- add the new genome
GCF_012559485.2 - add the new genome
bosTau9 - Switched T2T gene annotation from using
gencodeV35.bedtocatLiftOffGenesV1.bed
- add the
ballctrack support
- add
pixelsPaddingoption formodbedtrack while display in heatmap mode, used for padding the drawing tick pixel - add HPRC modbed hub for
hg38 - add the genome
rn5
- fix an bug that screenshot UI doesn't show axis label
- fix #319
- Fix Chrome 112 Navbar nested css problem (by Shane)
- small changes to modbed base pair level visualization
- new genome for Parhyale hawaiensis version 5 is added
- added an option to hide minimal annotations
- removed some old version gene annotation tracks (files can be requested if still need)
- added new track type
modbedfor display long read sequencing methylation data - added ensembl style for matplot and dynamic tracks
- fixed a screenshot bug when svg imported to Adobe Illustrator
- removed default highlight for gene/snp/region search
- added
highlightPositionurl parameter, used withposition, for example&position=chr3:181429711-181432223&highlightPosition=1will jump tochr3:181429711-181432223and highlight it at same time
- added light/dark theme setting
- added view sharing via email/embed/QR code
- added MANE select 1.0 for hg38
- updated gencode tracks for hg38, hg19, mm39 and mm10
- add opitimization for dropbox shared hubs
- 3D viewer can enable region set view from segments
- fixed a bug that MIN/MAX aggregator in numeric track crash
- added
t2t-chm13-v2.0 - fixed sceenshot issue with genome align track, reported by #252
- added highlights in screenshot by #251
rn7addedqBedadd secondary color config and horizontal line option per Slack user request- imporved highlighting functions
- changed circlet view UI slightly
- added chord view
- added Jaspar 2022 TF tracks to Annotation track sets
- new genomes
susScr3,calJac4,rheMac10added. - UI, button styles changes from Shane Liu
- added cookie consent notice
- new genomes
susScr11,oviAri4and Brapa (#228) added. - image tracks/hubs update
- add new feature to allow long range tracks with colors for each arc/heatmap diamond
- add spin utility for 3d model viewer
- add
clampHeightoption to chromatin interaction track forarcandheatmapstyle
- removed unnecessary re-render of numerical track
- adding push notification
- allow HiC track bin size and normalization options linked to the hic file itself
- add an option
italicizeTextto allow italic text/label on gene tracks - new genome
xenTro10added
- added
bigchainformat support bigchainandgenomealigncan be added from remote track UI with specify a query genome- layout header height bug fix
- region jumping following UCSC Browser: position coordinates
chr6:130129863-130129899(1 based, 37bp), bed coordinatechr6 130129863 130129899(0 based, 36bp)
- new T2T genome assembly
t2t-chm13-v1.1added - improved 3D module
- support Repeatmasker V2 format bigbed, track type
rmskv2
- for
bigwigtrack, specifyensemblStyleoption totruecan enable data with chromosome names as 1, 2, 3...work in the browser bedcolortrack type- Roadmap (hg38) hub update
- more genome alignment tracks
- pdf output label alignment improvement
- switched to
@gmod/bbilibrary for bigwig data fetching - removed zoom level configuration for bigWig track
- updated 4DN data hubs
- fixed a screenshot bug while tracks are grouped
- introduce
boxplottrack type show numerical data in small window as boxplots
vcftrack bug fix for certain tooltip clicking- updated SARS-CoV-2 data hubs by Changxu Fan
- 3D structure view module added, for more please check the 3D documentation
- basic support for
vcfformat
- add an option
queryEndpointtoTrackModelallow customization of gene/feature annotaion query - added 2 new Trypanosome genomes
TbruceiTREU927andTbruceiLister427per user's request
- fixed a bug that
dynseqtrack doesn't work correctly in Safari - fixed a bug that screenshot error on
hicheatmap mode - fixed a bug that
longrangetrack cannot change name/label - added a feature that sessions are sorted by date by dafault, can change to sorting by label
- add a new track type
rgbpeakto show the peaks inbigbedformat supportingitemRgbattribute - added a new virus genome
hpv16, ref sequence from https://www.ncbi.nlm.nih.gov/nuccore/NC_001526.4, gene annotation from https://www.ncbi.nlm.nih.gov/assembly/GCF_000863945.3/
- Dynamic sequence track type
dynseqadded, the track type is proposed and initially developped by Surag Nair from Anshul Kundaje's lab at Stanford University
- bug fix: numerical track y-scale threshold not working for negative values
- bug fix: local data hub missed track's
metadataattibute
- added percentile scale for interaction tracks (
hic,longrange) etc. - improved fixed scale config for numerical tracks
- video output for dynamic track
- added the highlight beams for interaction track at
heatmapmode
- fixed a bug that hic track fetches whole hic file when switch to other norm options with updated hic-straw package
- fixed a bug that numerical track shows wrong threshold for negative data values
- 6 new genomes added
- new
noDefaultTracksurl parameter remove the default tracks while loading a remote hub - removed limitation that querygenome of
genomealigntrack need be preconfigured - added default width and height for
withAutoDimensionsmodule - enhanced
genomeailgntrack by Silas Hsu
- add the API key and secret for accessing 4DN tracks
- added new genome
Plasmodium falciparum (Pfal3D7) - fixed one bug that bam track at density mode not showing y scale config
- fixed a bug when load
g3dtrack from datahub caused browser track facet table lost - fixed issue #111 and #176 for long range track data filtering
- add option to avoid use firebase database by remove session/live function (see doc for details)
- upgrade dependent packages: react, react-dom -> 16.13.1
- supports nodejs v12+
- SARS-CoV-2 hubs updated by Fan
- fixed a
smoothoption caused display issue (related to #163) - improved window/container resize listener using ResizeObserver API
- improved responsive layout of navigation menu and tools bar
- modified vendor
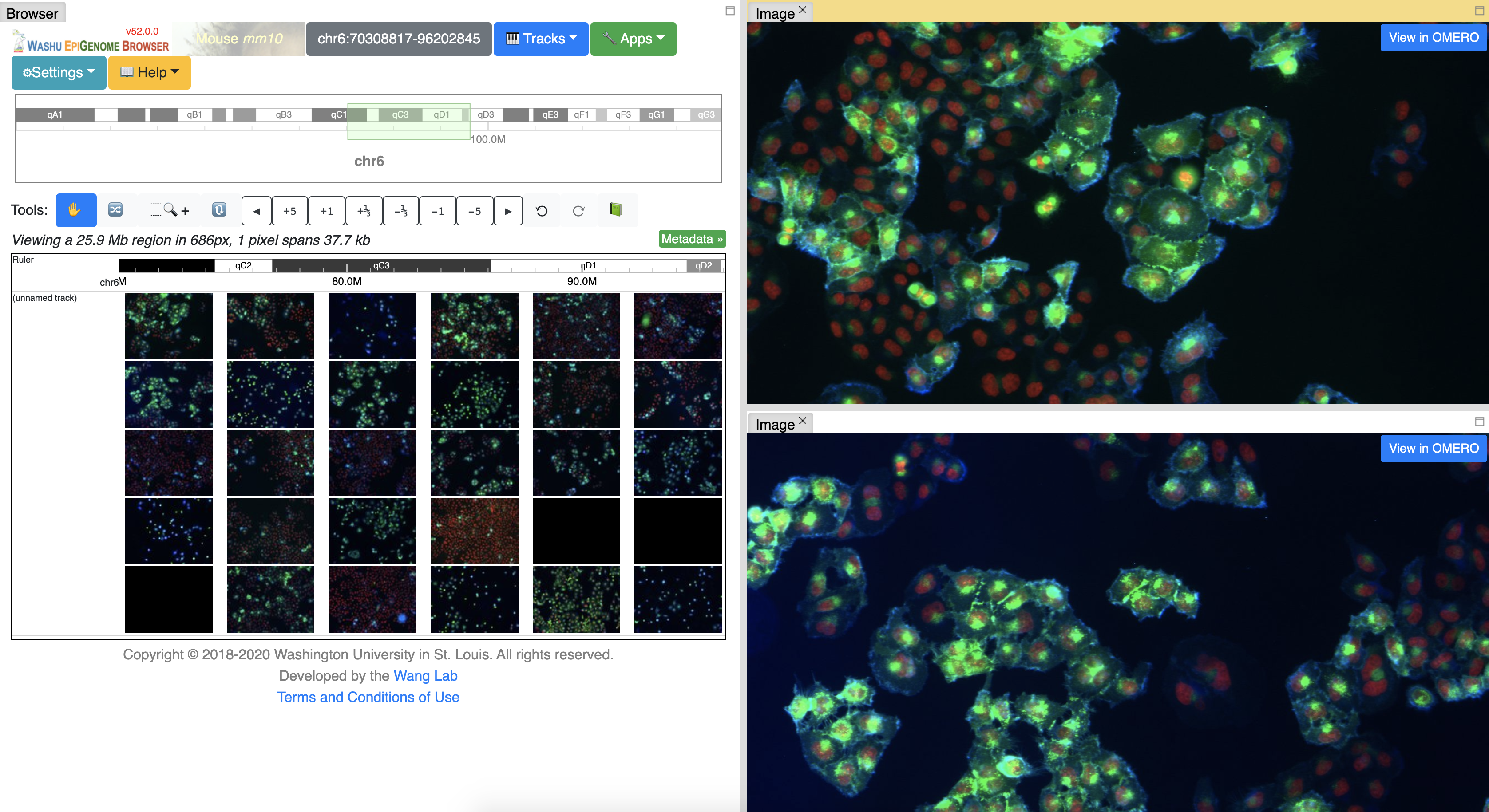
bigwig.jsto support chromosome names likechr1or1 - introduced image track:
- track file information can be displayed with track data (like resolution and normalization for
hic,g3dtrack etc) - new 'snv' track for display sequence variations from reference
- update hubs of virus genomes
- updated SARS-CoV-2 publib hubs and default tracks
- enabled tracks can be loaded from both hub url and previous session storage with new URL parameter
hubSessionStorage - fixed a
qBedwhen mouseover outside of the track caused TypeError bug - fixed a bug that remote hub load button not working in Firefox
- add dynamic arc view for dynamic hic tracks
- fixed a bug arc with negative score not show up in screenshot
- dynamic colors
dynamicColorsare supported for all dynamic tracks
- add a new option
greedyTooltipfor arc display of chromatin interaction data, this option will show more than one interaction from same 2 loci if exists - track type
callingcardrenamed to 'qBed' - update
hic-strawpackage
- fixed a crash bug when reize window with a dynamic bedgraph track
- added dynamic labels to dynamic tracks
- fixed a bug when zooming in a dynamic bedgraph data not show
- add tooltip for dynamic hic track
- fixed one bug when
bigwigfiles have just one zoomLevel causingdata fetch error - a new option
alwaysDrawLabeladded tobedandcategoricaltrack for telling the browser always render the label - update nCoV2019 public hubs
- added dynamic numerical track
- added dynamic plot menu for >=2 numerical tracks
- fixed a
virusBrowserModebug (redirected to main page) introduced in last few commits - fixed a bug long-range track data cannot have dot in chromosome names
- added an option
zoomLevelforbigwigtrack - changed custom track menu to remote track for better distinguish from local track
- added dynamic hic plot track/menu
- updated public hubs for nCoV2019
- added 4 virus genomes for aiding virus research
- global URL parameter
virusBrowserModeadded - VR freeze issue fixed
- VR mode supports
longrangetrack type now, thanks Silas for the help - certain public hubs added to 2019-nCoV by Changxu Fan
- added sea hare genome (aplCal3) by Xiaoyu
- added yeast sacCer3 genome
- calling card track update by Arnav
- multiple alignment track update by Xiaoyu
- this version we implemented the PWA design
- in case your device went offline, the local track and local text track function can still be used
- whenever there is a new version, a notice will show if user still use the old version
- fixed a bug for numerical track when there are negative values, set scale to fixed shifted values
- fixed a bug metadata terms were loaded when hub was loaded from URL
- fixed a small bug display view window in pixel
- fixed numerical track y-axis scale while setting minimal y value
- the
squaredisplay for hic track is added add all tracksbutton added for hub track table- text in
refbedformat supported
- introducing the new support for text tracks, text files can be visualized directly in the browser, check the documentation for more details
- text in
bedformat supported - text in
bedGraphformat supported - text in
longrangeformat supported - please contact us for customized format
- introduce a new track type
g3dfor genomic 3D structure display, see more at the docs - fixed link to slack invitation
- fixed a sequence fetch bug
- fixed a 'browser content empty' error for
Go Livefunction
- add
sessionFileURL param for loading a session file/state from URL - add new genome fruit fly
dm6and C.elegansce11
- the browser can respond to window resize event now, thanks to Silas who stopped by and helped debug this function
- add Zebrafish genome
danRer11 - from discussion with Arnav for
callingcardtrack, plot a single circle (per track) at the midpoint of the interval
- fixed a facet table bug when metadata is undefined, cannot distinguish track type with substrings like
bedandbedgraph - fixed a session saving bug due to updated firebase related packages
- when user define height in string like
"25"instead of25, the browser won't complain - fixed a bug that genome align track shows data fetch error
- this release is not an update for browser function but related to dependent package
- updated to use
webpack4 andtypescript3.4 - removed deprecated package
react-scripts-ts, use typescript support fromcreate-react-app - updated
react-app-rewiredto 2.1 - motivated by an issue that ArcDisplay is not working on build version but development version
- added chicken genome
galGal6andgalGal5 - a
flatarcmode added for chromatin interacion track: - fixed a metadata bug when define customized color
- fixed a bug that facet table shows error count of loaded tracks
- Rhesus
rheMac8genome is added - fixed bug #117
- enabled multiple rows for categorical track when categories overlap
- a new option
hiddenPixelsis added to control an item should be hidden for annotation tracks - fixed a bug that screenshot missed chromosome label in Ruler track
- user can input track options while submit a custom track or local track
urlin track of a custom datahub can use relative path
- track height and color configuration enabled for genome align track
- tooltip on heatmap and heatmap style of interaction track
- annotation tracks adding UI will be shown when corresponding genome align track is added
- fixed a bug that intra-region arcs are not displayed
- fixed an Unable to preventDefault inside passive event listener bug
- fixed the bug highlight region error when genomealign track is added, related to #104
- fixed an issue that tracks order in local hub doesn't follow order in
hub.config.jsonfile - local track supports BAM track now
- fixed a bug that categorical track error when category not exists in hub file
- tooltips on genome align track, both rough and fine mode (from Xiaoyu)
- annotation tracks UI for genome align track
- switched to latest bam-js from GMOD for bam file fetching
- free text search for track in track table using fuse.js
- pearson correlation value is added to scatter plot
- add YouTube channel link
- fix a bug for screenshot app missed background color for tracks
- cleaning package.json file
- screenshot can now generate pdf as well using svg2pdf.js, while the style may slight changed compared to SVG
hammocktrack type added with basic support- genome logo can now be clicked to change to another genome
- mouse
mm9and cowbosTau8added - arabidopsis
araTha1(same as TAIR10) added - session can either be downloaded as a session file or a datahub file
- highlight box color could be configured now
- color configurable for boxplot in geneplot
- marker size and color configurable for scatter plot
- heatmap type in geneplot switched back to using plotly.js as well
- fixed a bug for long-range track has negative score
- add line width config for interaction track when displayed as arcs
- updated to hic-straw 0.9.0 as suggested by Jim Robinson
- fixed a bug when reading local .hic file caused
data fetch error - removed timeout mechanism for bigwig tracks
- added the scatter plot app
- enabled canvas renderer for arc and heatmap display of chromatin interaction tracks
- enabled height config for chromatin interaction tracks
- use CND hosted plotly scripts to reduce script bundle size
- fix a bug that screenshot app shows left expanded region when there is interaction track
- use nivo library for heatmap in geneplot app
- fix a bug that values for geneplot missed strand information
- fix a bug certain gene symbol cannot be found in geneplot
- fix a bug strand for region sets by default sets to
-
- matplot function is added
- configuration for matplot track is added with
smooth,aggregate methods,line widthetc.
- retina screen display improved as track rendered in canvas
- SNP search through Ensembl API
- SNP track added as well, data comes from Ensembl API
- Aggregate method is configurable for numerical tracks,
MEAN,SUM,COUNT,MAX,MINare supported,MEANis default smoothoption added for numerical tracks
- new function to re-order many tracks at one time by drap and drop
- new hot key (Alt+G) to open the interface for re-order many function
- new track type
callingcardfor displaying Calling card data - new function geneplot for plot overall signal from a numerical track over a gene/region set
- function to download current session to local disk
- accept session files uploaded from local disk
- use hic-straw package for .hic data source, removed the dependency to igv.js and juicebox.js
- added
FetchSequenceApp to allow fetch sequence of current region or user specified regions - Fix a bug while upload file hub causing page freeze
- Rough alignment enhanced by Xiaoyu
- genome alignment track enhanced view, improved by Xiaoyu Zhuo
- allow users to upload local file as tracks
- allow users to upload entire folder or many files as a data hub by creating a
hub.config.jsonfile - HiC track normalization implemented by Silas Hsu
- first stable version since we start coding the new version of the browser code
- embedding browser function is available
- browser code also uploaded to npm
- (version number prior to 47 refers to the old browser codebase)