We need to first download SRA toolkit from NCBI's website:
http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=software
Select your appropriate binary distribution. For most modern Macs, MacOS 64 bit architecture would do the trick. The resulting file is ~ 51.5 mb
Double-click on sratoolkit.2.5.7-mac64.tar.gz to uncompress the file. Now you need to open a Terminal window (within Applications/Utilities/Terminal).
You should see a window like the following:
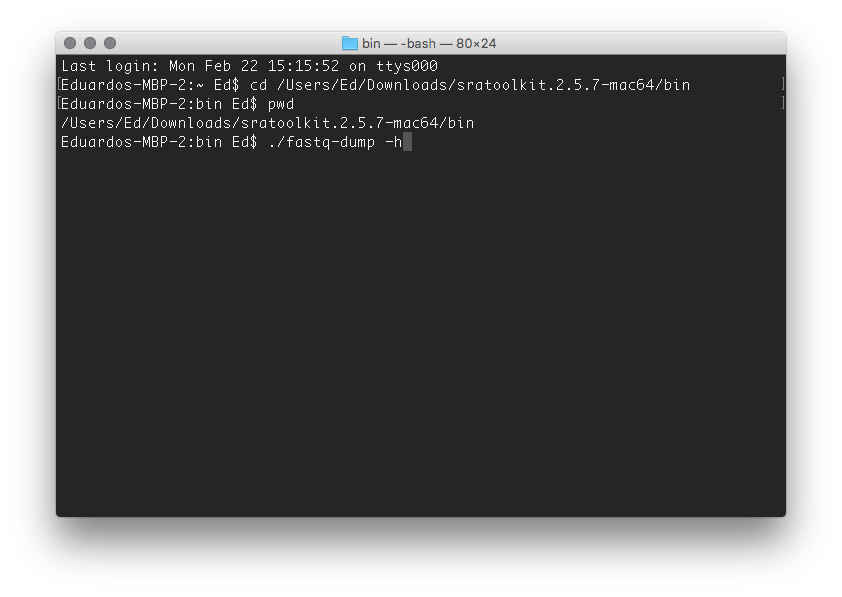
Now you need to "navigate" to the SRA toolkit folder by issueing the following command
cd /Users/Ed/Downloads/sratoolkit.2.5.7-mac64/bin
cd indicates that you want to change directory and the rest is simply the path to the destination directory. Make sure to replace Ed in the path above by your own home directory name. To confirm you are where you are suppossed to type pwd. The Terminal should return your current location, i.e., /Users/Ed/Downloads/sratoolkit.2.5.7-mac64/bin
Lastly, type ./fastq-dump -h to see the help menu.
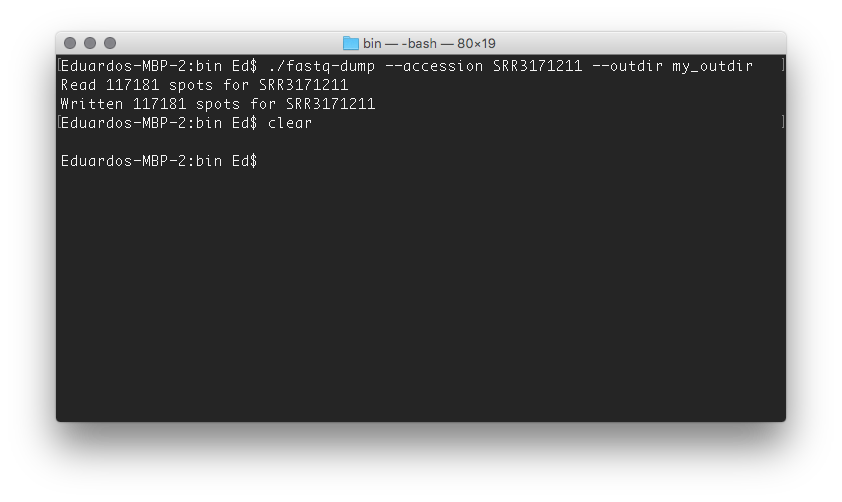
Next, we need to download actual data. We will use a somewhat small file hosted at SRA under the accession SRR3171211. Issue the following command in order to download the data:
./fastq-dump --accession SRR3171211 --outdir my_outdir
where SRR3171211 is the file we want and my_outdir is simply an arbitrary name for the output directory
and voilà! You should see a file named SRR3171211.fastq inside the output directory (takes about 10 minutes; 111.5 mb)