer.zero_grad()
+
+ with forward_context():
+ n_samples = self.n_objects_per_batch
+ n_samples = self._update_samples_for_gt_instances(y, n_samples)
+
+ n_pos, n_neg, get_boxes, multimask_output = self._get_prompt_and_multimasking_choices(self._iteration)
+
+ batched_inputs, sampled_ids = self.convert_inputs(x, y, n_pos, n_neg, get_boxes, n_samples)
+
+ assert len(y) == len(sampled_ids)
+ sampled_binary_y = []
+ for i in range(len(y)):
+ _sampled = [torch.isin(y[i], torch.tensor(idx)) for idx in sampled_ids[i]]
+ sampled_binary_y.append(_sampled)
+
+ # the steps below are done for one reason in a gist:
+ # to handle images where there aren't enough instances as expected
+ # (e.g. where one image has only one instance)
+ obj_lengths = [len(s) for s in sampled_binary_y]
+ sampled_binary_y = [s[:min(obj_lengths)] for s in sampled_binary_y]
+ sampled_binary_y = [torch.stack(s).to(torch.float32) for s in sampled_binary_y]
+ sampled_binary_y = torch.stack(sampled_binary_y)
+
+ # gist for below - while we find the mismatch, we need to update the batched inputs
+ # else it would still generate masks using mismatching prompts, and it doesn't help us
+ # with the subiterations again. hence we clip the number of input points as well
+ f_objs = sampled_binary_y.shape[1]
+ batched_inputs = [
+ {k: (v[:f_objs] if k in ("point_coords", "point_labels", "boxes") else v) for k, v in inp.items()}
+ for inp in batched_inputs
+ ]
+
+ loss, mask_loss, iou_regression_loss, model_iou = self._update_masks(batched_inputs, y,
+ sampled_binary_y, sampled_ids,
+ num_subiter=self.n_sub_iteration,
+ multimask_output=multimask_output)
+
+ backprop(loss)
+
+ if self.logger is not None:
+ lr = [pm["lr"] for pm in self.optimizer.param_groups][0]
+ samples = sampled_binary_y if self._iteration % self.log_image_interval == 0 else None
+ self.logger.log_train(self._iteration, loss, lr, x, y, samples,
+ mask_loss, iou_regression_loss, model_iou)
+
+ self._iteration += 1
+ n_iter += 1
+ if self._iteration >= self.max_iteration:
+ break
+ progress.update(1)
+
+ t_per_iter = (time.time() - t_per_iter) / n_iter
+ return t_per_iter
+
+ def _validate_impl(self, forward_context):
+ self.model.eval()
+
+ metric_val = 0.0
+ loss_val = 0.0
+ model_iou_val = 0.0
+ val_iteration = 0
+
+ with torch.no_grad():
+ for x, y in self.val_loader:
+ with forward_context():
+ n_samples = self.n_objects_per_batch
+ n_samples = self._update_samples_for_gt_instances(y, n_samples)
+
+ (n_pos, n_neg,
+ get_boxes, multimask_output) = self._get_prompt_and_multimasking_choices_for_val(val_iteration)
+
+ batched_inputs, sampled_ids = self.convert_inputs(x, y, n_pos, n_neg, get_boxes, n_samples)
+
+ batched_outputs = self.model(batched_inputs, multimask_output=multimask_output)
+
+ assert len(y) == len(sampled_ids)
+ sampled_binary_y = torch.stack(
+ [torch.isin(y[i], torch.tensor(sampled_ids[i])) for i in range(len(y))]
+ ).to(torch.float32)
+
+ loss, mask_loss, iou_regression_loss, model_iou = self._get_net_loss(batched_outputs,
+ y, sampled_ids)
+
+ metric = self._get_val_metric(batched_outputs, sampled_binary_y)
+
+ loss_val += loss.item()
+ metric_val += metric.item()
+ model_iou_val += model_iou.item()
+ val_iteration += 1
+
+ loss_val /= len(self.val_loader)
+ metric_val /= len(self.val_loader)
+ model_iou_val /= len(self.val_loader)
+ print()
+ print(f"The Average Dice Score for the Current Epoch is {1 - metric_val}")
+
+ if self.logger is not None:
+ self.logger.log_validation(
+ self._iteration, metric_val, loss_val, x, y,
+ sampled_binary_y, mask_loss, iou_regression_loss, model_iou_val
+ )
+
+ return metric_val
+
+
+class SamLogger(TorchEmLogger):
+ """@private"""
+ def __init__(self, trainer, save_root, **unused_kwargs):
+ super().__init__(trainer, save_root)
+ self.log_dir = f"./logs/{trainer.name}" if save_root is None else\
+ os.path.join(save_root, "logs", trainer.name)
+ os.makedirs(self.log_dir, exist_ok=True)
+
+ self.tb = torch.utils.tensorboard.SummaryWriter(self.log_dir)
+ self.log_image_interval = trainer.log_image_interval
+
+ def add_image(self, x, y, samples, name, step):
+ self.tb.add_image(tag=f"{name}/input", img_tensor=x[0], global_step=step)
+ self.tb.add_image(tag=f"{name}/target", img_tensor=y[0], global_step=step)
+ sample_grid = make_grid([sample[0] for sample in samples], nrow=4, padding=4)
+ self.tb.add_image(tag=f"{name}/samples", img_tensor=sample_grid, global_step=step)
+
+ def log_train(self, step, loss, lr, x, y, samples, mask_loss, iou_regression_loss, model_iou):
+ self.tb.add_scalar(tag="train/loss", scalar_value=loss, global_step=step)
+ self.tb.add_scalar(tag="train/mask_loss", scalar_value=mask_loss, global_step=step)
+ self.tb.add_scalar(tag="train/iou_loss", scalar_value=iou_regression_loss, global_step=step)
+ self.tb.add_scalar(tag="train/model_iou", scalar_value=model_iou, global_step=step)
+ self.tb.add_scalar(tag="train/learning_rate", scalar_value=lr, global_step=step)
+ if step % self.log_image_interval == 0:
+ self.add_image(x, y, samples, "train", step)
+
+ def log_validation(self, step, metric, loss, x, y, samples, mask_loss, iou_regression_loss, model_iou):
+ self.tb.add_scalar(tag="validation/loss", scalar_value=loss, global_step=step)
+ self.tb.add_scalar(tag="validation/mask_loss", scalar_value=mask_loss, global_step=step)
+ self.tb.add_scalar(tag="validation/iou_loss", scalar_value=iou_regression_loss, global_step=step)
+ self.tb.add_scalar(tag="validation/model_iou", scalar_value=model_iou, global_step=step)
+ self.tb.add_scalar(tag="validation/metric", scalar_value=metric, global_step=step)
+ self.add_image(x, y, samples, "validation", step)
diff --git a/micro_sam/training/trainable_sam.py b/micro_sam/training/trainable_sam.py
new file mode 100644
index 00000000..99728a1b
--- /dev/null
+++ b/micro_sam/training/trainable_sam.py
@@ -0,0 +1,117 @@
+from typing import Any, Dict, List, Optional, Union
+
+import torch
+from torch import nn
+from torch.nn import functional as F
+
+from segment_anything.modeling import Sam
+
+
+# simple wrapper around SAM in order to keep things trainable
+class TrainableSAM(nn.Module):
+ """Wrapper to make the SegmentAnything model trainable.
+
+ Args:
+ sam: The SegmentAnything Model.
+ device: The device for training.
+ """
+ def __init__(
+ self,
+ sam: Sam,
+ device: Union[str, torch.device],
+ ) -> None:
+ super().__init__()
+ self.sam = sam
+ self.device = device
+
+ def preprocess(self, x: torch.Tensor) -> torch.Tensor:
+ """Normalize pixel values and pad to a square input.
+
+ Args:
+ x: The input tensor.
+
+ Returns:
+ The normalized and padded tensor.
+ """
+ # Normalize colors
+ x = (x - self.sam.pixel_mean) / self.sam.pixel_std
+
+ # Pad
+ h, w = x.shape[-2:]
+ padh = self.sam.image_encoder.img_size - h
+ padw = self.sam.image_encoder.img_size - w
+ x = F.pad(x, (0, padw, 0, padh))
+ return x
+
+ def image_embeddings_oft(self, input_images):
+ """@private"""
+ image_embeddings = self.sam.image_encoder(input_images)
+ return image_embeddings
+
+ # batched inputs follow the same syntax as the input to sam.forward
+ def forward(
+ self,
+ batched_inputs: List[Dict[str, Any]],
+ multimask_output: bool = False,
+ image_embeddings: Optional[torch.Tensor] = None,
+ ) -> List[Dict[str, Any]]:
+ """Forward pass.
+
+ Args:
+ batched_inputs: The batched input images and prompts.
+ multimask_output: Whether to predict mutiple or just a single mask.
+ image_embeddings: The precompute image embeddings. If not passed then they will be computed.
+
+ Returns:
+ The predicted segmentation masks and iou values.
+ """
+ input_images = torch.stack([self.preprocess(x=x["image"].to(self.device)) for x in batched_inputs], dim=0)
+ if image_embeddings is None:
+ image_embeddings = self.sam.image_encoder(input_images)
+
+ outputs = []
+ for image_record, curr_embedding in zip(batched_inputs, image_embeddings):
+ if "point_coords" in image_record:
+ points = (image_record["point_coords"].to(self.device), image_record["point_labels"].to(self.device))
+ else:
+ points = None
+
+ if "boxes" in image_record:
+ boxes = image_record.get("boxes").to(self.device)
+ else:
+ boxes = None
+
+ if "mask_inputs" in image_record:
+ masks = image_record.get("mask_inputs").to(self.device)
+ else:
+ masks = None
+
+ sparse_embeddings, dense_embeddings = self.sam.prompt_encoder(
+ points=points,
+ boxes=boxes,
+ masks=masks,
+ )
+
+ low_res_masks, iou_predictions = self.sam.mask_decoder(
+ image_embeddings=curr_embedding.unsqueeze(0),
+ image_pe=self.sam.prompt_encoder.get_dense_pe(),
+ sparse_prompt_embeddings=sparse_embeddings,
+ dense_prompt_embeddings=dense_embeddings,
+ multimask_output=multimask_output,
+ )
+
+ masks = self.sam.postprocess_masks(
+ low_res_masks,
+ input_size=image_record["image"].shape[-2:],
+ original_size=image_record["original_size"],
+ )
+
+ outputs.append(
+ {
+ "low_res_masks": low_res_masks,
+ "masks": masks,
+ "iou_predictions": iou_predictions
+ }
+ )
+
+ return outputs
diff --git a/micro_sam/training/util.py b/micro_sam/training/util.py
new file mode 100644
index 00000000..3821ac7f
--- /dev/null
+++ b/micro_sam/training/util.py
@@ -0,0 +1,191 @@
+import os
+from typing import List, Optional, Union
+
+import torch
+import numpy as np
+
+from ..prompt_generators import PointAndBoxPromptGenerator
+from ..util import get_centers_and_bounding_boxes, get_sam_model
+from .trainable_sam import TrainableSAM
+
+
+def get_trainable_sam_model(
+ model_type: str = "vit_h",
+ checkpoint_path: Optional[Union[str, os.PathLike]] = None,
+ freeze: Optional[List[str]] = None,
+ device: Optional[Union[str, torch.device]] = None,
+) -> TrainableSAM:
+ """Get the trainable sam model.
+
+ Args:
+ model_type: The type of the segment anything model.
+ checkpoint_path: Path to a custom checkpoint from which to load the model weights.
+ freeze: Specify parts of the model that should be frozen.
+ By default nothing is frozen and the full model is updated.
+ device: The device to use for training.
+
+ Returns:
+ The trainable segment anything model.
+ """
+ # set the device here so that the correct one is passed to TrainableSAM below
+ if device is None:
+ device = "cuda" if torch.cuda.is_available() else "cpu"
+ _, sam = get_sam_model(device, model_type, checkpoint_path, return_sam=True)
+
+ # freeze components of the model if freeze was passed
+ # ideally we would want to add components in such a way that:
+ # - we would be able to freeze the choice of encoder/decoder blocks, yet be able to add components to the network
+ # (for e.g. encoder blocks to "image_encoder")
+ if freeze is not None:
+ for name, param in sam.named_parameters():
+ if isinstance(freeze, list):
+ # we would want to "freeze" all the components in the model if passed a list of parts
+ for l_item in freeze:
+ if name.startswith(f"{l_item}"):
+ param.requires_grad = False
+ else:
+ # we "freeze" only for one specific component when passed a "particular" part
+ if name.startswith(f"{freeze}"):
+ param.requires_grad = False
+
+ # convert to trainable sam
+ trainable_sam = TrainableSAM(sam, device)
+ return trainable_sam
+
+
+class ConvertToSamInputs:
+ """Convert outputs of data loader to the expected batched inputs of the SegmentAnything model.
+
+ Args:
+ dilation_strength: The dilation factor.
+ It determines a "safety" border from which prompts are not sampled to avoid ambiguous prompts
+ due to imprecise groundtruth masks.
+ box_distortion_factor: Factor for distorting the box annotations derived from the groundtruth masks.
+ Not yet implemented.
+ """
+ def __init__(
+ self,
+ dilation_strength: int = 10,
+ box_distortion_factor: Optional[float] = None,
+ ) -> None:
+ self.dilation_strength = dilation_strength
+ # TODO implement the box distortion logic
+ if box_distortion_factor is not None:
+ raise NotImplementedError
+
+ def _get_prompt_generator(self, n_positive_points, n_negative_points, get_boxes, get_points):
+ """Returns the prompt generator w.r.t. the "random" attributes inputed."""
+
+ # the class initialization below gets the random choice of n_positive and n_negative points as inputs
+ # (done in the trainer)
+ # in case of dynamic choice while choosing between points and/or box, it gets those as well
+ prompt_generator = PointAndBoxPromptGenerator(n_positive_points=n_positive_points,
+ n_negative_points=n_negative_points,
+ dilation_strength=self.dilation_strength,
+ get_box_prompts=get_boxes,
+ get_point_prompts=get_points)
+ return prompt_generator
+
+ def _get_prompt_lists(self, gt, n_samples, n_positive_points, n_negative_points, get_boxes,
+ get_points, prompt_generator, point_coordinates, bbox_coordinates):
+ """Returns a list of "expected" prompts subjected to the random input attributes for prompting."""
+ box_prompts = []
+ point_prompts = []
+ point_label_prompts = []
+
+ # getting the cell instance except the bg
+ cell_ids = np.unique(gt)[1:]
+
+ accepted_cell_ids = []
+ sampled_cell_ids = []
+
+ # while conditions gets all the prompts until it satisfies the requirement
+ while len(accepted_cell_ids) < min(n_samples, len(cell_ids)):
+ if len(sampled_cell_ids) == len(cell_ids): # we did not find enough cells
+ break
+
+ my_cell_id = np.random.choice(np.setdiff1d(cell_ids, sampled_cell_ids))
+ sampled_cell_ids.append(my_cell_id)
+
+ bbox = bbox_coordinates[my_cell_id]
+ # points = point_coordinates[my_cell_id]
+ # removed "points" to randomly choose fg points
+ coord_list, label_list, bbox_list, _ = prompt_generator(gt, my_cell_id, bbox)
+
+ if get_boxes is True and get_points is False: # only box
+ bbox_list = bbox_list[0]
+ box_prompts.append([bbox_list[1], bbox_list[0],
+ bbox_list[3], bbox_list[2]])
+ accepted_cell_ids.append(my_cell_id)
+
+ if get_points: # one with points expected
+ # check for the minimum point requirement per object in the batch
+ if len(label_list) == n_negative_points + n_positive_points:
+ point_prompts.append(np.array([ip[::-1] for ip in coord_list]))
+ point_label_prompts.append(np.array(label_list))
+ accepted_cell_ids.append(my_cell_id)
+ if get_boxes: # one with boxes expected with points as well
+ bbox_list = bbox_list[0]
+ box_prompts.append([bbox_list[1], bbox_list[0],
+ bbox_list[3], bbox_list[2]])
+
+ point_prompts = np.array(point_prompts)
+ point_label_prompts = np.array(point_label_prompts)
+ return box_prompts, point_prompts, point_label_prompts, accepted_cell_ids
+
+ def __call__(self, x, y, n_pos, n_neg, get_boxes=False, n_samples=None):
+ """Convert the outputs of dataloader and prompt settings to the batch format expected by SAM.
+ """
+
+ # condition to see if we get point prompts, then we (ofc) use point-prompting
+ # else we don't use point prompting
+ if n_pos == 0 and n_neg == 0:
+ get_points = False
+ else:
+ get_points = True
+
+ # keeping the solution open by checking for deterministic/dynamic choice of point prompts
+ prompt_generator = self._get_prompt_generator(n_pos, n_neg, get_boxes, get_points)
+
+ batched_inputs = []
+ batched_sampled_cell_ids_list = []
+ for i, gt in enumerate(y):
+ gt = gt.squeeze().numpy().astype(np.int32)
+ point_coordinates, bbox_coordinates = get_centers_and_bounding_boxes(gt)
+
+ this_n_samples = len(point_coordinates) if n_samples is None else n_samples
+ box_prompts, point_prompts, point_label_prompts, sampled_cell_ids = self._get_prompt_lists(
+ gt, this_n_samples,
+ n_pos, n_neg,
+ get_boxes,
+ get_points,
+ prompt_generator,
+ point_coordinates,
+ bbox_coordinates
+ )
+
+ # check to be sure about the expected size of the no. of elements in different settings
+ if get_boxes is True and get_points is False:
+ assert len(sampled_cell_ids) == len(box_prompts), \
+ print(len(sampled_cell_ids), len(box_prompts))
+
+ elif get_boxes is False and get_points is True:
+ assert len(sampled_cell_ids) == len(point_prompts) == len(point_label_prompts), \
+ print(len(sampled_cell_ids), len(point_prompts), len(point_label_prompts))
+
+ elif get_boxes is True and get_points is True:
+ assert len(sampled_cell_ids) == len(box_prompts) == len(point_prompts) == len(point_label_prompts), \
+ print(len(sampled_cell_ids), len(box_prompts), len(point_prompts), len(point_label_prompts))
+
+ batched_sampled_cell_ids_list.append(sampled_cell_ids)
+
+ batched_input = {"image": x[i], "original_size": x[i].shape[1:]}
+ if get_boxes:
+ batched_input["boxes"] = torch.tensor(box_prompts)
+ if get_points:
+ batched_input["point_coords"] = torch.tensor(point_prompts)
+ batched_input["point_labels"] = torch.tensor(point_label_prompts)
+
+ batched_inputs.append(batched_input)
+
+ return batched_inputs, batched_sampled_cell_ids_list
diff --git a/micro_sam/util.py b/micro_sam/util.py
index 7578c153..1ea21303 100644
--- a/micro_sam/util.py
+++ b/micro_sam/util.py
@@ -4,9 +4,11 @@
import hashlib
import os
+import pickle
import warnings
+from collections import OrderedDict
from shutil import copyfileobj
-from typing import Any, Callable, Dict, Optional, Tuple, Iterable
+from typing import Any, Callable, Dict, Iterable, Optional, Tuple, Union
import imageio.v3 as imageio
import numpy as np
@@ -27,25 +29,27 @@
from tqdm import tqdm
_MODEL_URLS = {
+ # the default segment anything models
"vit_h": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_h_4b8939.pth",
"vit_l": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_l_0b3195.pth",
"vit_b": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_b_01ec64.pth",
- # preliminary finetuned models
- "vit_h_lm": "https://owncloud.gwdg.de/index.php/s/CnxBvsdGPN0TD3A/download",
- "vit_b_lm": "https://owncloud.gwdg.de/index.php/s/gGlR1LFsav0eQ2k/download",
- "vit_h_em": "https://owncloud.gwdg.de/index.php/s/VcHoLC6AM0CrRpM/download",
- "vit_b_em": "https://owncloud.gwdg.de/index.php/s/BWupWhG1HRflI97/download",
+ # first version of finetuned models on zenodo
+ "vit_h_lm": "https://zenodo.org/record/8250299/files/vit_h_lm.pth?download=1",
+ "vit_b_lm": "https://zenodo.org/record/8250281/files/vit_b_lm.pth?download=1",
+ "vit_h_em": "https://zenodo.org/record/8250291/files/vit_h_em.pth?download=1",
+ "vit_b_em": "https://zenodo.org/record/8250260/files/vit_b_em.pth?download=1",
}
_CHECKPOINT_FOLDER = os.environ.get("SAM_MODELS", os.path.expanduser("~/.sam_models"))
_CHECKSUMS = {
+ # the default segment anything models
"vit_h": "a7bf3b02f3ebf1267aba913ff637d9a2d5c33d3173bb679e46d9f338c26f262e",
"vit_l": "3adcc4315b642a4d2101128f611684e8734c41232a17c648ed1693702a49a622",
"vit_b": "ec2df62732614e57411cdcf32a23ffdf28910380d03139ee0f4fcbe91eb8c912",
- # preliminary finetuned models
- "vit_h_lm": "c30a580e6ccaff2f4f0fbaf9cad10cee615a915cdd8c7bc4cb50ea9bdba3fc09",
- "vit_b_lm": "f2b8676f92a123f6f8ac998818118bd7269a559381ec60af4ac4be5c86024a1b",
- "vit_h_em": "652f70acad89ab855502bc10965e7d0baf7ef5f38fef063dd74f1787061d3919",
- "vit_b_em": "9eb783e538bb287c7086f825f1e1dc5d5681bd116541a0b98cab85f1e7f4dd62",
+ # first version of finetuned models on zenodo
+ "vit_h_lm": "9a65ee0cddc05a98d60469a12a058859c89dc3ea3ba39fed9b90d786253fbf26",
+ "vit_b_lm": "5a59cc4064092d54cd4d92cd967e39168f3760905431e868e474d60fe5464ecd",
+ "vit_h_em": "ae3798a0646c8df1d4db147998a2d37e402ff57d3aa4e571792fbb911d8a979c",
+ "vit_b_em": "c04a714a4e14a110f0eec055a65f7409d54e6bf733164d2933a0ce556f7d6f81",
}
# this is required so that the downloaded file is not called 'download'
_DOWNLOAD_NAMES = {
@@ -54,10 +58,19 @@
"vit_h_em": "vit_h_em.pth",
"vit_b_em": "vit_b_em.pth",
}
+# this is the default model used in micro_sam
+# currently set to the default vit_h
+_DEFAULT_MODEL = "vit_h"
# TODO define the proper type for image embeddings
ImageEmbeddings = Dict[str, Any]
+"""@private"""
+
+
+#
+# Functionality for model download and export
+#
def _download(url, path, model_type):
@@ -105,9 +118,9 @@ def _get_checkpoint(model_type, checkpoint_path=None):
def get_sam_model(
device: Optional[str] = None,
- model_type: str = "vit_h",
- checkpoint_path: Optional[str] = None,
- return_sam: bool = False
+ model_type: str = _DEFAULT_MODEL,
+ checkpoint_path: Optional[Union[str, os.PathLike]] = None,
+ return_sam: bool = False,
) -> SamPredictor:
"""Get the SegmentAnything Predictor.
@@ -117,7 +130,7 @@ def get_sam_model(
Args:
device: The device for the model. If none is given will use GPU if available.
- model_type: The SegmentAnything model to use.
+ model_type: The SegmentAnything model to use. Will use the standard vit_h model by default.
checkpoint_path: The path to the corresponding checkpoint if not in the default model folder.
return_sam: Return the sam model object as well as the predictor.
@@ -125,7 +138,8 @@ def get_sam_model(
The segment anything predictor.
"""
checkpoint = _get_checkpoint(model_type, checkpoint_path)
- device = "cuda" if torch.cuda.is_available() else "cpu"
+ if device is None:
+ device = "cuda" if torch.cuda.is_available() else "cpu"
# Our custom model types have a suffix "_...". This suffix needs to be stripped
# before calling sam_model_registry.
@@ -141,10 +155,107 @@ def get_sam_model(
return predictor
+# We write a custom unpickler that skips objects that cannot be found instead of
+# throwing an AttributeError or ModueNotFoundError.
+# NOTE: since we just want to unpickle the model to load its weights these errors don't matter.
+# See also https://stackoverflow.com/questions/27732354/unable-to-load-files-using-pickle-and-multiple-modules
+class _CustomUnpickler(pickle.Unpickler):
+ def find_class(self, module, name):
+ try:
+ return super().find_class(module, name)
+ except (AttributeError, ModuleNotFoundError) as e:
+ warnings.warn(f"Did not find {module}:{name} and will skip it, due to error {e}")
+ return None
+
+
+def get_custom_sam_model(
+ checkpoint_path: Union[str, os.PathLike],
+ device: Optional[str] = None,
+ model_type: str = "vit_h",
+ return_sam: bool = False,
+ return_state: bool = False,
+) -> SamPredictor:
+ """Load a SAM model from a torch_em checkpoint.
+
+ This function enables loading from the checkpoints saved by
+ the functionality in `micro_sam.training`.
+
+ Args:
+ checkpoint_path: The path to the corresponding checkpoint if not in the default model folder.
+ device: The device for the model. If none is given will use GPU if available.
+ model_type: The SegmentAnything model to use.
+ return_sam: Return the sam model object as well as the predictor.
+ return_state: Return the full state of the checkpoint in addition to the predictor.
+
+ Returns:
+ The segment anything predictor.
+ """
+ assert not (return_sam and return_state)
+
+ # over-ride the unpickler with our custom one
+ custom_pickle = pickle
+ custom_pickle.Unpickler = _CustomUnpickler
+
+ if device is None:
+ device = "cuda" if torch.cuda.is_available() else "cpu"
+ sam = sam_model_registry[model_type]()
+
+ # load the model state, ignoring any attributes that can't be found by pickle
+ state = torch.load(checkpoint_path, map_location=device, pickle_module=custom_pickle)
+ model_state = state["model_state"]
+
+ # copy the model weights from torch_em's training format
+ sam_prefix = "sam."
+ model_state = OrderedDict(
+ [(k[len(sam_prefix):] if k.startswith(sam_prefix) else k, v) for k, v in model_state.items()]
+ )
+ sam.load_state_dict(model_state)
+ sam.to(device)
+
+ predictor = SamPredictor(sam)
+ predictor.model_type = model_type
+
+ if return_sam:
+ return predictor, sam
+ if return_state:
+ return predictor, state
+ return predictor
+
+
+def export_custom_sam_model(
+ checkpoint_path: Union[str, os.PathLike],
+ model_type: str,
+ save_path: Union[str, os.PathLike],
+) -> None:
+ """Export a finetuned segment anything model to the standard model format.
+
+ The exported model can be used by the interactive annotation tools in `micro_sam.annotator`.
+
+ Args:
+ checkpoint_path: The path to the corresponding checkpoint if not in the default model folder.
+ model_type: The SegmentAnything model type to use (vit_h, vit_b or vit_l).
+ save_path: Where to save the exported model.
+ """
+ _, state = get_custom_sam_model(
+ checkpoint_path, model_type=model_type, return_state=True, device=torch.device("cpu"),
+ )
+ model_state = state["model_state"]
+ prefix = "sam."
+ model_state = OrderedDict(
+ [(k[len(prefix):] if k.startswith(prefix) else k, v) for k, v in model_state.items()]
+ )
+ torch.save(model_state, save_path)
+
+
def get_model_names() -> Iterable:
return _MODEL_URLS.keys()
+#
+# Functionality for precomputing embeddings and other state
+#
+
+
def _to_image(input_):
# we require the input to be uint8
if input_.dtype != np.dtype("uint8"):
@@ -375,7 +486,7 @@ def precompute_image_embeddings(
If 'save_path' is given the embeddings will be loaded/saved in a zarr container.
Args:
- predictor: The SegmentAnything predictor
+ predictor: The SegmentAnything predictor.
input_: The input data. Can be 2 or 3 dimensional, corresponding to an image, volume or timeseries.
save_path: Path to save the embeddings in a zarr container.
lazy_loading: Whether to load all embeddings into memory or return an
@@ -397,14 +508,22 @@ def precompute_image_embeddings(
data_signature = _compute_data_signature(input_)
f = zarr.open(save_path, "a")
- key_vals = [("data_signature", data_signature),
- ("tile_shape", tile_shape), ("model_type", predictor.model_type)]
- for key, val in key_vals:
- if "input_size" in f.attrs: # we have computed the embeddings already
- # key signature does not match or is not in the file
+ key_vals = [
+ ("data_signature", data_signature),
+ ("tile_shape", tile_shape if tile_shape is None else list(tile_shape)),
+ ("halo", halo if halo is None else list(halo)),
+ ("model_type", predictor.model_type)
+ ]
+ if "input_size" in f.attrs: # we have computed the embeddings already and perform checks
+ for key, val in key_vals:
+ if val is None:
+ continue
+ # check whether the key signature does not match or is not in the file
if key not in f.attrs or f.attrs[key] != val:
- warnings.warn(f"Embeddings file is invalid due to unmatching {key}. \
- Please recompute embeddings in a new file.")
+ warnings.warn(
+ f"Embeddings file {save_path} is invalid due to unmatching {key}: "
+ f"{f.attrs.get(key)} != {val}.Please recompute embeddings in a new file."
+ )
if wrong_file_callback is not None:
save_path = wrong_file_callback(save_path)
f = zarr.open(save_path, "a")
@@ -441,7 +560,7 @@ def set_precomputed(
i: Index for the image data. Required if `image` has three spatial dimensions
or a time dimension and two spatial dimensions.
"""
- device = "cuda" if torch.cuda.is_available() else "cpu"
+ device = predictor.device
features = image_embeddings["features"]
assert features.ndim in (4, 5)
@@ -463,6 +582,11 @@ def set_precomputed(
return predictor
+#
+# Misc functionality
+#
+
+
def compute_iou(mask1: np.ndarray, mask2: np.ndarray) -> float:
"""Compute the intersection over union of two masks.
@@ -508,7 +632,7 @@ def get_centers_and_bounding_boxes(
bbox_coordinates = {prop.label: prop.bbox for prop in properties}
- assert len(bbox_coordinates) == len(center_coordinates)
+ assert len(bbox_coordinates) == len(center_coordinates), f"{len(bbox_coordinates)}, {len(center_coordinates)}"
return center_coordinates, bbox_coordinates
@@ -535,25 +659,3 @@ def load_image_data(
if not lazy_loading:
image_data = image_data[:]
return image_data
-
-
-def main():
- """@private"""
- import argparse
-
- parser = argparse.ArgumentParser(description="Compute the embeddings for an image.")
- parser.add_argument("-i", "--input_path", required=True)
- parser.add_argument("-o", "--output_path", required=True)
- parser.add_argument("-m", "--model_type", default="vit_h")
- parser.add_argument("-c", "--checkpoint_path", default=None)
- parser.add_argument("-k", "--key")
- args = parser.parse_args()
-
- predictor = get_sam_model(model_type=args.model_type, checkpoint_path=args.checkpoint_path)
- with open_file(args.input_path, mode="r") as f:
- data = f[args.key]
- precompute_image_embeddings(predictor, data, save_path=args.output_path)
-
-
-if __name__ == "__main__":
- main()
diff --git a/test/test_instance_segmentation.py b/test/test_instance_segmentation.py
index 2cb932d7..8ee81614 100644
--- a/test/test_instance_segmentation.py
+++ b/test/test_instance_segmentation.py
@@ -91,10 +91,11 @@ def test_embedding_mask_generator(self):
mask, image = self.mask, self.image
predictor, image_embeddings = self.predictor, self.image_embeddings
+ pred_iou_thresh, stability_score_thresh = 0.95, 0.75
amg = EmbeddingMaskGenerator(predictor)
amg.initialize(image, image_embeddings=image_embeddings, verbose=False)
- predicted = amg.generate(pred_iou_thresh=0.96)
+ predicted = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
predicted = mask_data_to_segmentation(predicted, image.shape, with_background=True)
self.assertGreater(matching(predicted, mask, threshold=0.75)["segmentation_accuracy"], 0.99)
@@ -103,7 +104,7 @@ def test_embedding_mask_generator(self):
self.assertEqual(initial_seg.shape, image.shape)
# check that regenerating the segmentation works
- predicted2 = amg.generate(pred_iou_thresh=0.96)
+ predicted2 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
predicted2 = mask_data_to_segmentation(predicted2, image.shape, with_background=True)
self.assertTrue(np.array_equal(predicted, predicted2))
@@ -111,7 +112,7 @@ def test_embedding_mask_generator(self):
state = amg.get_state()
amg = EmbeddingMaskGenerator(predictor)
amg.set_state(state)
- predicted3 = amg.generate(pred_iou_thresh=0.96)
+ predicted3 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
predicted3 = mask_data_to_segmentation(predicted3, image.shape, with_background=True)
self.assertTrue(np.array_equal(predicted, predicted3))
@@ -120,23 +121,24 @@ def test_tiled_embedding_mask_generator(self):
mask, image = self.large_mask, self.large_image
predictor, image_embeddings = self.predictor, self.tiled_embeddings
+ pred_iou_thresh, stability_score_thresh = 0.90, 0.60
- amg = TiledEmbeddingMaskGenerator(predictor)
+ amg = TiledEmbeddingMaskGenerator(predictor, box_extension=0.1)
amg.initialize(image, image_embeddings=image_embeddings)
- predicted = amg.generate(pred_iou_thresh=0.96)
+ predicted = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
initial_seg = amg.get_initial_segmentation()
self.assertGreater(matching(predicted, mask, threshold=0.75)["segmentation_accuracy"], 0.99)
self.assertEqual(initial_seg.shape, image.shape)
- predicted2 = amg.generate(pred_iou_thresh=0.96)
+ predicted2 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
self.assertTrue(np.array_equal(predicted, predicted2))
# check that serializing and reserializing the state works
state = amg.get_state()
amg = TiledEmbeddingMaskGenerator(predictor)
amg.set_state(state)
- predicted3 = amg.generate(pred_iou_thresh=0.96)
+ predicted3 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
self.assertTrue(np.array_equal(predicted, predicted3))
def test_tiled_automatic_mask_generator(self):
+The training logic is implemented in `micro_sam.training` and is based on [torch-em](https://github.com/constantinpape/torch-em). Please check out [examples/finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/examples/finetuning) to see how you can finetune on your own data with it. The script `finetune_hela.py` contains an example for finetuning on a small custom microscopy dataset and `use_finetuned_model.py` shows how this model can then be used in the interactive annotatin tools.
+More advanced examples, including quantitative and qualitative evaluation, of finetuned models can be found in [finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/finetuning), which contains the code for training and evaluating our microscopy models.
diff --git a/doc/start_page.md b/doc/start_page.md
index b5fab2a9..840a650b 100644
--- a/doc/start_page.md
+++ b/doc/start_page.md
@@ -2,7 +2,7 @@
Segment Anything for Microscopy implements automatic and interactive annotation for microscopy data. It is built on top of [Segment Anything](https://segment-anything.com/) by Meta AI and specializes it for microscopy and other bio-imaging data.
Its core components are:
-- The `micro_sam` annotator tools for interactive data annotation with [napari](https://napari.org/stable/).
+- The `micro_sam` tools for interactive data annotation with [napari](https://napari.org/stable/).
- The `micro_sam` library to apply Segment Anything to 2d and 3d data or fine-tune it on your data.
- The `micro_sam` models that are fine-tuned on publicly available microscopy data.
@@ -19,20 +19,18 @@ On our roadmap for more functionality are:
If you run into any problems or have questions please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
-
## Quickstart
You can install `micro_sam` via conda:
```
-$ conda install -c conda-forge micro_sam
+$ conda install -c conda-forge micro_sam napari pyqt
```
-For more installation options check out [Installation](#installation)
+We also provide experimental installers for all operating systems.
+For more details on the available installation options check out [the installation section](#installation).
After installing `micro_sam` you can run the annotation tool via `$ micro_sam.annotator`, which opens a menu for selecting the annotation tool and its inputs.
-See [Annotation Tools](#annotation-tools) for an overview and explanation of the annotation functionality.
+See [the annotation tool section](#annotation-tools) for an overview and explanation of the annotation functionality.
The `micro_sam` python library can be used via
```python
@@ -47,7 +45,5 @@ For now, check out [the example script for the 2d annotator](https://github.com/
## Citation
If you are using `micro_sam` in your research please cite
-- [SegmentAnything](https://arxiv.org/abs/2304.02643)
-- and our repository on [zenodo](https://doi.org/10.5281/zenodo.7919746)
-
-We will release a preprint soon that describes this work and can be cited instead of zenodo.
+- Our [preprint](https://doi.org/10.1101/2023.08.21.554208)
+- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643)
diff --git a/environment_cpu.yaml b/environment_cpu.yaml
index 4d5f8ea7..ad91dab9 100644
--- a/environment_cpu.yaml
+++ b/environment_cpu.yaml
@@ -11,6 +11,7 @@ dependencies:
- pytorch
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/environment_gpu.yaml b/environment_gpu.yaml
index 21d5df53..57700759 100644
--- a/environment_gpu.yaml
+++ b/environment_gpu.yaml
@@ -12,6 +12,7 @@ dependencies:
- pytorch-cuda>=11.7 # you may need to update the cuda version to match your system
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/examples/README.md b/examples/README.md
index 87b3a963..4844167f 100644
--- a/examples/README.md
+++ b/examples/README.md
@@ -6,5 +6,8 @@ Examples for using the micro_sam annotation tools:
- `annotator_tracking.py`: run the interactive tracking annotation tool
- `image_series_annotator.py`: run the annotation tool for a series of images
+The folder `finetuning` contains example scripts that show how a Segment Anything model can be fine-tuned
+on custom data with the `micro_sam.train` library, and how the finetuned models can then be used within the annotatin tools.
+
The folder `use_as_library` contains example scripts that show how `micro_sam` can be used as a python
-library to apply Segment Anything on mult-dimensional data.
+library to apply Segment Anything to mult-dimensional data.
diff --git a/examples/annotator_2d.py b/examples/annotator_2d.py
index f0d6d44b..8f4930f2 100644
--- a/examples/annotator_2d.py
+++ b/examples/annotator_2d.py
@@ -34,7 +34,7 @@ def hela_2d_annotator(use_finetuned_model):
embedding_path = "./embeddings/embeddings-hela2d.zarr"
model_type = "vit_h"
- annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type)
+ annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type, precompute_amg_state=True)
def wholeslide_annotator(use_finetuned_model):
diff --git a/examples/annotator_with_custom_model.py b/examples/annotator_with_custom_model.py
new file mode 100644
index 00000000..ceb8b2cb
--- /dev/null
+++ b/examples/annotator_with_custom_model.py
@@ -0,0 +1,23 @@
+import h5py
+import micro_sam.sam_annotator as annotator
+from micro_sam.util import get_sam_model
+
+# TODO add an example for the 2d annotator with a custom model
+
+
+def annotator_3d_with_custom_model():
+ with h5py.File("./data/gut1_block_1.h5") as f:
+ raw = f["raw"][:]
+
+ custom_model = "/home/pape/Work/data/models/sam/user-study/vit_h_nuclei_em_finetuned.pt"
+ embedding_path = "./embeddings/nuclei3d-custom-vit-h.zarr"
+ predictor = get_sam_model(checkpoint_path=custom_model, model_type="vit_h")
+ annotator.annotator_3d(raw, embedding_path, predictor=predictor)
+
+
+def main():
+ annotator_3d_with_custom_model()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/finetuning/.gitignore b/examples/finetuning/.gitignore
new file mode 100644
index 00000000..ec28c5ea
--- /dev/null
+++ b/examples/finetuning/.gitignore
@@ -0,0 +1,2 @@
+checkpoints/
+logs/
diff --git a/examples/finetuning/finetune_hela.py b/examples/finetuning/finetune_hela.py
new file mode 100644
index 00000000..f83bd8cb
--- /dev/null
+++ b/examples/finetuning/finetune_hela.py
@@ -0,0 +1,140 @@
+import os
+
+import numpy as np
+import torch
+import torch_em
+
+import micro_sam.training as sam_training
+from micro_sam.sample_data import fetch_tracking_example_data, fetch_tracking_segmentation_data
+from micro_sam.util import export_custom_sam_model
+
+DATA_FOLDER = "data"
+
+
+def get_dataloader(split, patch_shape, batch_size):
+ """Return train or val data loader for finetuning SAM.
+
+ The data loader must be a torch data loader that retuns `x, y` tensors,
+ where `x` is the image data and `y` are the labels.
+ The labels have to be in a label mask instance segmentation format.
+ I.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
+ Important: the ID 0 is reseved for background, and the IDs must be consecutive
+
+ Here, we use `torch_em.default_segmentation_loader` for creating a suitable data loader from
+ the example hela data. You can either adapt this for your own data (see comments below)
+ or write a suitable torch dataloader yourself.
+ """
+ assert split in ("train", "val")
+ os.makedirs(DATA_FOLDER, exist_ok=True)
+
+ # This will download the image and segmentation data for training.
+ image_dir = fetch_tracking_example_data(DATA_FOLDER)
+ segmentation_dir = fetch_tracking_segmentation_data(DATA_FOLDER)
+
+ # torch_em.default_segmentation_loader is a convenience function to build a torch dataloader
+ # from image data and labels for training segmentation models.
+ # It supports image data in various formats. Here, we load image data and labels from the two
+ # folders with tif images that were downloaded by the example data functionality, by specifying
+ # `raw_key` and `label_key` as `*.tif`. This means all images in the respective folders that end with
+ # .tif will be loadded.
+ # The function supports many other file formats. For example, if you have tif stacks with multiple slices
+ # instead of multiple tif images in a foldder, then you can pass raw_key=label_key=None.
+
+ # Load images from multiple files in folder via pattern (here: all tif files)
+ raw_key, label_key = "*.tif", "*.tif"
+ # Alternative: if you have tif stacks you can just set raw_key and label_key to None
+ # raw_key, label_key= None, None
+
+ # The 'roi' argument can be used to subselect parts of the data.
+ # Here, we use it to select the first 70 frames fro the test split and the other frames for the val split.
+ if split == "train":
+ roi = np.s_[:70, :, :]
+ else:
+ roi = np.s_[70:, :, :]
+
+ loader = torch_em.default_segmentation_loader(
+ raw_paths=image_dir, raw_key=raw_key,
+ label_paths=segmentation_dir, label_key=label_key,
+ patch_shape=patch_shape, batch_size=batch_size,
+ ndim=2, is_seg_dataset=True, rois=roi,
+ label_transform=torch_em.transform.label.connected_components,
+ )
+ return loader
+
+
+def run_training(checkpoint_name, model_type):
+ """Run the actual model training."""
+
+ # All hyperparameters for training.
+ batch_size = 1 # the training batch size
+ patch_shape = (1, 512, 512) # the size of patches for training
+ n_objects_per_batch = 25 # the number of objects per batch that will be sampled
+ device = torch.device("cuda") # the device/GPU used for training
+ n_iterations = 10000 # how long we train (in iterations)
+
+ # Get the dataloaders.
+ train_loader = get_dataloader("train", patch_shape, batch_size)
+ val_loader = get_dataloader("val", patch_shape, batch_size)
+
+ # Get the segment anything model, the optimizer and the LR scheduler
+ model = sam_training.get_trainable_sam_model(model_type=model_type, device=device)
+ optimizer = torch.optim.Adam(model.parameters(), lr=1e-5)
+ scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode="min", factor=0.9, patience=10, verbose=True)
+
+ # This class creates all the training data for a batch (inputs, prompts and labels).
+ convert_inputs = sam_training.ConvertToSamInputs()
+
+ # the trainer which performs training and validation (implemented using "torch_em")
+ trainer = sam_training.SamTrainer(
+ name=checkpoint_name,
+ train_loader=train_loader,
+ val_loader=val_loader,
+ model=model,
+ optimizer=optimizer,
+ # currently we compute loss batch-wise, else we pass channelwise True
+ loss=torch_em.loss.DiceLoss(channelwise=False),
+ metric=torch_em.loss.DiceLoss(),
+ device=device,
+ lr_scheduler=scheduler,
+ logger=sam_training.SamLogger,
+ log_image_interval=10,
+ mixed_precision=True,
+ convert_inputs=convert_inputs,
+ n_objects_per_batch=n_objects_per_batch,
+ n_sub_iteration=8,
+ compile_model=False
+ )
+ trainer.fit(n_iterations)
+
+
+def export_model(checkpoint_name, model_type):
+ """Export the trained model."""
+ # export the model after training so that it can be used by the rest of the micro_sam library
+ export_path = "./finetuned_hela_model.pth"
+ checkpoint_path = os.path.join("checkpoints", checkpoint_name, "best.pt")
+ export_custom_sam_model(
+ checkpoint_path=checkpoint_path,
+ model_type=model_type,
+ save_path=export_path,
+ )
+
+
+def main():
+ """Finetune a Segment Anything model.
+
+ This example uses image data and segmentations from the cell tracking challenge,
+ but can easily be adapted for other data (including data you have annoated with micro_sam beforehand).
+ """
+ # The model_type determines which base model is used to initialize the weights that are finetuned.
+ # We use vit_b here because it can be trained faster. Note that vit_h usually yields higher quality results.
+ model_type = "vit_b"
+
+ # The name of the checkpoint. The checkpoints will be stored in './checkpoints/'
+ checkpoint_name = "sam_hela"
+
+ run_training(checkpoint_name, model_type)
+ export_model(checkpoint_name, model_type)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/finetuning/use_finetuned_model.py b/examples/finetuning/use_finetuned_model.py
new file mode 100644
index 00000000..19600241
--- /dev/null
+++ b/examples/finetuning/use_finetuned_model.py
@@ -0,0 +1,33 @@
+import imageio.v3 as imageio
+
+import micro_sam.util as util
+from micro_sam.sam_annotator import annotator_2d
+
+
+def run_annotator_with_custom_model():
+ """Run the 2d anntator with a custom (finetuned) model.
+
+ Here, we use the model that is produced by `finetuned_hela.py` and apply it
+ for an image from the validation set.
+ """
+ # take the last frame, which is part of the val set, so the model was not directly trained on it
+ im = imageio.imread("./data/DIC-C2DH-HeLa.zip.unzip/DIC-C2DH-HeLa/01/t083.tif")
+
+ # set the checkpoint and the path for caching the embeddings
+ checkpoint = "./finetuned_hela_model.pth"
+ embedding_path = "./embeddings/embeddings-finetuned.zarr"
+
+ model_type = "vit_b" # We finetune a vit_b in the example script.
+ # Adapt this if you finetune a different model type, e.g. vit_h.
+
+ # Load the custom model.
+ predictor = util.get_sam_model(model_type=model_type, checkpoint_path=checkpoint)
+
+ # Run the 2d annotator with the custom model.
+ annotator_2d(
+ im, embedding_path=embedding_path, predictor=predictor, precompute_amg_state=True,
+ )
+
+
+if __name__ == "__main__":
+ run_annotator_with_custom_model()
diff --git a/examples/image_series_annotator.py b/examples/image_series_annotator.py
index 2a1b65e7..1632d793 100644
--- a/examples/image_series_annotator.py
+++ b/examples/image_series_annotator.py
@@ -16,7 +16,8 @@ def series_annotation(use_finetuned_model):
example_data = fetch_image_series_example_data("./data")
image_folder_annotator(
example_data, "./data/series-segmentation-result", embedding_path=embedding_path,
- pattern="*.tif", model_type=model_type
+ pattern="*.tif", model_type=model_type,
+ precompute_amg_state=True,
)
diff --git a/examples/use_as_library/instance_segmentation.py b/examples/use_as_library/instance_segmentation.py
index 89b95900..be9300d3 100644
--- a/examples/use_as_library/instance_segmentation.py
+++ b/examples/use_as_library/instance_segmentation.py
@@ -50,7 +50,8 @@ def cell_segmentation():
# Generate the instance segmentation. You can call this again for different values of 'pred_iou_thresh'
# without having to call initialize again.
- # NOTE: the main advantage of this method is that it's considerably faster than the original implementation.
+ # NOTE: the main advantage of this method is that it's faster than the original implementation,
+ # however the quality is not as high as the original instance segmentation quality yet.
instances_mws = amg_mws.generate(pred_iou_thresh=0.88)
instances_mws = instance_segmentation.mask_data_to_segmentation(
instances_mws, shape=image.shape, with_background=True
@@ -64,7 +65,7 @@ def cell_segmentation():
napari.run()
-def segmentation_with_tiling():
+def cell_segmentation_with_tiling():
"""Run the instance segmentation functionality from micro_sam for segmentation of
cells in a large image. You need to run examples/annotator_2d.py:wholeslide_annotator once before
running this script so that all required data is downloaded and pre-computed.
@@ -111,20 +112,21 @@ def segmentation_with_tiling():
# Generate the instance segmentation. You can call this again for different values of 'pred_iou_thresh'
# without having to call initialize again.
- # NOTE: the main advantage of this method is that it's considerably faster than the original implementation.
+ # NOTE: the main advantage of this method is that it's faster than the original implementation.
+ # however the quality is not as high as the original instance segmentation quality yet.
instances_mws = amg_mws.generate(pred_iou_thresh=0.88)
# Show the results.
v = napari.Viewer()
v.add_image(image)
- # v.add_labels(instances_amg)
+ v.add_labels(instances_amg)
v.add_labels(instances_mws)
napari.run()
def main():
cell_segmentation()
- # segmentation_with_tiling()
+ # cell_segmentation_with_tiling()
if __name__ == "__main__":
diff --git a/finetuning/.gitignore b/finetuning/.gitignore
new file mode 100644
index 00000000..60fd41c2
--- /dev/null
+++ b/finetuning/.gitignore
@@ -0,0 +1,4 @@
+checkpoints/
+logs/
+sam_embeddings/
+results/
diff --git a/finetuning/README.md b/finetuning/README.md
new file mode 100644
index 00000000..6164837d
--- /dev/null
+++ b/finetuning/README.md
@@ -0,0 +1,58 @@
+# Segment Anything Finetuning
+
+Code for finetuning segment anything data on microscopy data and evaluating the finetuned models.
+
+## Example: LiveCELL
+
+**Finetuning**
+
+Run the script `livecell_finetuning.py` for fine-tuning a model on LiveCELL.
+
+**Inference**
+
+The script `livecell_inference.py` can be used to run inference on the test set. It supports different arguments for inference with different configurations.
+For example run
+```
+python livecell_inference.py -c checkpoints/livecell_sam/best.pt -m vit_b -e experiment -i /scratch/projects/nim00007/data/LiveCELL --points --positive 1 --negative 0
+```
+for inference with 1 positive point prompt and no negative point prompt (the prompts are derived from ground-truth).
+
+The arguments `-c`, `-e` and `-i` specify where the checkpoint for the model is, where the predictions from the model and other experiment data will be saved, and where the input dataset (LiveCELL) is stored.
+
+To run the default set of experiments from our publication use the command
+```
+python livecell_inference.py -c checkpoints/livecell_sam/best.pt -m vit_b -e experiment -i /scratch/projects/nim00007/data/LiveCELL -d --prompt_folder /scratch/projects/nim00007/sam/experiments/prompts/livecell
+```
+
+Here `-d` automatically runs the evaluation for these settings:
+- `--points --positive 1 --negative 0` (using point prompts with a single positive point)
+- `--points --positive 2 --negative 4` (using point prompts with two positive points and four negative points)
+- `--points --positive 4 --negative 8` (using point prompts with four positive points and eight negative points)
+- `--box` (using box prompts)
+
+In addition `--prompt_folder` specifies a folder with precomputed prompts. Using pre-computed prompts significantly speeds up the experiments and enables running them in a reproducible manner. (Without it the prompts will be recalculated each time.)
+
+You can also evaluate the automatic instance segmentation functionality, by running
+```
+python livecell_inference.py -c checkpoints/livecell_sam/best.pt -m vit_b -e experiment -i /scratch/projects/nim00007/data/LiveCELL -a
+```
+
+This will first perform a grid-search for the best parameters on a subset of the validation set and then run inference on the test set. This can take up to a day.
+
+**Evaluation**
+
+The script `livecell_evaluation.py` can then be used to evaluate the results from the inference runs.
+E.g. run the script like below to evaluate the previous predictions.
+```
+python livecell_evaluation.py -i /scratch/projects/nim00007/data/LiveCELL -e experiment
+```
+This will create a folder `experiment/results` with csv tables with the results per cell type and averaged over all images.
+
+
+## Finetuning and evaluation code
+
+The subfolders contain the code for different finetuning and evaluation experiments for microscopy data:
+- `livecell`: TODO
+- `generalist`: TODO
+
+Note: we still need to clean up most of this code and will add it later.
diff --git a/finetuning/generalists/cellpose_baseline.py b/finetuning/generalists/cellpose_baseline.py
new file mode 100644
index 00000000..f1a294b0
--- /dev/null
+++ b/finetuning/generalists/cellpose_baseline.py
@@ -0,0 +1,124 @@
+import argparse
+import os
+from glob import glob
+from subprocess import run
+
+import imageio.v3 as imageio
+
+from tqdm import tqdm
+
+DATA_ROOT = "/scratch/projects/nim00007/sam/datasets"
+EXP_ROOT = "/scratch/projects/nim00007/sam/experiments/cellpose"
+
+DATASETS = (
+ "covid-if",
+ "deepbacs",
+ "hpa",
+ "livecell",
+ "lizard",
+ "mouse-embryo",
+ "plantseg-ovules",
+ "plantseg-root",
+ "tissuenet",
+)
+
+
+def load_cellpose_model():
+ from cellpose import models
+
+ device, gpu = models.assign_device(True, True)
+ model = models.Cellpose(gpu=gpu, model_type="cyto", device=device)
+ return model
+
+
+def run_cellpose_segmentation(datasets, job_id):
+ dataset = datasets[job_id]
+ experiment_folder = os.path.join(EXP_ROOT, dataset)
+
+ prediction_folder = os.path.join(experiment_folder, "predictions")
+ os.makedirs(prediction_folder, exist_ok=True)
+
+ image_paths = sorted(glob(os.path.join(DATA_ROOT, dataset, "test", "image*.tif")))
+ model = load_cellpose_model()
+
+ for path in tqdm(image_paths, desc=f"Segmenting {dataset} with cellpose"):
+ fname = os.path.basename(path)
+ out_path = os.path.join(prediction_folder, fname)
+ if os.path.exists(out_path):
+ continue
+ image = imageio.imread(path)
+ if image.ndim == 3:
+ assert image.shape[-1] == 3
+ image = image.mean(axis=-1)
+ assert image.ndim == 2
+ seg = model.eval(image, diameter=None, flow_threshold=None, channels=[0, 0])[0]
+ assert seg.shape == image.shape
+ imageio.imwrite(out_path, seg, compression=5)
+
+
+def submit_array_job(datasets):
+ n_datasets = len(datasets)
+ cmd = ["sbatch", "-a", f"0-{n_datasets-1}", "cellpose_baseline.sbatch"]
+ run(cmd)
+
+
+def evaluate_dataset(dataset):
+ from micro_sam.evaluation.evaluation import run_evaluation
+
+ gt_paths = sorted(glob(os.path.join(DATA_ROOT, dataset, "test", "label*.tif")))
+ experiment_folder = os.path.join(EXP_ROOT, dataset)
+ pred_paths = sorted(glob(os.path.join(experiment_folder, "predictions", "*.tif")))
+ assert len(gt_paths) == len(pred_paths), f"{len(gt_paths)}, {len(pred_paths)}"
+ result_path = os.path.join(experiment_folder, "cellpose.csv")
+ run_evaluation(gt_paths, pred_paths, result_path)
+
+
+def evaluate_segmentations(datasets):
+ for dataset in datasets:
+ # we skip livecell, which has already been processed by cellpose
+ if dataset == "livecell":
+ continue
+ evaluate_dataset(dataset)
+
+
+def check_results(datasets):
+ for ds in datasets:
+ # we skip livecell, which has already been processed by cellpose
+ if ds == "livecell":
+ continue
+ result_path = os.path.join(EXP_ROOT, ds, "cellpose.csv")
+ if not os.path.exists(result_path):
+ print("Cellpose results missing for", ds)
+ print("All checks passed")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("--segment", "-s", action="store_true")
+ parser.add_argument("--evaluate", "-e", action="store_true")
+ parser.add_argument("--check", "-c", action="store_true")
+ parser.add_argument("--datasets", nargs="+")
+ args = parser.parse_args()
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+
+ if args.datasets is None:
+ datasets = DATASETS
+ else:
+ datasets = args.datasets
+ assert all(ds in DATASETS for ds in datasets)
+
+ if job_id is not None:
+ run_cellpose_segmentation(datasets, int(job_id))
+ elif args.segment:
+ submit_array_job(datasets)
+ elif args.evaluate:
+ evaluate_segmentations(datasets)
+ elif args.check:
+ check_results(datasets)
+ else:
+ raise ValueError("Doing nothing")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/cellpose_baseline.sbatch b/finetuning/generalists/cellpose_baseline.sbatch
new file mode 100755
index 00000000..c839abab
--- /dev/null
+++ b/finetuning/generalists/cellpose_baseline.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 300
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate cellpose
+python cellpose_baseline.py $@
diff --git a/finetuning/generalists/compile_results.py b/finetuning/generalists/compile_results.py
new file mode 100644
index 00000000..06165a75
--- /dev/null
+++ b/finetuning/generalists/compile_results.py
@@ -0,0 +1,104 @@
+import os
+from glob import glob
+
+import pandas as pd
+
+from evaluate_generalist import EXPERIMENT_ROOT
+from util import EM_DATASETS, LM_DATASETS
+
+
+def get_results(model, ds):
+ res_folder = os.path.join(EXPERIMENT_ROOT, model, ds, "results")
+ res_paths = sorted(glob(os.path.join(res_folder, "box", "*.csv"))) +\
+ sorted(glob(os.path.join(res_folder, "points", "*.csv")))
+
+ amg_res = os.path.join(res_folder, "amg.csv")
+ if os.path.exists(amg_res):
+ res_paths.append(amg_res)
+
+ results = []
+ for path in res_paths:
+ prompt_res = pd.read_csv(path)
+ prompt_name = os.path.splitext(os.path.relpath(path, res_folder))[0]
+ prompt_res.insert(0, "prompt", [prompt_name])
+ results.append(prompt_res)
+ results = pd.concat(results)
+ results.insert(0, "dataset", results.shape[0] * [ds])
+
+ return results
+
+
+def compile_results(models, datasets, out_path, load_results=False):
+ results = []
+
+ for model in models:
+ model_results = []
+
+ for ds in datasets:
+ ds_results = get_results(model, ds)
+ model_results.append(ds_results)
+
+ model_results = pd.concat(model_results)
+ model_results.insert(0, "model", [model] * model_results.shape[0])
+ results.append(model_results)
+
+ results = pd.concat(results)
+ if load_results:
+ assert os.path.exists(out_path)
+ all_results = pd.read_csv(out_path)
+ results = pd.concat([all_results, results])
+
+ results.to_csv(out_path, index=False)
+
+
+def compile_em():
+ compile_results(
+ ["vit_h", "vit_h_em", "vit_b", "vit_b_em"],
+ EM_DATASETS,
+ os.path.join(EXPERIMENT_ROOT, "evaluation-em.csv")
+ )
+
+
+def add_cellpose_results(datasets, out_path):
+ cp_root = "/scratch/projects/nim00007/sam/experiments/cellpose"
+
+ results = []
+ for dataset in datasets:
+ if dataset == "livecell":
+ continue
+ res_path = os.path.join(cp_root, dataset, "cellpose.csv")
+ ds_res = pd.read_csv(res_path)
+ ds_res.insert(0, "prompt", ["cellpose"] * ds_res.shape[0])
+ ds_res.insert(0, "dataset", [dataset] * ds_res.shape[0])
+ results.append(ds_res)
+
+ results = pd.concat(results)
+ results.insert(0, "model", ["cellpose"] * results.shape[0])
+
+ all_results = pd.read_csv(out_path)
+ results = pd.concat([all_results, results])
+ results.to_csv(out_path, index=False)
+
+
+def compile_lm():
+ res_path = os.path.join(EXPERIMENT_ROOT, "evaluation-lm.csv")
+ compile_results(
+ ["vit_h", "vit_h_lm", "vit_b", "vit_b_lm"], LM_DATASETS, res_path
+ )
+
+ # add the deepbacs and tissuenet specialist results
+ assert os.path.exists(res_path)
+ compile_results(["vit_h_tissuenet", "vit_b_tissuenet"], ["tissuenet"], res_path, True)
+ compile_results(["vit_h_deepbacs", "vit_b_deepbacs"], ["deepbacs"], res_path, True)
+
+ # add the cellpose results
+ add_cellpose_results(LM_DATASETS, res_path)
+
+
+def main():
+ # compile_em()
+ compile_lm()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/create_tissuenet_data.py b/finetuning/generalists/create_tissuenet_data.py
new file mode 100644
index 00000000..18114973
--- /dev/null
+++ b/finetuning/generalists/create_tissuenet_data.py
@@ -0,0 +1,63 @@
+
+import os
+from tqdm import tqdm
+import imageio.v2 as imageio
+import numpy as np
+
+from torch_em.data import MinInstanceSampler
+from torch_em.transform.label import label_consecutive
+from torch_em.data.datasets import get_tissuenet_loader
+from torch_em.transform.raw import standardize, normalize_percentile
+
+
+def rgb_to_gray_transform(raw):
+ raw = normalize_percentile(raw, axis=(1, 2))
+ raw = np.mean(raw, axis=0)
+ raw = standardize(raw)
+ return raw
+
+
+def get_tissuenet_loaders(input_path):
+ sampler = MinInstanceSampler()
+ label_transform = label_consecutive
+ raw_transform = rgb_to_gray_transform
+ val_loader = get_tissuenet_loader(path=input_path, split="val", raw_channel="rgb", label_channel="cell",
+ batch_size=1, patch_shape=(256, 256), num_workers=0,
+ sampler=sampler, label_transform=label_transform, raw_transform=raw_transform)
+ test_loader = get_tissuenet_loader(path=input_path, split="test", raw_channel="rgb", label_channel="cell",
+ batch_size=1, patch_shape=(256, 256), num_workers=0,
+ sampler=sampler, label_transform=label_transform, raw_transform=raw_transform)
+ return val_loader, test_loader
+
+
+def extract_images(loader, out_folder):
+ os.makedirs(out_folder, exist_ok=True)
+ for i, (x, y) in tqdm(enumerate(loader), total=len(loader)):
+ img_path = os.path.join(out_folder, "image_{:04d}.tif".format(i))
+ gt_path = os.path.join(out_folder, "label_{:04d}.tif".format(i))
+
+ img = x.squeeze().detach().cpu().numpy()
+ gt = y.squeeze().detach().cpu().numpy()
+
+ imageio.imwrite(img_path, img)
+ imageio.imwrite(gt_path, gt)
+
+
+def main():
+ val_loader, test_loader = get_tissuenet_loaders("/scratch-grete/projects/nim00007/data/tissuenet")
+ print("Length of val loader is:", len(val_loader))
+ print("Length of test loader is:", len(test_loader))
+
+ root_save_dir = "/scratch/projects/nim00007/sam/datasets/tissuenet"
+
+ # we use the val set for test because there are some issues with the test set
+ # out_folder = os.path.join(root_save_dir, "test")
+ # extract_images(val_loader, out_folder)
+
+ # we use the test folder for val and just use as many images as we can sample
+ out_folder = os.path.join(root_save_dir, "val")
+ extract_images(test_loader, out_folder)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/evaluate_generalist.py b/finetuning/generalists/evaluate_generalist.py
new file mode 100644
index 00000000..af217c0e
--- /dev/null
+++ b/finetuning/generalists/evaluate_generalist.py
@@ -0,0 +1,109 @@
+import argparse
+import os
+from subprocess import run
+
+from util import evaluate_checkpoint_for_dataset, ALL_DATASETS, EM_DATASETS, LM_DATASETS
+from micro_sam.evaluation import default_experiment_settings, get_experiment_setting_name
+
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/generalists"
+CHECKPOINTS = {
+ # Vanilla models
+ "vit_b": "/home/nimcpape/.sam_models/sam_vit_b_01ec64.pth",
+ "vit_h": "/home/nimcpape/.sam_models/sam_vit_h_4b8939.pth",
+ # Generalist LM models
+ "vit_b_lm": "/scratch/projects/nim00007/sam/models/LM/generalist/v2/vit_b/best.pt",
+ "vit_h_lm": "/scratch/projects/nim00007/sam/models/LM/generalist/v2/vit_h/best.pt",
+ # Generalist EM models
+ "vit_b_em": "/scratch/projects/nim00007/sam/models/EM/generalist/v2/vit_b/best.pt",
+ "vit_h_em": "/scratch/projects/nim00007/sam/models/EM/generalist/v2/vit_h/best.pt",
+ # Specialist Models (we don't add livecell, because these results are all computed already)
+ "vit_b_tissuenet": "/scratch/projects/nim00007/sam/models/LM/TissueNet/vit_b/best.pt",
+ "vit_h_tissuenet": "/scratch/projects/nim00007/sam/models/LM/TissueNet/vit_h/best.pt",
+ "vit_b_deepbacs": "/scratch/projects/nim00007/sam/models/LM/DeepBacs/vit_b/best.pt",
+ "vit_h_deepbacs": "/scratch/projects/nim00007/sam/models/LM/DeepBacs/vit_h/best.pt",
+}
+
+
+def submit_array_job(model_name, datasets):
+ n_datasets = len(datasets)
+ cmd = ["sbatch", "-a", f"0-{n_datasets-1}", "evaluate_generalist.sbatch", model_name, "--datasets"]
+ cmd.extend(datasets)
+ run(cmd)
+
+
+def evaluate_dataset_slurm(model_name, dataset):
+ if dataset in EM_DATASETS:
+ run_amg = False
+ max_num_val_images = None
+ else:
+ run_amg = True
+ max_num_val_images = 64
+
+ is_custom_model = model_name not in ("vit_h", "vit_b")
+ checkpoint = CHECKPOINTS[model_name]
+ model_type = model_name[:5]
+
+ experiment_folder = os.path.join(EXPERIMENT_ROOT, model_name, dataset)
+ evaluate_checkpoint_for_dataset(
+ checkpoint, model_type, dataset, experiment_folder,
+ run_default_evaluation=True, run_amg=run_amg,
+ is_custom_model=is_custom_model,
+ max_num_val_images=max_num_val_images,
+ )
+
+
+def _get_datasets(lm, em):
+ assert lm or em
+ datasets = []
+ if lm:
+ datasets.extend(LM_DATASETS)
+ if em:
+ datasets.extend(EM_DATASETS)
+ return datasets
+
+
+def check_computation(model_name, datasets):
+ prompt_settings = default_experiment_settings()
+ for ds in datasets:
+ experiment_folder = os.path.join(EXPERIMENT_ROOT, model_name, ds)
+ for setting in prompt_settings:
+ setting_name = get_experiment_setting_name(setting)
+ expected_path = os.path.join(experiment_folder, "results", f"{setting_name}.csv")
+ if not os.path.exists(expected_path):
+ print("Missing results for:", expected_path)
+ if ds in LM_DATASETS:
+ expected_path = os.path.join(experiment_folder, "results", "amg.csv")
+ if not os.path.exists(expected_path):
+ print("Missing results for:", expected_path)
+ print("All checks_run")
+
+
+# evaluation on slurm
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("model_name")
+ parser.add_argument("--check", "-c", action="store_true")
+ parser.add_argument("--lm", action="store_true")
+ parser.add_argument("--em", action="store_true")
+ parser.add_argument("--datasets", nargs="+")
+ args = parser.parse_args()
+
+ datasets = args.datasets
+ if datasets is None or len(datasets) == 0:
+ datasets = _get_datasets(args.lm, args.em)
+ assert all(ds in ALL_DATASETS for ds in datasets)
+
+ if args.check:
+ check_computation(args.model_name, datasets)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(args.model_name, datasets)
+ else: # we're in a slurm job
+ job_id = int(job_id)
+ evaluate_dataset_slurm(args.model_name, datasets[job_id])
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/evaluate_generalist.sbatch b/finetuning/generalists/evaluate_generalist.sbatch
new file mode 100755
index 00000000..121ba55f
--- /dev/null
+++ b/finetuning/generalists/evaluate_generalist.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 2800
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python evaluate_generalist.py $@
diff --git a/finetuning/generalists/evaluate_training_evolution.py b/finetuning/generalists/evaluate_training_evolution.py
new file mode 100644
index 00000000..c1435593
--- /dev/null
+++ b/finetuning/generalists/evaluate_training_evolution.py
@@ -0,0 +1,78 @@
+import argparse
+import os
+
+from glob import glob
+from subprocess import run
+
+import pandas as pd
+from util import evaluate_checkpoint_for_datasets, get_generalist_predictor
+
+CHECKPOINT_ROOT = "/scratch/projects/nim00007/sam/models/LM/generalist/v2"
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/training-evolution"
+# We evaluate these three datasets for the training evolution.
+# These are chosen based on observations from preliminary experiments.
+# - covid-if: out-of-domain dataset that shows the expected improvement (over vanilla).
+# - deepbacs: in domain dataset where we see the biggest gap to the specialist.
+# - lizard: out-of-domain that is furthest from the training data.
+EVAL_DATASETS = ("covid-if", "deepbacs", "lizard")
+
+
+def evaluate_checkpoint_slurm(model_type, job_id, checkpoints):
+ checkpoint = checkpoints[job_id]
+
+ predictor, state = get_generalist_predictor(
+ checkpoint, model_type, is_custom_model=True, return_state=True
+ )
+ epoch = state["epoch"] + 1
+
+ print("Run evaluation for", model_type, "epoch", epoch)
+ experiment_root = os.path.join(EXPERIMENT_ROOT, f"{model_type}-epoch-{epoch}")
+ result = evaluate_checkpoint_for_datasets(
+ None, None, experiment_root, EVAL_DATASETS,
+ run_default_evaluation=True, run_amg=False,
+ is_custom_model=True, predictor=predictor,
+ )
+
+ result.insert(0, "epoch", [epoch] * result.shape[0])
+ return result
+
+
+def evaluate_training_evolution(model_type, checkpoints):
+ results = []
+ for i in range(len(checkpoints)):
+ result = evaluate_checkpoint_slurm(model_type, i, checkpoints)
+ results.append(result)
+ results = pd.concat(results)
+ save_path = os.path.join(EXPERIMENT_ROOT, f"{model_type}.csv")
+ results.to_csv(save_path, index=False)
+
+
+def submit_array_job(model_type, checkpoints):
+ n_checkpoints = len(checkpoints)
+ cmd = ["sbatch", "-a", f"0-{n_checkpoints-1}", "evaluate_training_evolution.sbatch", model_type]
+ run(cmd)
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("model_type")
+ parser.add_argument("-e", "--evaluate", action="store_true")
+ args = parser.parse_args()
+
+ checkpoints = sorted(glob(os.path.join(CHECKPOINT_ROOT, args.model_type, "epoch-*.pt")))
+ assert len(checkpoints) > 0
+
+ if args.evaluate:
+ evaluate_training_evolution(args.model_type, checkpoints)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(args.model_type, checkpoints)
+ else: # we're in a slurm job
+ job_id = int(job_id)
+ evaluate_checkpoint_slurm(args.model_type, job_id, checkpoints)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/evaluate_training_evolution.sbatch b/finetuning/generalists/evaluate_training_evolution.sbatch
new file mode 100755
index 00000000..e628f44f
--- /dev/null
+++ b/finetuning/generalists/evaluate_training_evolution.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 96G
+#SBATCH -t 240
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python evaluate_training_evolution.py $@
diff --git a/finetuning/generalists/export_generalist_model.py b/finetuning/generalists/export_generalist_model.py
new file mode 100644
index 00000000..11073364
--- /dev/null
+++ b/finetuning/generalists/export_generalist_model.py
@@ -0,0 +1,24 @@
+import os
+from micro_sam.util import export_custom_sam_model
+from evaluate_generalist import CHECKPOINTS, EXPERIMENT_ROOT
+
+OUT_ROOT = os.path.join(EXPERIMENT_ROOT, "exported")
+os.makedirs(OUT_ROOT, exist_ok=True)
+
+
+def export_generalist(model):
+ checkpoint_path = CHECKPOINTS[model]
+ model_type = model[:5]
+ save_path = os.path.join(OUT_ROOT, f"{model}.pth")
+ export_custom_sam_model(checkpoint_path, model_type, save_path)
+
+
+def main():
+ export_generalist("vit_b_em")
+ export_generalist("vit_h_em")
+ export_generalist("vit_b_lm")
+ export_generalist("vit_h_lm")
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/generate_model_comparison.py b/finetuning/generalists/generate_model_comparison.py
new file mode 100644
index 00000000..f0ed3a9a
--- /dev/null
+++ b/finetuning/generalists/generate_model_comparison.py
@@ -0,0 +1,82 @@
+import os
+
+import imageio.v3 as imageio
+import micro_sam.evaluation.model_comparison as comparison
+import torch_em
+
+from util import get_data_paths, EM_DATASETS, LM_DATASETS
+
+OUTPUT_ROOT = "/scratch-grete/projects/nim00007/sam/experiments/model_comparison"
+
+
+def _get_patch_shape(path):
+ im_shape = imageio.imread(path).shape[:2]
+ patch_shape = tuple(min(sh, 512) for sh in im_shape)
+ return patch_shape
+
+
+def raw_trafo(raw):
+ raw = raw.transpose((2, 0, 1))
+ print(raw.shape)
+ return raw
+
+
+def get_loader(dataset):
+ image_paths, gt_paths = get_data_paths(dataset, split="test")
+ image_paths, gt_paths = image_paths[:100], gt_paths[:100]
+
+ with_channels = dataset in ("hpa", "lizard")
+
+ label_transform = torch_em.transform.label.connected_components
+ loader = torch_em.default_segmentation_loader(
+ image_paths, None, gt_paths, None,
+ batch_size=1, patch_shape=_get_patch_shape(image_paths[0]),
+ shuffle=True, n_samples=25, label_transform=label_transform,
+ with_channels=with_channels, is_seg_dataset=not with_channels
+ )
+ return loader
+
+
+def generate_comparison_for_dataset(dataset, model1, model2):
+ output_folder = os.path.join(OUTPUT_ROOT, dataset)
+ if os.path.exists(output_folder):
+ return output_folder
+ print("Generate model comparison data for", dataset)
+ loader = get_loader(dataset)
+ comparison.generate_data_for_model_comparison(loader, output_folder, model1, model2, n_samples=25)
+ return output_folder
+
+
+def create_comparison_images(output_folder, dataset):
+ plot_folder = os.path.join(OUTPUT_ROOT, "images", dataset)
+ if os.path.exists(plot_folder):
+ return
+ comparison.model_comparison(
+ output_folder, n_images_per_sample=25, min_size=100,
+ plot_folder=plot_folder, outline_dilation=1
+ )
+
+
+def generate_comparison_em():
+ model1 = "vit_h"
+ model2 = "vit_h_em"
+ for dataset in EM_DATASETS:
+ folder = generate_comparison_for_dataset(dataset, model1, model2)
+ create_comparison_images(folder, dataset)
+
+
+def generate_comparison_lm():
+ model1 = "vit_h"
+ model2 = "vit_h_lm"
+ for dataset in LM_DATASETS:
+ folder = generate_comparison_for_dataset(dataset, model1, model2)
+ create_comparison_images(folder, dataset)
+
+
+def main():
+ generate_comparison_lm()
+ # generate_comparison_em()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/precompute_prompts.py b/finetuning/generalists/precompute_prompts.py
new file mode 100644
index 00000000..73d5e4ed
--- /dev/null
+++ b/finetuning/generalists/precompute_prompts.py
@@ -0,0 +1,102 @@
+import argparse
+import os
+import pickle
+
+from subprocess import run
+
+import micro_sam.evaluation as evaluation
+from util import get_data_paths, ALL_DATASETS, LM_DATASETS
+from tqdm import tqdm
+
+PROMPT_ROOT = "/scratch/projects/nim00007/sam/experiments/prompts"
+
+
+def precompute_prompts(dataset):
+ # everything for livecell has been computed already
+ if dataset == "livecell":
+ return
+
+ prompt_folder = os.path.join(PROMPT_ROOT, dataset)
+ _, gt_paths = get_data_paths(dataset, "test")
+
+ settings = evaluation.default_experiment_settings()
+ evaluation.precompute_all_prompts(gt_paths, prompt_folder, settings)
+
+
+def precompute_prompts_slurm(job_id):
+ dataset = ALL_DATASETS[job_id]
+ precompute_prompts(dataset)
+
+
+def submit_array_job():
+ n_datasets = len(ALL_DATASETS)
+ cmd = ["sbatch", "-a", f"0-{n_datasets-1}", "precompute_prompts.sbatch"]
+ run(cmd)
+
+
+def _check_prompts(dataset, settings, expected_len):
+ prompt_folder = os.path.join(PROMPT_ROOT, dataset)
+
+ def check_prompt_file(prompt_file):
+ assert os.path.exists(prompt_file), prompt_file
+ with open(prompt_file, "rb") as f:
+ prompts = pickle.load(f)
+ assert len(prompts) == expected_len, f"{len(prompts)}, {expected_len}"
+
+ for setting in settings:
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ prompt_file = os.path.join(prompt_folder, f"points-p{pos}-n{neg}.pkl")
+ if pos == 0 and neg == 0:
+ prompt_file = os.path.join(prompt_folder, "boxes.pkl")
+ check_prompt_file(prompt_file)
+
+
+def check_prompts_and_datasets():
+
+ def check_dataset(dataset):
+ try:
+ images, _ = get_data_paths(dataset, "test")
+ except AssertionError as e:
+ print("Checking test split failed for datasset", dataset, "due to", e)
+
+ if dataset not in LM_DATASETS:
+ return len(images)
+
+ try:
+ get_data_paths(dataset, "val")
+ except AssertionError as e:
+ print("Checking val split failed for datasset", dataset, "due to", e)
+
+ return len(images)
+
+ settings = evaluation.default_experiment_settings()
+ for ds in tqdm(ALL_DATASETS, desc="Checking datasets"):
+ n_images = check_dataset(ds)
+ _check_prompts(ds, settings, n_images)
+ print("All checks done!")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-d", "--dataset")
+ parser.add_argument("--check", "-c", action="store_true")
+ args = parser.parse_args()
+
+ if args.check:
+ check_prompts_and_datasets()
+ return
+
+ if args.dataset is not None:
+ precompute_prompts(args.dataset)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job()
+ else: # we're in a slurm job and precompute a setting
+ job_id = int(job_id)
+ precompute_prompts_slurm(job_id)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/generalists/precompute_prompts.sbatch b/finetuning/generalists/precompute_prompts.sbatch
new file mode 100755
index 00000000..2b04cc96
--- /dev/null
+++ b/finetuning/generalists/precompute_prompts.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 2000
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python precompute_prompts.py $@
diff --git a/finetuning/generalists/util.py b/finetuning/generalists/util.py
new file mode 100644
index 00000000..d354d882
--- /dev/null
+++ b/finetuning/generalists/util.py
@@ -0,0 +1,195 @@
+import json
+import os
+import warnings
+
+from glob import glob
+from pathlib import Path
+
+import pandas as pd
+from micro_sam.evaluation import (
+ automatic_mask_generation, inference, evaluation,
+ default_experiment_settings, get_experiment_setting_name
+)
+from micro_sam.evaluation.livecell import _get_livecell_paths
+
+DATA_ROOT = "/scratch/projects/nim00007/sam/datasets"
+LIVECELL_ROOT = "/scratch/projects/nim00007/data/LiveCELL"
+PROMPT_ROOT = "/scratch-grete/projects/nim00007/sam/experiments/prompts"
+
+LM_DATASETS = (
+ "covid-if",
+ "deepbacs",
+ "hpa",
+ "livecell",
+ "lizard",
+ "mouse-embryo",
+ "plantseg-ovules",
+ "plantseg-root",
+ "tissuenet",
+)
+
+EM_DATASETS = (
+ "cremi",
+ "lucchi",
+ "mitoem",
+ "nuc_mm/mouse",
+ "nuc_mm/zebrafish",
+ "platy-cell",
+ "platy-cuticle",
+ "platy-nuclei",
+ "snemi",
+ "sponge-em",
+ "vnc",
+)
+ALL_DATASETS = EM_DATASETS + LM_DATASETS
+
+
+###
+# Dataset functionality
+###
+
+
+def get_data_paths(dataset, split, max_num_images=None):
+ if dataset == "livecell":
+ n_val_per_cell_type = None if max_num_images is None else int(max_num_images / 8)
+ return _get_livecell_paths(LIVECELL_ROOT, split=split, n_val_per_cell_type=n_val_per_cell_type)
+
+ image_pattern = os.path.join(DATA_ROOT, dataset, split, "image*.tif")
+ image_paths = sorted(glob(image_pattern))
+ gt_paths = sorted(glob(os.path.join(DATA_ROOT, dataset, split, "label*.tif")))
+ assert len(image_paths) == len(gt_paths)
+ assert len(image_paths) > 0, image_pattern
+ if max_num_images is not None:
+ image_paths, gt_paths = image_paths[:max_num_images], gt_paths[:max_num_images]
+ return image_paths, gt_paths
+
+
+###
+# Evaluation functionality
+###
+
+
+def get_generalist_predictor(checkpoint, model_type, is_custom_model, return_state=False):
+ with warnings.catch_warnings():
+ warnings.simplefilter("ignore")
+ return inference.get_predictor(
+ checkpoint, model_type=model_type,
+ return_state=return_state, is_custom_model=is_custom_model
+ )
+
+
+def evaluate_checkpoint_for_dataset(
+ checkpoint, model_type, dataset, experiment_folder,
+ run_default_evaluation, run_amg, is_custom_model,
+ predictor=None, max_num_val_images=None,
+):
+ """Evaluate a generalist checkpoint for a given dataset.
+ """
+ assert run_default_evaluation or run_amg
+
+ prompt_dir = os.path.join(PROMPT_ROOT, dataset)
+
+ if predictor is None:
+ predictor = get_generalist_predictor(checkpoint, model_type, is_custom_model)
+ test_image_paths, test_gt_paths = get_data_paths(dataset, "test")
+
+ embedding_dir = os.path.join(experiment_folder, "test", "embeddings")
+ os.makedirs(embedding_dir, exist_ok=True)
+ result_dir = os.path.join(experiment_folder, "results")
+
+ results = []
+ if run_default_evaluation:
+ prompt_settings = default_experiment_settings()
+ for setting in prompt_settings:
+
+ setting_name = get_experiment_setting_name(setting)
+ prediction_dir = os.path.join(experiment_folder, "test", setting_name)
+ os.makedirs(prediction_dir, exist_ok=True)
+
+ inference.run_inference_with_prompts(
+ predictor, test_image_paths, test_gt_paths,
+ embedding_dir, prediction_dir,
+ use_points=setting["use_points"], use_boxes=setting["use_boxes"],
+ n_positives=setting["n_positives"], n_negatives=setting["n_negatives"],
+ prompt_save_dir=prompt_dir,
+ )
+
+ if dataset == "livecell":
+ pred_paths = [
+ os.path.join(prediction_dir, os.path.basename(gt_path)) for gt_path in test_gt_paths
+ ]
+ assert all(os.path.exists(pred_path) for pred_path in pred_paths)
+ else:
+ pred_paths = sorted(glob(os.path.join(prediction_dir, "*.tif")))
+ result_path = os.path.join(result_dir, f"{setting_name}.csv")
+ os.makedirs(Path(result_path).parent, exist_ok=True)
+
+ result = evaluation.run_evaluation(test_gt_paths, pred_paths, result_path)
+ result.insert(0, "setting", [setting_name])
+ results.append(result)
+
+ if run_amg:
+ val_embedding_dir = os.path.join(experiment_folder, "val", "embeddings")
+ val_result_dir = os.path.join(experiment_folder, "val", "results")
+ os.makedirs(val_embedding_dir, exist_ok=True)
+
+ val_image_paths, val_gt_paths = get_data_paths(dataset, "val", max_num_images=max_num_val_images)
+ automatic_mask_generation.run_amg_grid_search(
+ predictor, val_image_paths, val_gt_paths, val_embedding_dir,
+ val_result_dir, verbose_gs=True,
+ )
+
+ best_iou_thresh, best_stability_thresh, _ = automatic_mask_generation.evaluate_amg_grid_search(val_result_dir)
+ best_settings = {"pred_iou_thresh": best_iou_thresh, "stability_score_thresh": best_stability_thresh}
+ gs_result_path = os.path.join(experiment_folder, "best_gs_params.json")
+ with open(gs_result_path, "w") as f:
+ json.dump(best_settings, f)
+
+ prediction_dir = os.path.join(experiment_folder, "test", "amg")
+ os.makedirs(prediction_dir, exist_ok=True)
+ automatic_mask_generation.run_amg_inference(
+ predictor, test_image_paths, embedding_dir, prediction_dir,
+ amg_generate_kwargs=best_settings,
+ )
+
+ if dataset == "livecell":
+ pred_paths = [
+ os.path.join(prediction_dir, os.path.basename(gt_path)) for gt_path in test_gt_paths
+ ]
+ assert all(os.path.exists(pred_path) for pred_path in pred_paths)
+ else:
+ pred_paths = sorted(glob(os.path.join(prediction_dir, "*.tif")))
+
+ result_path = os.path.join(result_dir, "amg.csv")
+ os.makedirs(Path(result_path).parent, exist_ok=True)
+
+ result = evaluation.run_evaluation(test_gt_paths, pred_paths, result_path)
+ result.insert(0, "setting", ["amg"])
+ results.append(result)
+
+ results = pd.concat(results)
+ results.insert(0, "dataset", [dataset] * results.shape[0])
+ return results
+
+
+def evaluate_checkpoint_for_datasets(
+ checkpoint, model_type, experiment_root, datasets,
+ run_default_evaluation, run_amg, is_custom_model,
+ predictor=None, max_num_val_images=None,
+):
+ if predictor is None:
+ predictor = get_generalist_predictor(checkpoint, model_type, is_custom_model)
+
+ results = []
+ for dataset in datasets:
+ experiment_folder = os.path.join(experiment_root, dataset)
+ os.makedirs(experiment_folder, exist_ok=True)
+ result = evaluate_checkpoint_for_dataset(
+ None, None, dataset, experiment_folder,
+ run_default_evaluation=run_default_evaluation,
+ run_amg=run_amg, is_custom_model=is_custom_model,
+ predictor=predictor, max_num_val_images=max_num_val_images,
+ )
+ results.append(result)
+
+ return pd.concat(results)
diff --git a/finetuning/livecell/README.md b/finetuning/livecell/README.md
new file mode 100644
index 00000000..fceedcb7
--- /dev/null
+++ b/finetuning/livecell/README.md
@@ -0,0 +1,27 @@
+# Finetuning Segment Anything for LiveCELL
+
+TODO: explain the set-up
+
+These experiments are implemented for a slurm cluster with access to GPUs (and you ideally need access to A100s or H100s with 80GB of memory, if you only use ViT-b then using a GPU with 32 GB or 40 GB should suffice.)
+
+## Training
+
+TODO: add training code and explain how to run it
+
+## Evaluation
+
+To run the evaluation experiments for the Segment Anything Models on LiveCELL follow these steps:
+
+- Preparation:
+ - Go to the `evaluation` directory.
+ - Adapt the path to the models, the data folder and the experiment folder in `util.py`
+ - Make sure you have the LiveCELL data downloaded in the data folder. If not you can run `python util.py` and it will automatically downloaded.
+ - Adapt the settings in `util/precompute_embeddings.sh` and `util/precompute_prompts.sh` to your slurm set-up.
+- Precompute the embeddings by running `sbatch precompute_embeddings.sbatch ` for all the models you want to evaluate.
+ - This will submit a slurm job that needs access to a GPU.
+- Precompute the prompts by running `python precompute_prompts.py -f`.
+ - This will submit a slurm array job that precomputes the prompts for all images. In total 31 jobs will be started. Note: these jobs do not need access to a GPU, the computation is purely CPU based.
+- Run inference via `python inference.py -n `.
+ - This will submit a slurm array job that runs the prediction for all prompt settings for this model. In total 61 jobs will be started, but each should only take 10-20 minutes (on a A100 and depending on the model type).
+- Run the evaluation of inference results via `sbatch evaluation.sbatch -n `.
+ - This will submit a single slurm job that does not need a GPU.
diff --git a/finetuning/livecell/amg/grid_search_and_inference.py b/finetuning/livecell/amg/grid_search_and_inference.py
new file mode 100644
index 00000000..1a04d763
--- /dev/null
+++ b/finetuning/livecell/amg/grid_search_and_inference.py
@@ -0,0 +1,40 @@
+import argparse
+from micro_sam.evaluation.livecell import run_livecell_amg
+from util import DATA_ROOT, get_checkpoint, get_experiment_folder, check_model
+
+
+def run_job(model_name, use_mws):
+ checkpoint, model_type = get_checkpoint(model_name)
+ experiment_folder = get_experiment_folder(model_name)
+ input_folder = DATA_ROOT
+
+ run_livecell_amg(
+ checkpoint, model_type, input_folder, experiment_folder,
+ n_val_per_cell_type=25, use_mws=use_mws,
+ )
+
+
+# TODO
+def check_amg(model_name, use_mws):
+ pass
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ parser.add_argument("--mws", action="store_true")
+ parser.add_argument("-c", "--check", action="store_true")
+ args = parser.parse_args()
+
+ model_name = args.name
+ use_mws = args.mws
+ check_model(model_name)
+
+ if args.check:
+ check_amg(model_name, use_mws)
+ else:
+ run_job(model_name, use_mws)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/amg/grid_search_and_inference.sbatch b/finetuning/livecell/amg/grid_search_and_inference.sbatch
new file mode 100755
index 00000000..a3692182
--- /dev/null
+++ b/finetuning/livecell/amg/grid_search_and_inference.sbatch
@@ -0,0 +1,9 @@
+#! /bin/bash
+#SBATCH -c 8
+#SBATCH --mem 96G
+#SBATCH -t 2880
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python grid_search_and_inference.py $@
diff --git a/finetuning/livecell/amg/util.py b/finetuning/livecell/amg/util.py
new file mode 100644
index 00000000..eff50eac
--- /dev/null
+++ b/finetuning/livecell/amg/util.py
@@ -0,0 +1,30 @@
+import os
+
+DATA_ROOT = "/scratch/projects/nim00007/data/LiveCELL"
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/livecell"
+MODELS = {
+ "vit_b": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_b_01ec64.pth",
+ "vit_h": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_h_4b8939.pth",
+ "vit_b_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_b/best.pt",
+ "vit_h_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_h/best.pt",
+ "vit_b_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_b/best.pt",
+ "vit_h_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_h/best.pt",
+}
+
+
+def get_checkpoint(name):
+ assert name in MODELS, name
+ ckpt = MODELS[name]
+ assert os.path.exists(ckpt), ckpt
+ model_type = name[:5]
+ assert model_type in ("vit_b", "vit_h"), model_type
+ return ckpt, model_type
+
+
+def get_experiment_folder(name):
+ return os.path.join(EXPERIMENT_ROOT, name)
+
+
+def check_model(name):
+ if name not in MODELS:
+ raise ValueError(f"Invalid model {name}, expect one of {MODELS.keys()}")
diff --git a/finetuning/livecell/evaluation/evaluation.py b/finetuning/livecell/evaluation/evaluation.py
new file mode 100644
index 00000000..18e93eb5
--- /dev/null
+++ b/finetuning/livecell/evaluation/evaluation.py
@@ -0,0 +1,58 @@
+import argparse
+import os
+from glob import glob
+
+import pandas as pd
+from tqdm import tqdm
+
+from micro_sam.evaluation.livecell import evaluate_livecell_predictions
+from util import get_experiment_folder, DATA_ROOT
+
+
+def run_eval(gt_dir, experiment_folder, prompt_prefix):
+ result_dir = os.path.join(experiment_folder, "results", prompt_prefix)
+ os.makedirs(result_dir, exist_ok=True)
+
+ pred_dirs = sorted(glob(os.path.join(experiment_folder, prompt_prefix, "*")))
+
+ results = []
+ for pred_dir in tqdm(pred_dirs, desc=f"Run livecell evaluation for all {prompt_prefix}-prompt settings"):
+ setting_name = os.path.basename(pred_dir)
+ save_path = os.path.join(result_dir, f"{setting_name}.csv")
+ if os.path.exists(save_path):
+ result = pd.read_csv(save_path)
+ else:
+ result = evaluate_livecell_predictions(gt_dir, pred_dir, verbose=False)
+ result.to_csv(save_path, index=False)
+ result.insert(0, "setting", [setting_name] * result.shape[0])
+ results.append(result)
+
+ results = pd.concat(results)
+ return results
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ args = parser.parse_args()
+
+ name = args.name
+
+ gt_dir = os.path.join(DATA_ROOT, "annotations", "livecell_test_images")
+ assert os.path.exists(gt_dir), "The LiveCELL Dataset is incomplete"
+
+ experiment_folder = get_experiment_folder(name)
+
+ result_box = run_eval(gt_dir, experiment_folder, "box")
+ result_box.insert(0, "prompt", ["box"] * result_box.shape[0])
+
+ result_point = run_eval(gt_dir, experiment_folder, "points")
+ result_point.insert(0, "prompt", ["points"] * result_point.shape[0])
+
+ result = pd.concat([result_box, result_point])
+ save_path = os.path.join(experiment_folder, f"result_{name}.csv")
+ result.to_csv(save_path, index=False)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/evaluation.sbatch b/finetuning/livecell/evaluation/evaluation.sbatch
new file mode 100755
index 00000000..8306a1df
--- /dev/null
+++ b/finetuning/livecell/evaluation/evaluation.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -N 1
+#SBATCH -c 4
+#SBATCH --mem 32G
+#SBATCH -t 720
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python evaluation.py $@
diff --git a/finetuning/livecell/evaluation/inference.py b/finetuning/livecell/evaluation/inference.py
new file mode 100644
index 00000000..00e1b894
--- /dev/null
+++ b/finetuning/livecell/evaluation/inference.py
@@ -0,0 +1,79 @@
+import argparse
+import os
+from glob import glob
+from subprocess import run
+
+import micro_sam.evaluation as evaluation
+from micro_sam.evaluation.livecell import livecell_inference
+from util import check_model, get_checkpoint, get_experiment_folder, DATA_ROOT, PROMPT_FOLDER
+
+
+def inference_job(prompt_settings, model_name):
+ experiment_folder = get_experiment_folder(model_name)
+ checkpoint, model_type = get_checkpoint(model_name)
+ livecell_inference(
+ checkpoint,
+ input_folder=DATA_ROOT,
+ model_type=model_type,
+ experiment_folder=experiment_folder,
+ use_points=prompt_settings["use_points"],
+ use_boxes=prompt_settings["use_boxes"],
+ n_positives=prompt_settings["n_positives"],
+ n_negatives=prompt_settings["n_negatives"],
+ prompt_folder=PROMPT_FOLDER,
+ )
+
+
+def submit_array_job(prompt_settings, model_name, test_run):
+ n_settings = len(prompt_settings)
+ cmd = ["sbatch", "-a", f"0-{n_settings-1}", "inference.sbatch", "-n", model_name]
+ if test_run:
+ cmd.append("-t")
+ run(cmd)
+
+
+def check_inference(settings, model_name):
+ experiment_folder = get_experiment_folder(model_name)
+ for setting in settings:
+ prefix = "box" if setting["use_boxes"] else "points"
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ pred_folder = os.path.join(experiment_folder, prefix, f"p{pos}-n{neg}")
+ assert os.path.exists(pred_folder), pred_folder
+ n_files = len(glob(os.path.join(pred_folder, "*.tif")))
+ assert n_files == 1512, str(n_files)
+
+ print("Inference checks successful!")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ parser.add_argument("-t", "--test_run", action="store_true")
+ parser.add_argument("-c", "--check", action="store_true")
+ args = parser.parse_args()
+
+ if args.test_run: # test run with only the three default experiment settings
+ settings = evaluation.default_experiment_settings()
+ else: # all experiment settings
+ settings = evaluation.full_experiment_settings()
+ settings.extend(evaluation.full_experiment_settings(use_boxes=True))
+
+ model_name = args.name
+ check_model(model_name)
+
+ if args.check:
+ check_inference(settings, model_name)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(settings, model_name, args.test_run)
+ else: # we're in a slurm job and run inference for a setting
+ job_id = int(job_id)
+ this_settings = settings[job_id]
+ inference_job(this_settings, model_name)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/inference.sbatch b/finetuning/livecell/evaluation/inference.sbatch
new file mode 100755
index 00000000..62d926eb
--- /dev/null
+++ b/finetuning/livecell/evaluation/inference.sbatch
@@ -0,0 +1,9 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 48G
+#SBATCH -t 60
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python inference.py $@
diff --git a/finetuning/livecell/evaluation/iterative_prompting.py b/finetuning/livecell/evaluation/iterative_prompting.py
new file mode 100644
index 00000000..0278ff38
--- /dev/null
+++ b/finetuning/livecell/evaluation/iterative_prompting.py
@@ -0,0 +1,52 @@
+import os
+from glob import glob
+
+from micro_sam.evaluation.inference import run_inference_with_iterative_prompting
+from micro_sam.evaluation.evaluation import run_evaluation
+
+from util import get_checkpoint, get_paths
+
+LIVECELL_GT_ROOT = "/scratch-grete/projects/nim00007/data/LiveCELL/annotations_corrected/livecell_test_images"
+# TODO update to make fit other models
+PREDICTION_ROOT = "./pred_interactive_prompting"
+
+
+def run_interactive_prompting():
+ prediction_root = PREDICTION_ROOT
+
+ checkpoint, model_type = get_checkpoint("vit_b")
+ image_paths, gt_paths = get_paths()
+
+ run_inference_with_iterative_prompting(
+ checkpoint, model_type, image_paths, gt_paths,
+ prediction_root, use_boxes=False, batch_size=16,
+ )
+
+
+def get_pg_paths(pred_folder):
+ pred_paths = sorted(glob(os.path.join(pred_folder, "*.tif")))
+ names = [os.path.split(path)[1] for path in pred_paths]
+ gt_paths = [
+ os.path.join(LIVECELL_GT_ROOT, name.split("_")[0], name) for name in names
+ ]
+ assert all(os.path.exists(pp) for pp in gt_paths)
+ return pred_paths, gt_paths
+
+
+def evaluate_interactive_prompting():
+ prediction_root = PREDICTION_ROOT
+ prediction_folders = sorted(glob(os.path.join(prediction_root, "iteration*")))
+ for pred_folder in prediction_folders:
+ print("Evaluating", pred_folder)
+ pred_paths, gt_paths = get_pg_paths(pred_folder)
+ res = run_evaluation(gt_paths, pred_paths, save_path=None)
+ print(res)
+
+
+def main():
+ # run_interactive_prompting()
+ evaluate_interactive_prompting()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/precompute_embeddings.py b/finetuning/livecell/evaluation/precompute_embeddings.py
new file mode 100644
index 00000000..3d53ed83
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_embeddings.py
@@ -0,0 +1,24 @@
+import argparse
+import os
+
+from micro_sam.evaluation import precompute_all_embeddings
+from util import get_paths, get_model, get_experiment_folder
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-n", "--name", required=True)
+ args = parser.parse_args()
+
+ name = args.name
+
+ image_paths, _ = get_paths()
+ predictor = get_model(name)
+ exp_folder = get_experiment_folder(name)
+ embedding_dir = os.path.join(exp_folder, "embeddings")
+ os.makedirs(embedding_dir, exist_ok=True)
+ precompute_all_embeddings(predictor, image_paths, embedding_dir)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/precompute_embeddings.sbatch b/finetuning/livecell/evaluation/precompute_embeddings.sbatch
new file mode 100755
index 00000000..70faec19
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_embeddings.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -N 1
+#SBATCH -c 4
+#SBATCH --mem 64G
+#SBATCH -t 120
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+
+source activate sam
+python precompute_embeddings.py -n $1
diff --git a/finetuning/livecell/evaluation/precompute_prompts.py b/finetuning/livecell/evaluation/precompute_prompts.py
new file mode 100644
index 00000000..f22ccdc6
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_prompts.py
@@ -0,0 +1,68 @@
+import argparse
+import os
+import pickle
+from subprocess import run
+
+import micro_sam.evaluation as evaluation
+from tqdm import tqdm
+from util import get_paths, PROMPT_FOLDER
+
+
+def precompute_setting(prompt_settings):
+ _, gt_paths = get_paths()
+ evaluation.precompute_all_prompts(gt_paths, PROMPT_FOLDER, prompt_settings)
+
+
+def submit_array_job(prompt_settings, full_settings):
+ n_settings = len(prompt_settings)
+ cmd = ["sbatch", "-a", f"0-{n_settings-1}", "precompute_prompts.sbatch"]
+ if full_settings:
+ cmd.append("-f")
+ run(cmd)
+
+
+def check_settings(settings):
+
+ def check_prompt_file(prompt_file):
+ assert os.path.exists(prompt_file), prompt_file
+ with open(prompt_file, "rb") as f:
+ prompts = pickle.load(f)
+ assert len(prompts) == 1512, f"{len(prompts)}"
+
+ for setting in tqdm(settings, desc="Check prompt files"):
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ prompt_file = os.path.join(PROMPT_FOLDER, f"points-p{pos}-n{neg}.pkl")
+ if pos == 0 and neg == 0:
+ prompt_file = os.path.join(PROMPT_FOLDER, "boxes.pkl")
+ check_prompt_file(prompt_file)
+
+ print("All files checked!")
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-f", "--full_settings", action="store_true")
+ parser.add_argument("-c", "--check", action="store_true")
+ args = parser.parse_args()
+
+ if args.full_settings:
+ settings = evaluation.full_experiment_settings(use_boxes=True)
+ else:
+ settings = evaluation.default_experiment_settings()
+
+ if args.check:
+ check_settings(settings)
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+
+ if job_id is None: # this is the main script that submits slurm jobs
+ submit_array_job(settings, args.full_settings)
+ else: # we're in a slurm job and precompute a setting
+ job_id = int(job_id)
+ this_settings = [settings[job_id]]
+ precompute_setting(this_settings)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/evaluation/precompute_prompts.sbatch b/finetuning/livecell/evaluation/precompute_prompts.sbatch
new file mode 100755
index 00000000..2a9e2df0
--- /dev/null
+++ b/finetuning/livecell/evaluation/precompute_prompts.sbatch
@@ -0,0 +1,8 @@
+#! /bin/bash
+#SBATCH -p standard96
+#SBATCH -c 4
+#SBATCH --mem 64G
+#SBATCH -t 720
+
+source activate sam
+python precompute_prompts.py $@
diff --git a/finetuning/livecell/evaluation/util.py b/finetuning/livecell/evaluation/util.py
new file mode 100644
index 00000000..48767a38
--- /dev/null
+++ b/finetuning/livecell/evaluation/util.py
@@ -0,0 +1,55 @@
+import os
+
+from micro_sam.evaluation import get_predictor
+from micro_sam.evaluation.livecell import _get_livecell_paths
+
+DATA_ROOT = "/scratch/projects/nim00007/data/LiveCELL"
+EXPERIMENT_ROOT = "/scratch/projects/nim00007/sam/experiments/livecell"
+PROMPT_FOLDER = "/scratch-grete/projects/nim00007/sam/experiments/prompts/livecell"
+MODELS = {
+ "vit_b": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_b_01ec64.pth",
+ "vit_h": "/scratch-grete/projects/nim00007/sam/vanilla/sam_vit_h_4b8939.pth",
+ "vit_b_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_b/best.pt",
+ "vit_h_specialist": "/scratch-grete/projects/nim00007/sam/LM/LiveCELL/vit_h/best.pt",
+ "vit_b_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_b/best.pt",
+ "vit_h_generalist": "/scratch-grete/projects/nim00007/sam/LM/generalist/vit_h/best.pt",
+}
+
+
+def get_paths():
+ return _get_livecell_paths(DATA_ROOT)
+
+
+def get_checkpoint(name):
+ assert name in MODELS, name
+ ckpt = MODELS[name]
+ assert os.path.exists(ckpt), ckpt
+ model_type = name[:5]
+ assert model_type in ("vit_b", "vit_h"), model_type
+ return ckpt, model_type
+
+
+def get_model(name):
+ ckpt, model_type = get_checkpoint(name)
+ predictor = get_predictor(ckpt, model_type)
+ return predictor
+
+
+def get_experiment_folder(name):
+ return os.path.join(EXPERIMENT_ROOT, name)
+
+
+def check_model(name):
+ if name not in MODELS:
+ raise ValueError(f"Invalid model {name}, expect one of {MODELS.keys()}")
+
+
+def download_livecell():
+ from torch_em.data.datasets import get_livecell_loader
+ get_livecell_loader(DATA_ROOT, "train", (512, 512), 1, download=True)
+ get_livecell_loader(DATA_ROOT, "val", (512, 512), 1, download=True)
+ get_livecell_loader(DATA_ROOT, "test", (512, 512), 1, download=True)
+
+
+if __name__ == "__main__":
+ download_livecell()
diff --git a/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.py b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.py
new file mode 100644
index 00000000..031984dd
--- /dev/null
+++ b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.py
@@ -0,0 +1,119 @@
+import argparse
+import os
+import warnings
+from subprocess import run
+
+import pandas as pd
+
+from micro_sam.evaluation import (
+ inference,
+ evaluation,
+ default_experiment_settings,
+ get_experiment_setting_name
+)
+from micro_sam.evaluation.livecell import _get_livecell_paths
+
+DATA_ROOT = "/scratch-grete/projects/nim00007/data/LiveCELL"
+EXPERIMENT_ROOT = "/scratch-grete/projects/nim00007/sam/experiments/livecell/partial-finetuning"
+PROMPT_DIR = "/scratch-grete/projects/nim00007/sam/experiments/prompts/livecell"
+MODELS = {
+ "freeze-image_encoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-image_encoder",
+ "freeze-image_encoder-mask_decoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-image_encoder-mask_decoder",
+ "freeze-image_encoder-prompt_encoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-image_encoder-prompt_encoder",
+ "freeze-mask_decoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-mask_decoder",
+ "freeze-None": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-None",
+ "freeze-prompt_encoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-prompt_encoder",
+ "freeze-prompt_encoder-mask_decoder": "/scratch-grete/projects/nim00007/sam/partial_finetuning/checkpoints/livecell_sam-freeze-prompt_encoder-mask_decoder",
+ "vanilla": "/home/nimcpape/.sam_models/sam_vit_b_01ec64.pth",
+}
+
+
+def evaluate_model(model_id):
+ model_name = list(MODELS.keys())[model_id]
+ print("Evaluating", model_name)
+
+ try:
+ checkpoint = os.path.join(MODELS[model_name], "best.pt")
+ assert os.path.exists(checkpoint)
+ except AssertionError:
+ checkpoint = MODELS[model_name]
+
+ print("Evalute", model_name, "from", checkpoint)
+ model_type = "vit_b"
+ with warnings.catch_warnings():
+ warnings.simplefilter("ignore")
+ predictor = inference.get_predictor(checkpoint, model_type=model_type)
+
+ experiment_dir = os.path.join(EXPERIMENT_ROOT, model_name)
+
+ embedding_dir = os.path.join(experiment_dir, "embeddings")
+ os.makedirs(embedding_dir, exist_ok=True)
+
+ result_dir = os.path.join(experiment_dir, "results")
+ os.makedirs(result_dir, exist_ok=True)
+
+ image_paths, gt_paths = _get_livecell_paths(DATA_ROOT)
+ experiment_settings = default_experiment_settings()
+
+ results = []
+ for setting in experiment_settings:
+ setting_name = get_experiment_setting_name(setting)
+ prediction_dir = os.path.join(experiment_dir, setting_name)
+
+ os.makedirs(prediction_dir, exist_ok=True)
+ inference.run_inference_with_prompts(
+ predictor, image_paths, gt_paths,
+ embedding_dir, prediction_dir,
+ prompt_save_dir=PROMPT_DIR, **setting
+ )
+
+ pred_paths = [os.path.join(prediction_dir, os.path.basename(gt_path)) for gt_path in gt_paths]
+ assert len(pred_paths) == len(gt_paths)
+ result_path = os.path.join(result_dir, f"{setting_name}.csv")
+
+ if os.path.exists(result_path):
+ result = pd.read_csv(result_path)
+ else:
+ result = evaluation.run_evaluation(gt_paths, pred_paths, result_path)
+ result.insert(0, "setting", [setting_name])
+ results.append(result)
+
+ results = pd.concat(results)
+ return results
+
+
+def combine_results():
+ results = []
+ for model_id, model_name in enumerate(MODELS):
+ res = evaluate_model(model_id)
+ res.insert(0, "frozen", res.shape[0] * [model_name.lstrip("freeze-")])
+ results.append(res)
+ results = pd.concat(results)
+ res_path = os.path.join(EXPERIMENT_ROOT, "partial_finetuning_results.csv")
+ results.to_csv(res_path, index=False)
+
+
+def submit_array_job():
+ n_models = len(MODELS)
+ cmd = ["sbatch", "-a", f"0-{n_models-1}", "evaluate_partially_finetuned.sbatch"]
+ run(cmd)
+
+
+def main():
+ parser = argparse.ArgumentParser()
+ parser.add_argument("-e", "--evaluate", action="store_true")
+ args = parser.parse_args()
+
+ if args.evaluate:
+ combine_results()
+ return
+
+ job_id = os.environ.get("SLURM_ARRAY_TASK_ID", None)
+ if job_id is None:
+ submit_array_job()
+ else:
+ evaluate_model(int(job_id))
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.sbatch b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.sbatch
new file mode 100755
index 00000000..68fee4fa
--- /dev/null
+++ b/finetuning/livecell/partial_finetuning/evaluate_partially_finetuned.sbatch
@@ -0,0 +1,10 @@
+#! /bin/bash
+#SBATCH -c 4
+#SBATCH --mem 96G
+#SBATCH -t 2880
+#SBATCH -p grete:shared
+#SBATCH -G A100:1
+#SBATCH -A nim00007
+
+source activate sam
+python evaluate_partially_finetuned.py $@
diff --git a/finetuning/livecell_evaluation.py b/finetuning/livecell_evaluation.py
new file mode 100644
index 00000000..3f545304
--- /dev/null
+++ b/finetuning/livecell_evaluation.py
@@ -0,0 +1,3 @@
+from micro_sam.evaluation.livecell import run_livecell_evaluation
+
+run_livecell_evaluation()
diff --git a/finetuning/livecell_finetuning.py b/finetuning/livecell_finetuning.py
new file mode 100644
index 00000000..3521db4b
--- /dev/null
+++ b/finetuning/livecell_finetuning.py
@@ -0,0 +1,115 @@
+import argparse
+import os
+
+import micro_sam.training as sam_training
+import torch
+import torch_em
+
+from torch_em.data.datasets import get_livecell_loader
+from micro_sam.util import export_custom_sam_model
+
+
+def get_dataloaders(patch_shape, data_path, cell_type=None):
+ """This returns the livecell data loaders implemented in torch_em:
+ https://github.com/constantinpape/torch-em/blob/main/torch_em/data/datasets/livecell.py
+ It will automatically download the livecell data.
+
+ Note: to replace this with another data loader you need to return a torch data loader
+ that retuns `x, y` tensors, where `x` is the image data and `y` are the labels.
+ The labels have to be in a label mask instance segmentation format.
+ I.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
+ Important: the ID 0 is reseved for background, and the IDs must be consecutive
+ """
+ label_transform = torch_em.transform.label.label_consecutive # to ensure consecutive IDs
+ train_loader = get_livecell_loader(path=data_path, patch_shape=patch_shape, split="train", batch_size=2,
+ num_workers=8, cell_types=cell_type, download=True,
+ label_transform=label_transform)
+ val_loader = get_livecell_loader(path=data_path, patch_shape=patch_shape, split="val", batch_size=1,
+ num_workers=8, cell_types=cell_type, download=True,
+ label_transform=label_transform)
+ return train_loader, val_loader
+
+
+def finetune_livecell(args):
+ """Example code for finetuning SAM on LiveCELL"""
+
+ # training settings:
+ model_type = args.model_type
+ checkpoint_path = None # override this to start training from a custom checkpoint
+ device = "cuda" # override this if you have some more complex set-up and need to specify the exact gpu
+ patch_shape = (520, 740) # the patch shape for training
+ n_objects_per_batch = 25 # this is the number of objects per batch that will be sampled
+
+ # get the trainable segment anything model
+ model = sam_training.get_trainable_sam_model(model_type, checkpoint_path, device=device)
+
+ # all the stuff we need for training
+ optimizer = torch.optim.Adam(model.parameters(), lr=1e-5)
+ scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode="min", factor=0.9, patience=10, verbose=True)
+ train_loader, val_loader = get_dataloaders(patch_shape=patch_shape, data_path=args.input_path)
+
+ # this class creates all the training data for a batch (inputs, prompts and labels)
+ convert_inputs = sam_training.ConvertToSamInputs()
+
+ checkpoint_name = "livecell_sam"
+ # the trainer which performs training and validation (implemented using "torch_em")
+ trainer = sam_training.SamTrainer(
+ name=checkpoint_name,
+ save_root=args.save_root,
+ train_loader=train_loader,
+ val_loader=val_loader,
+ model=model,
+ optimizer=optimizer,
+ # currently we compute loss batch-wise, else we pass channelwise True
+ loss=torch_em.loss.DiceLoss(channelwise=False),
+ metric=torch_em.loss.DiceLoss(),
+ device=device,
+ lr_scheduler=scheduler,
+ logger=sam_training.SamLogger,
+ log_image_interval=10,
+ mixed_precision=True,
+ convert_inputs=convert_inputs,
+ n_objects_per_batch=n_objects_per_batch,
+ n_sub_iteration=8,
+ compile_model=False
+ )
+ trainer.fit(args.iterations)
+ if args.export_path is not None:
+ checkpoint_path = os.path.join(
+ "" if args.save_root is None else args.save_root, "checkpoints", checkpoint_name, "best.pt"
+ )
+ export_custom_sam_model(
+ checkpoint_path=checkpoint_path,
+ model_type=model_type,
+ save_path=args.export_path,
+ )
+
+
+def main():
+ parser = argparse.ArgumentParser(description="Finetune Segment Anything for the LiveCELL dataset.")
+ parser.add_argument(
+ "--input_path", "-i", default="",
+ help="The filepath to the LiveCELL data. If the data does not exist yet it will be downloaded."
+ )
+ parser.add_argument(
+ "--model_type", "-m", default="vit_b",
+ help="The model type to use for fine-tuning. Either vit_h, vit_b or vit_l."
+ )
+ parser.add_argument(
+ "--save_root", "-s",
+ help="Where to save the checkpoint and logs. By default they will be saved where this script is run."
+ )
+ parser.add_argument(
+ "--iterations", type=int, default=int(1e5),
+ help="For how many iterations should the model be trained? By default 100k."
+ )
+ parser.add_argument(
+ "--export_path", "-e",
+ help="Where to export the finetuned model to. The exported model can be use din the annotation tools."
+ )
+ args = parser.parse_args()
+ finetune_livecell(args)
+
+
+if __name__ == "__main__":
+ main()
diff --git a/finetuning/livecell_inference.py b/finetuning/livecell_inference.py
new file mode 100644
index 00000000..3ec2f6ce
--- /dev/null
+++ b/finetuning/livecell_inference.py
@@ -0,0 +1,3 @@
+from micro_sam.evaluation.livecell import run_livecell_inference
+
+run_livecell_inference()
diff --git a/micro_sam/__init__.py b/micro_sam/__init__.py
index 5e90fe5b..9a135385 100644
--- a/micro_sam/__init__.py
+++ b/micro_sam/__init__.py
@@ -3,6 +3,7 @@
.. include:: ../doc/installation.md
.. include:: ../doc/annotation_tools.md
.. include:: ../doc/python_library.md
+.. include:: ../doc/finetuned_models.md
"""
from .__version__ import __version__
diff --git a/micro_sam/__version__.py b/micro_sam/__version__.py
index 02e905d7..b5fdc753 100644
--- a/micro_sam/__version__.py
+++ b/micro_sam/__version__.py
@@ -1 +1 @@
-__version__ = "0.1.2.post1"
+__version__ = "0.2.2"
diff --git a/micro_sam/evaluation/__init__.py b/micro_sam/evaluation/__init__.py
new file mode 100644
index 00000000..b16a45c1
--- /dev/null
+++ b/micro_sam/evaluation/__init__.py
@@ -0,0 +1,19 @@
+"""Functionality for evaluating Segment Anything models on microscopy data.
+"""
+
+from .automatic_mask_generation import (
+ run_amg_inference,
+ run_amg_grid_search,
+ run_amg_grid_search_and_inference,
+)
+from .inference import (
+ get_predictor,
+ run_inference_with_prompts,
+ precompute_all_embeddings,
+ precompute_all_prompts,
+)
+from .experiments import (
+ default_experiment_settings,
+ full_experiment_settings,
+ get_experiment_setting_name,
+)
diff --git a/micro_sam/evaluation/automatic_mask_generation.py b/micro_sam/evaluation/automatic_mask_generation.py
new file mode 100644
index 00000000..46b12ef5
--- /dev/null
+++ b/micro_sam/evaluation/automatic_mask_generation.py
@@ -0,0 +1,272 @@
+"""Inference and evaluation for the automatic instance segmentation functionality.
+"""
+
+import os
+from glob import glob
+from pathlib import Path
+from typing import Any, Dict, List, Optional, Tuple, Union
+
+import imageio.v3 as imageio
+import numpy as np
+import pandas as pd
+
+from elf.evaluation import mean_segmentation_accuracy
+from segment_anything import SamPredictor
+from tqdm import tqdm
+
+from .. import instance_segmentation
+from .. import util
+
+
+def _get_range_of_search_values(input_vals, step):
+ if isinstance(input_vals, list):
+ search_range = np.arange(input_vals[0], input_vals[1] + step, step)
+ search_range = [round(e, 2) for e in search_range]
+ else:
+ search_range = [input_vals]
+ return search_range
+
+
+def _grid_search(
+ amg, gt, image_name, iou_thresh_values, stability_score_values, result_path, amg_generate_kwargs, verbose,
+):
+ net_list = []
+ gs_combinations = [(r1, r2) for r1 in iou_thresh_values for r2 in stability_score_values]
+
+ for iou_thresh, stability_thresh in tqdm(gs_combinations, disable=not verbose):
+ masks = amg.generate(
+ pred_iou_thresh=iou_thresh, stability_score_thresh=stability_thresh, **amg_generate_kwargs
+ )
+ instance_labels = instance_segmentation.mask_data_to_segmentation(
+ masks, gt.shape, with_background=True,
+ min_object_size=amg_generate_kwargs.get("min_mask_region_area", 0),
+ )
+ m_sas, sas = mean_segmentation_accuracy(instance_labels, gt, return_accuracies=True) # type: ignore
+
+ result_dict = {
+ "image_name": image_name,
+ "pred_iou_thresh": iou_thresh,
+ "stability_score_thresh": stability_thresh,
+ "mSA": m_sas,
+ "SA50": sas[0],
+ "SA75": sas[5]
+ }
+ tmp_df = pd.DataFrame([result_dict])
+ net_list.append(tmp_df)
+
+ img_gs_df = pd.concat(net_list)
+ img_gs_df.to_csv(result_path, index=False)
+
+
+# ideally we would generalize the parameters that GS runs over
+def run_amg_grid_search(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ gt_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ result_dir: Union[str, os.PathLike],
+ iou_thresh_values: Optional[List[float]] = None,
+ stability_score_values: Optional[List[float]] = None,
+ amg_kwargs: Optional[Dict[str, Any]] = None,
+ amg_generate_kwargs: Optional[Dict[str, Any]] = None,
+ AMG: instance_segmentation.AMGBase = instance_segmentation.AutomaticMaskGenerator,
+ verbose_gs: bool = False,
+) -> None:
+ """Run grid search for automatic mask generation.
+
+ The grid search goes over the two most important parameters:
+ - `pred_iou_thresh`, the threshold for keeping objects according to the IoU predicted by the model
+ - `stability_score_thresh`, the theshold for keepong objects according to their stability
+
+ Args:
+ predictor: The segment anything predictor.
+ image_paths: The input images for the grid search.
+ gt_paths: The ground-truth segmentation for the grid search.
+ embedding_dir: Folder to cache the image embeddings.
+ result_dir: Folder to cache the evaluation results per image.
+ iou_thresh_values: The values for `pred_iou_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ stability_score_values: The values for `stability_score_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ amg_kwargs: The keyword arguments for the automatic mask generator class.
+ amg_generate_kwargs: The keyword arguments for the `generate` method of the mask generator.
+ This must not contain `pred_iou_thresh` or `stability_score_thresh`.
+ AMG: The automatic mask generator. By default `micro_sam.instance_segmentation.AutomaticMaskGenerator`.
+ verbose_gs: Whether to run the gridsearch for individual images in a verbose mode.
+ """
+ assert len(image_paths) == len(gt_paths)
+ amg_kwargs = {} if amg_kwargs is None else amg_kwargs
+ amg_generate_kwargs = {} if amg_generate_kwargs is None else amg_generate_kwargs
+ if "pred_iou_thresh" in amg_generate_kwargs or "stability_score_thresh" in amg_generate_kwargs:
+ raise ValueError("The threshold parameters are optimized in the grid-search. You must not pass them as kwargs.")
+
+ if iou_thresh_values is None:
+ iou_thresh_values = _get_range_of_search_values([0.6, 0.9], step=0.025)
+ if stability_score_values is None:
+ stability_score_values = _get_range_of_search_values([0.6, 0.95], step=0.025)
+
+ os.makedirs(result_dir, exist_ok=True)
+ amg = AMG(predictor, **amg_kwargs)
+
+ for image_path, gt_path in tqdm(
+ zip(image_paths, gt_paths), desc="Run grid search for AMG", total=len(image_paths)
+ ):
+ image_name = Path(image_path).stem
+ result_path = os.path.join(result_dir, f"{image_name}.csv")
+
+ # We skip images for which the grid search was done already.
+ if os.path.exists(result_path):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ assert os.path.exists(gt_path), gt_path
+
+ image = imageio.imread(image_path)
+ gt = imageio.imread(gt_path)
+
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ image_embeddings = util.precompute_image_embeddings(predictor, image, embedding_path, ndim=2)
+ amg.initialize(image, image_embeddings)
+
+ _grid_search(
+ amg, gt, image_name,
+ iou_thresh_values, stability_score_values,
+ result_path, amg_generate_kwargs, verbose=verbose_gs,
+ )
+
+
+def run_amg_inference(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ prediction_dir: Union[str, os.PathLike],
+ amg_kwargs: Optional[Dict[str, Any]] = None,
+ amg_generate_kwargs: Optional[Dict[str, Any]] = None,
+ AMG: instance_segmentation.AMGBase = instance_segmentation.AutomaticMaskGenerator,
+) -> None:
+ """Run inference for automatic mask generation.
+
+ Args:
+ predictor: The segment anything predictor.
+ image_paths: The input images.
+ embedding_dir: Folder to cache the image embeddings.
+ prediction_dir: Folder to save the predictions.
+ amg_kwargs: The keyword arguments for the automatic mask generator class.
+ amg_generate_kwargs: The keyword arguments for the `generate` method of the mask generator.
+ This must not contain `pred_iou_thresh` or `stability_score_thresh`.
+ AMG: The automatic mask generator. By default `micro_sam.instance_segmentation.AutomaticMaskGenerator`.
+ """
+ amg_kwargs = {} if amg_kwargs is None else amg_kwargs
+ amg_generate_kwargs = {} if amg_generate_kwargs is None else amg_generate_kwargs
+
+ amg = AMG(predictor, **amg_kwargs)
+
+ for image_path in tqdm(image_paths, desc="Run inference for automatic mask generation"):
+ image_name = os.path.basename(image_path)
+
+ # We skip the images that already have been segmented.
+ prediction_path = os.path.join(prediction_dir, image_name)
+ if os.path.exists(prediction_path):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ image = imageio.imread(image_path)
+
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ image_embeddings = util.precompute_image_embeddings(predictor, image, embedding_path, ndim=2)
+
+ amg.initialize(image, image_embeddings)
+ masks = amg.generate(**amg_generate_kwargs)
+ instances = instance_segmentation.mask_data_to_segmentation(
+ masks, image.shape, with_background=True, min_object_size=amg_generate_kwargs.get("min_mask_region_area", 0)
+ )
+
+ # It's important to compress here, otherwise the predictions would take up a lot of space.
+ imageio.imwrite(prediction_path, instances, compression=5)
+
+
+def evaluate_amg_grid_search(result_dir: Union[str, os.PathLike], criterion: str = "mSA") -> Tuple[float, float, float]:
+ """Evaluate gridsearch results.
+
+ Args:
+ result_dir: The folder with the gridsearch results.
+ criterion: The metric to use for determining the best parameters.
+
+ Returns:
+ - The best value for `pred_iou_thresh`.
+ - The best value for `stability_score_thresh`.
+ - The evaluation score for the best setting.
+ """
+
+ # load all the grid search results
+ gs_files = glob(os.path.join(result_dir, "*.csv"))
+ gs_result = pd.concat([pd.read_csv(gs_file) for gs_file in gs_files])
+
+ # contain only the relevant columns and group by the gridsearch columns
+ gs_col1 = "pred_iou_thresh"
+ gs_col2 = "stability_score_thresh"
+ gs_result = gs_result[[gs_col1, gs_col2, criterion]]
+
+ # compute the mean over the grouped columns
+ grouped_result = gs_result.groupby([gs_col1, gs_col2]).mean()
+
+ # find the best grouped result and return the corresponding thresholds
+ best_score = grouped_result.max().values[0]
+ best_result = grouped_result.idxmax()
+ best_iou_thresh, best_stability_score = best_result.values[0]
+ return best_iou_thresh, best_stability_score, best_score
+
+
+def run_amg_grid_search_and_inference(
+ predictor: SamPredictor,
+ val_image_paths: List[Union[str, os.PathLike]],
+ val_gt_paths: List[Union[str, os.PathLike]],
+ test_image_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ prediction_dir: Union[str, os.PathLike],
+ result_dir: Union[str, os.PathLike],
+ iou_thresh_values: Optional[List[float]] = None,
+ stability_score_values: Optional[List[float]] = None,
+ amg_kwargs: Optional[Dict[str, Any]] = None,
+ amg_generate_kwargs: Optional[Dict[str, Any]] = None,
+ AMG: instance_segmentation.AMGBase = instance_segmentation.AutomaticMaskGenerator,
+ verbose_gs: bool = True,
+) -> None:
+ """Run grid search and inference for automatic mask generation.
+
+ Args:
+ predictor: The segment anything predictor.
+ val_image_paths: The input images for the grid search.
+ val_gt_paths: The ground-truth segmentation for the grid search.
+ test_image_paths: The input images for inference.
+ embedding_dir: Folder to cache the image embeddings.
+ prediction_dir: Folder to save the predictions.
+ result_dir: Folder to cache the evaluation results per image.
+ iou_thresh_values: The values for `pred_iou_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ stability_score_values: The values for `stability_score_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ amg_kwargs: The keyword arguments for the automatic mask generator class.
+ amg_generate_kwargs: The keyword arguments for the `generate` method of the mask generator.
+ This must not contain `pred_iou_thresh` or `stability_score_thresh`.
+ AMG: The automatic mask generator. By default `micro_sam.instance_segmentation.AutomaticMaskGenerator`.
+ verbose_gs: Whether to run the gridsearch for individual images in a verbose mode.
+ """
+ run_amg_grid_search(
+ predictor, val_image_paths, val_gt_paths, embedding_dir, result_dir,
+ iou_thresh_values=iou_thresh_values, stability_score_values=stability_score_values,
+ amg_kwargs=amg_kwargs, amg_generate_kwargs=amg_generate_kwargs, AMG=AMG, verbose_gs=verbose_gs,
+ )
+
+ amg_generate_kwargs = {} if amg_generate_kwargs is None else amg_generate_kwargs
+ best_iou_thresh, best_stability_score, best_msa = evaluate_amg_grid_search(result_dir)
+ print(
+ "Best grid-search result:", best_msa,
+ f"@ iou_thresh = {best_iou_thresh}, stability_score = {best_stability_score}"
+ )
+ amg_generate_kwargs["pred_iou_thresh"] = best_iou_thresh
+ amg_generate_kwargs["stability_score_thresh"] = best_stability_score
+
+ run_amg_inference(
+ predictor, test_image_paths, embedding_dir, prediction_dir, amg_kwargs, amg_generate_kwargs, AMG
+ )
diff --git a/micro_sam/evaluation/evaluation.py b/micro_sam/evaluation/evaluation.py
new file mode 100644
index 00000000..854804ae
--- /dev/null
+++ b/micro_sam/evaluation/evaluation.py
@@ -0,0 +1,78 @@
+"""Evaluation functionality for segmentation predictions from `micro_sam.evaluation.automatic_mask_generation`
+and `micro_sam.evaluation.inference`.
+"""
+
+import os
+from pathlib import Path
+from typing import List, Optional, Union
+
+import imageio.v3 as imageio
+import numpy as np
+import pandas as pd
+
+from elf.evaluation import mean_segmentation_accuracy
+from skimage.measure import label
+from tqdm import tqdm
+
+
+def _run_evaluation(gt_paths, prediction_paths, verbose=True):
+ assert len(gt_paths) == len(prediction_paths)
+ msas, sa50s, sa75s = [], [], []
+
+ for gt_path, pred_path in tqdm(
+ zip(gt_paths, prediction_paths), desc="Evaluate predictions", total=len(gt_paths), disable=not verbose
+ ):
+ assert os.path.exists(gt_path), gt_path
+ assert os.path.exists(pred_path), pred_path
+
+ gt = imageio.imread(gt_path)
+ gt = label(gt)
+ pred = imageio.imread(pred_path)
+
+ msa, scores = mean_segmentation_accuracy(pred, gt, return_accuracies=True)
+ sa50, sa75 = scores[0], scores[5]
+ msas.append(msa), sa50s.append(sa50), sa75s.append(sa75)
+
+ return msas, sa50s, sa75s
+
+
+def run_evaluation(
+ gt_paths: List[Union[os.PathLike, str]],
+ prediction_paths: List[Union[os.PathLike, str]],
+ save_path: Optional[Union[os.PathLike, str]] = None,
+ verbose: bool = True,
+) -> pd.DataFrame:
+ """Run evaluation for instance segmentation predictions.
+
+ Args:
+ gt_folder: The folder with ground-truth images.
+ prediction_folder: The folder with the instance segmentations to evaluate.
+ save_path: Optional path for saving the results.
+ pattern: Optional pattern for selecting the images to evaluate via glob.
+ By default all images with ending .tif will be evaluated.
+ verbose: Whether to print the progress.
+
+ Returns:
+ A DataFrame that contains the evaluation results.
+ """
+ assert len(gt_paths) == len(prediction_paths)
+ # if a save_path is given and it already exists then just load it instead of running the eval
+ if save_path is not None and os.path.exists(save_path):
+ return pd.read_csv(save_path)
+
+ msas, sa50s, sa75s = _run_evaluation(gt_paths, prediction_paths, verbose=verbose)
+
+ results = pd.DataFrame.from_dict({
+ "msa": [np.mean(msas)],
+ "sa50": [np.mean(sa50s)],
+ "sa75": [np.mean(sa75s)],
+ })
+
+ if save_path is not None:
+ os.makedirs(Path(save_path).parent, exist_ok=True)
+ results.to_csv(save_path, index=False)
+
+ return results
+
+
+# TODO function to evaluate full experiment and resave in one table
diff --git a/micro_sam/evaluation/experiments.py b/micro_sam/evaluation/experiments.py
new file mode 100644
index 00000000..4646af52
--- /dev/null
+++ b/micro_sam/evaluation/experiments.py
@@ -0,0 +1,83 @@
+"""Predefined experiment settings for experiments with different prompt strategies.
+"""
+
+from typing import Dict, List, Optional
+
+# TODO fully define the dict type
+ExperimentSetting = Dict
+ExperimentSettings = List[ExperimentSetting]
+"""@private"""
+
+
+def full_experiment_settings(
+ use_boxes: bool = False,
+ positive_range: Optional[List[int]] = None,
+ negative_range: Optional[List[int]] = None,
+) -> ExperimentSettings:
+ """The full experiment settings.
+
+ Args:
+ use_boxes: Whether to run the experiments with or without boxes.
+ positive_range: The different number of positive points that will be used.
+ By defaul the values are set to [1, 2, 4, 8, 16].
+ negative_range: The different number of negative points that will be used.
+ By defaul the values are set to [0, 1, 2, 4, 8, 16].
+
+ Returns:
+ The list of experiment settings.
+ """
+ experiment_settings = []
+ if use_boxes:
+ experiment_settings.append(
+ {"use_points": False, "use_boxes": True, "n_positives": 0, "n_negatives": 0}
+ )
+
+ # set default values for the ranges if none were passed
+ if positive_range is None:
+ positive_range = [1, 2, 4, 8, 16]
+ if negative_range is None:
+ negative_range = [0, 1, 2, 4, 8, 16]
+
+ for n_positives in positive_range:
+ for n_negatives in negative_range:
+ if n_positives == 0 and n_negatives == 0:
+ continue
+ experiment_settings.append(
+ {"use_points": True, "use_boxes": use_boxes, "n_positives": n_positives, "n_negatives": n_negatives}
+ )
+
+ return experiment_settings
+
+
+def default_experiment_settings() -> ExperimentSettings:
+ """The three default experiment settings.
+
+ For the default experiments we use a single positive prompt,
+ two positive and four negative prompts and box prompts.
+
+ Returns:
+ The list of experiment settings.
+ """
+ experiment_settings = [
+ {"use_points": True, "use_boxes": False, "n_positives": 1, "n_negatives": 0}, # p1-n0
+ {"use_points": True, "use_boxes": False, "n_positives": 2, "n_negatives": 4}, # p2-n4
+ {"use_points": True, "use_boxes": False, "n_positives": 4, "n_negatives": 8}, # p4-n8
+ {"use_points": False, "use_boxes": True, "n_positives": 0, "n_negatives": 0}, # only box prompts
+ ]
+ return experiment_settings
+
+
+def get_experiment_setting_name(setting: ExperimentSetting) -> str:
+ """Get the name for the given experiment setting.
+
+ Args:
+ setting: The experiment setting.
+ Returns:
+ The name for this experiment setting.
+ """
+ use_points, use_boxes = setting["use_points"], setting["use_boxes"]
+ assert use_points or use_boxes
+ prefix = "points" if use_points else "box"
+ pos, neg = setting["n_positives"], setting["n_negatives"]
+ name = f"p{pos}-n{neg}" if use_points else "p0-n0"
+ return f"{prefix}/{name}"
diff --git a/micro_sam/evaluation/inference.py b/micro_sam/evaluation/inference.py
new file mode 100644
index 00000000..e490b6c6
--- /dev/null
+++ b/micro_sam/evaluation/inference.py
@@ -0,0 +1,553 @@
+"""Inference with Segment Anything models and different prompt strategies.
+"""
+
+import os
+import pickle
+import warnings
+
+from copy import deepcopy
+from typing import Any, Dict, List, Optional, Union
+
+import imageio.v3 as imageio
+import numpy as np
+import torch
+
+from skimage.segmentation import relabel_sequential
+from tqdm import tqdm
+
+from segment_anything import SamPredictor
+from segment_anything.utils.transforms import ResizeLongestSide
+
+from .. import util as util
+from ..instance_segmentation import mask_data_to_segmentation
+from ..prompt_generators import PointAndBoxPromptGenerator, IterativePromptGenerator
+from ..training import get_trainable_sam_model, ConvertToSamInputs
+
+
+def _load_prompts(
+ cached_point_prompts, save_point_prompts,
+ cached_box_prompts, save_box_prompts,
+ image_name
+):
+
+ def load_prompt_type(cached_prompts, save_prompts):
+ # Check if we have saved prompts.
+ if cached_prompts is None or save_prompts: # we don't have cached prompts
+ return cached_prompts, None
+
+ # we have cached prompts, but they have not been loaded yet
+ if isinstance(cached_prompts, str):
+ with open(cached_prompts, "rb") as f:
+ cached_prompts = pickle.load(f)
+
+ prompts = cached_prompts[image_name]
+ return cached_prompts, prompts
+
+ cached_point_prompts, point_prompts = load_prompt_type(cached_point_prompts, save_point_prompts)
+ cached_box_prompts, box_prompts = load_prompt_type(cached_box_prompts, save_box_prompts)
+
+ # we don't have anything cached
+ if point_prompts is None and box_prompts is None:
+ return None, cached_point_prompts, cached_box_prompts
+
+ if point_prompts is None:
+ input_point, input_label = [], []
+ else:
+ input_point, input_label = point_prompts
+
+ if box_prompts is None:
+ input_box = []
+ else:
+ input_box = box_prompts
+
+ prompts = (input_point, input_label, input_box)
+ return prompts, cached_point_prompts, cached_box_prompts
+
+
+def _get_batched_prompts(
+ gt,
+ gt_ids,
+ use_points,
+ use_boxes,
+ n_positives,
+ n_negatives,
+ dilation,
+ transform_function,
+):
+ input_point, input_label, input_box = [], [], []
+
+ # Initialize the prompt generator.
+ center_coordinates, bbox_coordinates = util.get_centers_and_bounding_boxes(gt)
+ prompt_generator = PointAndBoxPromptGenerator(
+ n_positive_points=n_positives, n_negative_points=n_negatives,
+ dilation_strength=dilation, get_point_prompts=use_points,
+ get_box_prompts=use_boxes
+ )
+
+ # Iterate over the gt ids, generate the corresponding prompts and combine them to batched input.
+ for gt_id in gt_ids:
+ centers, bboxes = center_coordinates.get(gt_id), bbox_coordinates.get(gt_id)
+ input_point_list, input_label_list, input_box_list, objm = prompt_generator(gt, gt_id, bboxes, centers)
+
+ if use_boxes:
+ # indexes hard-coded to adapt with SAM's bbox format
+ # default format: [a, b, c, d] -> SAM's format: [b, a, d, c]
+ _ib = [input_box_list[0][1], input_box_list[0][0],
+ input_box_list[0][3], input_box_list[0][2]]
+ # transform boxes to the expected format - see predictor.predict function for details
+ _ib = transform_function.apply_boxes(np.array(_ib), gt.shape)
+ input_box.append(_ib)
+
+ if use_points:
+ assert len(input_point_list) == (n_positives + n_negatives)
+ _ip = [ip[::-1] for ip in input_point_list] # to match the coordinate system used by SAM
+
+ # transform coords to the expected format - see predictor.predict function for details
+ _ip = transform_function.apply_coords(np.array(_ip), gt.shape)
+ input_point.append(_ip)
+ input_label.append(input_label_list)
+
+ return input_point, input_label, input_box
+
+
+def _run_inference_with_prompts_for_image(
+ predictor,
+ gt,
+ use_points,
+ use_boxes,
+ n_positives,
+ n_negatives,
+ dilation,
+ batch_size,
+ cached_prompts,
+):
+ # We need the resize transformation for the expected model input size.
+ transform_function = ResizeLongestSide(1024)
+ gt_ids = np.unique(gt)[1:]
+
+ if cached_prompts is None:
+ input_point, input_label, input_box = _get_batched_prompts(
+ gt, gt_ids, use_points, use_boxes, n_positives, n_negatives, dilation, transform_function,
+ )
+ else:
+ input_point, input_label, input_box = cached_prompts
+
+ # Make a copy of the point prompts to return them at the end.
+ prompts = deepcopy((input_point, input_label, input_box))
+
+ # Transform the prompts into batches
+ device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
+ input_point = torch.tensor(np.array(input_point)).to(device) if len(input_point) > 0 else None
+ input_label = torch.tensor(np.array(input_label)).to(device) if len(input_label) > 0 else None
+ input_box = torch.tensor(np.array(input_box)).to(device) if len(input_box) > 0 else None
+
+ # Use multi-masking only if we have a single positive point without box
+ multimasking = False
+ if not use_boxes and (n_positives == 1 and n_negatives == 0):
+ multimasking = True
+
+ # Run the batched inference.
+ n_samples = input_box.shape[0] if input_point is None else input_point.shape[0]
+ n_batches = int(np.ceil(float(n_samples) / batch_size))
+ masks, ious = [], []
+ with torch.no_grad():
+ for batch_idx in range(n_batches):
+ batch_start = batch_idx * batch_size
+ batch_stop = min((batch_idx + 1) * batch_size, n_samples)
+
+ batch_points = None if input_point is None else input_point[batch_start:batch_stop]
+ batch_labels = None if input_label is None else input_label[batch_start:batch_stop]
+ batch_boxes = None if input_box is None else input_box[batch_start:batch_stop]
+
+ batch_masks, batch_ious, _ = predictor.predict_torch(
+ point_coords=batch_points, point_labels=batch_labels,
+ boxes=batch_boxes, multimask_output=multimasking
+ )
+ masks.append(batch_masks)
+ ious.append(batch_ious)
+ masks = torch.cat(masks)
+ ious = torch.cat(ious)
+ assert len(masks) == len(ious) == n_samples
+
+ # TODO we should actually use non-max suppression here
+ # I will implement it somewhere to have it refactored
+ instance_labels = np.zeros_like(gt, dtype=int)
+ for m, iou, gt_idx in zip(masks, ious, gt_ids):
+ best_idx = torch.argmax(iou)
+ best_mask = m[best_idx]
+ instance_labels[best_mask.detach().cpu().numpy()] = gt_idx
+
+ return instance_labels, prompts
+
+
+def get_predictor(
+ checkpoint_path: Union[str, os.PathLike],
+ model_type: str,
+ return_state: bool = False,
+ is_custom_model: Optional[bool] = None,
+) -> SamPredictor:
+ """Get the segment anything predictor from an exported or custom checkpoint.
+
+ Args:
+ checkpoint_path: The checkpoint filepath.
+ model_type: The type of the model, either vit_h, vit_b or vit_l.
+ return_state: Whether to return the complete state of the checkpoint in addtion to the predictor.
+ is_custom_model: Whether this is a custom model or not.
+ Returns:
+ The segment anything predictor.
+ """
+ # By default we check if the model follows the torch_em checkpint naming scheme to check whether it is a
+ # custom model or not. This can be over-ridden by passing True or False for is_custom_model.
+ is_custom_model = checkpoint_path.split("/")[-1] == "best.pt" if is_custom_model is None else is_custom_model
+
+ if is_custom_model: # Finetuned SAM model
+ predictor = util.get_custom_sam_model(
+ checkpoint_path=checkpoint_path, model_type=model_type, return_state=return_state
+ )
+ else: # Vanilla SAM model
+ assert not return_state
+ predictor = util.get_sam_model(model_type=model_type, checkpoint_path=checkpoint_path) # type: ignore
+ return predictor
+
+
+def precompute_all_embeddings(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+) -> None:
+ """Precompute all image embeddings.
+
+ To enable running different inference tasks in parallel afterwards.
+
+ Args:
+ predictor: The SegmentAnything predictor.
+ image_paths: The image file paths.
+ embedding_dir: The directory where the embeddings will be saved.
+ """
+ for image_path in tqdm(image_paths, desc="Precompute embeddings"):
+ image_name = os.path.basename(image_path)
+ im = imageio.imread(image_path)
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ util.precompute_image_embeddings(predictor, im, embedding_path, ndim=2)
+
+
+def _precompute_prompts(gt_path, use_points, use_boxes, n_positives, n_negatives, dilation, transform_function):
+ name = os.path.basename(gt_path)
+
+ gt = imageio.imread(gt_path).astype("uint32")
+ gt = relabel_sequential(gt)[0]
+ gt_ids = np.unique(gt)[1:]
+
+ input_point, input_label, input_box = _get_batched_prompts(
+ gt, gt_ids, use_points, use_boxes, n_positives, n_negatives, dilation, transform_function
+ )
+
+ if use_boxes and not use_points:
+ return name, input_box
+ return name, (input_point, input_label)
+
+
+def precompute_all_prompts(
+ gt_paths: List[Union[str, os.PathLike]],
+ prompt_save_dir: Union[str, os.PathLike],
+ prompt_settings: List[Dict[str, Any]],
+) -> None:
+ """Precompute all point prompts.
+
+ To enable running different inference tasks in parallel afterwards.
+
+ Args:
+ gt_paths: The file paths to the ground-truth segmentations.
+ prompt_save_dir: The directory where the prompt files will be saved.
+ prompt_settings: The settings for which the prompts will be computed.
+ """
+ os.makedirs(prompt_save_dir, exist_ok=True)
+ transform_function = ResizeLongestSide(1024)
+
+ for settings in tqdm(prompt_settings, desc="Precompute prompts"):
+
+ use_points, use_boxes = settings["use_points"], settings["use_boxes"]
+ n_positives, n_negatives = settings["n_positives"], settings["n_negatives"]
+ dilation = settings.get("dilation", 5)
+
+ # check if the prompts were already computed
+ if use_boxes and not use_points:
+ prompt_save_path = os.path.join(prompt_save_dir, "boxes.pkl")
+ else:
+ prompt_save_path = os.path.join(prompt_save_dir, f"points-p{n_positives}-n{n_negatives}.pkl")
+ if os.path.exists(prompt_save_path):
+ continue
+
+ results = []
+ for gt_path in tqdm(gt_paths, desc=f"Precompute prompts for p{n_positives}-n{n_negatives}"):
+ prompts = _precompute_prompts(
+ gt_path,
+ use_points=use_points,
+ use_boxes=use_boxes,
+ n_positives=n_positives,
+ n_negatives=n_negatives,
+ dilation=dilation,
+ transform_function=transform_function,
+ )
+ results.append(prompts)
+
+ saved_prompts = {res[0]: res[1] for res in results}
+ with open(prompt_save_path, "wb") as f:
+ pickle.dump(saved_prompts, f)
+
+
+def _get_prompt_caching(prompt_save_dir, use_points, use_boxes, n_positives, n_negatives):
+
+ def get_prompt_type_caching(use_type, save_name):
+ if not use_type:
+ return None, False, None
+
+ prompt_save_path = os.path.join(prompt_save_dir, save_name)
+ if os.path.exists(prompt_save_path):
+ print("Using precomputed prompts from", prompt_save_path)
+ # We delay loading the prompts, so we only have to load them once they're needed the first time.
+ # This avoids loading the prompts (which are in a big pickle file) if all predictions are done already.
+ cached_prompts = prompt_save_path
+ save_prompts = False
+ else:
+ print("Saving prompts in", prompt_save_path)
+ cached_prompts = {}
+ save_prompts = True
+ return cached_prompts, save_prompts, prompt_save_path
+
+ # Check if prompt serialization is enabled.
+ # If it is then load the prompts if they are already cached and otherwise store them.
+ if prompt_save_dir is None:
+ print("Prompts are not cached.")
+ cached_point_prompts, cached_box_prompts = None, None
+ save_point_prompts, save_box_prompts = False, False
+ point_prompt_save_path, box_prompt_save_path = None, None
+ else:
+ cached_point_prompts, save_point_prompts, point_prompt_save_path = get_prompt_type_caching(
+ use_points, f"points-p{n_positives}-n{n_negatives}.pkl"
+ )
+ cached_box_prompts, save_box_prompts, box_prompt_save_path = get_prompt_type_caching(
+ use_boxes, "boxes.pkl"
+ )
+
+ return (cached_point_prompts, save_point_prompts, point_prompt_save_path,
+ cached_box_prompts, save_box_prompts, box_prompt_save_path)
+
+
+def run_inference_with_prompts(
+ predictor: SamPredictor,
+ image_paths: List[Union[str, os.PathLike]],
+ gt_paths: List[Union[str, os.PathLike]],
+ embedding_dir: Union[str, os.PathLike],
+ prediction_dir: Union[str, os.PathLike],
+ use_points: bool,
+ use_boxes: bool,
+ n_positives: int,
+ n_negatives: int,
+ dilation: int = 5,
+ prompt_save_dir: Optional[Union[str, os.PathLike]] = None,
+ batch_size: int = 512,
+) -> None:
+ """Run segment anything inference for multiple images using prompts derived form groundtruth.
+
+ Args:
+ predictor: The SegmentAnything predictor.
+ image_paths: The image file paths.
+ gt_paths: The ground-truth segmentation file paths.
+ embedding_dir: The directory where the image embddings will be saved or are already saved.
+ use_points: Whether to use point prompts.
+ use_boxes: Whetehr to use box prompts
+ n_positives: The number of positive point prompts that will be sampled.
+ n_negativess: The number of negative point prompts that will be sampled.
+ dilation: The dilation factor for the radius around the ground-truth object
+ around which points will not be sampled.
+ prompt_save_dir: The directory where point prompts will be saved or are already saved.
+ This enables running multiple experiments in a reproducible manner.
+ batch_size: The batch size used for batched prediction.
+ """
+ if not (use_points or use_boxes):
+ raise ValueError("You need to use at least one of point or box prompts.")
+
+ if len(image_paths) != len(gt_paths):
+ raise ValueError(f"Expect same number of images and gt images, got {len(image_paths)}, {len(gt_paths)}")
+
+ (cached_point_prompts, save_point_prompts, point_prompt_save_path,
+ cached_box_prompts, save_box_prompts, box_prompt_save_path) = _get_prompt_caching(
+ prompt_save_dir, use_points, use_boxes, n_positives, n_negatives
+ )
+
+ for image_path, gt_path in tqdm(
+ zip(image_paths, gt_paths), total=len(image_paths), desc="Run inference with prompts"
+ ):
+ image_name = os.path.basename(image_path)
+ label_name = os.path.basename(gt_path)
+
+ # We skip the images that already have been segmented.
+ prediction_path = os.path.join(prediction_dir, image_name)
+ if os.path.exists(prediction_path):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ assert os.path.exists(gt_path), gt_path
+
+ im = imageio.imread(image_path)
+ gt = imageio.imread(gt_path).astype("uint32")
+ gt = relabel_sequential(gt)[0]
+
+ embedding_path = os.path.join(embedding_dir, f"{os.path.splitext(image_name)[0]}.zarr")
+ image_embeddings = util.precompute_image_embeddings(predictor, im, embedding_path, ndim=2)
+ util.set_precomputed(predictor, image_embeddings)
+
+ this_prompts, cached_point_prompts, cached_box_prompts = _load_prompts(
+ cached_point_prompts, save_point_prompts,
+ cached_box_prompts, save_box_prompts,
+ label_name
+ )
+ instances, this_prompts = _run_inference_with_prompts_for_image(
+ predictor, gt, n_positives=n_positives, n_negatives=n_negatives,
+ dilation=dilation, use_points=use_points, use_boxes=use_boxes,
+ batch_size=batch_size, cached_prompts=this_prompts
+ )
+
+ if save_point_prompts:
+ cached_point_prompts[label_name] = this_prompts[:2]
+ if save_box_prompts:
+ cached_box_prompts[label_name] = this_prompts[-1]
+
+ # It's important to compress here, otherwise the predictions would take up a lot of space.
+ imageio.imwrite(prediction_path, instances, compression=5)
+
+ # Save the prompts if we run experiments with prompt caching and have computed them
+ # for the first time.
+ if save_point_prompts:
+ with open(point_prompt_save_path, "wb") as f:
+ pickle.dump(cached_point_prompts, f)
+ if save_box_prompts:
+ with open(box_prompt_save_path, "wb") as f:
+ pickle.dump(cached_box_prompts, f)
+
+
+def _save_segmentation(masks, prediction_path):
+ # masks to segmentation
+ masks = masks.cpu().numpy().squeeze().astype("bool")
+ shape = masks.shape[-2:]
+ masks = [{"segmentation": mask, "area": mask.sum()} for mask in masks]
+ segmentation = mask_data_to_segmentation(masks, shape, with_background=True)
+ imageio.imwrite(prediction_path, segmentation)
+
+
+def _run_inference_with_iterative_prompting_for_image(
+ model,
+ image,
+ gt,
+ n_iterations,
+ device,
+ use_boxes,
+ prediction_paths,
+ batch_size,
+):
+ assert len(prediction_paths) == n_iterations, f"{len(prediction_paths)}, {n_iterations}"
+ to_sam_inputs = ConvertToSamInputs()
+
+ image = torch.from_numpy(
+ image[None, None] if image.ndim == 2 else image[None]
+ )
+ gt = torch.from_numpy(gt[None].astype("int32"))
+
+ n_pos = 0 if use_boxes else 1
+ batched_inputs, sampled_ids = to_sam_inputs(image, gt, n_pos=n_pos, n_neg=0, get_boxes=use_boxes)
+
+ input_images = torch.stack([model.preprocess(x=x["image"].to(device)) for x in batched_inputs], dim=0)
+ image_embeddings = model.image_embeddings_oft(input_images)
+
+ multimasking = n_pos == 1
+ prompt_generator = IterativePromptGenerator()
+
+ n_samples = len(sampled_ids[0])
+ n_batches = int(np.ceil(float(n_samples) / batch_size))
+
+ for iteration in range(n_iterations):
+ final_masks = []
+ for batch_idx in range(n_batches):
+ batch_start = batch_idx * batch_size
+ batch_stop = min((batch_idx + 1) * batch_size, n_samples)
+
+ this_batched_inputs = [{
+ k: v[batch_start:batch_stop] if k in ("point_coords", "point_labels") else v
+ for k, v in batched_inputs[0].items()
+ }]
+
+ sampled_binary_y = torch.stack([
+ torch.stack([_gt == idx for idx in sampled[batch_start:batch_stop]])[:, None]
+ for _gt, sampled in zip(gt, sampled_ids)
+ ]).to(torch.float32)
+
+ batched_outputs = model(
+ this_batched_inputs,
+ multimask_output=multimasking if iteration == 0 else False,
+ image_embeddings=image_embeddings
+ )
+
+ masks, logits_masks = [], []
+ for m in batched_outputs:
+ mask, l_mask = [], []
+ for _m, _l, _iou in zip(m["masks"], m["low_res_masks"], m["iou_predictions"]):
+ best_iou_idx = torch.argmax(_iou)
+ mask.append(torch.sigmoid(_m[best_iou_idx][None]))
+ l_mask.append(_l[best_iou_idx][None])
+ mask, l_mask = torch.stack(mask), torch.stack(l_mask)
+ masks.append(mask)
+ logits_masks.append(l_mask)
+
+ masks, logits_masks = torch.stack(masks), torch.stack(logits_masks)
+ masks = (masks > 0.5).to(torch.float32)
+ final_masks.append(masks)
+
+ for _pred, _gt, _inp, logits in zip(masks, sampled_binary_y, this_batched_inputs, logits_masks):
+ next_coords, next_labels = prompt_generator(_gt, _pred, _inp["point_coords"], _inp["point_labels"])
+ _inp["point_coords"], _inp["point_labels"], _inp["mask_inputs"] = next_coords, next_labels, logits
+
+ final_masks = torch.cat(final_masks, dim=1)
+ _save_segmentation(final_masks, prediction_paths[iteration])
+
+
+def run_inference_with_iterative_prompting(
+ checkpoint_path: Union[str, os.PathLike],
+ model_type: str,
+ image_paths: List[Union[str, os.PathLike]],
+ gt_paths: List[Union[str, os.PathLike]],
+ prediction_root: Union[str, os.PathLike],
+ use_boxes: bool,
+ n_iterations: int = 8,
+ batch_size: int = 32,
+) -> None:
+ """@private"""
+ warnings.warn("The iterative prompting functionality is not working correctly yet.")
+
+ device = torch.device("cuda")
+ model = get_trainable_sam_model(model_type, checkpoint_path)
+
+ # create all prediction folders
+ for i in range(n_iterations):
+ os.makedirs(os.path.join(prediction_root, f"iteration{i:02}"), exist_ok=True)
+
+ for image_path, gt_path in tqdm(
+ zip(image_paths, gt_paths), total=len(image_paths), desc="Run inference with prompts"
+ ):
+ image_name = os.path.basename(image_path)
+
+ prediction_paths = [os.path.join(prediction_root, f"iteration{i:02}", image_name) for i in range(n_iterations)]
+ if all(os.path.exists(prediction_path) for prediction_path in prediction_paths):
+ continue
+
+ assert os.path.exists(image_path), image_path
+ assert os.path.exists(gt_path), gt_path
+
+ image = imageio.imread(image_path)
+ gt = imageio.imread(gt_path).astype("uint32")
+ gt = relabel_sequential(gt)[0]
+
+ with torch.no_grad():
+ _run_inference_with_iterative_prompting_for_image(
+ model, image, gt, n_iterations, device, use_boxes, prediction_paths, batch_size,
+ )
diff --git a/micro_sam/evaluation/livecell.py b/micro_sam/evaluation/livecell.py
new file mode 100644
index 00000000..752a0359
--- /dev/null
+++ b/micro_sam/evaluation/livecell.py
@@ -0,0 +1,362 @@
+"""Inference and evaluation for the [LiveCELL dataset](https://www.nature.com/articles/s41592-021-01249-6) and
+the different cell lines contained in it.
+"""
+
+import argparse
+import json
+import os
+
+from glob import glob
+from typing import List, Optional, Union
+
+import numpy as np
+import pandas as pd
+
+from segment_anything import SamPredictor
+from tqdm import tqdm
+
+from ..instance_segmentation import AutomaticMaskGenerator, EmbeddingMaskGenerator
+from . import automatic_mask_generation, inference, evaluation
+from .experiments import default_experiment_settings, full_experiment_settings
+
+CELL_TYPES = ["A172", "BT474", "BV2", "Huh7", "MCF7", "SHSY5Y", "SkBr3", "SKOV3"]
+
+
+#
+# Inference
+#
+
+
+def _get_livecell_paths(input_folder, split="test", n_val_per_cell_type=None):
+ assert split in ["val", "test"]
+ assert os.path.exists(input_folder), "Please download the LIVECell Dataset"
+
+ if split == "test":
+
+ img_dir = os.path.join(input_folder, "images", "livecell_test_images")
+ assert os.path.exists(img_dir), "The LIVECell Dataset is incomplete"
+ gt_dir = os.path.join(input_folder, "annotations", "livecell_test_images")
+ assert os.path.exists(gt_dir), "The LIVECell Dataset is incomplete"
+ image_paths, gt_paths = [], []
+ for ctype in CELL_TYPES:
+ for img_path in glob(os.path.join(img_dir, f"{ctype}*")):
+ image_paths.append(img_path)
+ img_name = os.path.basename(img_path)
+ gt_path = os.path.join(gt_dir, ctype, img_name)
+ assert os.path.exists(gt_path), gt_path
+ gt_paths.append(gt_path)
+ else:
+
+ with open(os.path.join(input_folder, "val.json")) as f:
+ data = json.load(f)
+ livecell_val_ids = [i["file_name"] for i in data["images"]]
+
+ img_dir = os.path.join(input_folder, "images", "livecell_train_val_images")
+ assert os.path.exists(img_dir), "The LIVECell Dataset is incomplete"
+ gt_dir = os.path.join(input_folder, "annotations", "livecell_train_val_images")
+ assert os.path.exists(gt_dir), "The LIVECell Dataset is incomplete"
+
+ image_paths, gt_paths = [], []
+ count_per_cell_type = {ct: 0 for ct in CELL_TYPES}
+
+ for img_name in livecell_val_ids:
+ cell_type = img_name.split("_")[0]
+ if n_val_per_cell_type is not None and count_per_cell_type[cell_type] >= n_val_per_cell_type:
+ continue
+
+ image_paths.append(os.path.join(img_dir, img_name))
+ gt_paths.append(os.path.join(gt_dir, cell_type, img_name))
+ count_per_cell_type[cell_type] += 1
+
+ return image_paths, gt_paths
+
+
+def livecell_inference(
+ checkpoint: Union[str, os.PathLike],
+ input_folder: Union[str, os.PathLike],
+ model_type: str,
+ experiment_folder: Union[str, os.PathLike],
+ use_points: bool,
+ use_boxes: bool,
+ n_positives: Optional[int] = None,
+ n_negatives: Optional[int] = None,
+ prompt_folder: Optional[Union[str, os.PathLike]] = None,
+ predictor: Optional[SamPredictor] = None,
+) -> None:
+ """Run inference for livecell with a fixed prompt setting.
+
+ Args:
+ checkpoint: The segment anything model checkpoint.
+ input_folder: The folder with the livecell data.
+ model_type: The type of the segment anything model.
+ experiment_folder: The folder where to save all data associated with the experiment.
+ use_points: Whether to use point prompts.
+ use_boxes: Whether to use box prompts.
+ n_positives: The number of positive point prompts.
+ n_negatives: The number of negative point prompts.
+ prompt_folder: The folder where the prompts should be saved.
+ predictor: The segment anything predictor.
+ """
+ image_paths, gt_paths = _get_livecell_paths(input_folder)
+ if predictor is None:
+ predictor = inference.get_predictor(checkpoint, model_type)
+
+ if use_boxes and use_points:
+ assert (n_positives is not None) and (n_negatives is not None)
+ setting_name = f"box/p{n_positives}-n{n_negatives}"
+ elif use_boxes:
+ setting_name = "box/p0-n0"
+ elif use_points:
+ assert (n_positives is not None) and (n_negatives is not None)
+ setting_name = f"points/p{n_positives}-n{n_negatives}"
+ else:
+ raise ValueError("You need to use at least one of point or box prompts.")
+
+ # we organize all folders with data from this experiment beneath 'experiment_folder'
+ prediction_folder = os.path.join(experiment_folder, setting_name) # where the predicted segmentations are saved
+ os.makedirs(prediction_folder, exist_ok=True)
+ embedding_folder = os.path.join(experiment_folder, "embeddings") # where the precomputed embeddings are saved
+ os.makedirs(embedding_folder, exist_ok=True)
+
+ # NOTE: we can pass an external prompt folder, to make re-use prompts from another experiment
+ # for reproducibility / fair comparison of results
+ if prompt_folder is None:
+ prompt_folder = os.path.join(experiment_folder, "prompts")
+ os.makedirs(prompt_folder, exist_ok=True)
+
+ inference.run_inference_with_prompts(
+ predictor,
+ image_paths,
+ gt_paths,
+ embedding_dir=embedding_folder,
+ prediction_dir=prediction_folder,
+ prompt_save_dir=prompt_folder,
+ use_points=use_points,
+ use_boxes=use_boxes,
+ n_positives=n_positives,
+ n_negatives=n_negatives,
+ )
+
+
+def run_livecell_amg(
+ checkpoint: Union[str, os.PathLike],
+ input_folder: Union[str, os.PathLike],
+ model_type: str,
+ experiment_folder: Union[str, os.PathLike],
+ iou_thresh_values: Optional[List[float]] = None,
+ stability_score_values: Optional[List[float]] = None,
+ verbose_gs: bool = False,
+ n_val_per_cell_type: int = 25,
+ use_mws: bool = False,
+) -> None:
+ """Run automatic mask generation grid-search and inference for livecell.
+
+ Args:
+ checkpoint: The segment anything model checkpoint.
+ input_folder: The folder with the livecell data.
+ model_type: The type of the segmenta anything model.
+ experiment_folder: The folder where to save all data associated with the experiment.
+ iou_thresh_values: The values for `pred_iou_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ stability_score_values: The values for `stability_score_thresh` used in the gridsearch.
+ By default values in the range from 0.6 to 0.9 with a stepsize of 0.025 will be used.
+ verbose_gs: Whether to run the gridsearch for individual images in a verbose mode.
+ n_val_per_cell_type: The number of validation images per cell type.
+ use_mws: Whether to use the mutex watershed based automatic mask generator approach.
+ """
+ embedding_folder = os.path.join(experiment_folder, "embeddings") # where the precomputed embeddings are saved
+ os.makedirs(embedding_folder, exist_ok=True)
+
+ if use_mws:
+ amg_prefix = "amg_mws"
+ AMG = EmbeddingMaskGenerator
+ else:
+ amg_prefix = "amg"
+ AMG = AutomaticMaskGenerator
+
+ # where the predictions are saved
+ prediction_folder = os.path.join(experiment_folder, amg_prefix, "inference")
+ os.makedirs(prediction_folder, exist_ok=True)
+
+ # where the grid-search results are saved
+ gs_result_folder = os.path.join(experiment_folder, amg_prefix, "grid_search")
+ os.makedirs(gs_result_folder, exist_ok=True)
+
+ val_image_paths, val_gt_paths = _get_livecell_paths(input_folder, "val", n_val_per_cell_type=n_val_per_cell_type)
+ test_image_paths, _ = _get_livecell_paths(input_folder, "test")
+
+ predictor = inference.get_predictor(checkpoint, model_type)
+ automatic_mask_generation.run_amg_grid_search_and_inference(
+ predictor, val_image_paths, val_gt_paths, test_image_paths,
+ embedding_folder, prediction_folder, gs_result_folder,
+ iou_thresh_values=iou_thresh_values, stability_score_values=stability_score_values,
+ AMG=AMG, verbose_gs=verbose_gs,
+ )
+
+
+def _run_multiple_prompt_settings(args, prompt_settings):
+ predictor = inference.get_predictor(args.ckpt, args.model)
+ for settings in prompt_settings:
+ livecell_inference(
+ args.ckpt,
+ args.input,
+ args.model,
+ args.experiment_folder,
+ use_points=settings["use_points"],
+ use_boxes=settings["use_boxes"],
+ n_positives=settings["n_positives"],
+ n_negatives=settings["n_negatives"],
+ prompt_folder=args.prompt_folder,
+ predictor=predictor
+ )
+
+
+def run_livecell_inference() -> None:
+ """Run LiveCELL inference with command line tool."""
+ parser = argparse.ArgumentParser()
+
+ # the checkpoint, input and experiment folder
+ parser.add_argument("-c", "--ckpt", type=str, required=True,
+ help="Provide model checkpoints (vanilla / finetuned).")
+ parser.add_argument("-i", "--input", type=str, required=True,
+ help="Provide the data directory for LIVECell Dataset.")
+ parser.add_argument("-e", "--experiment_folder", type=str, required=True,
+ help="Provide the path where all data for the inference run will be stored.")
+ parser.add_argument("-m", "--model", type=str, required=True,
+ help="Pass the checkpoint-specific model name being used for inference.")
+
+ # the experiment type:
+ # - default settings (p1-n0, p2-n4, box)
+ # - full experiment (ranges: p:1-16, n:0-16)
+ # - automatic mask generation (auto)
+ # if none of the two are active then the prompt setting arguments will be parsed
+ # and used to run inference for a single prompt setting
+ parser.add_argument("-f", "--full_experiment", action="store_true")
+ parser.add_argument("-d", "--default_experiment", action="store_true")
+ parser.add_argument("-a", "--auto_mask_generation", action="store_true")
+
+ # the prompt settings for an individual inference run
+ parser.add_argument("--box", action="store_true", help="Activate box-prompted based inference")
+ parser.add_argument("--points", action="store_true", help="Activate point-prompt based inference")
+ parser.add_argument("-p", "--positive", type=int, default=1, help="No. of positive prompts")
+ parser.add_argument("-n", "--negative", type=int, default=0, help="No. of negative prompts")
+
+ # optional external prompt folder
+ parser.add_argument("--prompt_folder", help="")
+
+ args = parser.parse_args()
+ if sum([args.full_experiment, args.default_experiment, args.auto_mask_generation]) > 2:
+ raise ValueError("Can only run one of 'full_experiment', 'default_experiment' or 'auto_mask_generation'.")
+
+ if args.full_experiment:
+ prompt_settings = full_experiment_settings(args.box)
+ _run_multiple_prompt_settings(args, prompt_settings)
+ elif args.default_experiment:
+ prompt_settings = default_experiment_settings()
+ _run_multiple_prompt_settings(args, prompt_settings)
+ elif args.auto_mask_generation:
+ run_livecell_amg(args.ckpt, args.input, args.model, args.experiment_folder)
+ else:
+ livecell_inference(
+ args.ckpt, args.input, args.model, args.experiment_folder,
+ args.points, args.box, args.positive, args.negative, args.prompt_folder,
+ )
+
+
+#
+# Evaluation
+#
+
+
+def evaluate_livecell_predictions(
+ gt_dir: Union[os.PathLike, str],
+ pred_dir: Union[os.PathLike, str],
+ verbose: bool,
+) -> None:
+ """Evaluate LiveCELL predictions.
+
+ Args:
+ gt_dir: The folder with the groundtruth segmentations.
+ pred_dir: The folder with the segmentation predictions.
+ verbose: Whether to run the evaluation in verbose mode.
+ """
+ assert os.path.exists(gt_dir), gt_dir
+ assert os.path.exists(pred_dir), pred_dir
+
+ msas, sa50s, sa75s = [], [], []
+ msas_ct, sa50s_ct, sa75s_ct = [], [], []
+
+ for ct in tqdm(CELL_TYPES, desc="Evaluate livecell predictions", disable=not verbose):
+
+ gt_pattern = os.path.join(gt_dir, f"{ct}/*.tif")
+ gt_paths = glob(gt_pattern)
+ assert len(gt_paths) > 0, "gt_pattern"
+
+ pred_paths = [
+ os.path.join(pred_dir, os.path.basename(path)) for path in gt_paths
+ ]
+
+ this_msas, this_sa50s, this_sa75s = evaluation._run_evaluation(
+ gt_paths, pred_paths, False
+ )
+
+ msas.extend(this_msas), sa50s.extend(this_sa50s), sa75s.extend(this_sa75s)
+ msas_ct.append(np.mean(this_msas))
+ sa50s_ct.append(np.mean(this_sa50s))
+ sa75s_ct.append(np.mean(this_sa75s))
+
+ result_dict = {
+ "cell_type": CELL_TYPES + ["Total"],
+ "msa": msas_ct + [np.mean(msas)],
+ "sa50": sa50s_ct + [np.mean(sa50s_ct)],
+ "sa75": sa75s_ct + [np.mean(sa75s_ct)],
+ }
+ df = pd.DataFrame.from_dict(result_dict)
+ df = df.round(decimals=4)
+ return df
+
+
+def run_livecell_evaluation() -> None:
+ """Run LiveCELL evaluation with command line tool."""
+ parser = argparse.ArgumentParser()
+ parser.add_argument(
+ "-i", "--input", required=True, help="Provide the data directory for LIVECell Dataset"
+ )
+ parser.add_argument(
+ "-e", "--experiment_folder", required=True,
+ help="Provide the path where the inference data is stored."
+ )
+ parser.add_argument(
+ "-f", "--force", action="store_true",
+ help="Force recomputation of already cached eval results."
+ )
+ args = parser.parse_args()
+
+ gt_dir = os.path.join(args.input, "annotations", "livecell_test_images")
+ assert os.path.exists(gt_dir), "The LiveCELL Dataset is incomplete"
+
+ experiment_folder = args.experiment_folder
+ save_root = os.path.join(experiment_folder, "results")
+
+ inference_root_names = ["points", "box", "amg/inference"]
+ for inf_root in inference_root_names:
+
+ pred_root = os.path.join(experiment_folder, inf_root)
+ if inf_root.startswith("amg"):
+ pred_folders = [pred_root]
+ else:
+ pred_folders = sorted(glob(os.path.join(pred_root, "*")))
+
+ if inf_root == "amg/inference":
+ save_folder = os.path.join(save_root, "amg")
+ else:
+ save_folder = os.path.join(save_root, inf_root)
+ os.makedirs(save_folder, exist_ok=True)
+
+ for pred_folder in tqdm(pred_folders, desc=f"Evaluate predictions for {inf_root} prompt settings"):
+ exp_name = os.path.basename(pred_folder)
+ save_path = os.path.join(save_folder, f"{exp_name}.csv")
+ if os.path.exists(save_path) and not args.force:
+ continue
+ results = evaluate_livecell_predictions(gt_dir, pred_folder, verbose=False)
+ results.to_csv(save_path, index=False)
diff --git a/micro_sam/evaluation/model_comparison.py b/micro_sam/evaluation/model_comparison.py
new file mode 100644
index 00000000..a65437b7
--- /dev/null
+++ b/micro_sam/evaluation/model_comparison.py
@@ -0,0 +1,429 @@
+"""Functionality for qualitative comparison of Segment Anything models on microscopy data.
+"""
+
+import os
+from functools import partial
+from glob import glob
+from pathlib import Path
+
+import h5py
+import matplotlib.pyplot as plt
+import numpy as np
+import pandas as pd
+import torch
+
+import skimage.draw as draw
+from scipy.ndimage import binary_dilation
+from skimage import exposure
+from skimage.segmentation import relabel_sequential, find_boundaries
+
+from tqdm import tqdm
+from typing import Optional, Union
+
+from .. import util
+from ..prompt_generators import PointAndBoxPromptGenerator
+from ..prompt_based_segmentation import segment_from_box, segment_from_points
+
+
+#
+# Compute all required data for the model comparison
+#
+
+
+def _predict_models_with_loader(loader, n_samples, prompt_generator, predictor1, predictor2, output_folder):
+ i = 0
+ os.makedirs(output_folder, exist_ok=True)
+
+ for x, y in tqdm(loader, total=n_samples):
+ out_path = os.path.join(output_folder, f"sample_{i}.h5")
+
+ im = x.numpy().squeeze()
+ if im.ndim == 3 and im.shape[0] == 3:
+ im = im.transpose((1, 2, 0))
+
+ gt = y.numpy().squeeze().astype("uint32")
+ gt = relabel_sequential(gt)[0]
+
+ emb1 = util.precompute_image_embeddings(predictor1, im, ndim=2)
+ util.set_precomputed(predictor1, emb1)
+
+ emb2 = util.precompute_image_embeddings(predictor2, im, ndim=2)
+ util.set_precomputed(predictor2, emb2)
+
+ centers, boxes = util.get_centers_and_bounding_boxes(gt)
+
+ with h5py.File(out_path, "a") as f:
+ f.create_dataset("image", data=im, compression="gzip")
+
+ gt_ids = np.unique(gt)[1:]
+ for gt_id in tqdm(gt_ids):
+
+ gt_mask = (gt == gt_id).astype("uint8")
+ point_coords, point_labels, box, _ = prompt_generator(gt, gt_id, boxes[gt_id], centers[gt_id])
+
+ box = np.array(box[0])
+ mask1_box = segment_from_box(predictor1, box)
+ mask2_box = segment_from_box(predictor2, box)
+ mask1_box, mask2_box = mask1_box.squeeze(), mask2_box.squeeze()
+
+ point_coords, point_labels = np.array(point_coords), np.array(point_labels)
+ mask1_points = segment_from_points(predictor1, point_coords, point_labels)
+ mask2_points = segment_from_points(predictor2, point_coords, point_labels)
+ mask1_points, mask2_points = mask1_points.squeeze(), mask2_points.squeeze()
+
+ with h5py.File(out_path, "a") as f:
+ g = f.create_group(str(gt_id))
+ g.attrs["point_coords"] = point_coords
+ g.attrs["point_labels"] = point_labels
+ g.attrs["box"] = box
+
+ g.create_dataset("gt_mask", data=gt_mask, compression="gzip")
+ g.create_dataset("box/mask1", data=mask1_box.astype("uint8"), compression="gzip")
+ g.create_dataset("box/mask2", data=mask2_box.astype("uint8"), compression="gzip")
+ g.create_dataset("points/mask1", data=mask1_points.astype("uint8"), compression="gzip")
+ g.create_dataset("points/mask2", data=mask2_points.astype("uint8"), compression="gzip")
+
+ i += 1
+ if i >= n_samples:
+ return
+
+
+def generate_data_for_model_comparison(
+ loader: torch.utils.data.DataLoader,
+ output_folder: Union[str, os.PathLike],
+ model_type1: str,
+ model_type2: str,
+ n_samples: int,
+) -> None:
+ """Generate samples for qualitative model comparison.
+
+ This precomputes the input for `model_comparison` and `model_comparison_with_napari`.
+
+ Args:
+ loader: The torch dataloader from which samples are drawn.
+ output_folder: The folder where the samples will be saved.
+ model_type1: The first model to use for comparison.
+ The value needs to be a valid model_type for `micro_sam.util.get_sam_model`.
+ model_type1: The second model to use for comparison.
+ The value needs to be a valid model_type for `micro_sam.util.get_sam_model`.
+ n_samples: The number of samples to draw from the dataloader.
+ """
+ prompt_generator = PointAndBoxPromptGenerator(
+ n_positive_points=1,
+ n_negative_points=0,
+ dilation_strength=3,
+ get_point_prompts=True,
+ get_box_prompts=True,
+ )
+ predictor1 = util.get_sam_model(model_type=model_type1)
+ predictor2 = util.get_sam_model(model_type=model_type2)
+ _predict_models_with_loader(loader, n_samples, prompt_generator, predictor1, predictor2, output_folder)
+
+
+#
+# Visual evaluation accroding to metrics
+#
+
+
+def _evaluate_samples(f, prefix, min_size):
+ eval_result = {
+ "gt_id": [],
+ "score1": [],
+ "score2": [],
+ }
+ for name, group in f.items():
+ if name == "image":
+ continue
+
+ gt_mask = group["gt_mask"][:]
+
+ size = gt_mask.sum()
+ if size < min_size:
+ continue
+
+ m1 = group[f"{prefix}/mask1"][:]
+ m2 = group[f"{prefix}/mask2"][:]
+
+ score1 = util.compute_iou(gt_mask, m1)
+ score2 = util.compute_iou(gt_mask, m2)
+
+ eval_result["gt_id"].append(name)
+ eval_result["score1"].append(score1)
+ eval_result["score2"].append(score2)
+
+ eval_result = pd.DataFrame.from_dict(eval_result)
+ eval_result["advantage1"] = eval_result["score1"] - eval_result["score2"]
+ eval_result["advantage2"] = eval_result["score2"] - eval_result["score1"]
+ return eval_result
+
+
+def _overlay_mask(image, mask):
+ assert image.ndim in (2, 3)
+ # overlay the mask
+ if image.ndim == 2:
+ overlay = np.stack([image, image, image]).transpose((1, 2, 0))
+ else:
+ overlay = image
+ assert overlay.shape[-1] == 3
+ mask_overlay = np.zeros_like(overlay)
+ mask_overlay[mask == 1] = [255, 0, 0]
+ alpha = 0.6
+ overlay = alpha * overlay + (1.0 - alpha) * mask_overlay
+ return overlay.astype("uint8")
+
+
+def _enhance_image(im):
+ # apply CLAHE to improve the image quality
+ im -= im.min(axis=(0, 1), keepdims=True)
+ im /= (im.max(axis=(0, 1), keepdims=True) + 1e-6)
+ im = exposure.equalize_adapthist(im)
+ im *= 255
+ return im
+
+
+def _overlay_outline(im, mask, outline_dilation):
+ outline = find_boundaries(mask)
+ if outline_dilation > 0:
+ outline = binary_dilation(outline, iterations=outline_dilation)
+ overlay = im.copy()
+ overlay[outline] = [255, 255, 0]
+ return overlay
+
+
+def _overlay_box(im, prompt, outline_dilation):
+ start, end = prompt
+ rr, cc = draw.rectangle_perimeter(start, end=end, shape=im.shape[:2])
+
+ box_outline = np.zeros(im.shape[:2], dtype="bool")
+ box_outline[rr, cc] = 1
+ if outline_dilation > 0:
+ box_outline = binary_dilation(box_outline, iterations=outline_dilation)
+
+ overlay = im.copy()
+ overlay[box_outline] = [0, 255, 255]
+
+ return overlay
+
+
+# NOTE: we currently only support a single point
+def _overlay_points(im, prompt, radius):
+ coords, labels = prompt
+ # make sure we have a single positive prompt, other options are
+ # currently not supported
+ assert coords.shape[0] == labels.shape[0] == 1
+ assert labels[0] == 1
+
+ rr, cc = draw.disk(coords[0], radius, shape=im.shape[:2])
+ overlay = im.copy()
+ draw.set_color(overlay, (rr, cc), [0, 255, 255], alpha=1.0)
+
+ return overlay
+
+
+def _compare_eval(
+ f, eval_result, advantage_column,
+ n_images_per_sample, prefix,
+ sample_name, plot_folder,
+ point_radius, outline_dilation,
+):
+ result = eval_result.sort_values(advantage_column, ascending=False).iloc[:n_images_per_sample]
+ n_rows = result.shape[0]
+
+ image = f["image"][:]
+ is_box_prompt = prefix == "box"
+ overlay_prompts = partial(_overlay_box, outline_dilation=outline_dilation) if is_box_prompt else\
+ partial(_overlay_points, radius=point_radius)
+
+ def make_square(bb, shape):
+ box_shape = [b.stop - b.start for b in bb]
+ longest_side = max(box_shape)
+ padding = [(longest_side - sh) // 2 for sh in box_shape]
+ bb = tuple(
+ slice(max(b.start - pad, 0), min(b.stop + pad, sh)) for b, pad, sh in zip(bb, padding, shape)
+ )
+ return bb
+
+ def plot_ax(axis, i, row):
+ g = f[row.gt_id]
+
+ gt = g["gt_mask"][:]
+ mask1 = g[f"{prefix}/mask1"][:]
+ mask2 = g[f"{prefix}/mask2"][:]
+
+ fg_mask = (gt + mask1 + mask2) > 0
+ # if this is a box prompt we dilate the mask so that the bounding box
+ # can be seen
+ if is_box_prompt:
+ fg_mask = binary_dilation(fg_mask, iterations=5)
+ bb = np.where(fg_mask)
+ bb = tuple(
+ slice(int(b.min()), int(b.max() + 1)) for b in bb
+ )
+ bb = make_square(bb, fg_mask.shape)
+
+ offset = np.array([b.start for b in bb])
+ if is_box_prompt:
+ prompt = g.attrs["box"]
+ prompt = np.array(
+ [prompt[:2], prompt[2:]]
+ ) - offset
+ else:
+ prompt = (g.attrs["point_coords"] - offset, g.attrs["point_labels"])
+
+ im = _enhance_image(image[bb])
+ gt, mask1, mask2 = gt[bb], mask1[bb], mask2[bb]
+
+ im1 = _overlay_mask(im, mask1)
+ im1 = _overlay_outline(im1, gt, outline_dilation)
+ im1 = overlay_prompts(im1, prompt)
+ ax = axis[0] if i is None else axis[i, 0]
+ ax.axis("off")
+ ax.imshow(im1)
+
+ im2 = _overlay_mask(im, mask2)
+ im2 = _overlay_outline(im2, gt, outline_dilation)
+ im2 = overlay_prompts(im2, prompt)
+ ax = axis[1] if i is None else axis[i, 1]
+ ax.axis("off")
+ ax.imshow(im2)
+
+ if plot_folder is None:
+ fig, axis = plt.subplots(n_rows, 2)
+ for i, (_, row) in enumerate(result.iterrows()):
+ plot_ax(axis, i, row)
+ plt.show()
+ else:
+ for i, (_, row) in enumerate(result.iterrows()):
+ fig, axis = plt.subplots(1, 2)
+ plot_ax(axis, None, row)
+ plt.subplots_adjust(wspace=0.05, hspace=0)
+ plt.savefig(os.path.join(plot_folder, f"{sample_name}_{i}.png"), bbox_inches="tight")
+ plt.close()
+
+
+def _compare_prompts(
+ f, prefix, n_images_per_sample, min_size, sample_name, plot_folder,
+ point_radius, outline_dilation
+):
+ box_eval = _evaluate_samples(f, prefix, min_size)
+ if plot_folder is None:
+ plot_folder1, plot_folder2 = None, None
+ else:
+ plot_folder1 = os.path.join(plot_folder, "advantage1")
+ plot_folder2 = os.path.join(plot_folder, "advantage2")
+ os.makedirs(plot_folder1, exist_ok=True)
+ os.makedirs(plot_folder2, exist_ok=True)
+ _compare_eval(
+ f, box_eval, "advantage1", n_images_per_sample, prefix, sample_name, plot_folder1,
+ point_radius, outline_dilation
+ )
+ _compare_eval(
+ f, box_eval, "advantage2", n_images_per_sample, prefix, sample_name, plot_folder2,
+ point_radius, outline_dilation
+ )
+
+
+def _compare_models(
+ path, n_images_per_sample, min_size, plot_folder, point_radius, outline_dilation
+):
+ sample_name = Path(path).stem
+ with h5py.File(path, "r") as f:
+ if plot_folder is None:
+ plot_folder_points, plot_folder_box = None, None
+ else:
+ plot_folder_points = os.path.join(plot_folder, "points")
+ plot_folder_box = os.path.join(plot_folder, "box")
+ _compare_prompts(
+ f, "points", n_images_per_sample, min_size, sample_name, plot_folder_points,
+ point_radius, outline_dilation
+ )
+ _compare_prompts(
+ f, "box", n_images_per_sample, min_size, sample_name, plot_folder_box,
+ point_radius, outline_dilation
+ )
+
+
+def model_comparison(
+ output_folder: Union[str, os.PathLike],
+ n_images_per_sample: int,
+ min_size: int,
+ plot_folder: Optional[Union[str, os.PathLike]] = None,
+ point_radius: int = 4,
+ outline_dilation: int = 0,
+) -> None:
+ """Create images for a qualitative model comparision.
+
+ Args:
+ output_folder: The folder with the data precomputed by `generate_data_for_model_comparison`.
+ n_images_per_sample: The number of images to generate per precomputed sample.
+ min_size: The min size of ground-truth objects to take into account.
+ plot_folder: The folder where to save the plots. If not given the plots will be displayed.
+ point_radius: The radius of the point overlay.
+ outline_dilation: The dilation factor of the outline overlay.
+ """
+ files = glob(os.path.join(output_folder, "*.h5"))
+ for path in tqdm(files):
+ _compare_models(
+ path, n_images_per_sample, min_size, plot_folder, point_radius, outline_dilation
+ )
+
+
+#
+# Quick visual evaluation with napari
+#
+
+
+def _check_group(g, show_points):
+ import napari
+
+ image = g["image"][:]
+ gt = g["gt_mask"][:]
+ if show_points:
+ m1 = g["points/mask1"][:]
+ m2 = g["points/mask2"][:]
+ points = g.attrs["point_coords"]
+ else:
+ m1 = g["box/mask1"][:]
+ m2 = g["box/mask2"][:]
+ box = g.attrs["box"]
+ box = np.array([
+ [box[0], box[1]], [box[2], box[3]]
+ ])
+
+ v = napari.Viewer()
+ v.add_image(image)
+ v.add_labels(gt)
+ v.add_labels(m1)
+ v.add_labels(m2)
+ if show_points:
+ # TODO use point labels for coloring
+ v.add_points(
+ points,
+ edge_color="#00FF00",
+ symbol="o",
+ face_color="transparent",
+ edge_width=0.5,
+ size=12,
+ )
+ else:
+ v.add_shapes(
+ box, face_color="transparent", edge_color="green", edge_width=4,
+ )
+ napari.run()
+
+
+def model_comparison_with_napari(output_folder: Union[str, os.PathLike], show_points: bool = True) -> None:
+ """Use napari to display the qualtiative comparison results for two models.
+
+ Args:
+ output_folder: The folder with the data precomputed by `generate_data_for_model_comparison`.
+ show_points: Whether to show the results for point or for box prompts.
+ """
+ files = glob(os.path.join(output_folder, "*.h5"))
+ for path in files:
+ print("Comparing models in", path)
+ with h5py.File(path, "r") as f:
+ for name, g in f.items():
+ if name == "image":
+ continue
+ _check_group(g, show_points=show_points)
diff --git a/micro_sam/instance_segmentation.py b/micro_sam/instance_segmentation.py
index f21ad9fa..f1c5e644 100644
--- a/micro_sam/instance_segmentation.py
+++ b/micro_sam/instance_segmentation.py
@@ -54,6 +54,7 @@ def mask_data_to_segmentation(
masks: List[Dict[str, Any]],
shape: tuple[int, ...],
with_background: bool,
+ min_object_size: int = 0,
) -> np.ndarray:
"""Convert the output of the automatic mask generation to an instance segmentation.
@@ -63,6 +64,7 @@ def mask_data_to_segmentation(
shape: The image shape.
with_background: Whether the segmentation has background. If yes this function assures that the largest
object in the output will be mapped to zero (the background value).
+ min_object_size: The minimal size of an object in pixels.
Returns:
The instance segmentation.
"""
@@ -70,8 +72,12 @@ def mask_data_to_segmentation(
masks = sorted(masks, key=(lambda x: x["area"]), reverse=True)
segmentation = np.zeros(shape[:2], dtype="uint32")
- for seg_id, mask in enumerate(masks, 1):
+ seg_id = 1
+ for mask in masks:
+ if mask["area"] < min_object_size:
+ continue
segmentation[mask["segmentation"]] = seg_id
+ seg_id += 1
if with_background:
seg_ids, sizes = np.unique(segmentation, return_counts=True)
@@ -129,7 +135,6 @@ def _postprocess_batch(
original_size,
pred_iou_thresh,
stability_score_thresh,
- stability_score_offset,
box_nms_thresh,
):
orig_h, orig_w = original_size
@@ -139,28 +144,16 @@ def _postprocess_batch(
keep_mask = data["iou_preds"] > pred_iou_thresh
data.filter(keep_mask)
- # calculate stability score
- data["stability_score"] = amg_utils.calculate_stability_score(
- data["masks"], self._predictor.model.mask_threshold, stability_score_offset
- )
+ # filter by stability score
if stability_score_thresh > 0.0:
keep_mask = data["stability_score"] >= stability_score_thresh
data.filter(keep_mask)
- # threshold masks and calculate boxes
- data["masks"] = data["masks"] > self._predictor.model.mask_threshold
- data["boxes"] = amg_utils.batched_mask_to_box(data["masks"])
-
# filter boxes that touch crop boundaries
keep_mask = ~amg_utils.is_box_near_crop_edge(data["boxes"], crop_box, [0, 0, orig_w, orig_h])
if not torch.all(keep_mask):
data.filter(keep_mask)
- # compress to RLE
- data["masks"] = amg_utils.uncrop_masks(data["masks"], crop_box, orig_h, orig_w)
- data["rles"] = amg_utils.mask_to_rle_pytorch(data["masks"])
- del data["masks"]
-
# remove duplicates within this crop.
keep_by_nms = batched_nms(
data["boxes"].float(),
@@ -261,6 +254,32 @@ def _postprocess_masks(self, mask_data, min_mask_region_area, box_nms_thresh, cr
return curr_anns
+ def _to_mask_data(self, masks, iou_preds, crop_box, original_size, points=None):
+ orig_h, orig_w = original_size
+
+ # serialize predictions and store in MaskData
+ data = amg_utils.MaskData(masks=masks.flatten(0, 1), iou_preds=iou_preds.flatten(0, 1))
+ if points is not None:
+ data["points"] = torch.as_tensor(points.repeat(masks.shape[1], axis=0))
+
+ del masks
+
+ # calculate the stability scores
+ data["stability_score"] = amg_utils.calculate_stability_score(
+ data["masks"], self._predictor.model.mask_threshold, self._stability_score_offset
+ )
+
+ # threshold masks and calculate boxes
+ data["masks"] = data["masks"] > self._predictor.model.mask_threshold
+ data["boxes"] = amg_utils.batched_mask_to_box(data["masks"])
+
+ # compress to RLE
+ data["masks"] = amg_utils.uncrop_masks(data["masks"], crop_box, orig_h, orig_w)
+ data["rles"] = amg_utils.mask_to_rle_pytorch(data["masks"])
+ del data["masks"]
+
+ return data
+
def get_state(self) -> Dict[str, Any]:
"""Get the initialized state of the mask generator.
@@ -269,6 +288,7 @@ def get_state(self) -> Dict[str, Any]:
"""
if not self.is_initialized:
raise RuntimeError("The state has not been computed yet. Call initialize first.")
+
return {"crop_list": self.crop_list, "crop_boxes": self.crop_boxes, "original_size": self.original_size}
def set_state(self, state: Dict[str, Any]) -> None:
@@ -309,6 +329,7 @@ class AutomaticMaskGenerator(AMGBase):
crop_n_points_downscale_factor: How the number of points is downsampled when predicting with crops.
point_grids: A lisst over explicit grids of points used for sampling masks.
Normalized to [0, 1] with respect to the image coordinate system.
+ stability_score_offset: The amount to shift the cutoff when calculating the stability score.
"""
def __init__(
self,
@@ -319,6 +340,7 @@ def __init__(
crop_overlap_ratio: float = 512 / 1500,
crop_n_points_downscale_factor: int = 1,
point_grids: Optional[List[np.ndarray]] = None,
+ stability_score_offset: float = 1.0,
):
super().__init__()
@@ -339,8 +361,9 @@ def __init__(
self._crop_n_layers = crop_n_layers
self._crop_overlap_ratio = crop_overlap_ratio
self._crop_n_points_downscale_factor = crop_n_points_downscale_factor
+ self._stability_score_offset = stability_score_offset
- def _process_batch(self, points, im_size):
+ def _process_batch(self, points, im_size, crop_box, original_size):
# run model on this batch
transformed_points = self._predictor.transform.apply_coords(points, im_size)
in_points = torch.as_tensor(transformed_points, device=self._predictor.device)
@@ -351,24 +374,14 @@ def _process_batch(self, points, im_size):
multimask_output=True,
return_logits=True,
)
-
- # serialize predictions and store in MaskData
- data = amg_utils.MaskData(
- masks=masks.flatten(0, 1),
- iou_preds=iou_preds.flatten(0, 1),
- points=torch.as_tensor(points.repeat(masks.shape[1], axis=0)),
- )
+ data = self._to_mask_data(masks, iou_preds, crop_box, original_size, points=points)
del masks
-
return data
def _process_crop(self, image, crop_box, crop_layer_idx, verbose, precomputed_embeddings):
# crop the image and calculate embeddings
- if crop_box is None:
- cropped_im = image
- else:
- x0, y0, x1, y1 = crop_box
- cropped_im = image[y0:y1, x0:x1, :]
+ x0, y0, x1, y1 = crop_box
+ cropped_im = image[y0:y1, x0:x1, :]
cropped_im_size = cropped_im.shape[:2]
if not precomputed_embeddings:
@@ -387,7 +400,7 @@ def _process_crop(self, image, crop_box, crop_layer_idx, verbose, precomputed_em
disable=not verbose, total=n_batches,
desc="Predict masks for point grid prompts",
):
- batch_data = self._process_batch(points, cropped_im_size)
+ batch_data = self._process_batch(points, cropped_im_size, crop_box, self.original_size)
data.cat(batch_data)
del batch_data
@@ -415,6 +428,8 @@ def initialize(
verbose: Whether to print computation progress.
"""
original_size = image.shape[:2]
+ self._original_size = original_size
+
crop_boxes, layer_idxs = amg_utils.generate_crop_boxes(
original_size, self._crop_n_layers, self._crop_overlap_ratio
)
@@ -443,14 +458,12 @@ def initialize(
self._is_initialized = True
self._crop_list = crop_list
self._crop_boxes = crop_boxes
- self._original_size = original_size
@torch.no_grad()
def generate(
self,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- stability_score_offset: float = 1.0,
box_nms_thresh: float = 0.7,
crop_nms_thresh: float = 0.7,
min_mask_region_area: int = 0,
@@ -462,7 +475,6 @@ def generate(
pred_iou_thresh: Filter threshold in [0, 1], using the mask quality predicted by the model.
stability_score_thresh: Filter threshold in [0, 1], using the stability of the mask
under changes to the cutoff used to binarize the model prediction.
- stability_score_offset: The amount to shift the cutoff when calculating the stability score.
box_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks.
crop_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks between crops.
min_mask_region_area: Minimal size for the predicted masks.
@@ -481,7 +493,6 @@ def generate(
crop_box=crop_box, original_size=self.original_size,
pred_iou_thresh=pred_iou_thresh,
stability_score_thresh=stability_score_thresh,
- stability_score_offset=stability_score_offset,
box_nms_thresh=box_nms_thresh
)
data.cat(crop_data)
@@ -529,6 +540,7 @@ class EmbeddingMaskGenerator(AMGBase):
use_mask: Whether to use the initial segments as prompts.
use_points: Whether to use points derived from the initial segments as prompts.
box_extension: Factor for extending the bounding box prompts, given in the relative box size.
+ stability_score_offset: The amount to shift the cutoff when calculating the stability score.
"""
default_offsets = [[-1, 0], [0, -1], [-3, 0], [0, -3], [-9, 0], [0, -9]]
@@ -543,6 +555,7 @@ def __init__(
use_mask: bool = True,
use_points: bool = False,
box_extension: float = 0.05,
+ stability_score_offset: float = 1.0,
):
super().__init__()
@@ -555,6 +568,7 @@ def __init__(
self._use_mask = use_mask
self._use_points = use_points
self._box_extension = box_extension
+ self._stability_score_offset = stability_score_offset
# additional state that is set 'initialize'
self._initial_segmentation = None
@@ -581,7 +595,7 @@ def _compute_initial_segmentation(self):
return initial_segmentation
- def _compute_mask_data(self, initial_segmentation, original_size, verbose):
+ def _compute_mask_data(self, initial_segmentation, crop_box, original_size, verbose):
seg_ids = np.unique(initial_segmentation)
if seg_ids[0] == 0:
seg_ids = seg_ids[1:]
@@ -592,15 +606,13 @@ def _compute_mask_data(self, initial_segmentation, original_size, verbose):
mask = initial_segmentation == seg_id
masks, iou_preds, _ = segment_from_mask(
self._predictor, mask, original_size=original_size,
- multimask_output=True, return_logits=True, return_all=True,
+ multimask_output=False, return_logits=True, return_all=True,
use_box=self._use_box, use_mask=self._use_mask, use_points=self._use_points,
box_extension=self._box_extension,
)
- data = amg_utils.MaskData(
- masks=torch.from_numpy(masks),
- iou_preds=torch.from_numpy(iou_preds),
- seg_id=torch.from_numpy(np.full(len(masks), seg_id, dtype="int64")),
- )
+ # bring masks and iou_preds to a format compatible with _to_mask_data
+ masks, iou_preds = torch.from_numpy(masks[None]), torch.from_numpy(iou_preds[None])
+ data = self._to_mask_data(masks, iou_preds, crop_box, original_size)
del masks
mask_data.cat(data)
@@ -625,6 +637,11 @@ def initialize(
verbose: Whether to print computation progress.
"""
original_size = image.shape[:2]
+ self._original_size = original_size
+
+ # the crop box is always the full image
+ crop_box = [0, 0, original_size[1], original_size[0]]
+ self._crop_boxes = [crop_box]
if image_embeddings is None:
image_embeddings = util.precompute_image_embeddings(self._predictor, image,)
@@ -633,7 +650,7 @@ def initialize(
# compute the initial segmentation via embedding based MWS and then refine the masks
# with the segment anything model
initial_segmentation = self._compute_initial_segmentation()
- mask_data = self._compute_mask_data(initial_segmentation, original_size, verbose)
+ mask_data = self._compute_mask_data(initial_segmentation, crop_box, original_size, verbose)
# to be compatible with the file format of the super class we have to wrap the mask data in a list
crop_list = [mask_data]
@@ -641,18 +658,12 @@ def initialize(
self._is_initialized = True
self._initial_segmentation = initial_segmentation
self._crop_list = crop_list
- # the crop box is always the full image
- self._crop_boxes = [
- [0, 0, original_size[1], original_size[0]]
- ]
- self._original_size = original_size
@torch.no_grad()
def generate(
self,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- stability_score_offset: float = 1.0,
box_nms_thresh: float = 0.7,
min_mask_region_area: int = 0,
output_mode: str = "binary_mask",
@@ -663,7 +674,6 @@ def generate(
pred_iou_thresh: Filter threshold in [0, 1], using the mask quality predicted by the model.
stability_score_thresh: Filter threshold in [0, 1], using the stability of the mask
under changes to the cutoff used to binarize the model prediction.
- stability_score_offset: The amount to shift the cutoff when calculating the stability score.
box_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks.
min_mask_region_area: Minimal size for the predicted masks.
output_mode: The form masks are returned in.
@@ -679,7 +689,6 @@ def generate(
original_size=self.original_size,
pred_iou_thresh=pred_iou_thresh,
stability_score_thresh=stability_score_thresh,
- stability_score_offset=stability_score_offset,
box_nms_thresh=box_nms_thresh
)
@@ -771,6 +780,7 @@ class TiledAutomaticMaskGenerator(AutomaticMaskGenerator):
Higher numbers may be faster but use more GPU memory.
point_grids: A lisst over explicit grids of points used for sampling masks.
Normalized to [0, 1] with respect to the image coordinate system.
+ stability_score_offset: The amount to shift the cutoff when calculating the stability score.
"""
# We only expose the arguments that make sense for the tiled mask generator.
@@ -782,12 +792,14 @@ def __init__(
points_per_side: Optional[int] = 32,
points_per_batch: int = 64,
point_grids: Optional[List[np.ndarray]] = None,
+ stability_score_offset: float = 1.0,
) -> None:
super().__init__(
predictor=predictor,
points_per_side=points_per_side,
points_per_batch=points_per_batch,
point_grids=point_grids,
+ stability_score_offset=stability_score_offset,
)
@torch.no_grad()
@@ -815,6 +827,8 @@ def initialize(
embedding_save_path: Where to save the image embeddings.
"""
original_size = image.shape[:2]
+ self._original_size = original_size
+
image_embeddings, tile_shape, halo = _compute_tiled_embeddings(
self._predictor, image, image_embeddings, embedding_save_path, tile_shape, halo
)
@@ -822,13 +836,15 @@ def initialize(
tiling = blocking([0, 0], original_size, tile_shape)
n_tiles = tiling.numberOfBlocks
+ # the crop box is always the full local tile
+ tiles = [tiling.getBlockWithHalo(tile_id, list(halo)).outerBlock for tile_id in range(n_tiles)]
+ crop_boxes = [[tile.begin[1], tile.begin[0], tile.end[1], tile.end[0]] for tile in tiles]
+
+ # we need to cast to the image representation that is compatible with SAM
+ image = util._to_image(image)
+
mask_data = []
for tile_id in tqdm(range(n_tiles), total=n_tiles, desc="Compute masks for tile", disable=not verbose):
- # get the bounding box for this tile and crop the image data
- tile = tiling.getBlockWithHalo(tile_id, list(halo)).outerBlock
- tile_bb = tuple(slice(beg, end) for beg, end in zip(tile.begin, tile.end))
- tile_data = image[tile_bb]
-
# set the pre-computed embeddings for this tile
features = image_embeddings["features"][tile_id]
tile_embeddings = {
@@ -840,18 +856,14 @@ def initialize(
# compute the mask data for this tile and append it
this_mask_data = self._process_crop(
- tile_data, crop_box=None, crop_layer_idx=0, verbose=verbose, precomputed_embeddings=True
+ image, crop_box=crop_boxes[tile_id], crop_layer_idx=0, verbose=verbose, precomputed_embeddings=True
)
mask_data.append(this_mask_data)
# set the initialized data
self._is_initialized = True
self._crop_list = mask_data
- self._original_size = original_size
-
- # the crop box is always the full local tile
- tiles = [tiling.getBlockWithHalo(tile_id, list(halo)).outerBlock for tile_id in range(n_tiles)]
- self._crop_boxes = [[tile.begin[1], tile.begin[0], tile.end[1], tile.end[0]] for tile in tiles]
+ self._crop_boxes = crop_boxes
class TiledEmbeddingMaskGenerator(EmbeddingMaskGenerator):
@@ -918,7 +930,10 @@ def _compute_mask_data_tiled(self, image_embeddings, i, initial_segmentations, n
"original_size": this_tile_shape
}
util.set_precomputed(self._predictor, tile_image_embeddings, i)
- tile_data = self._compute_mask_data(initial_segmentations[tile_id], this_tile_shape, verbose=False)
+ this_crop_box = [0, 0, this_tile_shape[1], this_tile_shape[0]]
+ tile_data = self._compute_mask_data(
+ initial_segmentations[tile_id], this_crop_box, this_tile_shape, verbose=False
+ )
mask_data.append(tile_data)
return mask_data
@@ -976,7 +991,6 @@ def generate(
self,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- stability_score_offset: float = 1.0,
box_nms_thresh: float = 0.7,
min_mask_region_area: int = 0,
verbose: bool = False
@@ -987,7 +1001,6 @@ def generate(
pred_iou_thresh: Filter threshold in [0, 1], using the mask quality predicted by the model.
stability_score_thresh: Filter threshold in [0, 1], using the stability of the mask
under changes to the cutoff used to binarize the model prediction.
- stability_score_offset: The amount to shift the cutoff when calculating the stability score.
box_nms_thresh: The IoU threshold used by nonmax suppression to filter duplicate masks.
min_mask_region_area: Minimal size for the predicted masks.
verbose: Whether to print progress of the computation.
@@ -1008,7 +1021,6 @@ def segment_tile(_, tile_id):
data=mask_data, crop_box=crop_box, original_size=this_tile_shape,
pred_iou_thresh=pred_iou_thresh,
stability_score_thresh=stability_score_thresh,
- stability_score_offset=stability_score_offset,
box_nms_thresh=box_nms_thresh,
)
mask_data.to_numpy()
@@ -1080,6 +1092,35 @@ def set_state(self, state: Dict[str, Any]) -> None:
super().set_state(state)
+def get_amg(
+ predictor: SamPredictor,
+ is_tiled: bool,
+ embedding_based_amg: bool = False,
+ **kwargs,
+) -> AMGBase:
+ """Get the automatic mask generator class.
+
+ Args:
+ predictor: The segment anything predictor.
+ is_tiled: Whether tiled embeddings are used.
+ embedding_based_amg: Whether to use the embedding based instance segmentation functionality.
+ This functionality is still experimental.
+ kwargs: The keyword arguments for the amg class.
+
+ Returns:
+ The automatic mask generator.
+ """
+ if embedding_based_amg:
+ warnings.warn("The embedding based instance segmentation functionality is experimental.")
+ if is_tiled:
+ amg = TiledEmbeddingMaskGenerator(predictor, **kwargs) if embedding_based_amg else\
+ TiledAutomaticMaskGenerator(predictor, **kwargs)
+ else:
+ amg = EmbeddingMaskGenerator(predictor, **kwargs) if embedding_based_amg else\
+ AutomaticMaskGenerator(predictor, **kwargs)
+ return amg
+
+
#
# Experimental functionality
#
diff --git a/micro_sam/napari.yaml b/micro_sam/napari.yaml
index 6107c87e..f6cdc25b 100644
--- a/micro_sam/napari.yaml
+++ b/micro_sam/napari.yaml
@@ -2,6 +2,9 @@ name: micro-sam
display_name: SegmentAnything for Microscopy
contributions:
commands:
+ - id: micro-sam.sample_data_image_series
+ python_name: micro_sam.sample_data:sample_data_image_series
+ title: Load image series sample data from micro-sam plugin
- id: micro-sam.sample_data_wholeslide
python_name: micro_sam.sample_data:sample_data_wholeslide
title: Load WholeSlide sample data from micro-sam plugin
@@ -17,7 +20,13 @@ contributions:
- id: micro-sam.sample_data_tracking
python_name: micro_sam.sample_data:sample_data_tracking
title: Load tracking sample data from micro-sam plugin
+ - id: micro-sam.sample_data_segmentation
+ python_name: micro_sam.sample_data:sample_data_segmentation
+ title: Load segmentation sample data from micro-sam plugin
sample_data:
+ - command: micro-sam.sample_data_image_series
+ display_name: Image series example data
+ key: micro-sam-image-series
- command: micro-sam.sample_data_wholeslide
display_name: WholeSlide example data
key: micro-sam-wholeslide
@@ -33,3 +42,6 @@ contributions:
- command: micro-sam.sample_data_tracking
display_name: Tracking sample dataset
key: micro-sam-tracking
+ - command: micro-sam.sample_data_segmentation
+ display_name: Segmentation sample dataset
+ key: micro-sam-segmentation
diff --git a/micro_sam/precompute_state.py b/micro_sam/precompute_state.py
new file mode 100644
index 00000000..6844c0f9
--- /dev/null
+++ b/micro_sam/precompute_state.py
@@ -0,0 +1,173 @@
+"""Precompute image embeddings and automatic mask generator state for image data.
+"""
+
+import os
+import pickle
+
+from glob import glob
+from pathlib import Path
+from typing import Optional, Tuple, Union
+
+import numpy as np
+import torch
+from segment_anything.predictor import SamPredictor
+from tqdm import tqdm
+
+from . import instance_segmentation, util
+
+
+def cache_amg_state(
+ predictor: SamPredictor,
+ raw: np.ndarray,
+ image_embeddings: util.ImageEmbeddings,
+ save_path: Union[str, os.PathLike],
+ verbose: bool = True,
+ **kwargs,
+) -> instance_segmentation.AMGBase:
+ """Compute and cache or load the state for the automatic mask generator.
+
+ Args:
+ predictor: The segment anything predictor.
+ raw: The image data.
+ image_embeddings: The image embeddings.
+ save_path: The embedding save path. The AMG state will be stored in 'save_path/amg_state.pickle'.
+ verbose: Whether to run the computation verbose.
+ kwargs: The keyword arguments for the amg class.
+
+ Returns:
+ The automatic mask generator class with the cached state.
+ """
+ is_tiled = image_embeddings["input_size"] is None
+ amg = instance_segmentation.get_amg(predictor, is_tiled, **kwargs)
+
+ save_path_amg = os.path.join(save_path, "amg_state.pickle")
+ if os.path.exists(save_path_amg):
+ if verbose:
+ print("Load the AMG state from", save_path_amg)
+ with open(save_path_amg, "rb") as f:
+ amg_state = pickle.load(f)
+ amg.set_state(amg_state)
+ return amg
+
+ if verbose:
+ print("Precomputing the state for instance segmentation.")
+
+ amg.initialize(raw, image_embeddings=image_embeddings, verbose=verbose)
+ amg_state = amg.get_state()
+
+ # put all state onto the cpu so that the state can be deserialized without a gpu
+ new_crop_list = []
+ for mask_data in amg_state["crop_list"]:
+ for k, v in mask_data.items():
+ if torch.is_tensor(v):
+ mask_data[k] = v.cpu()
+ new_crop_list.append(mask_data)
+ amg_state["crop_list"] = new_crop_list
+
+ with open(save_path_amg, "wb") as f:
+ pickle.dump(amg_state, f)
+
+ return amg
+
+
+def _precompute_state_for_file(
+ predictor, input_path, output_path, key, ndim, tile_shape, halo, precompute_amg_state,
+):
+ image_data = util.load_image_data(input_path, key)
+ output_path = Path(output_path).with_suffix(".zarr")
+ embeddings = util.precompute_image_embeddings(
+ predictor, image_data, output_path, ndim=ndim, tile_shape=tile_shape, halo=halo,
+ )
+ if precompute_amg_state:
+ cache_amg_state(predictor, image_data, embeddings, output_path, verbose=True)
+
+
+def _precompute_state_for_files(
+ predictor, input_files, output_path, ndim, tile_shape, halo, precompute_amg_state,
+):
+ os.makedirs(output_path, exist_ok=True)
+ for file_path in tqdm(input_files, desc="Precompute state for files."):
+ out_path = os.path.join(output_path, os.path.basename(file_path))
+ _precompute_state_for_file(
+ predictor, file_path, out_path,
+ key=None, ndim=ndim, tile_shape=tile_shape, halo=halo,
+ precompute_amg_state=precompute_amg_state,
+ )
+
+
+def precompute_state(
+ input_path: Union[os.PathLike, str],
+ output_path: Union[os.PathLike, str],
+ model_type: str = util._DEFAULT_MODEL,
+ checkpoint_path: Optional[Union[os.PathLike, str]] = None,
+ key: Optional[str] = None,
+ ndim: Union[int] = None,
+ tile_shape: Optional[Tuple[int, int]] = None,
+ halo: Optional[Tuple[int, int]] = None,
+ precompute_amg_state: bool = False,
+) -> None:
+ """Precompute the image embeddings and other optional state for the input image(s).
+
+ Args:
+ input_path: The input image file(s). Can either be a single image file (e.g. tif or png),
+ a container file (e.g. hdf5 or zarr) or a folder with images files.
+ In case of a container file the argument `key` must be given. In case of a folder
+ it can be given to provide a glob pattern to subselect files from the folder.
+ output_path: The output path were the embeddings and other state will be saved.
+ model_type: The SegmentAnything model to use. Will use the standard vit_h model by default.
+ checkpoint_path: Path to a checkpoint for a custom model.
+ key: The key to the input file. This is needed for contaner files (e.g. hdf5 or zarr)
+ and can be used to provide a glob pattern if the input is a folder with image files.
+ ndim: The dimensionality of the data.
+ tile_shape: Shape of tiles for tiled prediction. By default prediction is run without tiling.
+ halo: Overlap of the tiles for tiled prediction.
+ precompute_amg_state: Whether to precompute the state for automatic instance segmentation
+ in addition to the image embeddings.
+ """
+ predictor = util.get_sam_model(model_type=model_type, checkpoint_path=checkpoint_path)
+ # check if we precompute the state for a single file or for a folder with image files
+ if os.path.isdir(input_path) and Path(input_path).suffix not in (".n5", ".zarr"):
+ pattern = "*" if key is None else key
+ input_files = glob(os.path.join(input_path, pattern))
+ _precompute_state_for_files(
+ predictor, input_files, output_path,
+ ndim=ndim, tile_shape=tile_shape, halo=halo,
+ precompute_amg_state=precompute_amg_state,
+ )
+ else:
+ _precompute_state_for_file(
+ predictor, input_path, output_path, key,
+ ndim=ndim, tile_shape=tile_shape, halo=halo,
+ precompute_amg_state=precompute_amg_state,
+ )
+
+
+def main():
+ """@private"""
+ import argparse
+
+ parser = argparse.ArgumentParser(description="Compute the embeddings for an image.")
+ parser.add_argument("-i", "--input_path", required=True)
+ parser.add_argument("-o", "--output_path", required=True)
+ parser.add_argument("-m", "--model_type", default="vit_h")
+ parser.add_argument("-c", "--checkpoint_path", default=None)
+ parser.add_argument("-k", "--key")
+ parser.add_argument(
+ "--tile_shape", nargs="+", type=int, help="The tile shape for using tiled prediction", default=None
+ )
+ parser.add_argument(
+ "--halo", nargs="+", type=int, help="The halo for using tiled prediction", default=None
+ )
+ parser.add_argument("-n", "--ndim")
+ parser.add_argument("-p", "--precompute_amg_state")
+
+ args = parser.parse_args()
+ precompute_state(
+ args.input_path, args.output_path, args.model_type, args.checkpoint_path,
+ key=args.key, tile_shape=args.tile_shape, halo=args.halo, ndim=args.ndim,
+ precompute_amg_state=args.precompute_amg_state,
+ )
+
+
+if __name__ == "__main__":
+ main()
diff --git a/micro_sam/prompt_generators.py b/micro_sam/prompt_generators.py
index 18c6e6fe..51849e3e 100644
--- a/micro_sam/prompt_generators.py
+++ b/micro_sam/prompt_generators.py
@@ -4,11 +4,14 @@
"""
from collections.abc import Mapping
-from typing import Optional
+from typing import Optional, Tuple
import numpy as np
from scipy.ndimage import binary_dilation
+import torch
+from kornia.morphology import dilation
+
class PointAndBoxPromptGenerator:
"""Generate point and/or box prompts from an instance segmentation.
@@ -52,34 +55,7 @@ def __init__(
if self.get_point_prompts is False and self.get_box_prompts is False:
raise ValueError("You need to request box prompts, point prompts or both.")
- def __call__(
- self,
- segmentation: np.ndarray,
- segmentation_id: int,
- bbox_coordinates: Mapping[int, tuple],
- center_coordinates: Optional[Mapping[int, np.ndarray]] = None
- ) -> tuple[
- Optional[list[tuple]], Optional[list[int]], Optional[list[tuple]], np.ndarray
- ]:
- """Generate the prompts for one object in the segmentation.
-
- Args:
- segmentation: The instance segmentation.
- segmentation_id: The ID of the instance.
- bbox_coordinates: The precomputed bounding boxes of all objects in the segmentation.
- center_coordinates: The precomputed center coordinates of all objects in the segmentation.
- If passed, these coordinates will be used as the first positive point prompt.
- If not passed a random point from within the object mask will be used.
-
- Returns:
- List of point coordinates. Returns None, if get_point_prompts is false.
- List of point labels. Returns None, if get_point_prompts is false.
- List containing the object bounding box. Returns None, if get_box_prompts is false.
- Object mask.
- """
- coord_list = []
- label_list = []
-
+ def _sample_positive_points(self, object_mask, center_coordinates, coord_list, label_list):
if center_coordinates is not None:
# getting the center coordinate as the first positive point (OPTIONAL)
coord_list.append(tuple(map(int, center_coordinates))) # to get int coords instead of float
@@ -93,11 +69,6 @@ def __call__(
# need to sample "self.n_positive_points" number of points
n_positive_remaining = self.n_positive_points
- if self.get_box_prompts:
- bbox_list = [bbox_coordinates]
-
- object_mask = segmentation == segmentation_id
-
if n_positive_remaining > 0:
# all coordinates of our current object
object_coordinates = np.where(object_mask)
@@ -117,11 +88,14 @@ def __call__(
coord_list.append(positive_coordinates)
label_list.append(1)
+ return coord_list, label_list
+
+ def _sample_negative_points(self, object_mask, bbox_coordinates, coord_list, label_list):
# getting the negative points
# for this we do the opposite and we set the mask to the bounding box - the object mask
# we need to dilate the object mask before doing this: we use scipy.ndimage.binary_dilation for this
dilated_object = binary_dilation(object_mask, iterations=self.dilation_strength)
- background_mask = np.zeros(segmentation.shape)
+ background_mask = np.zeros(object_mask.shape)
background_mask[bbox_coordinates[0]:bbox_coordinates[2], bbox_coordinates[1]:bbox_coordinates[3]] = 1
background_mask = binary_dilation(background_mask, iterations=self.dilation_strength)
background_mask = abs(
@@ -148,12 +122,175 @@ def __call__(
coord_list.append(negative_coordinates)
label_list.append(0)
- # returns object-level masks per instance for cross-verification (TODO: fix it later)
- if self.get_point_prompts is True and self.get_box_prompts is True: # we want points and box
- return coord_list, label_list, bbox_list, object_mask
+ return coord_list, label_list
+
+ def _ensure_num_points(self, object_mask, coord_list, label_list):
+ num_points = self.n_positive_points + self.n_negative_points
+
+ # fill up to the necessary number of points if we did not sample enough of them
+ if len(coord_list) != num_points:
+ # to stay consistent, we add random points in the background of an object
+ # if there's no neg region around the object - usually happens with small rois
+ needed_points = num_points - len(coord_list)
+ more_neg_points = np.where(object_mask == 0)
+ chosen_idx = np.random.choice(len(more_neg_points[0]), size=needed_points)
+
+ coord_list.extend([
+ (more_neg_points[0][idx], more_neg_points[1][idx]) for idx in chosen_idx
+ ])
+ label_list.extend([0] * needed_points)
+
+ assert len(coord_list) == len(label_list) == num_points
+ return coord_list, label_list
+
+ def _sample_points(self, object_mask, bbox_coordinates, center_coordinates):
+ coord_list, label_list = [], []
+
+ coord_list, label_list = self._sample_positive_points(object_mask, center_coordinates, coord_list, label_list)
+ coord_list, label_list = self._sample_negative_points(object_mask, bbox_coordinates, coord_list, label_list)
+ coord_list, label_list = self._ensure_num_points(object_mask, coord_list, label_list)
+
+ return coord_list, label_list
+
+ def __call__(
+ self,
+ segmentation: np.ndarray,
+ segmentation_id: int,
+ bbox_coordinates: Mapping[int, tuple],
+ center_coordinates: Optional[Mapping[int, np.ndarray]] = None
+ ) -> tuple[
+ Optional[list[tuple]], Optional[list[int]], Optional[list[tuple]], np.ndarray
+ ]:
+ """Generate the prompts for one object in the segmentation.
+
+ Args:
+ segmentation: The instance segmentation.
+ segmentation_id: The ID of the instance.
+ bbox_coordinates: The precomputed bounding boxes of all objects in the segmentation.
+ center_coordinates: The precomputed center coordinates of all objects in the segmentation.
+ If passed, these coordinates will be used as the first positive point prompt.
+ If not passed a random point from within the object mask will be used.
+
+ Returns:
+ List of point coordinates. Returns None, if get_point_prompts is false.
+ List of point labels. Returns None, if get_point_prompts is false.
+ List containing the object bounding box. Returns None, if get_box_prompts is false.
+ Object mask.
+ """
+ object_mask = segmentation == segmentation_id
+
+ if self.get_point_prompts:
+ coord_list, label_list = self._sample_points(object_mask, bbox_coordinates, center_coordinates)
+ else:
+ coord_list, label_list = None, None
+
+ if self.get_box_prompts:
+ bbox_list = [bbox_coordinates]
+ else:
+ bbox_list = None
+
+ return coord_list, label_list, bbox_list, object_mask
+
+
+class IterativePromptGenerator:
+ """Generate point prompts from an instance segmentation iteratively.
+ """
+ def _get_positive_points(self, pos_region, overlap_region):
+ positive_locations = [torch.where(pos_reg) for pos_reg in pos_region]
+ # we may have objects withput a positive region (= missing true foreground)
+ # in this case we just sample a point where the model was already correct
+ positive_locations = [
+ torch.where(ovlp_reg) if len(pos_loc[0]) == 0 else pos_loc
+ for pos_loc, ovlp_reg in zip(positive_locations, overlap_region)
+ ]
+ # we sample one location for each object in the batch
+ sampled_indices = [np.random.choice(len(pos_loc[0])) for pos_loc in positive_locations]
+ # get the corresponding coordinates (Note that we flip the axis order here due to the expected order of SAM)
+ pos_coordinates = [
+ [pos_loc[-1][idx], pos_loc[-2][idx]] for pos_loc, idx in zip(positive_locations, sampled_indices)
+ ]
+
+ # make sure that we still have the correct batch size
+ assert len(pos_coordinates) == pos_region.shape[0]
+ pos_labels = [1] * len(pos_coordinates)
+
+ return pos_coordinates, pos_labels
+
+ # TODO get rid of this looped implementation and use proper batched computation instead
+ def _get_negative_points(self, negative_region_batched, true_object_batched, gt_batched):
+ device = negative_region_batched.device
+
+ negative_coordinates, negative_labels = [], []
+ for neg_region, true_object, gt in zip(negative_region_batched, true_object_batched, gt_batched):
+
+ tmp_neg_loc = torch.where(neg_region)
+ if torch.stack(tmp_neg_loc).shape[-1] == 0:
+ tmp_true_loc = torch.where(true_object)
+ x_coords, y_coords = tmp_true_loc[1], tmp_true_loc[2]
+ bbox = torch.stack([torch.min(x_coords), torch.min(y_coords),
+ torch.max(x_coords) + 1, torch.max(y_coords) + 1])
+ bbox_mask = torch.zeros_like(true_object).squeeze(0)
+ bbox_mask[bbox[0]:bbox[2], bbox[1]:bbox[3]] = 1
+ bbox_mask = bbox_mask[None].to(device)
+
+ # NOTE: FIX: here we add dilation to the bbox because in some case we couldn't find objects at all
+ # TODO: just expand the pixels of bbox
+ dilated_bbox_mask = dilation(bbox_mask[None], torch.ones(3, 3).to(device)).squeeze(0)
+ background_mask = abs(dilated_bbox_mask - true_object)
+ tmp_neg_loc = torch.where(background_mask)
+
+ # there is a chance that the object is small to not return a decent-sized bounding box
+ # hence we might not find points sometimes there as well, hence we sample points from true background
+ if torch.stack(tmp_neg_loc).shape[-1] == 0:
+ tmp_neg_loc = torch.where(gt == 0)
+
+ neg_index = np.random.choice(len(tmp_neg_loc[1]))
+ neg_coordinates = [tmp_neg_loc[1][neg_index], tmp_neg_loc[2][neg_index]]
+ neg_coordinates = neg_coordinates[::-1]
+ neg_labels = 0
+
+ negative_coordinates.append(neg_coordinates)
+ negative_labels.append(neg_labels)
+
+ return negative_coordinates, negative_labels
+
+ def __call__(
+ self,
+ gt: torch.Tensor,
+ object_mask: torch.Tensor,
+ current_points: torch.Tensor,
+ current_labels: torch.Tensor
+ ) -> Tuple[torch.Tensor, torch.Tensor]:
+ """Generate the prompts for each object iteratively in the segmentation.
+
+ Args:
+ The groundtruth segmentation.
+ The predicted objects.
+ The current points.
+ Thr current labels.
+
+ Returns:
+ The updated point prompt coordinates.
+ The updated point prompt labels.
+ """
+ assert gt.shape == object_mask.shape
+ device = object_mask.device
+
+ true_object = gt.to(device)
+ expected_diff = (object_mask - true_object)
+ neg_region = (expected_diff == 1).to(torch.float)
+ pos_region = (expected_diff == -1)
+ overlap_region = torch.logical_and(object_mask == 1, true_object == 1).to(torch.float32)
+
+ pos_coordinates, pos_labels = self._get_positive_points(pos_region, overlap_region)
+ neg_coordinates, neg_labels = self._get_negative_points(neg_region, true_object, gt)
+ assert len(pos_coordinates) == len(pos_labels) == len(neg_coordinates) == len(neg_labels)
+
+ pos_coordinates = torch.tensor(pos_coordinates)[:, None]
+ neg_coordinates = torch.tensor(neg_coordinates)[:, None]
+ pos_labels, neg_labels = torch.tensor(pos_labels)[:, None], torch.tensor(neg_labels)[:, None]
- elif self.get_point_prompts is True and self.get_box_prompts is False: # we want only points
- return coord_list, label_list, None, object_mask
+ net_coords = torch.cat([current_points, pos_coordinates, neg_coordinates], dim=1)
+ net_labels = torch.cat([current_labels, pos_labels, neg_labels], dim=1)
- elif self.get_point_prompts is False and self.get_box_prompts is True: # we want only boxes
- return None, None, bbox_list, object_mask
+ return net_coords, net_labels
diff --git a/micro_sam/sam_annotator/annotator.py b/micro_sam/sam_annotator/annotator.py
index 0c8fae27..9be755f5 100644
--- a/micro_sam/sam_annotator/annotator.py
+++ b/micro_sam/sam_annotator/annotator.py
@@ -9,7 +9,7 @@
from magicgui.application import use_app
from PyQt5.QtWidgets import QFileDialog, QMessageBox
-from ..util import load_image_data, get_model_names
+from ..util import load_image_data, get_model_names, _DEFAULT_MODEL
from .annotator_2d import annotator_2d
from .annotator_3d import annotator_3d
from .image_series_annotator import image_folder_annotator
@@ -64,7 +64,7 @@ def _on_2d():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select image", labels=False)
def on_select_image():
@@ -156,7 +156,7 @@ def _on_3d():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select images", labels=False)
def on_select_image():
@@ -271,7 +271,7 @@ def _on_series():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select input directory", labels=False)
def on_select_input_dir():
@@ -364,7 +364,7 @@ def _on_tracking():
re_halo_y = SpinBox(value=0, max=10000, label="Halo y")
re_tile_x = SpinBox(value=0, max=10000, label="Tile x")
re_tile_y = SpinBox(value=0, max=10000, label="Tile y")
- cb_model = ComboBox(value="vit_h_lm", choices=get_model_names(), label="Model Type")
+ cb_model = ComboBox(value=_DEFAULT_MODEL, choices=get_model_names(), label="Model Type")
@magicgui.magicgui(call_button="Select images", labels=False)
def on_select_image():
diff --git a/micro_sam/sam_annotator/annotator_2d.py b/micro_sam/sam_annotator/annotator_2d.py
index 73fd5159..eceb19db 100644
--- a/micro_sam/sam_annotator/annotator_2d.py
+++ b/micro_sam/sam_annotator/annotator_2d.py
@@ -8,8 +8,8 @@
from napari import Viewer
from segment_anything import SamPredictor
-from .. import util
-from .. import instance_segmentation
+from .. import instance_segmentation, util
+from ..precompute_state import cache_amg_state
from ..visualization import project_embeddings_for_visualization
from . import util as vutil
from .gui_utils import show_wrong_file_warning
@@ -38,18 +38,6 @@ def _segment_widget(v: Viewer) -> None:
v.layers["current_object"].refresh()
-def _get_amg(is_tiled, with_background, min_initial_size, box_extension=0.05):
- if is_tiled:
- amg = instance_segmentation.TiledEmbeddingMaskGenerator(
- PREDICTOR, with_background=with_background, min_initial_size=min_initial_size, box_extension=box_extension,
- )
- else:
- amg = instance_segmentation.EmbeddingMaskGenerator(
- PREDICTOR, min_initial_size=min_initial_size, box_extension=box_extension,
- )
- return amg
-
-
def _changed_param(amg, **params):
if amg is None:
return None
@@ -62,29 +50,25 @@ def _changed_param(amg, **params):
@magicgui(call_button="Automatic Segmentation")
def _autosegment_widget(
v: Viewer,
- with_background: bool = True,
pred_iou_thresh: float = 0.88,
stability_score_thresh: float = 0.95,
- min_initial_size: int = 10,
- box_extension: float = 0.05,
+ min_object_size: int = 100,
+ with_background: bool = True,
) -> None:
global AMG
is_tiled = IMAGE_EMBEDDINGS["input_size"] is None
- param_changed = _changed_param(
- AMG, with_background=with_background, min_initial_size=min_initial_size, box_extension=box_extension
- )
- if AMG is None or param_changed:
- if param_changed:
- print(f"The parameter {param_changed} was changed, so the full instance segmentation has to be recomputed.")
- AMG = _get_amg(is_tiled, with_background, min_initial_size, box_extension)
+ if AMG is None:
+ AMG = instance_segmentation.get_amg(PREDICTOR, is_tiled)
if not AMG.is_initialized:
AMG.initialize(v.layers["raw"].data, image_embeddings=IMAGE_EMBEDDINGS, verbose=True)
seg = AMG.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
- if not is_tiled:
- shape = v.layers["raw"].data.shape[:2]
- seg = instance_segmentation.mask_data_to_segmentation(seg, shape, with_background)
+
+ shape = v.layers["raw"].data.shape[:2]
+ seg = instance_segmentation.mask_data_to_segmentation(
+ seg, shape, with_background=True, min_object_size=min_object_size
+ )
assert isinstance(seg, np.ndarray)
v.layers["auto_segmentation"].data = seg
@@ -197,12 +181,13 @@ def annotator_2d(
embedding_path: Optional[str] = None,
show_embeddings: bool = False,
segmentation_result: Optional[np.ndarray] = None,
- model_type: str = "vit_h",
+ model_type: str = util._DEFAULT_MODEL,
tile_shape: Optional[Tuple[int, int]] = None,
halo: Optional[Tuple[int, int]] = None,
return_viewer: bool = False,
v: Optional[Viewer] = None,
predictor: Optional[SamPredictor] = None,
+ precompute_amg_state: bool = False,
) -> Optional[Viewer]:
"""The 2d annotation tool.
@@ -225,6 +210,9 @@ def annotator_2d(
This enables using a pre-initialized viewer, for example in `sam_annotator.image_series_annotator`.
predictor: The Segment Anything model. Passing this enables using fully custom models.
If you pass `predictor` then `model_type` will be ignored.
+ precompute_amg_state: Whether to precompute the state for automatic mask generation.
+ This will take more time when precomputing embeddings, but will then make
+ automatic mask generation much faster.
Returns:
The napari viewer, only returned if `return_viewer=True`.
@@ -237,10 +225,13 @@ def annotator_2d(
PREDICTOR = util.get_sam_model(model_type=model_type)
else:
PREDICTOR = predictor
+
IMAGE_EMBEDDINGS = util.precompute_image_embeddings(
PREDICTOR, raw, save_path=embedding_path, ndim=2, tile_shape=tile_shape, halo=halo,
wrong_file_callback=show_wrong_file_warning
)
+ if precompute_amg_state and (embedding_path is not None):
+ AMG = cache_amg_state(PREDICTOR, raw, IMAGE_EMBEDDINGS, embedding_path)
# we set the pre-computed image embeddings if we don't use tiling
# (if we use tiling we cannot directly set it because the tile will be chosen dynamically)
@@ -268,6 +259,7 @@ def annotator_2d(
def main():
"""@private"""
parser = vutil._initialize_parser(description="Run interactive segmentation for an image.")
+ parser.add_argument("--precompute_amg_state", action="store_true")
args = parser.parse_args()
raw = util.load_image_data(args.input, key=args.key)
@@ -283,4 +275,5 @@ def main():
raw, embedding_path=args.embedding_path,
show_embeddings=args.show_embeddings, segmentation_result=segmentation_result,
model_type=args.model_type, tile_shape=args.tile_shape, halo=args.halo,
+ precompute_amg_state=args.precompute_amg_state,
)
diff --git a/micro_sam/sam_annotator/annotator_3d.py b/micro_sam/sam_annotator/annotator_3d.py
index 6518336b..ba1740fd 100644
--- a/micro_sam/sam_annotator/annotator_3d.py
+++ b/micro_sam/sam_annotator/annotator_3d.py
@@ -154,7 +154,7 @@ def _segment_slice_wigdet(v: Viewer) -> None:
@magicgui(call_button="Segment Volume [V]", projection={"choices": ["default", "bounding_box", "mask", "points"]})
def _segment_volume_widget(
- v: Viewer, iou_threshold: float = 0.8, projection: str = "default", box_extension: float = 0.1
+ v: Viewer, iou_threshold: float = 0.8, projection: str = "default", box_extension: float = 0.05
) -> None:
# step 1: segment all slices with prompts
shape = v.layers["raw"].data.shape
@@ -192,7 +192,7 @@ def annotator_3d(
embedding_path: Optional[str] = None,
show_embeddings: bool = False,
segmentation_result: Optional[np.ndarray] = None,
- model_type: str = "vit_h",
+ model_type: str = util._DEFAULT_MODEL,
tile_shape: Optional[Tuple[int, int]] = None,
halo: Optional[Tuple[int, int]] = None,
return_viewer: bool = False,
diff --git a/micro_sam/sam_annotator/annotator_tracking.py b/micro_sam/sam_annotator/annotator_tracking.py
index e22a9172..691cd6ae 100644
--- a/micro_sam/sam_annotator/annotator_tracking.py
+++ b/micro_sam/sam_annotator/annotator_tracking.py
@@ -1,4 +1,6 @@
+import json
import warnings
+from pathlib import Path
from typing import Optional, Tuple
import napari
@@ -24,6 +26,9 @@
STATE_COLOR_CYCLE = ["#00FFFF", "#FF00FF", ]
"""@private"""
+COMMITTED_LINEAGES = []
+"""@private"""
+
#
# util functionality
@@ -336,6 +341,8 @@ def _reset_tracking_state():
@magicgui(call_button="Commit [C]", layer={"choices": ["current_track"]})
def _commit_tracking_widget(v: Viewer, layer: str = "current_track") -> None:
+ global COMMITTED_LINEAGES
+
seg = v.layers[layer].data
id_offset = int(v.layers["committed_tracks"].data.max())
@@ -348,6 +355,11 @@ def _commit_tracking_widget(v: Viewer, layer: str = "current_track") -> None:
v.layers[layer].data = np.zeros(shape, dtype="uint32")
v.layers[layer].refresh()
+ updated_lineage = {
+ parent + id_offset: [child + id_offset for child in children] for parent, children in LINEAGE.items()
+ }
+ COMMITTED_LINEAGES.append(updated_lineage)
+
_reset_tracking_state()
vutil.clear_annotations(v, clear_segmentations=False)
@@ -358,12 +370,19 @@ def _clear_widget_tracking(v: Viewer) -> None:
vutil.clear_annotations(v)
+@magicgui(call_button="Save Lineage")
+def _save_lineage_widget(v: Viewer, path: Path) -> None:
+ path = path.with_suffix(".json")
+ with open(path, "w") as f:
+ json.dump(COMMITTED_LINEAGES, f)
+
+
def annotator_tracking(
raw: np.ndarray,
embedding_path: Optional[str] = None,
show_embeddings: bool = False,
tracking_result: Optional[str] = None,
- model_type: str = "vit_h",
+ model_type: str = util._DEFAULT_MODEL,
tile_shape: Optional[Tuple[int, int]] = None,
halo: Optional[Tuple[int, int]] = None,
return_viewer: bool = False,
@@ -486,6 +505,7 @@ def annotator_tracking(
v.window.add_dock_widget(_segment_frame_wigdet)
v.window.add_dock_widget(_track_objet_widget)
v.window.add_dock_widget(_commit_tracking_widget)
+ v.window.add_dock_widget(_save_lineage_widget)
v.window.add_dock_widget(_clear_widget_tracking)
#
diff --git a/micro_sam/sam_annotator/gui_utils.py b/micro_sam/sam_annotator/gui_utils.py
index 8965daf6..fb412429 100644
--- a/micro_sam/sam_annotator/gui_utils.py
+++ b/micro_sam/sam_annotator/gui_utils.py
@@ -47,36 +47,26 @@ def _overwrite():
@magicgui.magicgui(call_button="Create new file", labels=False)
def _create():
- msg_box.close()
# unfortunately there exists no dialog to create a directory so we have
# to use "create new file" dialog with some adjustments.
dialog = QtWidgets.QFileDialog(None)
dialog.setFileMode(QtWidgets.QFileDialog.AnyFile)
dialog.setOption(QtWidgets.QFileDialog.ShowDirsOnly)
dialog.setNameFilter("Archives (*.zarr)")
- try_cnt = 0
- while os.path.splitext(new_path["value"])[1] != ".zarr":
- if try_cnt > 3:
- new_path["value"] = file_path
- return
- dialog.exec_()
- res = dialog.selectedFiles()
- new_path["value"] = res[0] if len(res) > 0 else ""
- try_cnt += 1
+ dialog.exec_()
+ res = dialog.selectedFiles()
+ new_path["value"] = res[0] if len(res) > 0 else ""
+ if os.path.splitext(new_path["value"])[1] != ".zarr":
+ new_path["value"] += ".zarr"
os.makedirs(new_path["value"])
+ msg_box.close()
@magicgui.magicgui(call_button="Select different file", labels=False)
def _select():
+ new_path["value"] = QtWidgets.QFileDialog.getExistingDirectory(
+ None, "Open a folder", os.path.split(file_path)[0], QtWidgets.QFileDialog.ShowDirsOnly
+ )
msg_box.close()
- try_cnt = 0
- while not os.path.exists(new_path["value"]):
- if try_cnt > 3:
- new_path["value"] = file_path
- return
- new_path["value"] = QtWidgets.QFileDialog.getExistingDirectory(
- None, "Open a folder", os.path.split(file_path)[0], QtWidgets.QFileDialog.ShowDirsOnly
- )
- try_cnt += 1
msg_box = Container(widgets=[_select, _ignore, _overwrite, _create], layout='horizontal', labels=False)
msg_box.root_native_widget.setWindowTitle("The input data does not match the embeddings file")
diff --git a/micro_sam/sam_annotator/image_series_annotator.py b/micro_sam/sam_annotator/image_series_annotator.py
index 80fa7602..d561b7d5 100644
--- a/micro_sam/sam_annotator/image_series_annotator.py
+++ b/micro_sam/sam_annotator/image_series_annotator.py
@@ -1,33 +1,19 @@
import os
import warnings
+
from glob import glob
+from pathlib import Path
from typing import List, Optional, Union
import imageio.v3 as imageio
import napari
from magicgui import magicgui
-from napari.utils import progress as tqdm
from segment_anything import SamPredictor
-from .annotator_2d import annotator_2d
from .. import util
-
-
-def _precompute_embeddings_for_image_series(predictor, image_files, embedding_root, tile_shape, halo):
- os.makedirs(embedding_root, exist_ok=True)
- embedding_paths = []
- for image_file in tqdm(image_files, desc="Precompute embeddings"):
- fname = os.path.basename(image_file)
- fname = os.path.splitext(fname)[0] + ".zarr"
- embedding_path = os.path.join(embedding_root, fname)
- image = imageio.imread(image_file)
- util.precompute_image_embeddings(
- predictor, image, save_path=embedding_path, ndim=2,
- tile_shape=tile_shape, halo=halo
- )
- embedding_paths.append(embedding_path)
- return embedding_paths
+from ..precompute_state import _precompute_state_for_files
+from .annotator_2d import annotator_2d
def image_series_annotator(
@@ -47,6 +33,7 @@ def image_series_annotator(
embedding_path: Filepath where to save the embeddings.
predictor: The Segment Anything model. Passing this enables using fully custom models.
If you pass `predictor` then `model_type` will be ignored.
+ kwargs: The keywored arguments for `micro_sam.sam_annotator.annotator_2d`.
"""
# make sure we don't set incompatible kwargs
assert kwargs.get("show_embeddings", False) is False
@@ -58,13 +45,20 @@ def image_series_annotator(
next_image_id = 0
if predictor is None:
- predictor = util.get_sam_model(model_type=kwargs.get("model_type", "vit_h"))
+ predictor = util.get_sam_model(model_type=kwargs.get("model_type", util._DEFAULT_MODEL))
if embedding_path is None:
embedding_paths = None
else:
- embedding_paths = _precompute_embeddings_for_image_series(
- predictor, image_files, embedding_path, kwargs.get("tile_shape", None), kwargs.get("halo", None)
+ _precompute_state_for_files(
+ predictor, image_files, embedding_path, ndim=2,
+ tile_shape=kwargs.get("tile_shape", None),
+ halo=kwargs.get("halo", None),
+ precompute_amg_state=kwargs.get("precompute_amg_state", False),
)
+ embedding_paths = [
+ os.path.join(embedding_path, f"{Path(path).stem}.zarr") for path in image_files
+ ]
+ assert all(os.path.exists(emb_path) for emb_path in embedding_paths)
def _save_segmentation(image_path, segmentation):
fname = os.path.basename(image_path)
@@ -127,6 +121,7 @@ def image_folder_annotator(
embedding_path: Filepath where to save the embeddings.
predictor: The Segment Anything model. Passing this enables using fully custom models.
If you pass `predictor` then `model_type` will be ignored.
+ kwargs: The keywored arguments for `micro_sam.sam_annotator.annotator_2d`.
"""
image_files = sorted(glob(os.path.join(input_folder, pattern)))
image_series_annotator(image_files, output_folder, embedding_path, predictor, **kwargs)
@@ -136,6 +131,9 @@ def main():
"""@private"""
import argparse
+ available_models = list(util.get_model_names())
+ available_models = ", ".join(available_models)
+
parser = argparse.ArgumentParser(description="Annotate a series of images from a folder.")
parser.add_argument(
"-i", "--input_folder", required=True,
@@ -157,7 +155,8 @@ def main():
"otherwise they will be recomputed every time (which can take a long time)."
)
parser.add_argument(
- "--model_type", default="vit_h", help="The segment anything model that will be used, one of vit_h,l,b."
+ "--model_type", default=util._DEFAULT_MODEL,
+ help=f"The segment anything model that will be used, one of {available_models}."
)
parser.add_argument(
"--tile_shape", nargs="+", type=int, help="The tile shape for using tiled prediction", default=None
@@ -165,6 +164,7 @@ def main():
parser.add_argument(
"--halo", nargs="+", type=int, help="The halo for using tiled prediction", default=None
)
+ parser.add_argument("--precompute_amg_state", action="store_true")
args = parser.parse_args()
@@ -175,4 +175,5 @@ def main():
args.input_folder, args.output_folder, args.pattern,
embedding_path=args.embedding_path, model_type=args.model_type,
tile_shape=args.tile_shape, halo=args.halo,
+ precompute_amg_state=args.precompute_amg_state,
)
diff --git a/micro_sam/sam_annotator/util.py b/micro_sam/sam_annotator/util.py
index e4dbf20d..48a02622 100644
--- a/micro_sam/sam_annotator/util.py
+++ b/micro_sam/sam_annotator/util.py
@@ -1,4 +1,6 @@
import argparse
+import os
+import pickle
from typing import Optional, Tuple
import napari
@@ -7,6 +9,7 @@
from magicgui import magicgui
from magicgui.widgets import ComboBox, Container
+from .. import instance_segmentation, util
from ..prompt_based_segmentation import segment_from_box, segment_from_box_and_points, segment_from_points
# Green and Red
@@ -362,6 +365,10 @@ def toggle_label(prompts):
def _initialize_parser(description, with_segmentation_result=True, with_show_embeddings=True):
+
+ available_models = list(util.get_model_names())
+ available_models = ", ".join(available_models)
+
parser = argparse.ArgumentParser(description=description)
parser.add_argument(
@@ -401,7 +408,8 @@ def _initialize_parser(description, with_segmentation_result=True, with_show_emb
help="Visualize the embeddings computed by SegmentAnything. This can be helpful for debugging."
)
parser.add_argument(
- "--model_type", default="vit_h", help="The segment anything model that will be used, one of vit_h,l,b."
+ "--model_type", default=util._DEFAULT_MODEL,
+ help=f"The segment anything model that will be used, one of {available_models}."
)
parser.add_argument(
"--tile_shape", nargs="+", type=int, help="The tile shape for using tiled prediction", default=None
diff --git a/micro_sam/sample_data.py b/micro_sam/sample_data.py
index 533d97ae..8c9523c5 100644
--- a/micro_sam/sample_data.py
+++ b/micro_sam/sample_data.py
@@ -11,7 +11,57 @@
import pooch
-def fetch_wholeslide_example_data(save_directory: Union[str, os.PathLike]) -> Union[str, os.PathLike]:
+def fetch_image_series_example_data(save_directory: Union[str, os.PathLike]) -> str:
+ """Download the sample images for the image series annotator.
+
+ Args:
+ save_directory: Root folder to save the downloaded data.
+ Returns:
+ The folder that contains the downloaded data.
+ """
+ # This sample dataset is currently not provided to napari by the micro-sam
+ # plugin, because images are not all the same shape and cannot be combined
+ # into a single layer
+ save_directory = Path(save_directory)
+ os.makedirs(save_directory, exist_ok=True)
+ print("Example data directory is:", save_directory.resolve())
+ fname = "image-series.zip"
+ unpack_filenames = [os.path.join("series", f"im{i}.tif") for i in range(3)]
+ unpack = pooch.Unzip(members=unpack_filenames)
+ pooch.retrieve(
+ url="https://owncloud.gwdg.de/index.php/s/M1zGnfkulWoAhUG/download",
+ known_hash="92346ca9770bcaf55248efee590718d54c7135b6ebca15d669f3b77b6afc8706",
+ fname=fname,
+ path=save_directory,
+ progressbar=True,
+ processor=unpack,
+ )
+ data_folder = os.path.join(save_directory, f"{fname}.unzip", "series")
+ assert os.path.exists(data_folder)
+ return data_folder
+
+
+def sample_data_image_series():
+ """Provides image series example image to napari.
+
+ Opens as three separate image layers in napari (one per image in series).
+ The third image in the series has a different size and modality.
+ """
+ # Return list of tuples
+ # [(data1, add_image_kwargs1), (data2, add_image_kwargs2)]
+ # Check the documentation for more information about the
+ # add_image_kwargs
+ # https://napari.org/stable/api/napari.Viewer.html#napari.Viewer.add_image
+ default_base_data_dir = pooch.os_cache('micro-sam')
+ data_directory = fetch_image_series_example_data(default_base_data_dir)
+ fnames = os.listdir(data_directory)
+ full_filenames = [os.path.join(data_directory, f) for f in fnames]
+ full_filenames.sort()
+ data_and_image_kwargs = [(imageio.imread(f), {"name": f"img-{i}"}) for i, f in enumerate(full_filenames)]
+ return data_and_image_kwargs
+
+
+def fetch_wholeslide_example_data(save_directory: Union[str, os.PathLike]) -> str:
"""Download the sample data for the 2d annotator.
This downloads part of a whole-slide image from the NeurIPS Cell Segmentation Challenge.
@@ -220,3 +270,49 @@ def sample_data_tracking():
data = np.stack([imageio.imread(f) for f in full_filenames], axis=0)
add_image_kwargs = {"name": "tracking"}
return [(data, add_image_kwargs)]
+
+
+def fetch_tracking_segmentation_data(save_directory: Union[str, os.PathLike]) -> str:
+ """Download groundtruth segmentation for the tracking example data.
+
+ This downloads the groundtruth segmentation for the image data from `fetch_tracking_example_data`.
+
+ Args:
+ save_directory: Root folder to save the downloaded data.
+ Returns:
+ The folder that contains the downloaded data.
+ """
+ save_directory = Path(save_directory)
+ os.makedirs(save_directory, exist_ok=True)
+ print("Example data directory is:", save_directory.resolve())
+ unpack_filenames = [os.path.join("masks", f"mask_{str(i).zfill(4)}.tif") for i in range(84)]
+ unpack = pooch.Unzip(members=unpack_filenames)
+ fname = "hela-ctc-01-gt.zip"
+ pooch.retrieve(
+ url="https://owncloud.gwdg.de/index.php/s/AWxQMblxwR99OjC/download",
+ known_hash="c0644d8ebe1390fb60125560ba15aa2342caf44f50ff0667a0318ea0ac6c958b",
+ fname=fname,
+ path=save_directory,
+ progressbar=True,
+ processor=unpack,
+ )
+ cell_tracking_dir = save_directory.joinpath(f"{fname}.unzip", "masks")
+ assert os.path.exists(cell_tracking_dir)
+ return str(cell_tracking_dir)
+
+
+def sample_data_segmentation():
+ """Provides segmentation example dataset to napari."""
+ # Return list of tuples
+ # [(data1, add_image_kwargs1), (data2, add_image_kwargs2)]
+ # Check the documentation for more information about the
+ # add_image_kwargs
+ # https://napari.org/stable/api/napari.Viewer.html#napari.Viewer.add_image
+ default_base_data_dir = pooch.os_cache("micro-sam")
+ data_directory = fetch_tracking_segmentation_data(default_base_data_dir)
+ fnames = os.listdir(data_directory)
+ full_filenames = [os.path.join(data_directory, f) for f in fnames]
+ full_filenames.sort()
+ data = np.stack([imageio.imread(f) for f in full_filenames], axis=0)
+ add_image_kwargs = {"name": "segmentation"}
+ return [(data, add_image_kwargs)]
diff --git a/micro_sam/training/__init__.py b/micro_sam/training/__init__.py
new file mode 100644
index 00000000..225b6568
--- /dev/null
+++ b/micro_sam/training/__init__.py
@@ -0,0 +1,5 @@
+"""Functionality for training Segment Anything.
+"""
+
+from .sam_trainer import SamTrainer, SamLogger
+from .util import ConvertToSamInputs, get_trainable_sam_model
diff --git a/micro_sam/training/sam_trainer.py b/micro_sam/training/sam_trainer.py
new file mode 100644
index 00000000..9a30554b
--- /dev/null
+++ b/micro_sam/training/sam_trainer.py
@@ -0,0 +1,458 @@
+import os
+import time
+from typing import Optional
+
+import numpy as np
+import torch
+import torch_em
+
+from kornia.morphology import dilation
+from torchvision.utils import make_grid
+from torch_em.trainer.logger_base import TorchEmLogger
+
+
+class SamTrainer(torch_em.trainer.DefaultTrainer):
+ """Trainer class for training the Segment Anything model.
+
+ This class is derived from `torch_em.trainer.DefaultTrainer`.
+ Check out https://github.com/constantinpape/torch-em/blob/main/torch_em/trainer/default_trainer.py
+ for details on its usage and implementation.
+
+ Args:
+ convert_inputs: Class that converts the output of the dataloader to the expected input format of SAM.
+ The class `micro_sam.training.util.ConvertToSamInputs` can be used here.
+ n_sub_iteration: The number of iteration steps for which the masks predicted for one object are updated.
+ In each sub-iteration new point prompts are sampled where the model was wrong.
+ n_objects_per_batch: If not given, we compute the loss for all objects in a sample.
+ Otherwise the loss computation is limited to n_objects_per_batch, and the objects are randomly sampled.
+ mse_loss: The regression loss to compare the IoU predicted by the model with the true IoU.
+ sigmoid: The activation function for normalizing the model output.
+ **kwargs: The keyword arguments of the DefaultTrainer super class.
+ """
+
+ def __init__(
+ self,
+ convert_inputs,
+ n_sub_iteration: int,
+ n_objects_per_batch: Optional[int] = None,
+ mse_loss: torch.nn.Module = torch.nn.MSELoss(),
+ _sigmoid: torch.nn.Module = torch.nn.Sigmoid(),
+ **kwargs
+ ):
+ super().__init__(**kwargs)
+ self.convert_inputs = convert_inputs
+ self.mse_loss = mse_loss
+ self._sigmoid = _sigmoid
+ self.n_objects_per_batch = n_objects_per_batch
+ self.n_sub_iteration = n_sub_iteration
+ self._kwargs = kwargs
+
+ def _get_prompt_and_multimasking_choices(self, current_iteration):
+ """Choose the type of prompts we sample for training, and then we call
+ 'convert_inputs' with the correct prompting from here.
+ """
+ if current_iteration % 2 == 0: # sample only a single point per object
+ n_pos, n_neg = 1, 0
+ get_boxes = False
+ multimask_output = True
+
+ else: # sample only a single box per object
+ n_pos, n_neg = 0, 0
+ get_boxes = True
+ multimask_output = False
+
+ return n_pos, n_neg, get_boxes, multimask_output
+
+ def _get_prompt_and_multimasking_choices_for_val(self, current_iteration):
+ """Choose the type of prompts we sample for validation, and then we call
+ 'convert_inputs' with the correct prompting from here.
+ """
+ if current_iteration % 4 == 0: # sample only a single point per object
+ n_pos, n_neg = 1, 0
+ get_boxes = False
+ multimask_output = True
+
+ elif current_iteration % 4 == 1: # sample only a single box per object
+ n_pos, n_neg = 0, 0
+ get_boxes = True
+ multimask_output = False
+
+ elif current_iteration % 4 == 2: # sample a random no. of points
+ pos_range, neg_range = 4, 4
+
+ n_pos = np.random.randint(1, pos_range + 1)
+ if n_pos == 1: # to avoid (1, 0) combination for redundancy but still have (n_pos, 0)
+ n_neg = np.random.randint(1, neg_range + 1)
+ else:
+ n_neg = np.random.randint(0, neg_range + 1)
+ get_boxes = False
+ multimask_output = False
+
+ else: # sample boxes AND random no. of points
+ # here we can have (1, 0) because we also have box
+ pos_range, neg_range = 4, 4
+
+ n_pos = np.random.randint(1, pos_range + 1)
+ n_neg = np.random.randint(0, neg_range + 1)
+ get_boxes = True
+ multimask_output = False
+
+ return n_pos, n_neg, get_boxes, multimask_output
+
+ def _get_dice(self, input_, target):
+ """Using the default "DiceLoss" called by the trainer from "torch_em"
+ """
+ dice_loss = self.loss(input_, target)
+ return dice_loss
+
+ def _get_iou(self, pred, true, eps=1e-7):
+ """Getting the IoU score for the predicted and true labels
+ """
+ pred_mask = pred > 0.5 # binarizing the output predictions
+ overlap = pred_mask.logical_and(true).sum()
+ union = pred_mask.logical_or(true).sum()
+ iou = overlap / (union + eps)
+ return iou
+
+ def _get_net_loss(self, batched_outputs, y, sampled_ids):
+ """What do we do here? two **separate** things
+ 1. compute the mask loss: loss between the predicted and ground-truth masks
+ for this we just use the dice of the prediction vs. the gt (binary) mask
+ 2. compute the mask for the "IOU Regression Head": so we want the iou output from the decoder to
+ match the actual IOU between predicted and (binary) ground-truth mask. And we use L2Loss / MSE for this.
+ """
+ masks = [m["masks"] for m in batched_outputs]
+ predicted_iou_values = [m["iou_predictions"] for m in batched_outputs]
+ with torch.no_grad():
+ mean_model_iou = torch.mean(torch.stack([p.mean() for p in predicted_iou_values]))
+
+ mask_loss = 0.0 # this is the loss term for 1.
+ iou_regression_loss = 0.0 # this is the loss term for 2.
+
+ # outer loop is over the batch (different image/patch predictions)
+ for m_, y_, ids_, predicted_iou_ in zip(masks, y, sampled_ids, predicted_iou_values):
+ per_object_dice_scores = []
+ per_object_iou_scores = []
+
+ # inner loop is over the channels, this corresponds to the different predicted objects
+ for i, (predicted_obj, predicted_iou) in enumerate(zip(m_, predicted_iou_)):
+ predicted_obj = self._sigmoid(predicted_obj).to(self.device)
+ true_obj = (y_ == ids_[i]).to(self.device)
+
+ # this is computing the LOSS for 1.)
+ _dice_score = min([self._get_dice(p[None], true_obj) for p in predicted_obj])
+ per_object_dice_scores.append(_dice_score)
+
+ # now we need to compute the loss for 2.)
+ with torch.no_grad():
+ true_iou = torch.stack([self._get_iou(p[None], true_obj) for p in predicted_obj])
+ _iou_score = self.mse_loss(true_iou, predicted_iou)
+ per_object_iou_scores.append(_iou_score)
+
+ mask_loss = mask_loss + torch.mean(torch.stack(per_object_dice_scores))
+ iou_regression_loss = iou_regression_loss + torch.mean(torch.stack(per_object_iou_scores))
+
+ loss = mask_loss + iou_regression_loss
+
+ return loss, mask_loss, iou_regression_loss, mean_model_iou
+
+ def _postprocess_outputs(self, masks):
+ """ masks look like -> (B, 1, X, Y)
+ where, B is the number of objects, (X, Y) is the input image shape
+ """
+ instance_labels = []
+ for m in masks:
+ instance_list = [self._sigmoid(_val) for _val in m.squeeze(1)]
+ instance_label = torch.stack(instance_list, dim=0).sum(dim=0).clip(0, 1)
+ instance_labels.append(instance_label)
+ instance_labels = torch.stack(instance_labels).unsqueeze(1)
+ return instance_labels
+
+ def _get_val_metric(self, batched_outputs, sampled_binary_y):
+ """ Tracking the validation metric based on the DiceLoss
+ """
+ masks = [m["masks"] for m in batched_outputs]
+ pred_labels = self._postprocess_outputs(masks)
+
+ # we do the condition below to adapt w.r.t. the multimask output
+ # to select the "objectively" best response
+ if pred_labels.dim() == 5:
+ metric = min([self.metric(pred_labels[:, :, i, :, :], sampled_binary_y.to(self.device))
+ for i in range(pred_labels.shape[2])])
+ else:
+ metric = self.metric(pred_labels, sampled_binary_y.to(self.device))
+
+ return metric
+
+ #
+ # Update Masks Iteratively while Training
+ #
+ def _update_masks(self, batched_inputs, y, sampled_binary_y, sampled_ids, num_subiter, multimask_output):
+ # estimating the image inputs to make the computations faster for the decoder
+ input_images = torch.stack([self.model.preprocess(x=x["image"].to(self.device)) for x in batched_inputs], dim=0)
+ image_embeddings = self.model.image_embeddings_oft(input_images)
+
+ loss = 0.0
+ mask_loss = 0.0
+ iou_regression_loss = 0.0
+ mean_model_iou = 0.0
+
+ # this loop takes care of the idea of sub-iterations, i.e. the number of times we iterate over each batch
+ for i in range(0, num_subiter):
+ # we do multimasking only in the first sub-iteration as we then pass single prompt
+ # after the first sub-iteration, we don't do multimasking because we get multiple prompts
+ batched_outputs = self.model(batched_inputs,
+ multimask_output=multimask_output if i == 0 else False,
+ image_embeddings=image_embeddings)
+
+ # we want to average the loss and then backprop over the net sub-iterations
+ net_loss, net_mask_loss, net_iou_regression_loss, net_mean_model_iou = self._get_net_loss(batched_outputs,
+ y, sampled_ids)
+ loss += net_loss
+ mask_loss += net_mask_loss
+ iou_regression_loss += net_iou_regression_loss
+ mean_model_iou += net_mean_model_iou
+
+ masks, logits_masks = [], []
+ # the loop below gets us the masks and logits from the batch-level outputs
+ for m in batched_outputs:
+ mask, l_mask = [], []
+ for _m, _l, _iou in zip(m["masks"], m["low_res_masks"], m["iou_predictions"]):
+ best_iou_idx = torch.argmax(_iou)
+
+ best_mask, best_logits = _m[best_iou_idx], _l[best_iou_idx]
+ best_mask, best_logits = best_mask[None], best_logits[None]
+ mask.append(self._sigmoid(best_mask))
+ l_mask.append(best_logits)
+
+ mask, l_mask = torch.stack(mask), torch.stack(l_mask)
+ masks.append(mask)
+ logits_masks.append(l_mask)
+
+ masks, logits_masks = torch.stack(masks), torch.stack(logits_masks)
+ masks = (masks > 0.5).to(torch.float32)
+
+ self._get_updated_points_per_mask_per_subiter(masks, sampled_binary_y, batched_inputs, logits_masks)
+
+ loss = loss / num_subiter
+ mask_loss = mask_loss / num_subiter
+ iou_regression_loss = iou_regression_loss / num_subiter
+ mean_model_iou = mean_model_iou / num_subiter
+
+ return loss, mask_loss, iou_regression_loss, mean_model_iou
+
+ def _get_updated_points_per_mask_per_subiter(self, masks, sampled_binary_y, batched_inputs, logits_masks):
+ # here, we get the pair-per-batch of predicted and true elements (and also the "batched_inputs")
+ for x1, x2, _inp, logits in zip(masks, sampled_binary_y, batched_inputs, logits_masks):
+ net_coords, net_labels = [], []
+
+ # here, we get each object in the pairs and do the point choices per-object
+ for pred_obj, true_obj in zip(x1, x2):
+ true_obj = true_obj.to(self.device)
+
+ expected_diff = (pred_obj - true_obj)
+
+ neg_region = (expected_diff == 1).to(torch.float32)
+ pos_region = (expected_diff == -1)
+ overlap_region = torch.logical_and(pred_obj == 1, true_obj == 1).to(torch.float32)
+
+ # POSITIVE POINTS
+ tmp_pos_loc = torch.where(pos_region)
+ if torch.stack(tmp_pos_loc).shape[-1] == 0:
+ tmp_pos_loc = torch.where(overlap_region)
+
+ pos_index = np.random.choice(len(tmp_pos_loc[1]))
+ pos_coordinates = int(tmp_pos_loc[1][pos_index]), int(tmp_pos_loc[2][pos_index])
+ pos_coordinates = pos_coordinates[::-1]
+ pos_labels = 1
+
+ # NEGATIVE POINTS
+ tmp_neg_loc = torch.where(neg_region)
+ if torch.stack(tmp_neg_loc).shape[-1] == 0:
+ tmp_true_loc = torch.where(true_obj)
+ x_coords, y_coords = tmp_true_loc[1], tmp_true_loc[2]
+ bbox = torch.stack([torch.min(x_coords), torch.min(y_coords),
+ torch.max(x_coords) + 1, torch.max(y_coords) + 1])
+ bbox_mask = torch.zeros_like(true_obj).squeeze(0)
+ bbox_mask[bbox[0]:bbox[2], bbox[1]:bbox[3]] = 1
+ bbox_mask = bbox_mask[None].to(self.device)
+
+ dilated_bbox_mask = dilation(bbox_mask[None], torch.ones(3, 3).to(self.device)).squeeze(0)
+ background_mask = abs(dilated_bbox_mask - true_obj)
+ tmp_neg_loc = torch.where(background_mask)
+
+ neg_index = np.random.choice(len(tmp_neg_loc[1]))
+ neg_coordinates = int(tmp_neg_loc[1][neg_index]), int(tmp_neg_loc[2][neg_index])
+ neg_coordinates = neg_coordinates[::-1]
+ neg_labels = 0
+
+ net_coords.append([pos_coordinates, neg_coordinates])
+ net_labels.append([pos_labels, neg_labels])
+
+ if "point_labels" in _inp.keys():
+ updated_point_coords = torch.cat([_inp["point_coords"], torch.tensor(net_coords)], dim=1)
+ updated_point_labels = torch.cat([_inp["point_labels"], torch.tensor(net_labels)], dim=1)
+ else:
+ updated_point_coords = torch.tensor(net_coords)
+ updated_point_labels = torch.tensor(net_labels)
+
+ _inp["point_coords"] = updated_point_coords
+ _inp["point_labels"] = updated_point_labels
+ _inp["mask_inputs"] = logits
+
+ #
+ # Training Loop
+ #
+
+ def _update_samples_for_gt_instances(self, y, n_samples):
+ num_instances_gt = [len(torch.unique(_y)) for _y in y]
+ if n_samples > min(num_instances_gt):
+ n_samples = min(num_instances_gt) - 1
+ return n_samples
+
+ def _train_epoch_impl(self, progress, forward_context, backprop):
+ self.model.train()
+
+ n_iter = 0
+ t_per_iter = time.time()
+ for x, y in self.train_loader:
+
+ self.optimizer.zero_grad()
+
+ with forward_context():
+ n_samples = self.n_objects_per_batch
+ n_samples = self._update_samples_for_gt_instances(y, n_samples)
+
+ n_pos, n_neg, get_boxes, multimask_output = self._get_prompt_and_multimasking_choices(self._iteration)
+
+ batched_inputs, sampled_ids = self.convert_inputs(x, y, n_pos, n_neg, get_boxes, n_samples)
+
+ assert len(y) == len(sampled_ids)
+ sampled_binary_y = []
+ for i in range(len(y)):
+ _sampled = [torch.isin(y[i], torch.tensor(idx)) for idx in sampled_ids[i]]
+ sampled_binary_y.append(_sampled)
+
+ # the steps below are done for one reason in a gist:
+ # to handle images where there aren't enough instances as expected
+ # (e.g. where one image has only one instance)
+ obj_lengths = [len(s) for s in sampled_binary_y]
+ sampled_binary_y = [s[:min(obj_lengths)] for s in sampled_binary_y]
+ sampled_binary_y = [torch.stack(s).to(torch.float32) for s in sampled_binary_y]
+ sampled_binary_y = torch.stack(sampled_binary_y)
+
+ # gist for below - while we find the mismatch, we need to update the batched inputs
+ # else it would still generate masks using mismatching prompts, and it doesn't help us
+ # with the subiterations again. hence we clip the number of input points as well
+ f_objs = sampled_binary_y.shape[1]
+ batched_inputs = [
+ {k: (v[:f_objs] if k in ("point_coords", "point_labels", "boxes") else v) for k, v in inp.items()}
+ for inp in batched_inputs
+ ]
+
+ loss, mask_loss, iou_regression_loss, model_iou = self._update_masks(batched_inputs, y,
+ sampled_binary_y, sampled_ids,
+ num_subiter=self.n_sub_iteration,
+ multimask_output=multimask_output)
+
+ backprop(loss)
+
+ if self.logger is not None:
+ lr = [pm["lr"] for pm in self.optimizer.param_groups][0]
+ samples = sampled_binary_y if self._iteration % self.log_image_interval == 0 else None
+ self.logger.log_train(self._iteration, loss, lr, x, y, samples,
+ mask_loss, iou_regression_loss, model_iou)
+
+ self._iteration += 1
+ n_iter += 1
+ if self._iteration >= self.max_iteration:
+ break
+ progress.update(1)
+
+ t_per_iter = (time.time() - t_per_iter) / n_iter
+ return t_per_iter
+
+ def _validate_impl(self, forward_context):
+ self.model.eval()
+
+ metric_val = 0.0
+ loss_val = 0.0
+ model_iou_val = 0.0
+ val_iteration = 0
+
+ with torch.no_grad():
+ for x, y in self.val_loader:
+ with forward_context():
+ n_samples = self.n_objects_per_batch
+ n_samples = self._update_samples_for_gt_instances(y, n_samples)
+
+ (n_pos, n_neg,
+ get_boxes, multimask_output) = self._get_prompt_and_multimasking_choices_for_val(val_iteration)
+
+ batched_inputs, sampled_ids = self.convert_inputs(x, y, n_pos, n_neg, get_boxes, n_samples)
+
+ batched_outputs = self.model(batched_inputs, multimask_output=multimask_output)
+
+ assert len(y) == len(sampled_ids)
+ sampled_binary_y = torch.stack(
+ [torch.isin(y[i], torch.tensor(sampled_ids[i])) for i in range(len(y))]
+ ).to(torch.float32)
+
+ loss, mask_loss, iou_regression_loss, model_iou = self._get_net_loss(batched_outputs,
+ y, sampled_ids)
+
+ metric = self._get_val_metric(batched_outputs, sampled_binary_y)
+
+ loss_val += loss.item()
+ metric_val += metric.item()
+ model_iou_val += model_iou.item()
+ val_iteration += 1
+
+ loss_val /= len(self.val_loader)
+ metric_val /= len(self.val_loader)
+ model_iou_val /= len(self.val_loader)
+ print()
+ print(f"The Average Dice Score for the Current Epoch is {1 - metric_val}")
+
+ if self.logger is not None:
+ self.logger.log_validation(
+ self._iteration, metric_val, loss_val, x, y,
+ sampled_binary_y, mask_loss, iou_regression_loss, model_iou_val
+ )
+
+ return metric_val
+
+
+class SamLogger(TorchEmLogger):
+ """@private"""
+ def __init__(self, trainer, save_root, **unused_kwargs):
+ super().__init__(trainer, save_root)
+ self.log_dir = f"./logs/{trainer.name}" if save_root is None else\
+ os.path.join(save_root, "logs", trainer.name)
+ os.makedirs(self.log_dir, exist_ok=True)
+
+ self.tb = torch.utils.tensorboard.SummaryWriter(self.log_dir)
+ self.log_image_interval = trainer.log_image_interval
+
+ def add_image(self, x, y, samples, name, step):
+ self.tb.add_image(tag=f"{name}/input", img_tensor=x[0], global_step=step)
+ self.tb.add_image(tag=f"{name}/target", img_tensor=y[0], global_step=step)
+ sample_grid = make_grid([sample[0] for sample in samples], nrow=4, padding=4)
+ self.tb.add_image(tag=f"{name}/samples", img_tensor=sample_grid, global_step=step)
+
+ def log_train(self, step, loss, lr, x, y, samples, mask_loss, iou_regression_loss, model_iou):
+ self.tb.add_scalar(tag="train/loss", scalar_value=loss, global_step=step)
+ self.tb.add_scalar(tag="train/mask_loss", scalar_value=mask_loss, global_step=step)
+ self.tb.add_scalar(tag="train/iou_loss", scalar_value=iou_regression_loss, global_step=step)
+ self.tb.add_scalar(tag="train/model_iou", scalar_value=model_iou, global_step=step)
+ self.tb.add_scalar(tag="train/learning_rate", scalar_value=lr, global_step=step)
+ if step % self.log_image_interval == 0:
+ self.add_image(x, y, samples, "train", step)
+
+ def log_validation(self, step, metric, loss, x, y, samples, mask_loss, iou_regression_loss, model_iou):
+ self.tb.add_scalar(tag="validation/loss", scalar_value=loss, global_step=step)
+ self.tb.add_scalar(tag="validation/mask_loss", scalar_value=mask_loss, global_step=step)
+ self.tb.add_scalar(tag="validation/iou_loss", scalar_value=iou_regression_loss, global_step=step)
+ self.tb.add_scalar(tag="validation/model_iou", scalar_value=model_iou, global_step=step)
+ self.tb.add_scalar(tag="validation/metric", scalar_value=metric, global_step=step)
+ self.add_image(x, y, samples, "validation", step)
diff --git a/micro_sam/training/trainable_sam.py b/micro_sam/training/trainable_sam.py
new file mode 100644
index 00000000..99728a1b
--- /dev/null
+++ b/micro_sam/training/trainable_sam.py
@@ -0,0 +1,117 @@
+from typing import Any, Dict, List, Optional, Union
+
+import torch
+from torch import nn
+from torch.nn import functional as F
+
+from segment_anything.modeling import Sam
+
+
+# simple wrapper around SAM in order to keep things trainable
+class TrainableSAM(nn.Module):
+ """Wrapper to make the SegmentAnything model trainable.
+
+ Args:
+ sam: The SegmentAnything Model.
+ device: The device for training.
+ """
+ def __init__(
+ self,
+ sam: Sam,
+ device: Union[str, torch.device],
+ ) -> None:
+ super().__init__()
+ self.sam = sam
+ self.device = device
+
+ def preprocess(self, x: torch.Tensor) -> torch.Tensor:
+ """Normalize pixel values and pad to a square input.
+
+ Args:
+ x: The input tensor.
+
+ Returns:
+ The normalized and padded tensor.
+ """
+ # Normalize colors
+ x = (x - self.sam.pixel_mean) / self.sam.pixel_std
+
+ # Pad
+ h, w = x.shape[-2:]
+ padh = self.sam.image_encoder.img_size - h
+ padw = self.sam.image_encoder.img_size - w
+ x = F.pad(x, (0, padw, 0, padh))
+ return x
+
+ def image_embeddings_oft(self, input_images):
+ """@private"""
+ image_embeddings = self.sam.image_encoder(input_images)
+ return image_embeddings
+
+ # batched inputs follow the same syntax as the input to sam.forward
+ def forward(
+ self,
+ batched_inputs: List[Dict[str, Any]],
+ multimask_output: bool = False,
+ image_embeddings: Optional[torch.Tensor] = None,
+ ) -> List[Dict[str, Any]]:
+ """Forward pass.
+
+ Args:
+ batched_inputs: The batched input images and prompts.
+ multimask_output: Whether to predict mutiple or just a single mask.
+ image_embeddings: The precompute image embeddings. If not passed then they will be computed.
+
+ Returns:
+ The predicted segmentation masks and iou values.
+ """
+ input_images = torch.stack([self.preprocess(x=x["image"].to(self.device)) for x in batched_inputs], dim=0)
+ if image_embeddings is None:
+ image_embeddings = self.sam.image_encoder(input_images)
+
+ outputs = []
+ for image_record, curr_embedding in zip(batched_inputs, image_embeddings):
+ if "point_coords" in image_record:
+ points = (image_record["point_coords"].to(self.device), image_record["point_labels"].to(self.device))
+ else:
+ points = None
+
+ if "boxes" in image_record:
+ boxes = image_record.get("boxes").to(self.device)
+ else:
+ boxes = None
+
+ if "mask_inputs" in image_record:
+ masks = image_record.get("mask_inputs").to(self.device)
+ else:
+ masks = None
+
+ sparse_embeddings, dense_embeddings = self.sam.prompt_encoder(
+ points=points,
+ boxes=boxes,
+ masks=masks,
+ )
+
+ low_res_masks, iou_predictions = self.sam.mask_decoder(
+ image_embeddings=curr_embedding.unsqueeze(0),
+ image_pe=self.sam.prompt_encoder.get_dense_pe(),
+ sparse_prompt_embeddings=sparse_embeddings,
+ dense_prompt_embeddings=dense_embeddings,
+ multimask_output=multimask_output,
+ )
+
+ masks = self.sam.postprocess_masks(
+ low_res_masks,
+ input_size=image_record["image"].shape[-2:],
+ original_size=image_record["original_size"],
+ )
+
+ outputs.append(
+ {
+ "low_res_masks": low_res_masks,
+ "masks": masks,
+ "iou_predictions": iou_predictions
+ }
+ )
+
+ return outputs
diff --git a/micro_sam/training/util.py b/micro_sam/training/util.py
new file mode 100644
index 00000000..3821ac7f
--- /dev/null
+++ b/micro_sam/training/util.py
@@ -0,0 +1,191 @@
+import os
+from typing import List, Optional, Union
+
+import torch
+import numpy as np
+
+from ..prompt_generators import PointAndBoxPromptGenerator
+from ..util import get_centers_and_bounding_boxes, get_sam_model
+from .trainable_sam import TrainableSAM
+
+
+def get_trainable_sam_model(
+ model_type: str = "vit_h",
+ checkpoint_path: Optional[Union[str, os.PathLike]] = None,
+ freeze: Optional[List[str]] = None,
+ device: Optional[Union[str, torch.device]] = None,
+) -> TrainableSAM:
+ """Get the trainable sam model.
+
+ Args:
+ model_type: The type of the segment anything model.
+ checkpoint_path: Path to a custom checkpoint from which to load the model weights.
+ freeze: Specify parts of the model that should be frozen.
+ By default nothing is frozen and the full model is updated.
+ device: The device to use for training.
+
+ Returns:
+ The trainable segment anything model.
+ """
+ # set the device here so that the correct one is passed to TrainableSAM below
+ if device is None:
+ device = "cuda" if torch.cuda.is_available() else "cpu"
+ _, sam = get_sam_model(device, model_type, checkpoint_path, return_sam=True)
+
+ # freeze components of the model if freeze was passed
+ # ideally we would want to add components in such a way that:
+ # - we would be able to freeze the choice of encoder/decoder blocks, yet be able to add components to the network
+ # (for e.g. encoder blocks to "image_encoder")
+ if freeze is not None:
+ for name, param in sam.named_parameters():
+ if isinstance(freeze, list):
+ # we would want to "freeze" all the components in the model if passed a list of parts
+ for l_item in freeze:
+ if name.startswith(f"{l_item}"):
+ param.requires_grad = False
+ else:
+ # we "freeze" only for one specific component when passed a "particular" part
+ if name.startswith(f"{freeze}"):
+ param.requires_grad = False
+
+ # convert to trainable sam
+ trainable_sam = TrainableSAM(sam, device)
+ return trainable_sam
+
+
+class ConvertToSamInputs:
+ """Convert outputs of data loader to the expected batched inputs of the SegmentAnything model.
+
+ Args:
+ dilation_strength: The dilation factor.
+ It determines a "safety" border from which prompts are not sampled to avoid ambiguous prompts
+ due to imprecise groundtruth masks.
+ box_distortion_factor: Factor for distorting the box annotations derived from the groundtruth masks.
+ Not yet implemented.
+ """
+ def __init__(
+ self,
+ dilation_strength: int = 10,
+ box_distortion_factor: Optional[float] = None,
+ ) -> None:
+ self.dilation_strength = dilation_strength
+ # TODO implement the box distortion logic
+ if box_distortion_factor is not None:
+ raise NotImplementedError
+
+ def _get_prompt_generator(self, n_positive_points, n_negative_points, get_boxes, get_points):
+ """Returns the prompt generator w.r.t. the "random" attributes inputed."""
+
+ # the class initialization below gets the random choice of n_positive and n_negative points as inputs
+ # (done in the trainer)
+ # in case of dynamic choice while choosing between points and/or box, it gets those as well
+ prompt_generator = PointAndBoxPromptGenerator(n_positive_points=n_positive_points,
+ n_negative_points=n_negative_points,
+ dilation_strength=self.dilation_strength,
+ get_box_prompts=get_boxes,
+ get_point_prompts=get_points)
+ return prompt_generator
+
+ def _get_prompt_lists(self, gt, n_samples, n_positive_points, n_negative_points, get_boxes,
+ get_points, prompt_generator, point_coordinates, bbox_coordinates):
+ """Returns a list of "expected" prompts subjected to the random input attributes for prompting."""
+ box_prompts = []
+ point_prompts = []
+ point_label_prompts = []
+
+ # getting the cell instance except the bg
+ cell_ids = np.unique(gt)[1:]
+
+ accepted_cell_ids = []
+ sampled_cell_ids = []
+
+ # while conditions gets all the prompts until it satisfies the requirement
+ while len(accepted_cell_ids) < min(n_samples, len(cell_ids)):
+ if len(sampled_cell_ids) == len(cell_ids): # we did not find enough cells
+ break
+
+ my_cell_id = np.random.choice(np.setdiff1d(cell_ids, sampled_cell_ids))
+ sampled_cell_ids.append(my_cell_id)
+
+ bbox = bbox_coordinates[my_cell_id]
+ # points = point_coordinates[my_cell_id]
+ # removed "points" to randomly choose fg points
+ coord_list, label_list, bbox_list, _ = prompt_generator(gt, my_cell_id, bbox)
+
+ if get_boxes is True and get_points is False: # only box
+ bbox_list = bbox_list[0]
+ box_prompts.append([bbox_list[1], bbox_list[0],
+ bbox_list[3], bbox_list[2]])
+ accepted_cell_ids.append(my_cell_id)
+
+ if get_points: # one with points expected
+ # check for the minimum point requirement per object in the batch
+ if len(label_list) == n_negative_points + n_positive_points:
+ point_prompts.append(np.array([ip[::-1] for ip in coord_list]))
+ point_label_prompts.append(np.array(label_list))
+ accepted_cell_ids.append(my_cell_id)
+ if get_boxes: # one with boxes expected with points as well
+ bbox_list = bbox_list[0]
+ box_prompts.append([bbox_list[1], bbox_list[0],
+ bbox_list[3], bbox_list[2]])
+
+ point_prompts = np.array(point_prompts)
+ point_label_prompts = np.array(point_label_prompts)
+ return box_prompts, point_prompts, point_label_prompts, accepted_cell_ids
+
+ def __call__(self, x, y, n_pos, n_neg, get_boxes=False, n_samples=None):
+ """Convert the outputs of dataloader and prompt settings to the batch format expected by SAM.
+ """
+
+ # condition to see if we get point prompts, then we (ofc) use point-prompting
+ # else we don't use point prompting
+ if n_pos == 0 and n_neg == 0:
+ get_points = False
+ else:
+ get_points = True
+
+ # keeping the solution open by checking for deterministic/dynamic choice of point prompts
+ prompt_generator = self._get_prompt_generator(n_pos, n_neg, get_boxes, get_points)
+
+ batched_inputs = []
+ batched_sampled_cell_ids_list = []
+ for i, gt in enumerate(y):
+ gt = gt.squeeze().numpy().astype(np.int32)
+ point_coordinates, bbox_coordinates = get_centers_and_bounding_boxes(gt)
+
+ this_n_samples = len(point_coordinates) if n_samples is None else n_samples
+ box_prompts, point_prompts, point_label_prompts, sampled_cell_ids = self._get_prompt_lists(
+ gt, this_n_samples,
+ n_pos, n_neg,
+ get_boxes,
+ get_points,
+ prompt_generator,
+ point_coordinates,
+ bbox_coordinates
+ )
+
+ # check to be sure about the expected size of the no. of elements in different settings
+ if get_boxes is True and get_points is False:
+ assert len(sampled_cell_ids) == len(box_prompts), \
+ print(len(sampled_cell_ids), len(box_prompts))
+
+ elif get_boxes is False and get_points is True:
+ assert len(sampled_cell_ids) == len(point_prompts) == len(point_label_prompts), \
+ print(len(sampled_cell_ids), len(point_prompts), len(point_label_prompts))
+
+ elif get_boxes is True and get_points is True:
+ assert len(sampled_cell_ids) == len(box_prompts) == len(point_prompts) == len(point_label_prompts), \
+ print(len(sampled_cell_ids), len(box_prompts), len(point_prompts), len(point_label_prompts))
+
+ batched_sampled_cell_ids_list.append(sampled_cell_ids)
+
+ batched_input = {"image": x[i], "original_size": x[i].shape[1:]}
+ if get_boxes:
+ batched_input["boxes"] = torch.tensor(box_prompts)
+ if get_points:
+ batched_input["point_coords"] = torch.tensor(point_prompts)
+ batched_input["point_labels"] = torch.tensor(point_label_prompts)
+
+ batched_inputs.append(batched_input)
+
+ return batched_inputs, batched_sampled_cell_ids_list
diff --git a/micro_sam/util.py b/micro_sam/util.py
index 7578c153..1ea21303 100644
--- a/micro_sam/util.py
+++ b/micro_sam/util.py
@@ -4,9 +4,11 @@
import hashlib
import os
+import pickle
import warnings
+from collections import OrderedDict
from shutil import copyfileobj
-from typing import Any, Callable, Dict, Optional, Tuple, Iterable
+from typing import Any, Callable, Dict, Iterable, Optional, Tuple, Union
import imageio.v3 as imageio
import numpy as np
@@ -27,25 +29,27 @@
from tqdm import tqdm
_MODEL_URLS = {
+ # the default segment anything models
"vit_h": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_h_4b8939.pth",
"vit_l": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_l_0b3195.pth",
"vit_b": "https://dl.fbaipublicfiles.com/segment_anything/sam_vit_b_01ec64.pth",
- # preliminary finetuned models
- "vit_h_lm": "https://owncloud.gwdg.de/index.php/s/CnxBvsdGPN0TD3A/download",
- "vit_b_lm": "https://owncloud.gwdg.de/index.php/s/gGlR1LFsav0eQ2k/download",
- "vit_h_em": "https://owncloud.gwdg.de/index.php/s/VcHoLC6AM0CrRpM/download",
- "vit_b_em": "https://owncloud.gwdg.de/index.php/s/BWupWhG1HRflI97/download",
+ # first version of finetuned models on zenodo
+ "vit_h_lm": "https://zenodo.org/record/8250299/files/vit_h_lm.pth?download=1",
+ "vit_b_lm": "https://zenodo.org/record/8250281/files/vit_b_lm.pth?download=1",
+ "vit_h_em": "https://zenodo.org/record/8250291/files/vit_h_em.pth?download=1",
+ "vit_b_em": "https://zenodo.org/record/8250260/files/vit_b_em.pth?download=1",
}
_CHECKPOINT_FOLDER = os.environ.get("SAM_MODELS", os.path.expanduser("~/.sam_models"))
_CHECKSUMS = {
+ # the default segment anything models
"vit_h": "a7bf3b02f3ebf1267aba913ff637d9a2d5c33d3173bb679e46d9f338c26f262e",
"vit_l": "3adcc4315b642a4d2101128f611684e8734c41232a17c648ed1693702a49a622",
"vit_b": "ec2df62732614e57411cdcf32a23ffdf28910380d03139ee0f4fcbe91eb8c912",
- # preliminary finetuned models
- "vit_h_lm": "c30a580e6ccaff2f4f0fbaf9cad10cee615a915cdd8c7bc4cb50ea9bdba3fc09",
- "vit_b_lm": "f2b8676f92a123f6f8ac998818118bd7269a559381ec60af4ac4be5c86024a1b",
- "vit_h_em": "652f70acad89ab855502bc10965e7d0baf7ef5f38fef063dd74f1787061d3919",
- "vit_b_em": "9eb783e538bb287c7086f825f1e1dc5d5681bd116541a0b98cab85f1e7f4dd62",
+ # first version of finetuned models on zenodo
+ "vit_h_lm": "9a65ee0cddc05a98d60469a12a058859c89dc3ea3ba39fed9b90d786253fbf26",
+ "vit_b_lm": "5a59cc4064092d54cd4d92cd967e39168f3760905431e868e474d60fe5464ecd",
+ "vit_h_em": "ae3798a0646c8df1d4db147998a2d37e402ff57d3aa4e571792fbb911d8a979c",
+ "vit_b_em": "c04a714a4e14a110f0eec055a65f7409d54e6bf733164d2933a0ce556f7d6f81",
}
# this is required so that the downloaded file is not called 'download'
_DOWNLOAD_NAMES = {
@@ -54,10 +58,19 @@
"vit_h_em": "vit_h_em.pth",
"vit_b_em": "vit_b_em.pth",
}
+# this is the default model used in micro_sam
+# currently set to the default vit_h
+_DEFAULT_MODEL = "vit_h"
# TODO define the proper type for image embeddings
ImageEmbeddings = Dict[str, Any]
+"""@private"""
+
+
+#
+# Functionality for model download and export
+#
def _download(url, path, model_type):
@@ -105,9 +118,9 @@ def _get_checkpoint(model_type, checkpoint_path=None):
def get_sam_model(
device: Optional[str] = None,
- model_type: str = "vit_h",
- checkpoint_path: Optional[str] = None,
- return_sam: bool = False
+ model_type: str = _DEFAULT_MODEL,
+ checkpoint_path: Optional[Union[str, os.PathLike]] = None,
+ return_sam: bool = False,
) -> SamPredictor:
"""Get the SegmentAnything Predictor.
@@ -117,7 +130,7 @@ def get_sam_model(
Args:
device: The device for the model. If none is given will use GPU if available.
- model_type: The SegmentAnything model to use.
+ model_type: The SegmentAnything model to use. Will use the standard vit_h model by default.
checkpoint_path: The path to the corresponding checkpoint if not in the default model folder.
return_sam: Return the sam model object as well as the predictor.
@@ -125,7 +138,8 @@ def get_sam_model(
The segment anything predictor.
"""
checkpoint = _get_checkpoint(model_type, checkpoint_path)
- device = "cuda" if torch.cuda.is_available() else "cpu"
+ if device is None:
+ device = "cuda" if torch.cuda.is_available() else "cpu"
# Our custom model types have a suffix "_...". This suffix needs to be stripped
# before calling sam_model_registry.
@@ -141,10 +155,107 @@ def get_sam_model(
return predictor
+# We write a custom unpickler that skips objects that cannot be found instead of
+# throwing an AttributeError or ModueNotFoundError.
+# NOTE: since we just want to unpickle the model to load its weights these errors don't matter.
+# See also https://stackoverflow.com/questions/27732354/unable-to-load-files-using-pickle-and-multiple-modules
+class _CustomUnpickler(pickle.Unpickler):
+ def find_class(self, module, name):
+ try:
+ return super().find_class(module, name)
+ except (AttributeError, ModuleNotFoundError) as e:
+ warnings.warn(f"Did not find {module}:{name} and will skip it, due to error {e}")
+ return None
+
+
+def get_custom_sam_model(
+ checkpoint_path: Union[str, os.PathLike],
+ device: Optional[str] = None,
+ model_type: str = "vit_h",
+ return_sam: bool = False,
+ return_state: bool = False,
+) -> SamPredictor:
+ """Load a SAM model from a torch_em checkpoint.
+
+ This function enables loading from the checkpoints saved by
+ the functionality in `micro_sam.training`.
+
+ Args:
+ checkpoint_path: The path to the corresponding checkpoint if not in the default model folder.
+ device: The device for the model. If none is given will use GPU if available.
+ model_type: The SegmentAnything model to use.
+ return_sam: Return the sam model object as well as the predictor.
+ return_state: Return the full state of the checkpoint in addition to the predictor.
+
+ Returns:
+ The segment anything predictor.
+ """
+ assert not (return_sam and return_state)
+
+ # over-ride the unpickler with our custom one
+ custom_pickle = pickle
+ custom_pickle.Unpickler = _CustomUnpickler
+
+ if device is None:
+ device = "cuda" if torch.cuda.is_available() else "cpu"
+ sam = sam_model_registry[model_type]()
+
+ # load the model state, ignoring any attributes that can't be found by pickle
+ state = torch.load(checkpoint_path, map_location=device, pickle_module=custom_pickle)
+ model_state = state["model_state"]
+
+ # copy the model weights from torch_em's training format
+ sam_prefix = "sam."
+ model_state = OrderedDict(
+ [(k[len(sam_prefix):] if k.startswith(sam_prefix) else k, v) for k, v in model_state.items()]
+ )
+ sam.load_state_dict(model_state)
+ sam.to(device)
+
+ predictor = SamPredictor(sam)
+ predictor.model_type = model_type
+
+ if return_sam:
+ return predictor, sam
+ if return_state:
+ return predictor, state
+ return predictor
+
+
+def export_custom_sam_model(
+ checkpoint_path: Union[str, os.PathLike],
+ model_type: str,
+ save_path: Union[str, os.PathLike],
+) -> None:
+ """Export a finetuned segment anything model to the standard model format.
+
+ The exported model can be used by the interactive annotation tools in `micro_sam.annotator`.
+
+ Args:
+ checkpoint_path: The path to the corresponding checkpoint if not in the default model folder.
+ model_type: The SegmentAnything model type to use (vit_h, vit_b or vit_l).
+ save_path: Where to save the exported model.
+ """
+ _, state = get_custom_sam_model(
+ checkpoint_path, model_type=model_type, return_state=True, device=torch.device("cpu"),
+ )
+ model_state = state["model_state"]
+ prefix = "sam."
+ model_state = OrderedDict(
+ [(k[len(prefix):] if k.startswith(prefix) else k, v) for k, v in model_state.items()]
+ )
+ torch.save(model_state, save_path)
+
+
def get_model_names() -> Iterable:
return _MODEL_URLS.keys()
+#
+# Functionality for precomputing embeddings and other state
+#
+
+
def _to_image(input_):
# we require the input to be uint8
if input_.dtype != np.dtype("uint8"):
@@ -375,7 +486,7 @@ def precompute_image_embeddings(
If 'save_path' is given the embeddings will be loaded/saved in a zarr container.
Args:
- predictor: The SegmentAnything predictor
+ predictor: The SegmentAnything predictor.
input_: The input data. Can be 2 or 3 dimensional, corresponding to an image, volume or timeseries.
save_path: Path to save the embeddings in a zarr container.
lazy_loading: Whether to load all embeddings into memory or return an
@@ -397,14 +508,22 @@ def precompute_image_embeddings(
data_signature = _compute_data_signature(input_)
f = zarr.open(save_path, "a")
- key_vals = [("data_signature", data_signature),
- ("tile_shape", tile_shape), ("model_type", predictor.model_type)]
- for key, val in key_vals:
- if "input_size" in f.attrs: # we have computed the embeddings already
- # key signature does not match or is not in the file
+ key_vals = [
+ ("data_signature", data_signature),
+ ("tile_shape", tile_shape if tile_shape is None else list(tile_shape)),
+ ("halo", halo if halo is None else list(halo)),
+ ("model_type", predictor.model_type)
+ ]
+ if "input_size" in f.attrs: # we have computed the embeddings already and perform checks
+ for key, val in key_vals:
+ if val is None:
+ continue
+ # check whether the key signature does not match or is not in the file
if key not in f.attrs or f.attrs[key] != val:
- warnings.warn(f"Embeddings file is invalid due to unmatching {key}. \
- Please recompute embeddings in a new file.")
+ warnings.warn(
+ f"Embeddings file {save_path} is invalid due to unmatching {key}: "
+ f"{f.attrs.get(key)} != {val}.Please recompute embeddings in a new file."
+ )
if wrong_file_callback is not None:
save_path = wrong_file_callback(save_path)
f = zarr.open(save_path, "a")
@@ -441,7 +560,7 @@ def set_precomputed(
i: Index for the image data. Required if `image` has three spatial dimensions
or a time dimension and two spatial dimensions.
"""
- device = "cuda" if torch.cuda.is_available() else "cpu"
+ device = predictor.device
features = image_embeddings["features"]
assert features.ndim in (4, 5)
@@ -463,6 +582,11 @@ def set_precomputed(
return predictor
+#
+# Misc functionality
+#
+
+
def compute_iou(mask1: np.ndarray, mask2: np.ndarray) -> float:
"""Compute the intersection over union of two masks.
@@ -508,7 +632,7 @@ def get_centers_and_bounding_boxes(
bbox_coordinates = {prop.label: prop.bbox for prop in properties}
- assert len(bbox_coordinates) == len(center_coordinates)
+ assert len(bbox_coordinates) == len(center_coordinates), f"{len(bbox_coordinates)}, {len(center_coordinates)}"
return center_coordinates, bbox_coordinates
@@ -535,25 +659,3 @@ def load_image_data(
if not lazy_loading:
image_data = image_data[:]
return image_data
-
-
-def main():
- """@private"""
- import argparse
-
- parser = argparse.ArgumentParser(description="Compute the embeddings for an image.")
- parser.add_argument("-i", "--input_path", required=True)
- parser.add_argument("-o", "--output_path", required=True)
- parser.add_argument("-m", "--model_type", default="vit_h")
- parser.add_argument("-c", "--checkpoint_path", default=None)
- parser.add_argument("-k", "--key")
- args = parser.parse_args()
-
- predictor = get_sam_model(model_type=args.model_type, checkpoint_path=args.checkpoint_path)
- with open_file(args.input_path, mode="r") as f:
- data = f[args.key]
- precompute_image_embeddings(predictor, data, save_path=args.output_path)
-
-
-if __name__ == "__main__":
- main()
diff --git a/test/test_instance_segmentation.py b/test/test_instance_segmentation.py
index 2cb932d7..8ee81614 100644
--- a/test/test_instance_segmentation.py
+++ b/test/test_instance_segmentation.py
@@ -91,10 +91,11 @@ def test_embedding_mask_generator(self):
mask, image = self.mask, self.image
predictor, image_embeddings = self.predictor, self.image_embeddings
+ pred_iou_thresh, stability_score_thresh = 0.95, 0.75
amg = EmbeddingMaskGenerator(predictor)
amg.initialize(image, image_embeddings=image_embeddings, verbose=False)
- predicted = amg.generate(pred_iou_thresh=0.96)
+ predicted = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
predicted = mask_data_to_segmentation(predicted, image.shape, with_background=True)
self.assertGreater(matching(predicted, mask, threshold=0.75)["segmentation_accuracy"], 0.99)
@@ -103,7 +104,7 @@ def test_embedding_mask_generator(self):
self.assertEqual(initial_seg.shape, image.shape)
# check that regenerating the segmentation works
- predicted2 = amg.generate(pred_iou_thresh=0.96)
+ predicted2 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
predicted2 = mask_data_to_segmentation(predicted2, image.shape, with_background=True)
self.assertTrue(np.array_equal(predicted, predicted2))
@@ -111,7 +112,7 @@ def test_embedding_mask_generator(self):
state = amg.get_state()
amg = EmbeddingMaskGenerator(predictor)
amg.set_state(state)
- predicted3 = amg.generate(pred_iou_thresh=0.96)
+ predicted3 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
predicted3 = mask_data_to_segmentation(predicted3, image.shape, with_background=True)
self.assertTrue(np.array_equal(predicted, predicted3))
@@ -120,23 +121,24 @@ def test_tiled_embedding_mask_generator(self):
mask, image = self.large_mask, self.large_image
predictor, image_embeddings = self.predictor, self.tiled_embeddings
+ pred_iou_thresh, stability_score_thresh = 0.90, 0.60
- amg = TiledEmbeddingMaskGenerator(predictor)
+ amg = TiledEmbeddingMaskGenerator(predictor, box_extension=0.1)
amg.initialize(image, image_embeddings=image_embeddings)
- predicted = amg.generate(pred_iou_thresh=0.96)
+ predicted = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
initial_seg = amg.get_initial_segmentation()
self.assertGreater(matching(predicted, mask, threshold=0.75)["segmentation_accuracy"], 0.99)
self.assertEqual(initial_seg.shape, image.shape)
- predicted2 = amg.generate(pred_iou_thresh=0.96)
+ predicted2 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
self.assertTrue(np.array_equal(predicted, predicted2))
# check that serializing and reserializing the state works
state = amg.get_state()
amg = TiledEmbeddingMaskGenerator(predictor)
amg.set_state(state)
- predicted3 = amg.generate(pred_iou_thresh=0.96)
+ predicted3 = amg.generate(pred_iou_thresh=pred_iou_thresh, stability_score_thresh=stability_score_thresh)
self.assertTrue(np.array_equal(predicted, predicted3))
def test_tiled_automatic_mask_generator(self):
 +The training logic is implemented in `micro_sam.training` and is based on [torch-em](https://github.com/constantinpape/torch-em). Please check out [examples/finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/examples/finetuning) to see how you can finetune on your own data with it. The script `finetune_hela.py` contains an example for finetuning on a small custom microscopy dataset and `use_finetuned_model.py` shows how this model can then be used in the interactive annotatin tools.
+More advanced examples, including quantitative and qualitative evaluation, of finetuned models can be found in [finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/finetuning), which contains the code for training and evaluating our microscopy models.
diff --git a/doc/start_page.md b/doc/start_page.md
index b5fab2a9..840a650b 100644
--- a/doc/start_page.md
+++ b/doc/start_page.md
@@ -2,7 +2,7 @@
Segment Anything for Microscopy implements automatic and interactive annotation for microscopy data. It is built on top of [Segment Anything](https://segment-anything.com/) by Meta AI and specializes it for microscopy and other bio-imaging data.
Its core components are:
-- The `micro_sam` annotator tools for interactive data annotation with [napari](https://napari.org/stable/).
+- The `micro_sam` tools for interactive data annotation with [napari](https://napari.org/stable/).
- The `micro_sam` library to apply Segment Anything to 2d and 3d data or fine-tune it on your data.
- The `micro_sam` models that are fine-tuned on publicly available microscopy data.
@@ -19,20 +19,18 @@ On our roadmap for more functionality are:
If you run into any problems or have questions please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
-
## Quickstart
You can install `micro_sam` via conda:
```
-$ conda install -c conda-forge micro_sam
+$ conda install -c conda-forge micro_sam napari pyqt
```
-For more installation options check out [Installation](#installation)
+We also provide experimental installers for all operating systems.
+For more details on the available installation options check out [the installation section](#installation).
After installing `micro_sam` you can run the annotation tool via `$ micro_sam.annotator`, which opens a menu for selecting the annotation tool and its inputs.
-See [Annotation Tools](#annotation-tools) for an overview and explanation of the annotation functionality.
+See [the annotation tool section](#annotation-tools) for an overview and explanation of the annotation functionality.
The `micro_sam` python library can be used via
```python
@@ -47,7 +45,5 @@ For now, check out [the example script for the 2d annotator](https://github.com/
## Citation
If you are using `micro_sam` in your research please cite
-- [SegmentAnything](https://arxiv.org/abs/2304.02643)
-- and our repository on [zenodo](https://doi.org/10.5281/zenodo.7919746)
-
-We will release a preprint soon that describes this work and can be cited instead of zenodo.
+- Our [preprint](https://doi.org/10.1101/2023.08.21.554208)
+- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643)
diff --git a/environment_cpu.yaml b/environment_cpu.yaml
index 4d5f8ea7..ad91dab9 100644
--- a/environment_cpu.yaml
+++ b/environment_cpu.yaml
@@ -11,6 +11,7 @@ dependencies:
- pytorch
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/environment_gpu.yaml b/environment_gpu.yaml
index 21d5df53..57700759 100644
--- a/environment_gpu.yaml
+++ b/environment_gpu.yaml
@@ -12,6 +12,7 @@ dependencies:
- pytorch-cuda>=11.7 # you may need to update the cuda version to match your system
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/examples/README.md b/examples/README.md
index 87b3a963..4844167f 100644
--- a/examples/README.md
+++ b/examples/README.md
@@ -6,5 +6,8 @@ Examples for using the micro_sam annotation tools:
- `annotator_tracking.py`: run the interactive tracking annotation tool
- `image_series_annotator.py`: run the annotation tool for a series of images
+The folder `finetuning` contains example scripts that show how a Segment Anything model can be fine-tuned
+on custom data with the `micro_sam.train` library, and how the finetuned models can then be used within the annotatin tools.
+
The folder `use_as_library` contains example scripts that show how `micro_sam` can be used as a python
-library to apply Segment Anything on mult-dimensional data.
+library to apply Segment Anything to mult-dimensional data.
diff --git a/examples/annotator_2d.py b/examples/annotator_2d.py
index f0d6d44b..8f4930f2 100644
--- a/examples/annotator_2d.py
+++ b/examples/annotator_2d.py
@@ -34,7 +34,7 @@ def hela_2d_annotator(use_finetuned_model):
embedding_path = "./embeddings/embeddings-hela2d.zarr"
model_type = "vit_h"
- annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type)
+ annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type, precompute_amg_state=True)
def wholeslide_annotator(use_finetuned_model):
diff --git a/examples/annotator_with_custom_model.py b/examples/annotator_with_custom_model.py
new file mode 100644
index 00000000..ceb8b2cb
--- /dev/null
+++ b/examples/annotator_with_custom_model.py
@@ -0,0 +1,23 @@
+import h5py
+import micro_sam.sam_annotator as annotator
+from micro_sam.util import get_sam_model
+
+# TODO add an example for the 2d annotator with a custom model
+
+
+def annotator_3d_with_custom_model():
+ with h5py.File("./data/gut1_block_1.h5") as f:
+ raw = f["raw"][:]
+
+ custom_model = "/home/pape/Work/data/models/sam/user-study/vit_h_nuclei_em_finetuned.pt"
+ embedding_path = "./embeddings/nuclei3d-custom-vit-h.zarr"
+ predictor = get_sam_model(checkpoint_path=custom_model, model_type="vit_h")
+ annotator.annotator_3d(raw, embedding_path, predictor=predictor)
+
+
+def main():
+ annotator_3d_with_custom_model()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/finetuning/.gitignore b/examples/finetuning/.gitignore
new file mode 100644
index 00000000..ec28c5ea
--- /dev/null
+++ b/examples/finetuning/.gitignore
@@ -0,0 +1,2 @@
+checkpoints/
+logs/
diff --git a/examples/finetuning/finetune_hela.py b/examples/finetuning/finetune_hela.py
new file mode 100644
index 00000000..f83bd8cb
--- /dev/null
+++ b/examples/finetuning/finetune_hela.py
@@ -0,0 +1,140 @@
+import os
+
+import numpy as np
+import torch
+import torch_em
+
+import micro_sam.training as sam_training
+from micro_sam.sample_data import fetch_tracking_example_data, fetch_tracking_segmentation_data
+from micro_sam.util import export_custom_sam_model
+
+DATA_FOLDER = "data"
+
+
+def get_dataloader(split, patch_shape, batch_size):
+ """Return train or val data loader for finetuning SAM.
+
+ The data loader must be a torch data loader that retuns `x, y` tensors,
+ where `x` is the image data and `y` are the labels.
+ The labels have to be in a label mask instance segmentation format.
+ I.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
+ Important: the ID 0 is reseved for background, and the IDs must be consecutive
+
+ Here, we use `torch_em.default_segmentation_loader` for creating a suitable data loader from
+ the example hela data. You can either adapt this for your own data (see comments below)
+ or write a suitable torch dataloader yourself.
+ """
+ assert split in ("train", "val")
+ os.makedirs(DATA_FOLDER, exist_ok=True)
+
+ # This will download the image and segmentation data for training.
+ image_dir = fetch_tracking_example_data(DATA_FOLDER)
+ segmentation_dir = fetch_tracking_segmentation_data(DATA_FOLDER)
+
+ # torch_em.default_segmentation_loader is a convenience function to build a torch dataloader
+ # from image data and labels for training segmentation models.
+ # It supports image data in various formats. Here, we load image data and labels from the two
+ # folders with tif images that were downloaded by the example data functionality, by specifying
+ # `raw_key` and `label_key` as `*.tif`. This means all images in the respective folders that end with
+ # .tif will be loadded.
+ # The function supports many other file formats. For example, if you have tif stacks with multiple slices
+ # instead of multiple tif images in a foldder, then you can pass raw_key=label_key=None.
+
+ # Load images from multiple files in folder via pattern (here: all tif files)
+ raw_key, label_key = "*.tif", "*.tif"
+ # Alternative: if you have tif stacks you can just set raw_key and label_key to None
+ # raw_key, label_key= None, None
+
+ # The 'roi' argument can be used to subselect parts of the data.
+ # Here, we use it to select the first 70 frames fro the test split and the other frames for the val split.
+ if split == "train":
+ roi = np.s_[:70, :, :]
+ else:
+ roi = np.s_[70:, :, :]
+
+ loader = torch_em.default_segmentation_loader(
+ raw_paths=image_dir, raw_key=raw_key,
+ label_paths=segmentation_dir, label_key=label_key,
+ patch_shape=patch_shape, batch_size=batch_size,
+ ndim=2, is_seg_dataset=True, rois=roi,
+ label_transform=torch_em.transform.label.connected_components,
+ )
+ return loader
+
+
+def run_training(checkpoint_name, model_type):
+ """Run the actual model training."""
+
+ # All hyperparameters for training.
+ batch_size = 1 # the training batch size
+ patch_shape = (1, 512, 512) # the size of patches for training
+ n_objects_per_batch = 25 # the number of objects per batch that will be sampled
+ device = torch.device("cuda") # the device/GPU used for training
+ n_iterations = 10000 # how long we train (in iterations)
+
+ # Get the dataloaders.
+ train_loader = get_dataloader("train", patch_shape, batch_size)
+ val_loader = get_dataloader("val", patch_shape, batch_size)
+
+ # Get the segment anything model, the optimizer and the LR scheduler
+ model = sam_training.get_trainable_sam_model(model_type=model_type, device=device)
+ optimizer = torch.optim.Adam(model.parameters(), lr=1e-5)
+ scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode="min", factor=0.9, patience=10, verbose=True)
+
+ # This class creates all the training data for a batch (inputs, prompts and labels).
+ convert_inputs = sam_training.ConvertToSamInputs()
+
+ # the trainer which performs training and validation (implemented using "torch_em")
+ trainer = sam_training.SamTrainer(
+ name=checkpoint_name,
+ train_loader=train_loader,
+ val_loader=val_loader,
+ model=model,
+ optimizer=optimizer,
+ # currently we compute loss batch-wise, else we pass channelwise True
+ loss=torch_em.loss.DiceLoss(channelwise=False),
+ metric=torch_em.loss.DiceLoss(),
+ device=device,
+ lr_scheduler=scheduler,
+ logger=sam_training.SamLogger,
+ log_image_interval=10,
+ mixed_precision=True,
+ convert_inputs=convert_inputs,
+ n_objects_per_batch=n_objects_per_batch,
+ n_sub_iteration=8,
+ compile_model=False
+ )
+ trainer.fit(n_iterations)
+
+
+def export_model(checkpoint_name, model_type):
+ """Export the trained model."""
+ # export the model after training so that it can be used by the rest of the micro_sam library
+ export_path = "./finetuned_hela_model.pth"
+ checkpoint_path = os.path.join("checkpoints", checkpoint_name, "best.pt")
+ export_custom_sam_model(
+ checkpoint_path=checkpoint_path,
+ model_type=model_type,
+ save_path=export_path,
+ )
+
+
+def main():
+ """Finetune a Segment Anything model.
+
+ This example uses image data and segmentations from the cell tracking challenge,
+ but can easily be adapted for other data (including data you have annoated with micro_sam beforehand).
+ """
+ # The model_type determines which base model is used to initialize the weights that are finetuned.
+ # We use vit_b here because it can be trained faster. Note that vit_h usually yields higher quality results.
+ model_type = "vit_b"
+
+ # The name of the checkpoint. The checkpoints will be stored in './checkpoints/
+The training logic is implemented in `micro_sam.training` and is based on [torch-em](https://github.com/constantinpape/torch-em). Please check out [examples/finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/examples/finetuning) to see how you can finetune on your own data with it. The script `finetune_hela.py` contains an example for finetuning on a small custom microscopy dataset and `use_finetuned_model.py` shows how this model can then be used in the interactive annotatin tools.
+More advanced examples, including quantitative and qualitative evaluation, of finetuned models can be found in [finetuning](https://github.com/computational-cell-analytics/micro-sam/tree/master/finetuning), which contains the code for training and evaluating our microscopy models.
diff --git a/doc/start_page.md b/doc/start_page.md
index b5fab2a9..840a650b 100644
--- a/doc/start_page.md
+++ b/doc/start_page.md
@@ -2,7 +2,7 @@
Segment Anything for Microscopy implements automatic and interactive annotation for microscopy data. It is built on top of [Segment Anything](https://segment-anything.com/) by Meta AI and specializes it for microscopy and other bio-imaging data.
Its core components are:
-- The `micro_sam` annotator tools for interactive data annotation with [napari](https://napari.org/stable/).
+- The `micro_sam` tools for interactive data annotation with [napari](https://napari.org/stable/).
- The `micro_sam` library to apply Segment Anything to 2d and 3d data or fine-tune it on your data.
- The `micro_sam` models that are fine-tuned on publicly available microscopy data.
@@ -19,20 +19,18 @@ On our roadmap for more functionality are:
If you run into any problems or have questions please open an issue on Github or reach out via [image.sc](https://forum.image.sc/) using the tag `micro-sam` and tagging @constantinpape.
-
## Quickstart
You can install `micro_sam` via conda:
```
-$ conda install -c conda-forge micro_sam
+$ conda install -c conda-forge micro_sam napari pyqt
```
-For more installation options check out [Installation](#installation)
+We also provide experimental installers for all operating systems.
+For more details on the available installation options check out [the installation section](#installation).
After installing `micro_sam` you can run the annotation tool via `$ micro_sam.annotator`, which opens a menu for selecting the annotation tool and its inputs.
-See [Annotation Tools](#annotation-tools) for an overview and explanation of the annotation functionality.
+See [the annotation tool section](#annotation-tools) for an overview and explanation of the annotation functionality.
The `micro_sam` python library can be used via
```python
@@ -47,7 +45,5 @@ For now, check out [the example script for the 2d annotator](https://github.com/
## Citation
If you are using `micro_sam` in your research please cite
-- [SegmentAnything](https://arxiv.org/abs/2304.02643)
-- and our repository on [zenodo](https://doi.org/10.5281/zenodo.7919746)
-
-We will release a preprint soon that describes this work and can be cited instead of zenodo.
+- Our [preprint](https://doi.org/10.1101/2023.08.21.554208)
+- and the original [Segment Anything publication](https://arxiv.org/abs/2304.02643)
diff --git a/environment_cpu.yaml b/environment_cpu.yaml
index 4d5f8ea7..ad91dab9 100644
--- a/environment_cpu.yaml
+++ b/environment_cpu.yaml
@@ -11,6 +11,7 @@ dependencies:
- pytorch
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/environment_gpu.yaml b/environment_gpu.yaml
index 21d5df53..57700759 100644
--- a/environment_gpu.yaml
+++ b/environment_gpu.yaml
@@ -12,6 +12,7 @@ dependencies:
- pytorch-cuda>=11.7 # you may need to update the cuda version to match your system
- segment-anything
- torchvision
+ - torch_em >=0.5.1
- tqdm
# - pip:
# - git+https://github.com/facebookresearch/segment-anything.git
diff --git a/examples/README.md b/examples/README.md
index 87b3a963..4844167f 100644
--- a/examples/README.md
+++ b/examples/README.md
@@ -6,5 +6,8 @@ Examples for using the micro_sam annotation tools:
- `annotator_tracking.py`: run the interactive tracking annotation tool
- `image_series_annotator.py`: run the annotation tool for a series of images
+The folder `finetuning` contains example scripts that show how a Segment Anything model can be fine-tuned
+on custom data with the `micro_sam.train` library, and how the finetuned models can then be used within the annotatin tools.
+
The folder `use_as_library` contains example scripts that show how `micro_sam` can be used as a python
-library to apply Segment Anything on mult-dimensional data.
+library to apply Segment Anything to mult-dimensional data.
diff --git a/examples/annotator_2d.py b/examples/annotator_2d.py
index f0d6d44b..8f4930f2 100644
--- a/examples/annotator_2d.py
+++ b/examples/annotator_2d.py
@@ -34,7 +34,7 @@ def hela_2d_annotator(use_finetuned_model):
embedding_path = "./embeddings/embeddings-hela2d.zarr"
model_type = "vit_h"
- annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type)
+ annotator_2d(image, embedding_path, show_embeddings=False, model_type=model_type, precompute_amg_state=True)
def wholeslide_annotator(use_finetuned_model):
diff --git a/examples/annotator_with_custom_model.py b/examples/annotator_with_custom_model.py
new file mode 100644
index 00000000..ceb8b2cb
--- /dev/null
+++ b/examples/annotator_with_custom_model.py
@@ -0,0 +1,23 @@
+import h5py
+import micro_sam.sam_annotator as annotator
+from micro_sam.util import get_sam_model
+
+# TODO add an example for the 2d annotator with a custom model
+
+
+def annotator_3d_with_custom_model():
+ with h5py.File("./data/gut1_block_1.h5") as f:
+ raw = f["raw"][:]
+
+ custom_model = "/home/pape/Work/data/models/sam/user-study/vit_h_nuclei_em_finetuned.pt"
+ embedding_path = "./embeddings/nuclei3d-custom-vit-h.zarr"
+ predictor = get_sam_model(checkpoint_path=custom_model, model_type="vit_h")
+ annotator.annotator_3d(raw, embedding_path, predictor=predictor)
+
+
+def main():
+ annotator_3d_with_custom_model()
+
+
+if __name__ == "__main__":
+ main()
diff --git a/examples/finetuning/.gitignore b/examples/finetuning/.gitignore
new file mode 100644
index 00000000..ec28c5ea
--- /dev/null
+++ b/examples/finetuning/.gitignore
@@ -0,0 +1,2 @@
+checkpoints/
+logs/
diff --git a/examples/finetuning/finetune_hela.py b/examples/finetuning/finetune_hela.py
new file mode 100644
index 00000000..f83bd8cb
--- /dev/null
+++ b/examples/finetuning/finetune_hela.py
@@ -0,0 +1,140 @@
+import os
+
+import numpy as np
+import torch
+import torch_em
+
+import micro_sam.training as sam_training
+from micro_sam.sample_data import fetch_tracking_example_data, fetch_tracking_segmentation_data
+from micro_sam.util import export_custom_sam_model
+
+DATA_FOLDER = "data"
+
+
+def get_dataloader(split, patch_shape, batch_size):
+ """Return train or val data loader for finetuning SAM.
+
+ The data loader must be a torch data loader that retuns `x, y` tensors,
+ where `x` is the image data and `y` are the labels.
+ The labels have to be in a label mask instance segmentation format.
+ I.e. a tensor of the same spatial shape as `x`, with each object mask having its own ID.
+ Important: the ID 0 is reseved for background, and the IDs must be consecutive
+
+ Here, we use `torch_em.default_segmentation_loader` for creating a suitable data loader from
+ the example hela data. You can either adapt this for your own data (see comments below)
+ or write a suitable torch dataloader yourself.
+ """
+ assert split in ("train", "val")
+ os.makedirs(DATA_FOLDER, exist_ok=True)
+
+ # This will download the image and segmentation data for training.
+ image_dir = fetch_tracking_example_data(DATA_FOLDER)
+ segmentation_dir = fetch_tracking_segmentation_data(DATA_FOLDER)
+
+ # torch_em.default_segmentation_loader is a convenience function to build a torch dataloader
+ # from image data and labels for training segmentation models.
+ # It supports image data in various formats. Here, we load image data and labels from the two
+ # folders with tif images that were downloaded by the example data functionality, by specifying
+ # `raw_key` and `label_key` as `*.tif`. This means all images in the respective folders that end with
+ # .tif will be loadded.
+ # The function supports many other file formats. For example, if you have tif stacks with multiple slices
+ # instead of multiple tif images in a foldder, then you can pass raw_key=label_key=None.
+
+ # Load images from multiple files in folder via pattern (here: all tif files)
+ raw_key, label_key = "*.tif", "*.tif"
+ # Alternative: if you have tif stacks you can just set raw_key and label_key to None
+ # raw_key, label_key= None, None
+
+ # The 'roi' argument can be used to subselect parts of the data.
+ # Here, we use it to select the first 70 frames fro the test split and the other frames for the val split.
+ if split == "train":
+ roi = np.s_[:70, :, :]
+ else:
+ roi = np.s_[70:, :, :]
+
+ loader = torch_em.default_segmentation_loader(
+ raw_paths=image_dir, raw_key=raw_key,
+ label_paths=segmentation_dir, label_key=label_key,
+ patch_shape=patch_shape, batch_size=batch_size,
+ ndim=2, is_seg_dataset=True, rois=roi,
+ label_transform=torch_em.transform.label.connected_components,
+ )
+ return loader
+
+
+def run_training(checkpoint_name, model_type):
+ """Run the actual model training."""
+
+ # All hyperparameters for training.
+ batch_size = 1 # the training batch size
+ patch_shape = (1, 512, 512) # the size of patches for training
+ n_objects_per_batch = 25 # the number of objects per batch that will be sampled
+ device = torch.device("cuda") # the device/GPU used for training
+ n_iterations = 10000 # how long we train (in iterations)
+
+ # Get the dataloaders.
+ train_loader = get_dataloader("train", patch_shape, batch_size)
+ val_loader = get_dataloader("val", patch_shape, batch_size)
+
+ # Get the segment anything model, the optimizer and the LR scheduler
+ model = sam_training.get_trainable_sam_model(model_type=model_type, device=device)
+ optimizer = torch.optim.Adam(model.parameters(), lr=1e-5)
+ scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode="min", factor=0.9, patience=10, verbose=True)
+
+ # This class creates all the training data for a batch (inputs, prompts and labels).
+ convert_inputs = sam_training.ConvertToSamInputs()
+
+ # the trainer which performs training and validation (implemented using "torch_em")
+ trainer = sam_training.SamTrainer(
+ name=checkpoint_name,
+ train_loader=train_loader,
+ val_loader=val_loader,
+ model=model,
+ optimizer=optimizer,
+ # currently we compute loss batch-wise, else we pass channelwise True
+ loss=torch_em.loss.DiceLoss(channelwise=False),
+ metric=torch_em.loss.DiceLoss(),
+ device=device,
+ lr_scheduler=scheduler,
+ logger=sam_training.SamLogger,
+ log_image_interval=10,
+ mixed_precision=True,
+ convert_inputs=convert_inputs,
+ n_objects_per_batch=n_objects_per_batch,
+ n_sub_iteration=8,
+ compile_model=False
+ )
+ trainer.fit(n_iterations)
+
+
+def export_model(checkpoint_name, model_type):
+ """Export the trained model."""
+ # export the model after training so that it can be used by the rest of the micro_sam library
+ export_path = "./finetuned_hela_model.pth"
+ checkpoint_path = os.path.join("checkpoints", checkpoint_name, "best.pt")
+ export_custom_sam_model(
+ checkpoint_path=checkpoint_path,
+ model_type=model_type,
+ save_path=export_path,
+ )
+
+
+def main():
+ """Finetune a Segment Anything model.
+
+ This example uses image data and segmentations from the cell tracking challenge,
+ but can easily be adapted for other data (including data you have annoated with micro_sam beforehand).
+ """
+ # The model_type determines which base model is used to initialize the weights that are finetuned.
+ # We use vit_b here because it can be trained faster. Note that vit_h usually yields higher quality results.
+ model_type = "vit_b"
+
+ # The name of the checkpoint. The checkpoints will be stored in './checkpoints/ +
+And after selecting them a new window will open where you can select the input file path and other optional parameter. Then click the top button to start the tool. **Note: If you are not starting the annotation tool with a path to pre-computed embeddings then it can take several minutes to open napari after pressing the button because the embeddings are being computed.**
+
## Annotator 2D
@@ -44,7 +57,7 @@ It contains the following elements:
Note that point prompts and box prompts can be combined. When you're using point prompts you can only segment one object at a time. With box prompts you can segment several objects at once.
-Check out [this video](https://youtu.be/DfWE_XRcqN8) for an example of how to use the interactive 2d annotator.
+Check out [this video](https://youtu.be/ket7bDUP9tI) for a tutorial for the 2d annotation tool.
We also provide the `image series annotator`, which can be used for running the 2d annotator for several images in a folder. You can start by clicking `Image series annotator` in the GUI, running `micro_sam.image_series_annotator` in the command line or from a [python script](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/image_series_annotator.py).
@@ -69,7 +82,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that you can only segment one object at a time with the 3d annotator.
-Check out [this video](https://youtu.be/5Jo_CtIefTM) for an overview of the interactive 3d segmentation functionality.
+Check out [this video](https://youtu.be/PEy9-rTCdS4) for a tutorial for the 3d annotation tool.
## Annotator Tracking
@@ -93,7 +106,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that the tracking annotator only supports 2d image data, volumetric data is not supported.
-Check out [this video](https://youtu.be/PBPW0rDOn9w) for an overview of the interactive tracking functionality.
+Check out [this video](https://youtu.be/Xi5pRWMO6_w) for a tutorial for how to use the tracking annotation tool.
## Tips & Tricks
@@ -105,7 +118,7 @@ You can activate tiling by passing the parameters `tile_shape`, which determines
- If you're using the command line functions you can pass them via the options `--tile_shape 1024 1024 --halo 128 128`
- Note that prediction with tiling only works when the embeddings are cached to file, so you must specify an `embedding_path` (`-e` in the CLI).
- You should choose the `halo` such that it is larger than half of the maximal radius of the objects your segmenting.
-- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_embeddings` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
+- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_state` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
- Most other processing steps are very fast even on a CPU, so interactive annotation is possible. An exception is the automatic segmentation step (2d segmentation), which takes several minutes without a GPU (depending on the image size). For large volumes and timeseries segmenting an object in 3d / tracking across time can take a couple settings with a CPU (it is very fast with a GPU).
- You can also try using a smaller version of the SegmentAnything model to speed up the computations. For this you can pass the `model_type` argument and either set it to `vit_b` or to `vit_l` (default is `vit_h`). However, this may lead to worse results.
- You can save and load the results from the `committed_objects` / `committed_tracks` layer to correct segmentations you obtained from another tool (e.g. CellPose) or to save intermediate annotation results. The results can be saved via `File -> Save Selected Layer(s) ...` in the napari menu (see the tutorial videos for details). They can be loaded again by specifying the corresponding location via the `segmentation_result` (2d and 3d segmentation) or `tracking_result` (tracking) argument.
diff --git a/doc/finetuned_models.md b/doc/finetuned_models.md
new file mode 100644
index 00000000..4fc9fb13
--- /dev/null
+++ b/doc/finetuned_models.md
@@ -0,0 +1,36 @@
+# Finetuned models
+
+We provide models that were finetuned on microscopy data using `micro_sam.training`. They are hosted on zenodo. We currently offer the following models:
+- `vit_h`: Default Segment Anything model with vit-h backbone.
+- `vit_l`: Default Segment Anything model with vit-l backbone.
+- `vit_b`: Default Segment Anything model with vit-b backbone.
+- `vit_h_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-h backbone.
+- `vit_b_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-b backbone.
+- `vit_h_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-h backbone.
+- `vit_b_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-b backbone.
+
+See the two figures below of the improvements through the finetuned model for LM and EM data.
+
+
+
+And after selecting them a new window will open where you can select the input file path and other optional parameter. Then click the top button to start the tool. **Note: If you are not starting the annotation tool with a path to pre-computed embeddings then it can take several minutes to open napari after pressing the button because the embeddings are being computed.**
+
## Annotator 2D
@@ -44,7 +57,7 @@ It contains the following elements:
Note that point prompts and box prompts can be combined. When you're using point prompts you can only segment one object at a time. With box prompts you can segment several objects at once.
-Check out [this video](https://youtu.be/DfWE_XRcqN8) for an example of how to use the interactive 2d annotator.
+Check out [this video](https://youtu.be/ket7bDUP9tI) for a tutorial for the 2d annotation tool.
We also provide the `image series annotator`, which can be used for running the 2d annotator for several images in a folder. You can start by clicking `Image series annotator` in the GUI, running `micro_sam.image_series_annotator` in the command line or from a [python script](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/image_series_annotator.py).
@@ -69,7 +82,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that you can only segment one object at a time with the 3d annotator.
-Check out [this video](https://youtu.be/5Jo_CtIefTM) for an overview of the interactive 3d segmentation functionality.
+Check out [this video](https://youtu.be/PEy9-rTCdS4) for a tutorial for the 3d annotation tool.
## Annotator Tracking
@@ -93,7 +106,7 @@ Most elements are the same as in [the 2d annotator](#annotator-2d):
Note that the tracking annotator only supports 2d image data, volumetric data is not supported.
-Check out [this video](https://youtu.be/PBPW0rDOn9w) for an overview of the interactive tracking functionality.
+Check out [this video](https://youtu.be/Xi5pRWMO6_w) for a tutorial for how to use the tracking annotation tool.
## Tips & Tricks
@@ -105,7 +118,7 @@ You can activate tiling by passing the parameters `tile_shape`, which determines
- If you're using the command line functions you can pass them via the options `--tile_shape 1024 1024 --halo 128 128`
- Note that prediction with tiling only works when the embeddings are cached to file, so you must specify an `embedding_path` (`-e` in the CLI).
- You should choose the `halo` such that it is larger than half of the maximal radius of the objects your segmenting.
-- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_embeddings` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
+- The applications pre-compute the image embeddings produced by SegmentAnything and (optionally) store them on disc. If you are using a CPU this step can take a while for 3d data or timeseries (you will see a progress bar with a time estimate). If you have access to a GPU without graphical interface (e.g. via a local computer cluster or a cloud provider), you can also pre-compute the embeddings there and then copy them to your laptop / local machine to speed this up. You can use the command `micro_sam.precompute_state` for this (it is installed with the rest of the applications). You can specify the location of the precomputed embeddings via the `embedding_path` argument.
- Most other processing steps are very fast even on a CPU, so interactive annotation is possible. An exception is the automatic segmentation step (2d segmentation), which takes several minutes without a GPU (depending on the image size). For large volumes and timeseries segmenting an object in 3d / tracking across time can take a couple settings with a CPU (it is very fast with a GPU).
- You can also try using a smaller version of the SegmentAnything model to speed up the computations. For this you can pass the `model_type` argument and either set it to `vit_b` or to `vit_l` (default is `vit_h`). However, this may lead to worse results.
- You can save and load the results from the `committed_objects` / `committed_tracks` layer to correct segmentations you obtained from another tool (e.g. CellPose) or to save intermediate annotation results. The results can be saved via `File -> Save Selected Layer(s) ...` in the napari menu (see the tutorial videos for details). They can be loaded again by specifying the corresponding location via the `segmentation_result` (2d and 3d segmentation) or `tracking_result` (tracking) argument.
diff --git a/doc/finetuned_models.md b/doc/finetuned_models.md
new file mode 100644
index 00000000..4fc9fb13
--- /dev/null
+++ b/doc/finetuned_models.md
@@ -0,0 +1,36 @@
+# Finetuned models
+
+We provide models that were finetuned on microscopy data using `micro_sam.training`. They are hosted on zenodo. We currently offer the following models:
+- `vit_h`: Default Segment Anything model with vit-h backbone.
+- `vit_l`: Default Segment Anything model with vit-l backbone.
+- `vit_b`: Default Segment Anything model with vit-b backbone.
+- `vit_h_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-h backbone.
+- `vit_b_lm`: Finetuned Segment Anything model for cells and nuclei in light microscopy data with vit-b backbone.
+- `vit_h_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-h backbone.
+- `vit_b_em`: Finetuned Segment Anything model for neurites and cells in electron microscopy data with vit-b backbone.
+
+See the two figures below of the improvements through the finetuned model for LM and EM data.
+
+ +
+
+
+ +
+You can select which of the models is used in the annotation tools by selecting the corresponding name from the `Model Type` menu:
+
+
+
+You can select which of the models is used in the annotation tools by selecting the corresponding name from the `Model Type` menu:
+
+ +
+To use a specific model in the python library you need to pass the corresponding name as value to the `model_type` parameter exposed by all relevant functions.
+See for example the [2d annotator example](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/annotator_2d.py#L62) where `use_finetuned_model` can be set to `True` to use the `vit_h_lm` model.
+
+## Which model should I choose?
+
+As a rule of thumb:
+- Use the `_lm` models for segmenting cells or nuclei in light microscopy.
+- Use the `_em` models for segmenting ceells or neurites in electron microscopy.
+ - Note that this model does not work well for segmenting mitochondria or other organelles becuase it is biased towards segmenting the full cell / cellular compartment.
+- For other cases use the default models.
+
+See also the figures above for examples where the finetuned models work better than the vanilla models.
+Currently the model `vit_h` is used by default.
+
+We are working on releasing more fine-tuned models, in particular for mitochondria and other organelles in EM.
diff --git a/doc/images/em_comparison.png b/doc/images/em_comparison.png
new file mode 100644
index 00000000..86b2d66c
Binary files /dev/null and b/doc/images/em_comparison.png differ
diff --git a/doc/images/lm_comparison.png b/doc/images/lm_comparison.png
new file mode 100644
index 00000000..4e115d61
Binary files /dev/null and b/doc/images/lm_comparison.png differ
diff --git a/doc/images/micro-sam-gui.png b/doc/images/micro-sam-gui.png
new file mode 100644
index 00000000..26254490
Binary files /dev/null and b/doc/images/micro-sam-gui.png differ
diff --git a/doc/images/model-type-selector.png b/doc/images/model-type-selector.png
new file mode 100644
index 00000000..cab3b077
Binary files /dev/null and b/doc/images/model-type-selector.png differ
diff --git a/doc/images/vanilla-v-finetuned.png b/doc/images/vanilla-v-finetuned.png
deleted file mode 100644
index a72446a4..00000000
Binary files a/doc/images/vanilla-v-finetuned.png and /dev/null differ
diff --git a/doc/installation.md b/doc/installation.md
index 12bda71e..0b49edc5 100644
--- a/doc/installation.md
+++ b/doc/installation.md
@@ -1,16 +1,38 @@
# Installation
-`micro_sam` requires the following dependencies:
+We provide three different ways of installing `micro_sam`:
+- [From conda](#from-conda) is the recommended way if you want to use all functionality.
+- [From source](#from-source) for setting up a development environment to change and potentially contribute to our software.
+- [From installer](#from-installer) to install without having to use conda. This mode of installation is still experimental! It only provides the annotation tools and does not enable model finetuning.
+
+Our software requires the following dependencies:
- [PyTorch](https://pytorch.org/get-started/locally/)
- [SegmentAnything](https://github.com/facebookresearch/segment-anything#installation)
-- [napari](https://napari.org/stable/)
- [elf](https://github.com/constantinpape/elf)
+- [napari](https://napari.org/stable/) (for the interactive annotation tools)
+- [torch_em](https://github.com/constantinpape/torch-em) (for the training functionality)
+
+## From conda
-It is available as a conda package and can be installed via
+`micro_sam` is available as a conda package and can be installed via
```
$ conda install -c conda-forge micro_sam
```
+This command will not install the required dependencies for the annotation tools and for training / finetuning.
+To use the annotation functionality you also need to install `napari`:
+```
+$ conda install -c conda-forge napari pyqt
+```
+And to use the training functionality `torch_em`:
+```
+$ conda install -c conda-forge torch_em
+```
+
+In case the installation via conda takes too long consider using [mamba](https://mamba.readthedocs.io/en/latest/).
+Once you have it installed you can simply replace the `conda` commands with `mamba`.
+
+
## From source
To install `micro_sam` from source, we recommend to first set up a conda environment with the necessary requirements:
@@ -54,3 +76,48 @@ $ pip install -e .
- Install `micro_sam` by running `pip install -e .` in this folder.
- **Note:** we have seen many issues with the pytorch installation on MAC. If a wrong pytorch version is installed for you (which will cause pytorch errors once you run the application) please try again with a clean `mambaforge` installation. Please install the `OS X, arm64` version from [here](https://github.com/conda-forge/miniforge#mambaforge).
- Some MACs require a specific installation order of packages. If the steps layed out above don't work for you please check out the procedure described [in this github issue](https://github.com/computational-cell-analytics/micro-sam/issues/77).
+
+
+## From installer
+
+We also provide installers for Linuxand Windows:
+- [Linux](https://owncloud.gwdg.de/index.php/s/Cw9RmA3BlyqKJeU)
+- [Windows](https://owncloud.gwdg.de/index.php/s/1iD1eIcMZvEyE6d)
+
+
+**The installers are stil experimental and not fully tested.** Mac is not supported yet, but we are working on also providing an installer for it.
+
+If you encounter problems with them then please consider installing `micro_sam` via [conda](#from-conda) instead.
+
+**Linux Installer:**
+
+To use the installer:
+- Unpack the zip file you have downloaded.
+- Make the installer executable: `$ chmod +x micro_sam-0.2.0post1-Linux-x86_64.sh`
+- Run the installer: `$./micro_sam-0.2.0post1-Linux-x86_64.sh$`
+ - You can select where to install `micro_sam` during the installation. By default it will be installed in `$HOME/micro_sam`.
+ - The installer will unpack all `micro_sam` files to the installation directory.
+- After the installation you can start the annotator with the command `.../micro_sam/bin/micro_sam.annotator`.
+ - To make it easier to run the annotation tool you can add `.../micro_sam/bin` to your `PATH` or set a softlink to `.../micro_sam/bin/micro_sam.annotator`.
+
+
+
+**Windows Installer:**
+
+- Unpack the zip file you have downloaded.
+- Run the installer by double clicking on it.
+- Choose installation type: `Just Me(recommended)` or `All Users(requires admin privileges)`.
+- Choose installation path. By default it will be installed in `C:\Users\
+
+To use a specific model in the python library you need to pass the corresponding name as value to the `model_type` parameter exposed by all relevant functions.
+See for example the [2d annotator example](https://github.com/computational-cell-analytics/micro-sam/blob/master/examples/annotator_2d.py#L62) where `use_finetuned_model` can be set to `True` to use the `vit_h_lm` model.
+
+## Which model should I choose?
+
+As a rule of thumb:
+- Use the `_lm` models for segmenting cells or nuclei in light microscopy.
+- Use the `_em` models for segmenting ceells or neurites in electron microscopy.
+ - Note that this model does not work well for segmenting mitochondria or other organelles becuase it is biased towards segmenting the full cell / cellular compartment.
+- For other cases use the default models.
+
+See also the figures above for examples where the finetuned models work better than the vanilla models.
+Currently the model `vit_h` is used by default.
+
+We are working on releasing more fine-tuned models, in particular for mitochondria and other organelles in EM.
diff --git a/doc/images/em_comparison.png b/doc/images/em_comparison.png
new file mode 100644
index 00000000..86b2d66c
Binary files /dev/null and b/doc/images/em_comparison.png differ
diff --git a/doc/images/lm_comparison.png b/doc/images/lm_comparison.png
new file mode 100644
index 00000000..4e115d61
Binary files /dev/null and b/doc/images/lm_comparison.png differ
diff --git a/doc/images/micro-sam-gui.png b/doc/images/micro-sam-gui.png
new file mode 100644
index 00000000..26254490
Binary files /dev/null and b/doc/images/micro-sam-gui.png differ
diff --git a/doc/images/model-type-selector.png b/doc/images/model-type-selector.png
new file mode 100644
index 00000000..cab3b077
Binary files /dev/null and b/doc/images/model-type-selector.png differ
diff --git a/doc/images/vanilla-v-finetuned.png b/doc/images/vanilla-v-finetuned.png
deleted file mode 100644
index a72446a4..00000000
Binary files a/doc/images/vanilla-v-finetuned.png and /dev/null differ
diff --git a/doc/installation.md b/doc/installation.md
index 12bda71e..0b49edc5 100644
--- a/doc/installation.md
+++ b/doc/installation.md
@@ -1,16 +1,38 @@
# Installation
-`micro_sam` requires the following dependencies:
+We provide three different ways of installing `micro_sam`:
+- [From conda](#from-conda) is the recommended way if you want to use all functionality.
+- [From source](#from-source) for setting up a development environment to change and potentially contribute to our software.
+- [From installer](#from-installer) to install without having to use conda. This mode of installation is still experimental! It only provides the annotation tools and does not enable model finetuning.
+
+Our software requires the following dependencies:
- [PyTorch](https://pytorch.org/get-started/locally/)
- [SegmentAnything](https://github.com/facebookresearch/segment-anything#installation)
-- [napari](https://napari.org/stable/)
- [elf](https://github.com/constantinpape/elf)
+- [napari](https://napari.org/stable/) (for the interactive annotation tools)
+- [torch_em](https://github.com/constantinpape/torch-em) (for the training functionality)
+
+## From conda
-It is available as a conda package and can be installed via
+`micro_sam` is available as a conda package and can be installed via
```
$ conda install -c conda-forge micro_sam
```
+This command will not install the required dependencies for the annotation tools and for training / finetuning.
+To use the annotation functionality you also need to install `napari`:
+```
+$ conda install -c conda-forge napari pyqt
+```
+And to use the training functionality `torch_em`:
+```
+$ conda install -c conda-forge torch_em
+```
+
+In case the installation via conda takes too long consider using [mamba](https://mamba.readthedocs.io/en/latest/).
+Once you have it installed you can simply replace the `conda` commands with `mamba`.
+
+
## From source
To install `micro_sam` from source, we recommend to first set up a conda environment with the necessary requirements:
@@ -54,3 +76,48 @@ $ pip install -e .
- Install `micro_sam` by running `pip install -e .` in this folder.
- **Note:** we have seen many issues with the pytorch installation on MAC. If a wrong pytorch version is installed for you (which will cause pytorch errors once you run the application) please try again with a clean `mambaforge` installation. Please install the `OS X, arm64` version from [here](https://github.com/conda-forge/miniforge#mambaforge).
- Some MACs require a specific installation order of packages. If the steps layed out above don't work for you please check out the procedure described [in this github issue](https://github.com/computational-cell-analytics/micro-sam/issues/77).
+
+
+## From installer
+
+We also provide installers for Linuxand Windows:
+- [Linux](https://owncloud.gwdg.de/index.php/s/Cw9RmA3BlyqKJeU)
+- [Windows](https://owncloud.gwdg.de/index.php/s/1iD1eIcMZvEyE6d)
+
+
+**The installers are stil experimental and not fully tested.** Mac is not supported yet, but we are working on also providing an installer for it.
+
+If you encounter problems with them then please consider installing `micro_sam` via [conda](#from-conda) instead.
+
+**Linux Installer:**
+
+To use the installer:
+- Unpack the zip file you have downloaded.
+- Make the installer executable: `$ chmod +x micro_sam-0.2.0post1-Linux-x86_64.sh`
+- Run the installer: `$./micro_sam-0.2.0post1-Linux-x86_64.sh$`
+ - You can select where to install `micro_sam` during the installation. By default it will be installed in `$HOME/micro_sam`.
+ - The installer will unpack all `micro_sam` files to the installation directory.
+- After the installation you can start the annotator with the command `.../micro_sam/bin/micro_sam.annotator`.
+ - To make it easier to run the annotation tool you can add `.../micro_sam/bin` to your `PATH` or set a softlink to `.../micro_sam/bin/micro_sam.annotator`.
+
+
+
+**Windows Installer:**
+
+- Unpack the zip file you have downloaded.
+- Run the installer by double clicking on it.
+- Choose installation type: `Just Me(recommended)` or `All Users(requires admin privileges)`.
+- Choose installation path. By default it will be installed in `C:\Users\