diff --git a/access_controlled/CDS_access.md b/access_controlled/CDS_access.md
index 1294c736..7347bcf4 100644
--- a/access_controlled/CDS_access.md
+++ b/access_controlled/CDS_access.md
@@ -4,8 +4,44 @@ order: 997
# NCI's Cancer Data Service
-The [CDS Portal](https://dataservice.datacommons.cancer.gov/), within NCI's Cancer Research Data Commons (CRDC), provides access to both open and controlled access data.
+The [CDS Portal](https://dataservice.datacommons.cancer.gov/), within NCI's Cancer Research Data Commons (CRDC), provides an interface to filter and select data from a variety of NCI programs, including controlled-access, primary sequence data from the Human Tumor Atlas Network (HTAN).
-CDS currently hosts a variety of data types from NCI projects, including those from the [Human Tumor Atlas Network](https://dataservice.datacommons.cancer.gov/#/study/phs002371).
+In order to access these HTAN data within the [CDS Portal](https://dataservice.datacommons.cancer.gov/), navigate to the portal in a web browser and click on the **Explore CDS Data** button on the landing page.
+
+ +
+
+
+
+
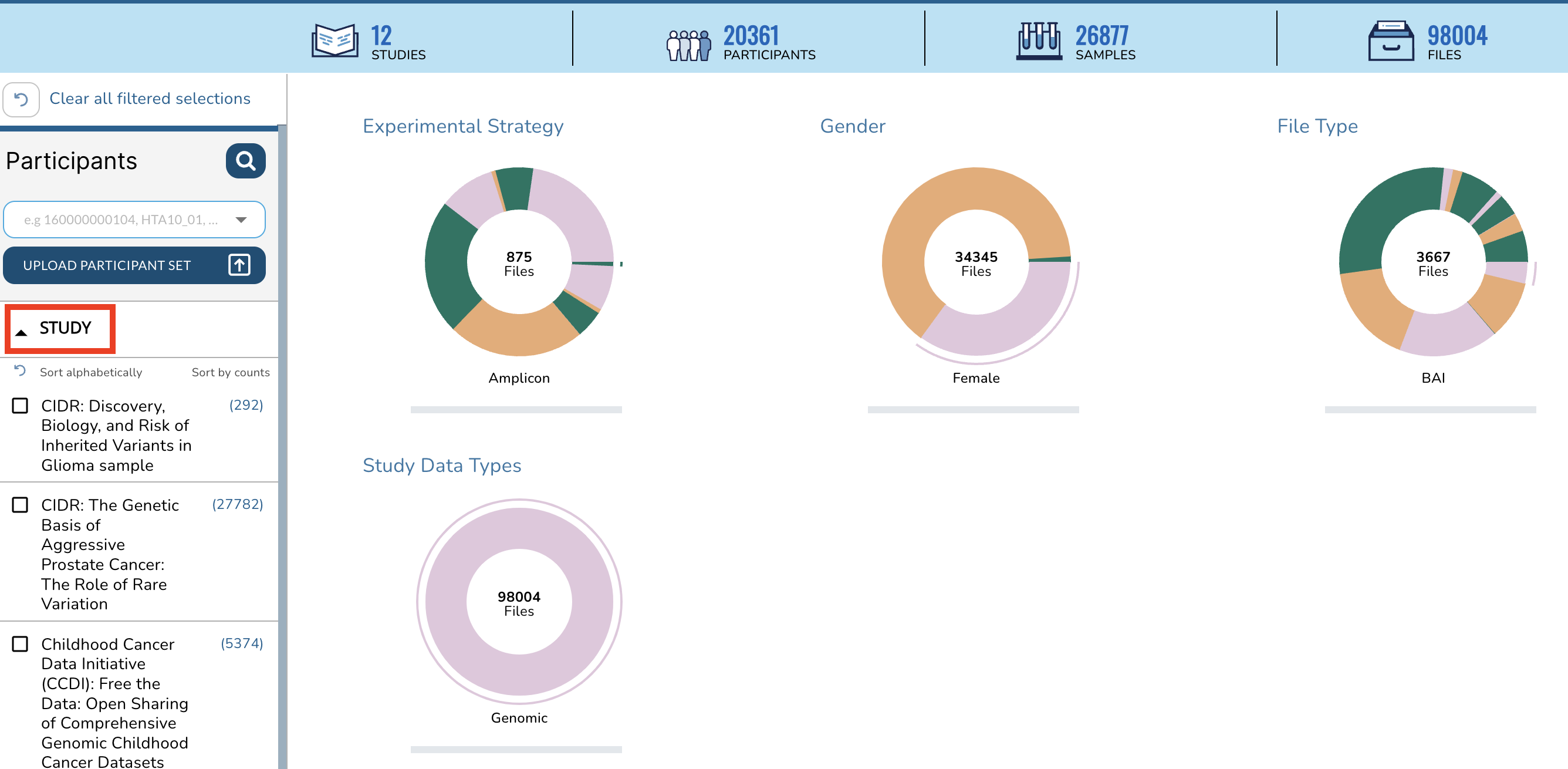
+On the Data Explorer page, expand the STUDY section on the left sidebar, scroll down, and check the box next to **Human Tumor Atlas (HTAN) primary sequence data**.
+
+
+
+
+
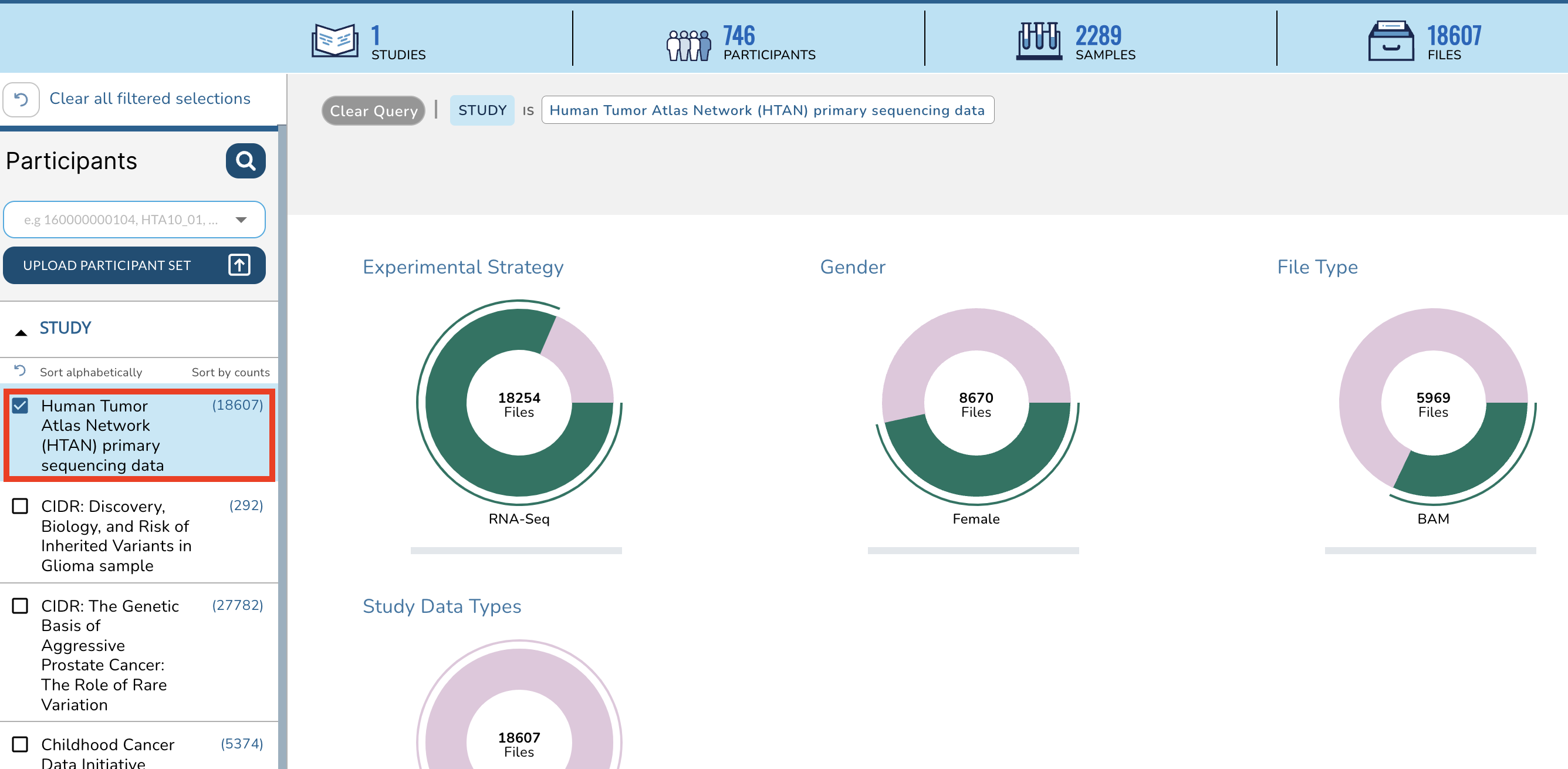
+This action will change the summary panel to reflect selecting HTAN data only.
+
+
+
+
+
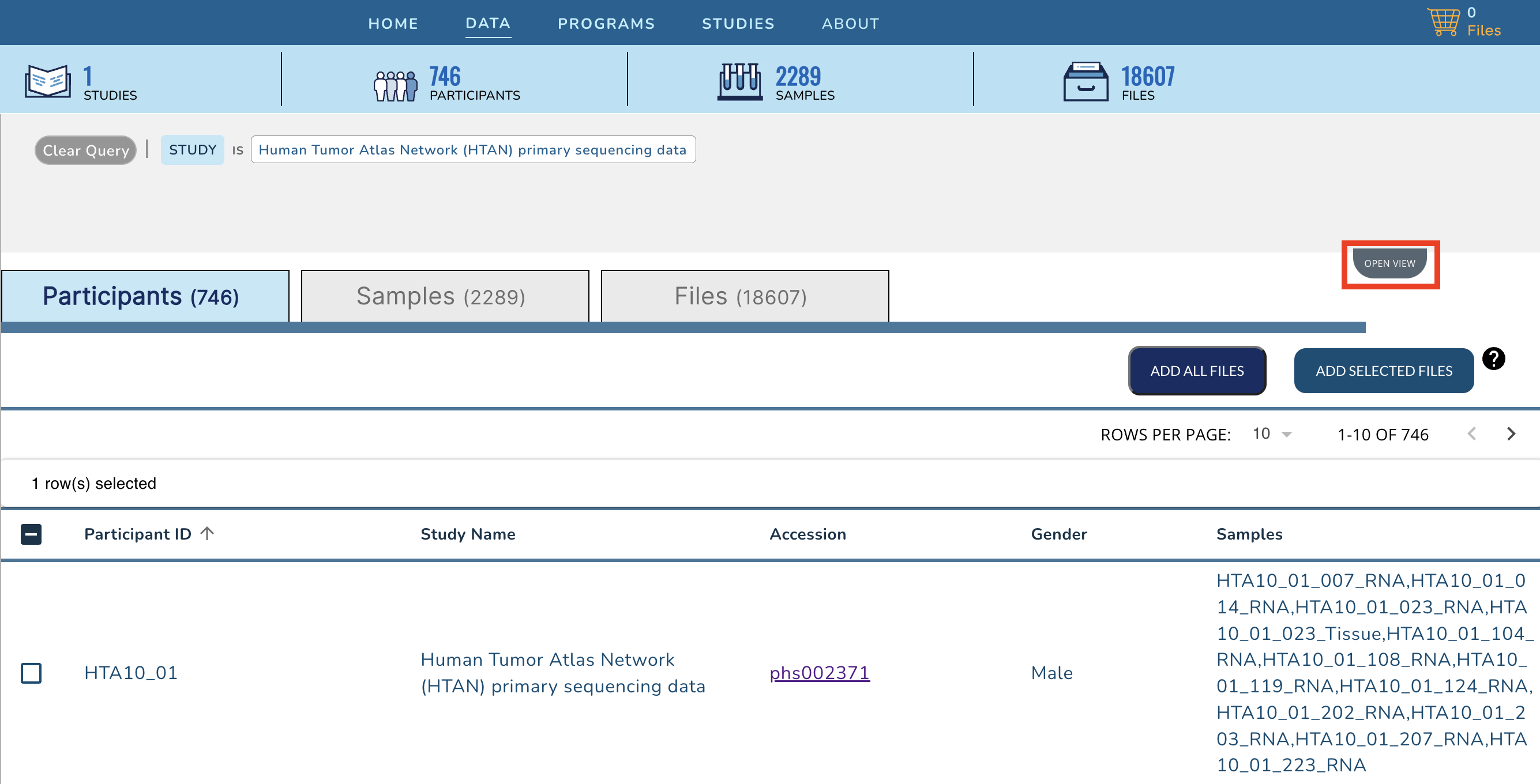
+Scroll down, or click on the **Collapse View** tab on the upper right just below the query summary line in order to see the tabulated view of all of the participants, samples or files in HTAN.
+
+
+
+
+
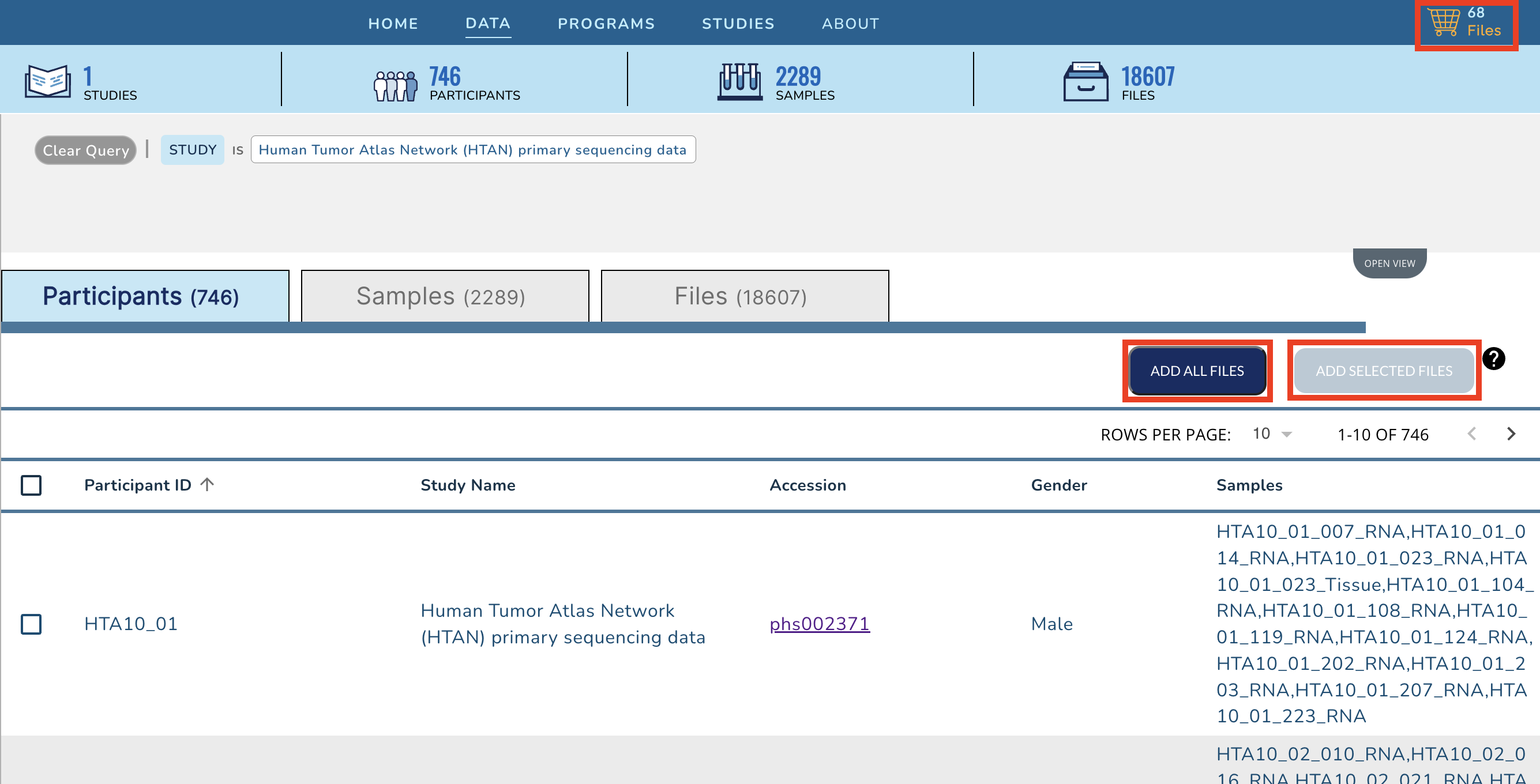
+Click on the **Add All Files** button, or select the check boxes next to all Participants, Samples or Files for a subselection and then click on the **Add Selected** button. This action will update your cart icon in the upper right corner.
+
+
+
+
+
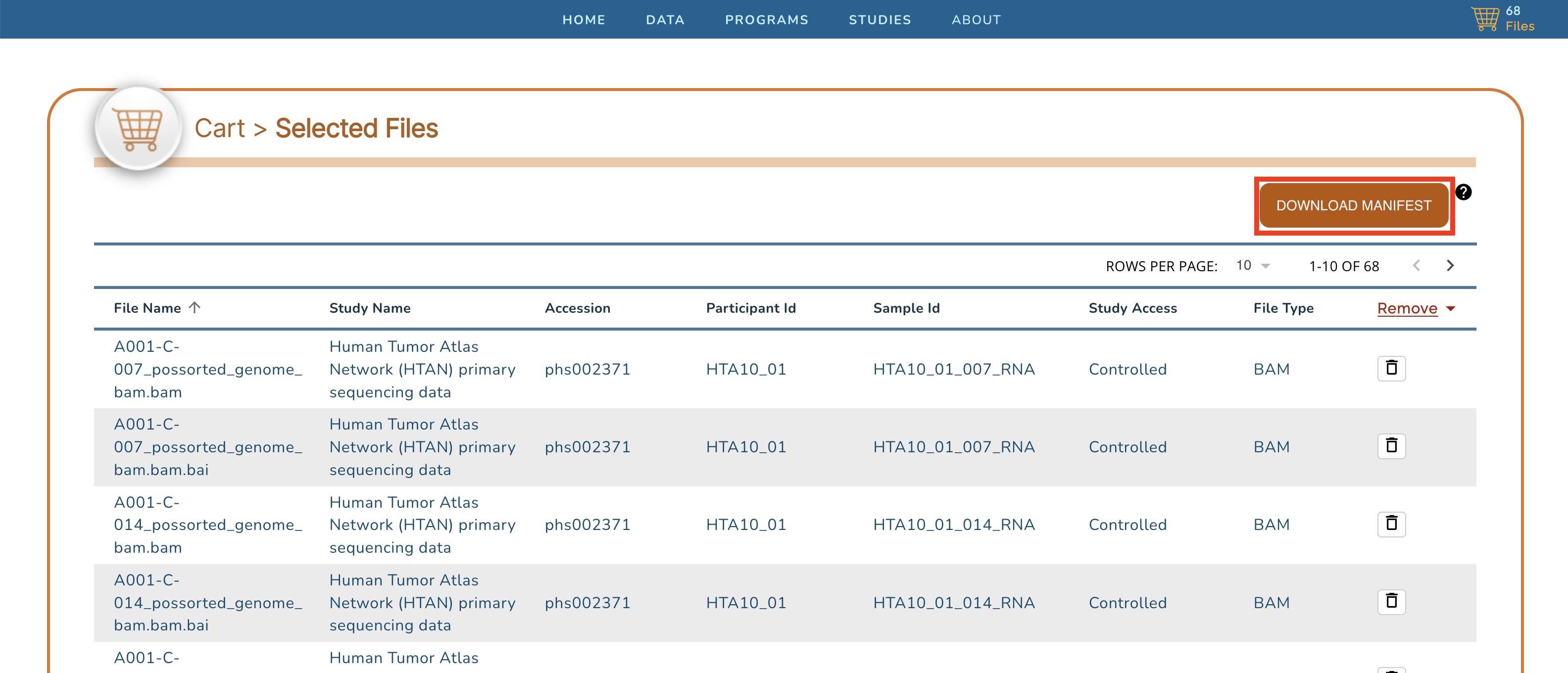
+Clicking on the cart icon, will bring up a list of the selected files. Click on the **Download Manifest** button in the upper right to download a CSV-formated (Excel compatible) file of this file list.
+
+
+
+
+
+Once this file manifest is downloaded, it will have to be uploaded into your [Seven Bridges Cancer Genomics Cloud](https://www.cancergenomicscloud.org/) account, in order for you to be able to download, or otherwise compute on, these data. **NOTE**: dbGaP approval for HTAN study [phs002371](https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002371.v3.p1) is required in order to access HTAN lower-level genomics data, such as RNAseq FASTQ and BAM files.
-dbGaP approval for HTAN study [phs002371](https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs002371.v3.p1) is required in order to access HTAN lower-level genomics data, such as RNAseq FASTQ and BAM files.
+