diff --git a/README.md b/README.md
index 23ca757..5a03be9 100644
--- a/README.md
+++ b/README.md
@@ -34,6 +34,8 @@ git clone https://github.com/arnodelorme/roiconnect.git
```
That's it! If you want to run the plugin, please start [EEGLAB](https://github.com/sccn/eeglab#to-use-eeglab) first. You may need to add EEGLAB to the [MATLAB path](https://de.mathworks.com/help/matlab/ref/addpath.html).
+📌 `test_pipes/` includes some test pipelines which can be used to get started.
+
# Key features
The features of the toolbox are implemented in the following three main functions: `pop_roi_activity`, `pop_roi_connect` and `pop_roi_connectplot`.
@@ -66,12 +68,12 @@ The function computes all FC metrics in a frequency-resolved way, i.e., the outp
You can visualize power and FC in different modes by calling `pop_roi_connectplot`. Below, we show results of a single subject from the real data example in [[1]](#1). You can find the MATLAB code and corresponding analyses [here](https://github.com/fpellegrini/MotorImag). The plots show power or FC in left motor imagery condition. Due to the nature of the task, we show results in the 8 to 13 Hz frequency band but you are free to choose any frequency or frequency band you want.
:pushpin: If any of the images are too small for you, simply click on them, they will open in full size in another tab.
-:round_pushpin: Plotting is particularly optimized for PSD, MIM/MIC and GC/TRGC. The matrix plots are only available for the Desikan-Killiany atlas (68 ROIs). We are currently working on a generalized solution for all atlases.
+:round_pushpin: Plotting is particularly optimized for PSD, MIM/MIC and GC/TRGC.
### Power as a region-wise bar plot
If you wish to visualize power as a barplot only, please make sure to explicitely turn `plotcortex` off because it is turned on by default.
```matlab
-EEG = pop_roi_connectplot(EEG, 'measure', 'roipsd', 'plotcortex', 'off', 'plotbarplot', on, 'freqrange', [8 13]) % alpha band;
+EEG = pop_roi_connectplot(EEG, 'measure', 'roipsd', 'plotcortex', 'off', 'plotbarplot', 'on', 'freqrange', [8 13]) % alpha band;
```
 @@ -93,7 +95,7 @@ Again, if you do not wish to see the cortex plot, you should explicitely turn `p
EEG = pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [8 13]);
```
@@ -93,7 +95,7 @@ Again, if you do not wish to see the cortex plot, you should explicitely turn `p
EEG = pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [8 13]);
```
-  +
+ 
@@ -103,17 +105,17 @@ pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'o
```
-  +
+ 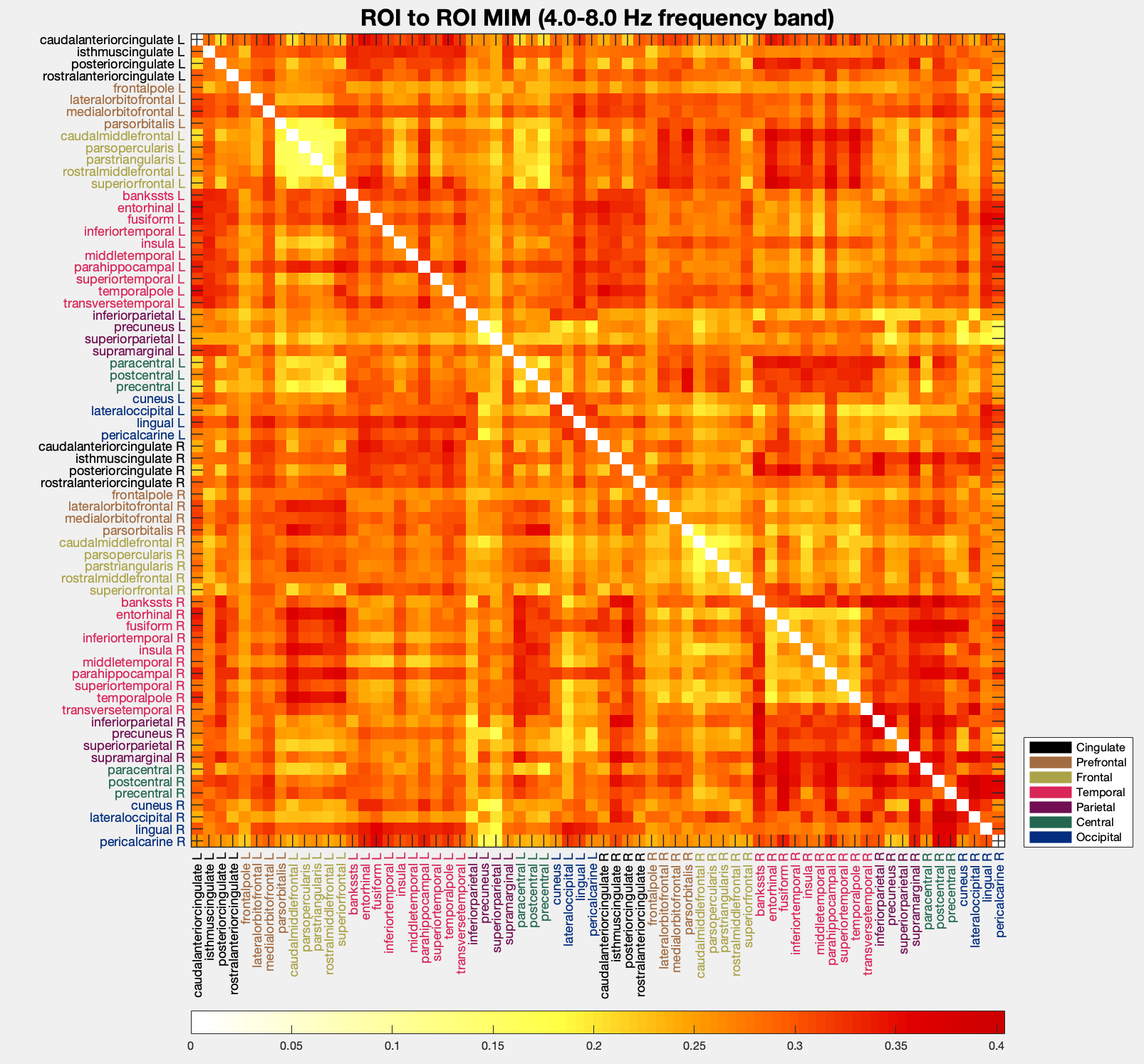
You can additionally filter by hemispheres and regions belonging to specific brain lobes. As an example, let us see how FC of the left hemisphere looks like.
```matlab
-pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [8 13], 'hemisphere', 'left') % left hemisphere, left motor imagery;
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [8 13], 'hemisphere', 'left');
```
-  +
+ 
diff --git a/pop_roi_connectplot.m b/pop_roi_connectplot.m
index 109b664..7bf7a91 100644
--- a/pop_roi_connectplot.m
+++ b/pop_roi_connectplot.m
@@ -416,13 +416,14 @@
% plot on matrix
if strcmpi(g.plotmatrix, 'on') && ~isempty(matrix)
- matrix = matrix.*seedMask;
- try
- roi_plotcoloredlobes(EEG, matrix, titleStr, g.measure, g.hemisphere, g.grouphemispheres, g.region);
- catch
- warning('Functionalities only available for the Desikan-Killiany atlas (68 ROIs).')
- figure; imagesc(matrix);
- end
+ matrix = matrix.*seedMask;
+ roi_plotcoloredlobes(EEG, matrix, titleStr, g.measure, g.hemisphere, g.grouphemispheres, g.region);
+% try
+% roi_plotcoloredlobes(EEG, matrix, titleStr, g.measure, g.hemisphere, g.grouphemispheres, g.region);
+% catch
+% warning('Functionalities only available for the Desikan-Killiany atlas (68 ROIs).')
+% figure; imagesc(matrix);
+% end
end
% plot on cortical surface
@@ -440,6 +441,8 @@
end
h = textsc(cortexTitle, 'title');
set(h, 'fontsize', 20);
+ elseif cortexFlag == -1
+ warning('EEG.roi.cortex does not contain the field "Faces" required to plot surface topographies.')
end
% plot 3D
@@ -477,15 +480,24 @@
end
end
-function [colors, color_idxx, roi_idxx, labels_dk_cell_idx, roi_loc] = get_colored_labels(EEG)
- % retrieve labels from atlas
+function labels = get_labels(EEG)
+% retrieve labels from atlas
labels = strings(1,length(EEG.roi.atlas.Scouts));
for i = 1:length(labels)
scout = struct2cell(EEG.roi.atlas.Scouts(i));
labels(i) = char(scout(1));
end
labels = cellstr(labels);
-
+end
+
+function new_labels = replace_underscores(labels)
+ % remove underscores in label names to avoid bug
+ new_labels = strrep(labels, '_', ' ');
+end
+
+function [colors, color_idxx, roi_idxx, labels_sorted, roi_loc] = get_colored_labels(EEG)
+ labels = get_labels(EEG);
+
colors = {[0,0,0]/255, [163, 107, 64]/255, [171, 163, 71]/255, [217, 37, 88]/255, [113, 15, 82]/255,[35, 103, 81]/255,[2, 45, 126]/255,};
% assign labels to colors
roi_loc ={'LT';'RT';'LL';'RL';'LF';'RF';'LO';'RO';'LT';'RT';'LPF';'RPF';'LT';'RT';'LP';'RP';'LT';'RT';'LT';'RT';'LL';'RL';'LO';'RO';'LPF';'RPF';'LO';'RO';'LPF';'RPF';'LT';'RT';'LC';'RC';'LT';'RT';'LF';'RF';'LPF';'RPF';'LF';'RF';'LO';'RO';'LC';'RC';'LL';'RL';'LC';'RC';'LP';'RP';'LL';'RL';'LF';'RF';'LF';'RF';'LP';'RP';'LT';'RT';'LP';'RP';'LT';'RT';'LT';'RT'};
@@ -502,21 +514,21 @@
roi_loc = strrep(roi_loc, 'R', '');
try
[color_idxx,roi_idxx] = sort(str2double(roi_loc));
- labels_dk_cell_idx = labels(roi_idxx);
+ labels_sorted = labels(roi_idxx);
catch
roi_idxx = 1:length(labels);
color_idxx = mod(roi_idxx, length(colors))+1;
- labels_dk_cell_idx = labels;
+ labels_sorted = labels;
end
end
function roi_plotpower(EEG, source_roi_power_norm_dB, titleStr)
- [colors, color_idxx, roi_idxx, labels_dk_cell_idx, ~] = get_colored_labels(EEG);
- n_roi_labels = size(labels_dk_cell_idx, 2);
+ [colors, color_idxx, roi_idxx, labels_sorted, ~] = get_colored_labels(EEG);
+ n_roi_labels = size(labels_sorted, 2);
barh(source_roi_power_norm_dB(roi_idxx));
set(gca, 'YDir', 'reverse');
- set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_dk_cell_idx(1:end), 'fontweight','bold','fontsize', 9, 'TickLength',[0.015, 0.02], 'LineWidth',0.7);
+ set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_sorted(1:end), 'fontweight','bold','fontsize', 9, 'TickLength',[0.015, 0.02], 'LineWidth',0.7);
h = title([ 'ROI source power' ' (' titleStr ')' ]);
set(h, 'fontsize', 16);
ylabel('power [dB]')
@@ -542,7 +554,7 @@ function roi_plotcoloredlobes( EEG, matrix, titleStr, measure, hemisphere, group
error('Region plotting is only supported for the Desikan-Killiany atlas.');
end
- % plot matrix with colored labels sorted by region according to the Desikan-Killiany atlas
+ % plot matrix with colored labels
load cm18
switch lower(measure)
case {'mim', 'mic', 'coh'}
@@ -550,35 +562,54 @@ function roi_plotcoloredlobes( EEG, matrix, titleStr, measure, hemisphere, group
otherwise
cmap = cm18;
end
- [colors, color_idxx, roi_idxx, labels_dk_cell_idx, roi_loc] = get_colored_labels(EEG);
-
clim_min = min(matrix, [], 'all');
clim_max = max(matrix, [], 'all');
-
- % assign region input to an index
- [GC, GR] = groupcounts(roi_loc);
- switch lower(region)
- case 'cingulate'
- region_idx = 1;
- case 'prefrontal'
- region_idx = 2;
- case 'frontal'
- region_idx = 3;
- case 'temporal'
- region_idx = 4;
- case 'parietal'
- region_idx = 5;
- case 'central'
- region_idx = 6;
- case 'occipital'
- region_idx = 7;
- otherwise
- region_idx = 99;
+
+ % hemisphere parameters to determine which labels to use
+ last_char = EEG.roi.atlas.Scouts(1).Label(end);
+ if strcmpi(hemisphere, 'left')
+ if strcmpi(last_char, 'R')
+ hem_idx = {2 2 2}; % use labels 2:2:end (first two values), only use 1/2 of the labels (3rd value)
+ else
+ hem_idx = {1 2 2}; % use labels 1:2:end (first two values), only use 1/2 of the labels (3rd value)
+ end
+ elseif strcmpi(hemisphere, 'right')
+ if strcmpi(last_char, 'L')
+ hem_idx = {1 2 2};
+ else
+ hem_idx = {2 2 2};
+ end
+ else
+ hem_idx = {1 1 1};
end
% sort matrix according to color scheme
% reduce matrix to only keep components corresponding to selected region
if isDKatlas == 1
+ [colors, color_idxx, roi_idxx, labels_sorted, roi_loc] = get_colored_labels(EEG);
+ labels = labels_sorted;
+
+ % assign region input to an index
+ [GC, ~] = groupcounts(roi_loc);
+ switch lower(region)
+ case 'cingulate'
+ region_idx = 1;
+ case 'prefrontal'
+ region_idx = 2;
+ case 'frontal'
+ region_idx = 3;
+ case 'temporal'
+ region_idx = 4;
+ case 'parietal'
+ region_idx = 5;
+ case 'central'
+ region_idx = 6;
+ case 'occipital'
+ region_idx = 7;
+ otherwise
+ region_idx = 99;
+ end
+
matrix = matrix(roi_idxx, roi_idxx);
if not(region_idx == 99)
if region_idx == 1
@@ -588,44 +619,40 @@ function roi_plotcoloredlobes( EEG, matrix, titleStr, measure, hemisphere, group
end
end_idx = start_idx + GC(region_idx) - 1;
matrix = matrix(start_idx:end_idx, start_idx:end_idx);
- labels_dk_cell_idx = labels_dk_cell_idx(start_idx:end_idx);
+ labels = labels(start_idx:end_idx);
color_idxx = color_idxx(start_idx:end_idx);
end
n_roi_labels = size(matrix, 1);
- end
-
- % hemisphere parameters to determine which labels to use
- if strcmpi(hemisphere, 'left')
- hem_idx = {1 2 2}; % use labels 1:2:end (first two values), only use 1/2 of the labels (3rd value)
- elseif strcmpi(hemisphere, 'right')
- hem_idx = {2 2 2}; % use labels 2:2:end (first two values), only use 1/2 of the labels (3rd value)
else
- hem_idx = {1 1 1}; % use labels 1:1:end (first two values, all labels), use 1/1 of the labels (3rd value, all labels)
+ labels = get_labels(EEG);
end
+ % remove underscores in labels to avoid plotting bug
+ labels = replace_underscores(labels);
+
% create dummy plot and add custom legend
f = figure();
%f.WindowState = 'maximized';
hold on
n_dummy_labels = 7;
x = 1:10;
- for k=1:n_dummy_labels
- plot(x, x*k, '-', 'LineWidth', 9, 'Color', colors{k});
- end
% labels on dummy plot for positioning
xlim([0 n_roi_labels])
ylim([0 n_roi_labels])
- set(gca,'xtick',[1:n_roi_labels],'xticklabel',labels_dk_cell_idx(hem_idx{1}:hem_idx{2}:n_roi_labels));%, 'TickLabelInterpreter','none');
+ set(gca,'xtick',1:n_roi_labels,'xticklabel',labels(hem_idx{1}:hem_idx{2}:n_roi_labels));%, 'TickLabelInterpreter','none');
ax = gca;
- for i=hem_idx{1}:hem_idx{2}:n_roi_labels
- ax.XTickLabel{ceil(i/hem_idx{3})} = sprintf('\\color[rgb]{%f,%f,%f}%s', colors{color_idxx(i)}, ax.XTickLabel{ceil(i/2)});
- end
- xtickangle(90)
- pos = get(gca, 'Position');
if isDKatlas == 1
+ for k=1:n_dummy_labels
+ plot(x, x*k, '-', 'LineWidth', 9, 'Color', colors{k});
+ end
+ for i=hem_idx{1}:hem_idx{2}:n_roi_labels
+ ax.XTickLabel{ceil(i/hem_idx{3})} = sprintf('\\color[rgb]{%f,%f,%f}%s', colors{color_idxx(i)}, ax.XTickLabel{ceil(i/2)});
+ end
legend('Cingulate', 'Prefrontal', 'Frontal', 'Temporal', 'Parietal', 'Central', 'Occipital', 'Location', 'southeastoutside'); % modify legend position
end
+ xtickangle(90)
+ pos = get(gca, 'Position');
set(gca, 'Position', pos, 'DataAspectRatio',[1 1 1], 'visible', 'off')
axes('pos', [pos(1) pos(2) pos(3) pos(4)]) % plot matrix over the dummy plot and keep the legend
@@ -640,14 +667,20 @@ function roi_plotcoloredlobes( EEG, matrix, titleStr, measure, hemisphere, group
matrix = horzcat(mat_left_col, mat_right_col); % sort columns
% sort labels
- lc = {labels_dk_cell_idx; transpose(color_idxx)};
- for i = 1:length(lc)
- left = lc{i}(1:2:end);
- right = lc{i}(2:2:end);
- lc{i} = [left right];
+ try % if color can be assigned
+ lc = {labels; transpose(color_idxx)};
+ for i = 1:length(lc)
+ left = lc{i}(1:2:end);
+ right = lc{i}(2:2:end);
+ lc{i} = [left right];
+ end
+ labels = lc{1};
+ color_idxx = transpose(lc{2});
+ catch
+ left = labels(1:2:end);
+ right = labels(2:2:end);
+ labels = [left right];
end
- labels_dk_cell_idx = lc{1};
- color_idxx = transpose(lc{2});
end
% reduce matrix to keep only one hemisphere
@@ -677,16 +710,16 @@ function roi_plotcoloredlobes( EEG, matrix, titleStr, measure, hemisphere, group
set(gca, 'Position', pos, 'DataAspectRatio',[1 1 1], 'visible', 'on')
% add colored labels with display option
+ ax = gca;
+ set(gca,'xtick',1:n_roi_labels,'xticklabel',labels(hem_idx{1}:hem_idx{2}:n_roi_labels));
if isDKatlas == 1
- set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_dk_cell_idx(hem_idx{1}:hem_idx{2}:n_roi_labels), 'fontweight','bold', 'fontsize', 9, 'TickLength',[0.015, 0.02], 'LineWidth',0.75);
+ set(gca,'ytick',1:n_roi_labels,'yticklabel',labels(hem_idx{1}:hem_idx{2}:n_roi_labels), 'fontsize', 9, 'TickLength',[0.015, 0.02], 'LineWidth',0.75);
+ for i=hem_idx{1}:hem_idx{2}:n_roi_labels
+ ax.XTickLabel{ceil(i/hem_idx{3})} = sprintf('\\color[rgb]{%f,%f,%f}%s', colors{color_idxx(i)}, ax.XTickLabel{ceil(i/hem_idx{3})});
+ ax.YTickLabel{ceil(i/hem_idx{3})} = sprintf('\\color[rgb]{%f,%f,%f}%s', colors{color_idxx(i)}, ax.YTickLabel{ceil(i/hem_idx{3})});
+ end
else
- set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_dk_cell_idx(hem_idx{1}:hem_idx{2}:n_roi_labels), 'fontsize', 7, 'TickLength',[0.015, 0.02], 'LineWidth',0.75);
- end
- set(gca,'xtick',[1:n_roi_labels],'xticklabel',labels_dk_cell_idx(hem_idx{1}:hem_idx{2}:n_roi_labels));
- ax = gca;
- for i=hem_idx{1}:hem_idx{2}:n_roi_labels
- ax.XTickLabel{ceil(i/hem_idx{3})} = sprintf('\\color[rgb]{%f,%f,%f}%s', colors{color_idxx(i)}, ax.XTickLabel{ceil(i/hem_idx{3})});
- ax.YTickLabel{ceil(i/hem_idx{3})} = sprintf('\\color[rgb]{%f,%f,%f}%s', colors{color_idxx(i)}, ax.YTickLabel{ceil(i/hem_idx{3})});
+ set(gca,'ytick',1:n_roi_labels,'yticklabel',labels(hem_idx{1}:hem_idx{2}:n_roi_labels), 'fontsize', 7, 'TickLength',[0.015, 0.02], 'LineWidth',0.75);
end
h = title([ 'ROI to ROI ' upper(measure) ' (' titleStr ')' ]);
set(h, 'fontsize', 16);
@@ -696,8 +729,8 @@ function roi_plotcoloredlobes( EEG, matrix, titleStr, measure, hemisphere, group
function roi_largeplot(EEG, mim, trgc, roipsd, titleStr)
% plot MIM, TRGC and power (barplot) in a single large figure
load cm18
- [colors, color_idxx, roi_idxx, labels_dk_cell_idx, ~] = get_colored_labels(EEG);
- n_roi_labels = size(labels_dk_cell_idx, 2);
+ [colors, color_idxx, roi_idxx, labels_sorted, ~] = get_colored_labels(EEG);
+ n_roi_labels = size(labels_sorted, 2);
f = figure();
f.WindowState = 'maximized';
@@ -713,8 +746,8 @@ function roi_largeplot(EEG, mim, trgc, roipsd, titleStr)
img_sorted = img(roi_idxx, roi_idxx);
imagesc(img_sorted)
- set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_dk_cell_idx(1:end), 'fontsize', 5, 'TickLength',[0.015, 0.02], 'LineWidth',0.75);
- set(gca,'xtick',[1:n_roi_labels],'xticklabel',labels_dk_cell_idx(1:end));
+ set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_sorted(1:end), 'fontsize', 5, 'TickLength',[0.015, 0.02], 'LineWidth',0.75);
+ set(gca,'xtick',[1:n_roi_labels],'xticklabel',labels_sorted(1:end));
h = title([ 'ROI to ROI ' fc_names{k} ' (' titleStr ')' ]);
set(h, 'fontsize', 16);
hcb = colorbar;
@@ -735,7 +768,7 @@ function roi_largeplot(EEG, mim, trgc, roipsd, titleStr)
barh(roipsd(roi_idxx));
set(gca, 'YDir', 'reverse');
- set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_dk_cell_idx(1:end), 'fontweight','bold','fontsize', 9, 'TickLength',[0.015, 0.02], 'LineWidth',0.7);
+ set(gca,'ytick',[1:n_roi_labels],'yticklabel',labels_sorted(1:end), 'fontsize', 9, 'TickLength',[0.015, 0.02], 'LineWidth',0.7);
h = title([ 'ROI source power' ' (' titleStr ')' ]);
set(h, 'fontsize', 16);
ylabel('power [dB]')
diff --git a/resources/FC_MIM_matrix.png b/resources/FC_MIM_matrix.png
new file mode 100644
index 0000000..9c4495c
Binary files /dev/null and b/resources/FC_MIM_matrix.png differ
diff --git a/resources/FC_MIM_matrix_groupedhems.png b/resources/FC_MIM_matrix_groupedhems.png
new file mode 100644
index 0000000..02b233c
Binary files /dev/null and b/resources/FC_MIM_matrix_groupedhems.png differ
diff --git a/resources/FC_MIM_matrix_left.png b/resources/FC_MIM_matrix_left.png
new file mode 100644
index 0000000..4ea9399
Binary files /dev/null and b/resources/FC_MIM_matrix_left.png differ
diff --git a/pipeline_connectivity.m b/test_pipes/pipeline_connectivity.m
similarity index 96%
rename from pipeline_connectivity.m
rename to test_pipes/pipeline_connectivity.m
index 2fe5d0e..a4b5159 100644
--- a/pipeline_connectivity.m
+++ b/test_pipes/pipeline_connectivity.m
@@ -46,6 +46,4 @@
tic
EEG = pop_roi_connect(EEG, 'methods', measures(iMeasure));
t(iMeasure) = toc;
-end
-
-pop_roi_connectplot(EEG, 'measure', 'MIM', 'plotmatrix', 'on', 'plotcortex', 'on');
\ No newline at end of file
+end
\ No newline at end of file
diff --git a/test_pipes/test_brainplots_dkatlas.m b/test_pipes/test_brainplots_dkatlas.m
new file mode 100644
index 0000000..247ddc3
--- /dev/null
+++ b/test_pipes/test_brainplots_dkatlas.m
@@ -0,0 +1,33 @@
+% Test cortical surface topographies with different parameters. Here, the Desikan-Killiany atlas with 68 ROIs is used as the source model.
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 1.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'functions','supportfiles','head_modelColin27_5003_Standard-10-5-Cap339.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','LORETA-Talairach-BAs','nPCA',3);
+EEG = pop_roi_connect(EEG, 'methods', {'MIM'});
+
+%% Plot brain plot with different parameters
+% brain plot without any filters
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'on');
+
+% frequency band specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'on', 'freqrange', [8 13]);
+
+% seed voxel specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'on', 'plotcortexseedregion', 49);
\ No newline at end of file
diff --git a/test_pipes/test_brainplots_loretaatlas.m b/test_pipes/test_brainplots_loretaatlas.m
new file mode 100644
index 0000000..7c9a490
--- /dev/null
+++ b/test_pipes/test_brainplots_loretaatlas.m
@@ -0,0 +1,33 @@
+% Test cortical surface topographies with different parameters. Here, the LORETA-Talairach-BAs atlas with 90 ROIs is used as the source model.
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 1.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'plugins','dipfit','LORETA-Talairach-BAs.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','LORETA-Talairach-BAs','nPCA',3);
+EEG = pop_roi_connect(EEG, 'methods', {'MIM'});
+
+%% Plot brain plot with different parameters
+% brain plot without any filters
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'on');
+
+% frequency band specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'on', 'freqrange', [8 13]);
+
+% seed voxel specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'on', 'plotcortexseedregion', 49);
\ No newline at end of file
diff --git a/test_pipes/test_flex_epochlength.m b/test_pipes/test_flex_epochlength.m
new file mode 100644
index 0000000..ca06458
--- /dev/null
+++ b/test_pipes/test_flex_epochlength.m
@@ -0,0 +1,31 @@
+% Test pipeline with an epoch length other than 2 seconds (test with 1 second epoch length).
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 0.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'functions','supportfiles','head_modelColin27_5003_Standard-10-5-Cap339.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','LORETA-Talairach-BAs','nPCA',1);
+
+measures = { 'MIM' 'wPLI' };
+for iMeasure = 1:length(measures)
+ tic
+ EEG = pop_roi_connect(EEG, 'methods', measures(iMeasure));
+ t(iMeasure) = toc;
+end
+
+
diff --git a/test_pipes/test_largebarplot_dkatlas.m b/test_pipes/test_largebarplot_dkatlas.m
new file mode 100644
index 0000000..a9b177f
--- /dev/null
+++ b/test_pipes/test_largebarplot_dkatlas.m
@@ -0,0 +1,27 @@
+% Test region-to-region power bar plot. Here, the Desikan-Killiany atlas with 68 ROIs is used as the source model.
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 1.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'functions','supportfiles','head_modelColin27_5003_Standard-10-5-Cap339.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','LORETA-Talairach-BAs','nPCA',3);
+EEG = pop_roi_connect(EEG, 'methods', {'MIM', 'TRGC'});
+
+%% Plot barplot
+% plot barplot with specified frequency band
+EEG = pop_roi_connectplot(EEG, 'measure', 'roipsd', 'plotcortex', 'off', 'plotbarplot', 'on', 'freqrange', [4 8]);
\ No newline at end of file
diff --git a/test_pipes/test_matrixplots_brainregions.m b/test_pipes/test_matrixplots_brainregions.m
new file mode 100644
index 0000000..39b89a2
--- /dev/null
+++ b/test_pipes/test_matrixplots_brainregions.m
@@ -0,0 +1,45 @@
+% Test region-to-region FC matrices (MIM) with different parameters. Here, the grouped LORETA-Talairach-BAs ('Brain-Regions') atlas with 34 ROIs is used as the source model.
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 1.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'plugins','dipfit','LORETA-Talairach-BAs.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','Brain-Regions','nPCA',3);
+EEG = pop_roi_connect(EEG, 'methods', {'MIM'});
+
+%% Plot matrix with different parameters
+% matrix without any filters
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on');
+
+% group by hemispheres
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'grouphemispheres', 'on');
+
+% frequency band specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [4 8]');
+
+% hemisphere specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'hemisphere', 'right');
+
+% cortical region specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'region', 'occipital');
+
+
+
+
+
+
diff --git a/test_pipes/test_matrixplots_dkatlas.m b/test_pipes/test_matrixplots_dkatlas.m
new file mode 100644
index 0000000..c9ae221
--- /dev/null
+++ b/test_pipes/test_matrixplots_dkatlas.m
@@ -0,0 +1,45 @@
+% Test region-to-region FC matrices (MIM) with different parameters. Here, the Desikan-Killiany atlas with 68 ROIs is used as the source model.
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 1.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'functions','supportfiles','head_modelColin27_5003_Standard-10-5-Cap339.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','LORETA-Talairach-BAs','nPCA',3);
+EEG = pop_roi_connect(EEG, 'methods', {'MIM'});
+
+%% Plot matrix with different parameters
+% matrix without any filters
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on');
+
+% group by hemispheres
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'grouphemispheres', 'on');
+
+% frequency band specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [4 8]');
+
+% hemisphere specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'hemisphere', 'right');
+
+% cortical region specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'region', 'occipital');
+
+
+
+
+
+
diff --git a/test_pipes/test_matrixplots_loretaatlas.m b/test_pipes/test_matrixplots_loretaatlas.m
new file mode 100644
index 0000000..9ab5d24
--- /dev/null
+++ b/test_pipes/test_matrixplots_loretaatlas.m
@@ -0,0 +1,45 @@
+% Test region-to-region FC matrices (MIM) with different parameters. Here, the LORETA-Talairach-BAs atlas with 90 ROIs is used as the source model.
+%% Run pipeline
+clear
+eeglab
+
+eeglabp = fileparts(which('eeglab.m'));
+EEG = pop_loadset('filename','eeglab_data_epochs_ica.set','filepath',fullfile(eeglabp, 'sample_data/'));
+EEG = pop_resample( EEG, 100);
+EEG = pop_epoch( EEG, { }, [-0.5 1.5], 'newname', 'EEG Data epochs epochs', 'epochinfo', 'yes');
+EEG = pop_select( EEG, 'trial',1:30);
+[ALLEEG, EEG, CURRENTSET] = eeg_store(ALLEEG, EEG);
+eeglab redraw;
+
+EEG = pop_dipfit_settings( EEG, 'hdmfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_vol.mat'), ...
+ 'coordformat','MNI','mrifile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','standard_mri.mat'),...
+ 'chanfile',fullfile(eeglabp, 'plugins','dipfit','standard_BEM','elec', 'standard_1005.elc'),...
+ 'coord_transform',[0.83215 -15.6287 2.4114 0.081214 0.00093739 -1.5732 1.1742 1.0601 1.1485] ,'chansel',[1:32] );
+
+EEG = pop_leadfield(EEG, 'sourcemodel',fullfile(eeglabp,'plugins','dipfit','LORETA-Talairach-BAs.mat'), ...
+ 'sourcemodel2mni',[0 -24 -45 0 0 -1.5708 1000 1000 1000] ,'downsample',1);
+
+EEG = pop_roi_activity(EEG, 'leadfield',EEG.dipfit.sourcemodel,'model','LCMV','modelparams',{0.05},'atlas','LORETA-Talairach-BAs','nPCA',3);
+EEG = pop_roi_connect(EEG, 'methods', {'MIM'});
+
+%% Plot matrix with different parameters
+% matrix without any filters
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on');
+
+% group by hemispheres
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'grouphemispheres', 'on');
+
+% frequency band specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [4 8]');
+
+% hemisphere specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'hemisphere', 'right');
+
+% cortical region specified
+pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'region', 'occipital');
+
+
+
+
+
+
![]() @@ -93,7 +95,7 @@ Again, if you do not wish to see the cortex plot, you should explicitely turn `p
EEG = pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [8 13]);
```
@@ -93,7 +95,7 @@ Again, if you do not wish to see the cortex plot, you should explicitely turn `p
EEG = pop_roi_connectplot(EEG, 'measure', 'mim', 'plotcortex', 'off', 'plotmatrix', 'on', 'freqrange', [8 13]);
```
![]() +
+ ![]()
![]() +
+ ![]()
![]() +
+ ![]()