You signed in with another tab or window. Reload to refresh your session.You signed out in another tab or window. Reload to refresh your session.You switched accounts on another tab or window. Reload to refresh your session.Dismiss alert
{{ message }}
This repository was archived by the owner on Jul 10, 2024. It is now read-only.
In certain cases, explicit hydrogens seem to cause trouble for the atom-labelling layer of the hash. In these cases, it seems that the smiles generated by the standardizer produces a different hash than the input molfile itself.
Consider the following poorly layed-out structure:
[molfile below]
This example isn't so much about explicit H's but more about their parities. To fix this we need to be able to define a canonical set of parity flags for the specified stereocenters. This is far from a trivial fix.
It can't just be parity though, because this messes up the atom label layer, not just the stereo layer. If it only had a problem with stereo, that'd be less concerning.
In certain cases, explicit hydrogens seem to cause trouble for the atom-labelling layer of the hash. In these cases, it seems that the smiles generated by the standardizer produces a different hash than the input molfile itself.
Consider the following poorly layed-out structure:
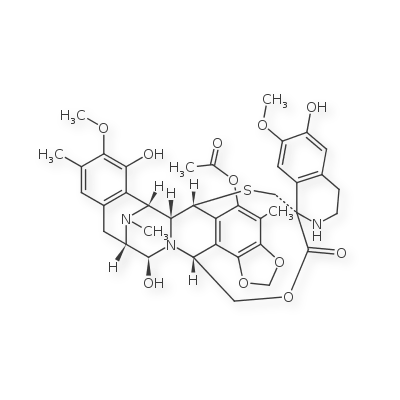
[molfile below]
Direct generation of hash from this Std_SMILES:
And this hash:
However, when that same smiles is fed into the standardizer, I get:
If the explicit hydrogens are removed entirely:
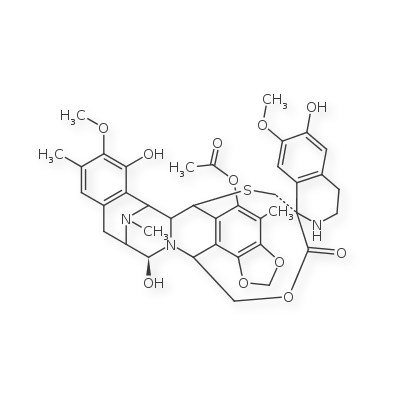
The output hash is now compatible with the smiles.
Molfile for explicit hydrogen version:
The text was updated successfully, but these errors were encountered: