diff --git a/latest/index.bs b/0.5/index.bs
similarity index 79%
rename from latest/index.bs
rename to 0.5/index.bs
index 26e27b3c..3f326ffa 100644
--- a/latest/index.bs
+++ b/0.5/index.bs
@@ -1,42 +1,33 @@
-Title: Next-generation file formats (NGFF)
-Shortname: ome-ngff
+Title: OME-Zarr specification
+Shortname: ome-zarr
Level: 1
-Status: LS-COMMIT
-Status: w3c/ED
-Group: ome
-URL: https://ngff.openmicroscopy.org/latest/
+Status: w3c/CG-FINAL
+TR: https://ngff.openmicroscopy.org/0.5/
+URL: https://ngff.openmicroscopy.org/0.5/
Repository: https://github.com/ome/ngff
Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff
Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg
-Local Boilerplate: header yes
-Local Boilerplate: copyright yes
+Local Boilerplate: header yes, copyright yes
Boilerplate: style-darkmode off
Markup Shorthands: markdown yes
-Editor: Josh Moore, University of Dundee (UoD) https://www.dundee.ac.uk, https://orcid.org/0000-0003-4028-811X
-Text Macro: NGFFVERSION 0.5-dev
+Editor: Josh Moore, German BioImaging e.V., https://gerbi-gmb.de/, https://orcid.org/0000-0003-4028-811X
+Text Macro: NGFFVERSION 0.5
Abstract: This document contains next-generation file format (NGFF)
Abstract: specifications for storing bioimaging data in the cloud.
Abstract: All specifications are submitted to the https://image.sc community for review.
-Status Text: The current released version of this specification is
-Status Text: 0.4. Migration scripts
+Status Text: The current released version of this specification is 0.5. Migration scripts
Status Text: will be provided between numbered versions. Data written with these latest changes
Status Text: (an "editor's draft") will not necessarily be supported.
-OME-NGFF {#ome-ngff}
---------------------
+OME-Zarr specification {#ome-zarr}
+----------------------------------
The conventions and specifications defined in this document are designed to
enable next-generation file formats to represent the same bioimaging data
that can be represented in \[OME-TIFF](http://www.openmicroscopy.org/ome-files/)
-and beyond. However, the conventions will also be usable by HDF5 and other sufficiently advanced
-binary containers. Eventually, we hope, the moniker "next-generation" will no longer be
-applicable, and this will simply be the most efficient, common, and useful representation
-of bioimaging data, whether during acquisition or sharing in the cloud.
-
-Note: The following text makes use of OME-Zarr [[ome-zarr-py]], the current prototype implementation,
-for all examples.
+and beyond.
Document conventions
--------------------
@@ -56,8 +47,14 @@ implementations that are later submitted as a formal specification. (See [[#bf2r
Some of the JSON examples in this document include comments. However, these are only for
clarity purposes and comments MUST NOT be included in JSON objects.
-On-disk (or in-cloud) layout {#on-disk}
-=======================================
+Storage format {#storage-format}
+================================
+
+OME-Zarr is implemented using the Zarr format as defined by the
+[version 3 of the Zarr specification](https://zarr-specs.readthedocs.io/en/latest/v3/core/v3.0.html).
+All features of the Zarr format including codecs, chunk grids, chunk
+key encodings, data types and storage transformers may be used with
+OME-Zarr unless explicitly disallowed in this specification.
An overview of the layout of an OME-Zarr fileset should make
understanding the following metadata sections easier. The hierarchy
@@ -65,66 +62,51 @@ is represented here as it would appear locally but could equally
be stored on a web server to be accessed via HTTP or in object storage
like S3 or GCS.
-OME-Zarr is an implementation of the OME-NGFF specification using the Zarr
-format. Arrays MUST be defined and stored in a hierarchical organization as
-defined by the
-[version 2 of the Zarr specification ](https://zarr.readthedocs.io/en/stable/spec/v2.html).
-OME-NGFF metadata MUST be stored as attributes in the corresponding Zarr
-groups.
-
Images {#image-layout}
----------------------
The following layout describes the expected Zarr hierarchy for images with
multiple levels of resolutions and optionally associated labels.
Note that the number of dimensions is variable between 2 and 5 and that axis names are arbitrary, see [[#multiscale-md]] for details.
-For this example we assume an image with 5 dimensions and axes called `t,c,z,y,x`.
-. # Root folder, potentially in S3,
-│ # with a flat list of images by image ID.
-│
-├── 123.zarr # One image (id=123) converted to Zarr.
+├── 123.zarr # One OME-Zarr image (id=123).
+│ ...
│
-└── 456.zarr # Another image (id=456) converted to Zarr.
+└── 456.zarr # Another OME-Zarr image (id=456).
│
- ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays.
- ├── .zattrs # Group level attributes are stored in the .zattrs file and include
- │ # "multiscales" and "omero" (see below). In addition, the group level attributes
- │ # may also contain "_ARRAY_DIMENSIONS" for compatibility with xarray if this group directly contains multi-scale arrays.
+ ├── zarr.json # Each image is a Zarr group of other groups and arrays.
+ │ # Group level attributes are stored in the `zarr.json` file and include
+ │ # "multiscales" and "omero" (see below).
│
├── 0 # Each multiscale level is stored as a separate Zarr array,
│ ... # which is a folder containing chunk files which compose the array.
├── n # The name of the array is arbitrary with the ordering defined by
│ │ # by the "multiscales" metadata, but is often a sequence starting at 0.
│ │
- │ ├── .zarray # All image arrays must be up to 5-dimensional
+ │ ├── zarr.json # All image arrays must be up to 5-dimensional
│ │ # with the axis of type time before type channel, before spatial axes.
│ │
- │ └─ t # Chunks are stored with the nested directory layout.
- │ └─ c # All but the last chunk element are stored as directories.
- │ └─ z # The terminal chunk is a file. Together the directory and file names
- │ └─ y # provide the "chunk coordinate" (t, c, z, y, x), where the maximum coordinate
- │ └─ x # will be `dimension_size / chunk_size`.
+ │ └─ ... # Chunks are stored conforming to the Zarr array specification and
+ │ # metadata as specified in the array's `zarr.json`.
│
└── labels
│
- ├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable
- │
- ├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }`
- │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the
+ ├── zarr.json # The labels group is a container which holds a list of labels to make the objects easily discoverable
+ │ # All labels will be listed in `zarr.json` e.g. `{ "labels": [ "original/0" ] }`
+ │ # Each dimension of the label should be either the same as the
│ # corresponding dimension of the image, or `1` if that dimension of the label
│ # is irrelevant.
│
└── original # Intermediate folders are permitted but not necessary and currently contain no extra metadata.
│
└── 0 # Multiscale, labeled image. The name is unimportant but is registered in the "labels" group above.
- ├── .zgroup # Zarr Group which is both a multiscaled image as well as a labeled image.
- ├── .zattrs # Metadata of the related image and as well as display information under the "image-label" key.
+ ├── zarr.json # Zarr Group which is both a multiscaled image as well as a labeled image.
+ │ # Metadata of the related image and as well as display information under the "image-label" key.
│
├── 0 # Each multiscale level is stored as a separate Zarr array, as above, but only integer values
- │ ... # are supported.
- └── n
+ └── ... # are supported.
+
@@ -148,51 +130,66 @@ A well group SHOULD NOT be present if there are no images in the well.
-. # Root folder, potentially in S3,
+.
│
-└── 5966.zarr # One plate (id=5966) converted to Zarr
- ├── .zgroup
- ├── .zattrs # Implements "plate" specification
+└── 5966.zarr # One OME-Zarr plate (id=5966)
+ ├── zarr.json # Implements "plate" specification
├── A # First row of the plate
- │ ├── .zgroup
+ │ ├── zarr.json
│ │
│ ├── 1 # First column of row A
- │ │ ├── .zgroup
- │ │ ├── .zattrs # Implements "well" specification
+ │ │ ├── zarr.json # Implements "well" specification
│ │ │
│ │ ├── 0 # First field of view of well A1
│ │ │ │
- │ │ │ ├── .zgroup
- │ │ │ ├── .zattrs # Implements "multiscales", "omero"
- │ │ │ ├── 0
- │ │ │ │ ... # Resolution levels
- │ │ │ ├── n
+ │ │ │ ├── zarr.json # Implements "multiscales", "omero"
+ │ │ │ ├── 0 # Resolution levels
+ │ │ │ ├── ...
│ │ │ └── labels # Labels (optional)
- │ │ ├── ... # Fields of view
- │ │ └── m
- │ ├── ... # Columns
- │ └── 12
- ├── ... # Rows
- └── H
+ │ │ └── ... # Other fields of view
+ │ └── ... # Other columns
+ └── ... # Other rows
-Metadata {#metadata}
-====================
+OME-Zarr Metadata {#metadata}
+=============================
+
+The "OME-Zarr Metadata" contains metadata keys as specified below
+for discovering certain types of data, especially images.
-The various `.zattrs` files throughout the above array hierarchy may contain metadata
-keys as specified below for discovering certain types of data, especially images.
+The OME-Zarr Metadata is stored in the various `zarr.json` files throughout the above array
+hierarchy. In this file, the metadata is stored under the namespaced key
+`ome` in `attributes`.
+The version of the OME-Zarr Metadata is denoted as a string in the `version` attribute within the `ome` namespace.
+
+The OME-Zarr Metadata version MUST be consistent within a hierarchy.
+
+```json
+{
+ ...
+ "attributes": {
+ "ome": {
+ "version": "0.5",
+ ...
+ }
+ }
+}
+```
"axes" metadata {#axes-md}
--------------------------
"axes" describes the dimensions of a physical coordinate space. It is a list of dictionaries, where each dictionary describes a dimension (axis) and:
- MUST contain the field "name" that gives the name for this dimension. The values MUST be unique across all "name" fields.
-- SHOULD contain the field "type". It SHOULD be one of "space", "time" or "channel", but MAY take other values for custom axis types that are not part of this specification yet.
+- SHOULD contain the field "type". It SHOULD be one of "space", "time" or "channel", but MAY take other string values for custom axis types that are not part of this specification yet.
- SHOULD contain the field "unit" to specify the physical unit of this dimension. The value SHOULD be one of the following strings, which are valid units according to UDUNITS-2.
- Units for "space" axes: 'angstrom', 'attometer', 'centimeter', 'decimeter', 'exameter', 'femtometer', 'foot', 'gigameter', 'hectometer', 'inch', 'kilometer', 'megameter', 'meter', 'micrometer', 'mile', 'millimeter', 'nanometer', 'parsec', 'petameter', 'picometer', 'terameter', 'yard', 'yoctometer', 'yottameter', 'zeptometer', 'zettameter'
- Units for "time" axes: 'attosecond', 'centisecond', 'day', 'decisecond', 'exasecond', 'femtosecond', 'gigasecond', 'hectosecond', 'hour', 'kilosecond', 'megasecond', 'microsecond', 'millisecond', 'minute', 'nanosecond', 'petasecond', 'picosecond', 'second', 'terasecond', 'yoctosecond', 'yottasecond', 'zeptosecond', 'zettasecond'
-If part of [[#multiscale-md]], the length of "axes" MUST be equal to the number of dimensions of the arrays that contain the image data.
+The "axes" are used as part of [[#multiscale-md]]. The length of "axes" MUST be equal to the number of dimensions of the arrays that contain the image data.
+
+The "dimension_names" attribute in the `zarr.json` of the Zarr array of a multiscale level MUST match the names in the "axes" metadata.
+
"bioformats2raw.layout" (transitional) {#bf2raw}
------------------------------------------------
@@ -201,7 +198,7 @@ If part of [[#multiscale-md]], the length of "axes" MUST be equal to the number
The need for the collection stems from the common "multi-image file" scenario in microscopy. Parsers like Bio-Formats
define a strict, stable ordering of the images in a single container that can be used to refer to them by other tools.
-In order to capture that information within an OME-NGFF dataset, `bioformats2raw` internally introduced a wrapping layer.
+In order to capture that information within an OME-Zarr dataset, `bioformats2raw` internally introduced a wrapping layer.
The bioformats2raw layout has been added to v0.4 as a transitional specification to specify filesets that already exist
in the wild. An upcoming NGFF specification will replace this layout with explicit metadata.
@@ -211,11 +208,9 @@ Typical Zarr layout produced by running `bioformats2raw` on a fileset that conta
series.ome.zarr # One converted fileset from bioformats2raw
- ├── .zgroup
- ├── .zattrs # Contains "bioformats2raw.layout" metadata
+ ├── zarr.json # Contains "bioformats2raw.layout" metadata
├── OME # Special group for containing OME metadata
- │ ├── .zgroup
- │ ├── .zattrs # Contains "series" metadata
+ │ ├── zarr.json # Contains "series" metadata
│ └── METADATA.ome.xml # OME-XML file stored within the Zarr fileset
├── 0 # First image in the collection
├── 1 # Second image in the collection
@@ -224,7 +219,7 @@ series.ome.zarr # One converted fileset from bioformats2raw
Attributes
-The top-level `.zattrs` file must contain the `bioformats2raw.layout` key:
+The OME-Zarr Metadata in the top-level `zarr.json` file must contain the `bioformats2raw.layout` key:
path: examples/bf2raw/image.json
highlight: json
@@ -239,7 +234,7 @@ path: examples/bf2raw/plate.json
highlight: json
-The `.zattrs` file within the OME group may contain the "series" key:
+The OME-Zarr Metadata in the `zarr.json` file within the OME group may contain the "series" key:
path: examples/ome/series-2.json
@@ -250,7 +245,7 @@ highlight: json
Conforming groups:
-- MUST have the value "3" for the "bioformats2raw.layout" key in their `.zattrs` metadata at the top of the hierarchy;
+- MUST have the value "3" for the "bioformats2raw.layout" key in their OME-Zarr Metadata in the `zarr.json` at the top of the hierarchy;
- SHOULD have OME metadata representing the entire collection of images in a file named "OME/METADATA.ome.xml" which:
- MUST adhere to the OME-XML specification but
- MUST use `` elements as opposed to ``, `` or ``;
@@ -298,7 +293,9 @@ The transformations in the list are applied sequentially and in order.
"multiscales" metadata {#multiscale-md}
---------------------------------------
-Metadata about an image can be found under the "multiscales" key in the group-level metadata. Here, image refers to 2 to 5 dimensional data representing image or volumetric data with optional time or channel axes. It is stored in a multiple resolution representation.
+Metadata about an image can be found under the "multiscales" key in the group-level OME-Zarr Metadata.
+Here, image refers to 2 to 5 dimensional data representing image or volumetric data with optional time or channel axes.
+It is stored in a multiple resolution representation.
"multiscales" contains a list of dictionaries where each entry describes a multiscale image.
@@ -324,7 +321,7 @@ Each "multiscales" dictionary MAY contain the field "coordinateTransformations",
The transformations MUST follow the same rules about allowed types, order, etc. as in "datasets:coordinateTransformations" and are applied after them.
They can for example be used to specify the `scale` for a dimension that is the same for all resolutions.
-Each "multiscales" dictionary SHOULD contain the field "name". It MUST contain the field "version", which indicates the version of the multiscale metadata of this image (current version is [NGFFVERSION]).
+Each "multiscales" dictionary SHOULD contain the field "name".
Each "multiscales" dictionary SHOULD contain the field "type", which gives the type of downscaling method used to generate the multiscale image pyramid.
It SHOULD contain the field "metadata", which contains a dictionary with additional information about the downscaling method.
@@ -358,7 +355,6 @@ can be found under the "omero" key in the group-level metadata:
```json
"id": 1, # ID in OMERO
"name": "example.tif", # Name as shown in the UI
-"version": "0.5-dev", # Current version
"channels": [ # Array matching the c dimension size
{
"active": true,
@@ -399,25 +395,32 @@ The "labels" group is not itself an image; it contains images. The pixels of the
[`uint8`, `int8`, `uint16`, `int16`, `uint32`, `int32`, `uint64`, `int64`]. Intermediate groups between "labels" and the images within it are allowed,
but these MUST NOT contain metadata. Names of the images in the "labels" group are arbitrary.
-The `.zattrs` file associated with the "labels" group MUST contain a JSON object with the key `labels`, whose value is a JSON array of paths to the
+The OME-Zarr Metadata in the `zarr.json` file associated with the "labels" group MUST contain a JSON object with the key `labels`, whose value is a JSON array of paths to the
labeled multiscale image(s). All label images SHOULD be listed within this metadata file. For example:
```json
{
- "labels": [
- "cell_space_segmentation"
- ]
+ ...
+ "attributes": {
+ "ome": {
+ "version": "0.5",
+ "schema_url": "https://ngff.openmicroscopy.org/latest/schemas/ome_zarr.schema",
+ "labels": [
+ "cell_space_segmentation"
+ ]
+ }
+ }
}
```
-The `.zattrs` file for the label image MUST implement the multiscales specification. Within the `multiscales` object, the JSON array
+The `zarr.json` file for the label image MUST implement the multiscales specification. Within the `multiscales` object, the JSON array
associated with the `datasets` key MUST have the same number of entries (scale levels) as the original unlabeled image.
-In addition to the `multiscales` key, the JSON object in this image-level `.zattrs` file SHOULD contain another key, `image-label`,
+In addition to the `multiscales` key, the OME-Zarr Metadata in this image-level `zarr.json` file SHOULD contain another key, `image-label`,
whose value is also a JSON object. The `image-label` object stores information about the display colors, source image, and optionally,
further arbitrary properties of the label image. That `image-label` object SHOULD contain the following keys: first, a `colors` key,
whose value MUST be a JSON array describing color information for the unique label values. Second, a `version` key, whose value MUST be a
-string specifying the version of the OME-NGFF `image-label` schema.
+string specifying the version of the OME-Zarr `image-label` schema.
Conforming readers SHOULD display labels using the colors specified by the `colors` JSON array, as follows. This array contains one
JSON object for each unique custom label. Each of these objects MUST contain the `label-value` key, whose value MUST be the integer
@@ -565,50 +568,14 @@ were added before this was adopted, but they should be updated in due course.
Implementations {#implementations}
==================================
-Projects which support reading and/or writing OME-NGFF data include:
-
-
-
- - [bigdataviewer-ome-zarr](https://github.com/mobie/bigdataviewer-ome-zarr)
- - Fiji-plugin for reading OME-Zarr.
-
- - [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw)
- - A performant, Bio-Formats image file format converter.
-
- - [omero-ms-zarr](https://github.com/ome/omero-ms-zarr)
- - A microservice for OMERO.server that converts images stored in OMERO to OME-Zarr files on the fly, served via a web API.
-
- - [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools)
- - A full workflow demonstrating the conversion of IDR images to OME-Zarr images on S3.
-
- - [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr)
- - An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file.
-
- - [ome-zarr-py](https://github.com/ome/ome-zarr-py)
- - A napari plugin for reading ome-zarr files.
-
- - [vizarr](https://github.com/hms-dbmi/vizarr/)
- - A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy.
-
- - [ITKIOOMEZarrNGFF](https://github.com/InsightSoftwareConsortium/ITKIOOMEZarrNGFF/)
- - ITK IO for images stored in OME-NGFF format.
-
-
-
-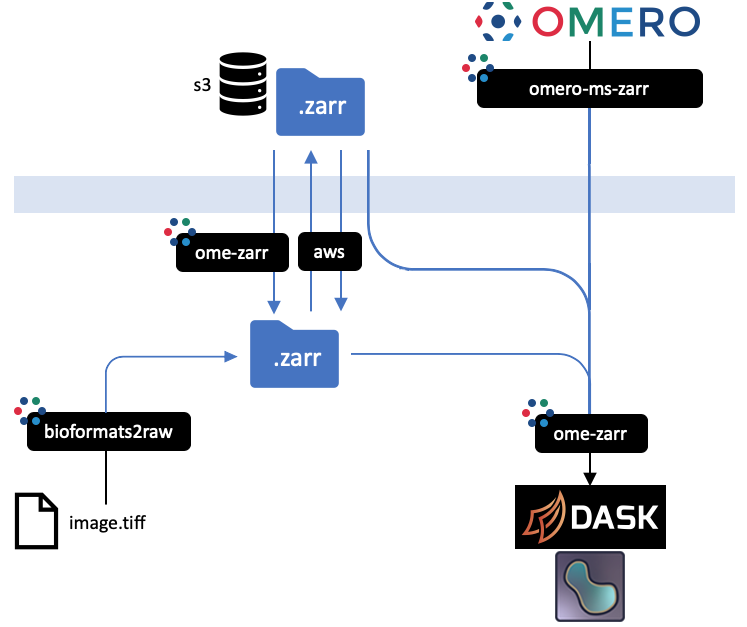 -
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+See [Tools](https://ngff.openmicroscopy.org/tools/index.html).
Citing {#citing}
================
[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.4)
J. Moore, *et al*. Open Microscopy Environment Consortium, 8 February 2022.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.4/](https://ngff.openmicroscopy.org/0.4/]).
+This edition of the specification is [https://ngff.openmicroscopy.org/0.5/](https://ngff.openmicroscopy.org/0.5/]).
The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
@@ -623,6 +590,11 @@ Version History {#history}
-
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+See [Tools](https://ngff.openmicroscopy.org/tools/index.html).
Citing {#citing}
================
[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.4)
J. Moore, *et al*. Open Microscopy Environment Consortium, 8 February 2022.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.4/](https://ngff.openmicroscopy.org/0.4/]).
+This edition of the specification is [https://ngff.openmicroscopy.org/0.5/](https://ngff.openmicroscopy.org/0.5/]).
The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
@@ -623,6 +590,11 @@ Version History {#history}
Description |
+
+ | 0.5 |
+ 2024-11-21 |
+ use Zarr v3 in OME-Zarr, see RFC-2. |
+
| 0.4.1 |
2023-02-09 |
diff --git a/0.5/schemas/_version.schema b/0.5/schemas/_version.schema
new file mode 100644
index 00000000..1da06a6d
--- /dev/null
+++ b/0.5/schemas/_version.schema
@@ -0,0 +1,10 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema",
+ "title": "OME-Zarr Metadata version",
+ "description": "The version of the OME-Zarr Metadata",
+ "type": "string",
+ "enum": [
+ "0.5"
+ ]
+}
diff --git a/0.5/schemas/bf2raw.schema b/0.5/schemas/bf2raw.schema
new file mode 100644
index 00000000..d3c87253
--- /dev/null
+++ b/0.5/schemas/bf2raw.schema
@@ -0,0 +1,32 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/bf2raw.schema",
+ "title": "OME-Zarr container produced by bioformats2raw",
+ "description": "The zarr.json attributes key",
+ "type": "object",
+ "properties": {
+ "ome": {
+ "description": "The versioned OME-Zarr Metadata namespace",
+ "type": "object",
+ "properties": {
+ "bioformats2raw.layout": {
+ "description": "The top-level identifier metadata added by bioformats2raw",
+ "type": "number",
+ "enum": [
+ 3
+ ]
+ },
+ "version": {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema"
+ }
+ },
+ "required": [
+ "bioformats2raw.layout",
+ "version"
+ ]
+ }
+ },
+ "required": [
+ "ome"
+ ]
+}
diff --git a/latest/schemas/image.schema b/0.5/schemas/image.schema
similarity index 77%
rename from latest/schemas/image.schema
rename to 0.5/schemas/image.schema
index cf2b477f..f94fd942 100644
--- a/latest/schemas/image.schema
+++ b/0.5/schemas/image.schema
@@ -1,10 +1,34 @@
{
"$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/image.schema",
- "title": "NGFF Image",
- "description": "JSON from OME-NGFF .zattrs",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/image.schema",
+ "title": "OME-Zarr Image",
+ "description": "The zarr.json attributes key",
"type": "object",
"properties": {
+ "ome": {
+ "description": "The versioned OME-Zarr Metadata namespace",
+ "type": "object",
+ "properties": {
+ "multiscales": {
+ "$ref": "#/$defs/multiscales"
+ },
+ "omero": {
+ "$ref": "#/$defs/omero"
+ },
+ "version": {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema"
+ }
+ },
+ "required": [
+ "multiscales",
+ "version"
+ ]
+ }
+ },
+ "required": [
+ "ome"
+ ],
+ "$defs": {
"multiscales": {
"description": "The multiscale datasets for this image",
"type": "array",
@@ -27,24 +51,22 @@
"$ref": "#/$defs/coordinateTransformations"
}
},
- "required": ["path", "coordinateTransformations"]
+ "required": [
+ "path",
+ "coordinateTransformations"
+ ]
}
},
- "version": {
- "type": "string",
- "enum": [
- "0.5-dev"
- ]
- },
"axes": {
"$ref": "#/$defs/axes"
},
"coordinateTransformations": {
"$ref": "#/$defs/coordinateTransformations"
- }
+ }
},
"required": [
- "datasets", "axes", "version"
+ "datasets",
+ "axes"
]
},
"minItems": 1,
@@ -100,11 +122,7 @@
"required": [
"channels"
]
- }
- },
- "required": [ "multiscales" ],
-
- "$defs": {
+ },
"axes": {
"type": "array",
"uniqueItems": true,
@@ -118,7 +136,9 @@
},
"type": {
"type": "string",
- "enum": ["space"]
+ "enum": [
+ "space"
+ ]
},
"unit": {
"type": "string"
@@ -137,10 +157,17 @@
},
"type": {
"type": "string",
- "enum": ["channel", "time", "space"]
+ "enum": [
+ "channel",
+ "time",
+ "space"
+ ]
}
},
- "required": ["name", "type"]
+ "required": [
+ "name",
+ "type"
+ ]
},
{
"type": "object",
@@ -151,11 +178,17 @@
"type": {
"type": "string",
"not": {
- "enum": ["space", "time", "channel"]
+ "enum": [
+ "space",
+ "time",
+ "channel"
+ ]
}
}
},
- "required": ["name"]
+ "required": [
+ "name"
+ ]
}
]
}
@@ -201,7 +234,10 @@
}
}
},
- "required": ["type", "scale"]
+ "required": [
+ "type",
+ "scale"
+ ]
},
{
"type": "object",
@@ -220,7 +256,10 @@
}
}
},
- "required": ["type", "translation"]
+ "required": [
+ "type",
+ "translation"
+ ]
}
]
}
diff --git a/latest/schemas/label.schema b/0.5/schemas/label.schema
similarity index 75%
rename from latest/schemas/label.schema
rename to 0.5/schemas/label.schema
index 8b23d577..dd5cb6d6 100644
--- a/latest/schemas/label.schema
+++ b/0.5/schemas/label.schema
@@ -1,10 +1,31 @@
{
"$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/label.schema",
- "title": "OME-NGFF labelled image schema",
- "description": "JSON from OME-NGFF .zattrs",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/label.schema",
+ "title": "OME-Zarr labelled image schema",
+ "description": "The zarr.json attributes key",
"type": "object",
"properties": {
+ "ome": {
+ "description": "The versioned OME-Zarr Metadata namespace",
+ "type": "object",
+ "properties": {
+ "image-label": {
+ "$ref": "#/$defs/image-label"
+ },
+ "version": {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema"
+ }
+ },
+ "required": [
+ "image-label",
+ "version"
+ ]
+ }
+ },
+ "required": [
+ "ome"
+ ],
+ "$defs": {
"image-label": {
"type": "object",
"properties": {
@@ -63,13 +84,6 @@
"type": "string"
}
}
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.5-dev"
- ]
}
}
}
diff --git a/0.5/schemas/ome.schema b/0.5/schemas/ome.schema
new file mode 100644
index 00000000..36569280
--- /dev/null
+++ b/0.5/schemas/ome.schema
@@ -0,0 +1,33 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/ome.schema",
+ "title": "OME-Zarr group produced by bioformats2raw to contain OME metadata",
+ "description": "The zarr.json attributes key",
+ "type": "object",
+ "properties": {
+ "ome": {
+ "description": "The versioned OME-Zarr Metadata namespace",
+ "type": "object",
+ "properties": {
+ "series": {
+ "description": "An array of the same length and the same order as the images defined in the OME-XML",
+ "type": "array",
+ "items": {
+ "type": "string"
+ },
+ "minContains": 1
+ },
+ "version": {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema"
+ }
+ },
+ "required": [
+ "series",
+ "version"
+ ]
+ }
+ },
+ "required": [
+ "ome"
+ ]
+}
diff --git a/0.5/schemas/ome_zarr.schema b/0.5/schemas/ome_zarr.schema
new file mode 100644
index 00000000..9b22bba6
--- /dev/null
+++ b/0.5/schemas/ome_zarr.schema
@@ -0,0 +1,24 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/ome_zarr.schema",
+ "anyOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/bf2raw.schema"
+ },
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/image.schema"
+ },
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/label.schema"
+ },
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/ome.schema"
+ },
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/plate.schema"
+ },
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/well.schema"
+ }
+ ]
+}
diff --git a/0.5/schemas/plate.schema b/0.5/schemas/plate.schema
new file mode 100644
index 00000000..3c8a1b97
--- /dev/null
+++ b/0.5/schemas/plate.schema
@@ -0,0 +1,153 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/plate.schema",
+ "title": "OME-Zarr plate schema",
+ "description": "The zarr.json attributes key",
+ "type": "object",
+ "properties": {
+ "ome": {
+ "description": "The versioned OME-Zarr Metadata namespace",
+ "type": "object",
+ "properties": {
+ "plate": {
+ "type": "object",
+ "properties": {
+ "acquisitions": {
+ "description": "The acquisitions for this plate",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "id": {
+ "description": "A unique identifier within the context of the plate",
+ "type": "integer",
+ "minimum": 0
+ },
+ "maximumfieldcount": {
+ "description": "The maximum number of fields of view for the acquisition",
+ "type": "integer",
+ "exclusiveMinimum": 0
+ },
+ "name": {
+ "description": "The name of the acquisition",
+ "type": "string"
+ },
+ "description": {
+ "description": "The description of the acquisition",
+ "type": "string"
+ },
+ "starttime": {
+ "description": "The start timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
+ "type": "integer",
+ "minimum": 0
+ },
+ "endtime": {
+ "description": "The end timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
+ "type": "integer",
+ "minimum": 0
+ }
+ },
+ "required": [
+ "id"
+ ]
+ }
+ },
+ "field_count": {
+ "description": "The maximum number of fields per view across all wells",
+ "type": "integer",
+ "exclusiveMinimum": 0
+ },
+ "name": {
+ "description": "The name of the plate",
+ "type": "string"
+ },
+ "columns": {
+ "description": "The columns of the plate",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "description": "The column name",
+ "type": "string",
+ "pattern": "^[A-Za-z0-9]+$"
+ }
+ },
+ "required": [
+ "name"
+ ]
+ },
+ "minItems": 1,
+ "uniqueItems": true
+ },
+ "rows": {
+ "description": "The rows of the plate",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "name": {
+ "description": "The row name",
+ "type": "string",
+ "pattern": "^[A-Za-z0-9]+$"
+ }
+ },
+ "required": [
+ "name"
+ ]
+ },
+ "minItems": 1,
+ "uniqueItems": true
+ },
+ "wells": {
+ "description": "The wells of the plate",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "path": {
+ "description": "The path to the well subgroup",
+ "type": "string",
+ "pattern": "^[A-Za-z0-9]+/[A-Za-z0-9]+$"
+ },
+ "rowIndex": {
+ "description": "The index of the well in the rows list",
+ "type": "integer",
+ "minimum": 0
+ },
+ "columnIndex": {
+ "description": "The index of the well in the columns list",
+ "type": "integer",
+ "minimum": 0
+ }
+ },
+ "required": [
+ "path",
+ "rowIndex",
+ "columnIndex"
+ ]
+ },
+ "minItems": 1,
+ "uniqueItems": true
+ }
+ },
+ "required": [
+ "columns",
+ "rows",
+ "wells"
+ ]
+ },
+ "version": {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema"
+ }
+ },
+ "required": [
+ "plate",
+ "version"
+ ]
+ }
+ },
+ "required": [
+ "ome"
+ ]
+}
diff --git a/0.5/schemas/strict_image.schema b/0.5/schemas/strict_image.schema
new file mode 100644
index 00000000..042790e7
--- /dev/null
+++ b/0.5/schemas/strict_image.schema
@@ -0,0 +1,25 @@
+{
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/strict_image.schema",
+ "allOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/image.schema"
+ },
+ {
+ "properties": {
+ "ome": {
+ "properties": {
+ "multiscales": {
+ "items": {
+ "required": [
+ "metadata",
+ "type",
+ "name"
+ ]
+ }
+ }
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/0.5/schemas/strict_label.schema b/0.5/schemas/strict_label.schema
new file mode 100644
index 00000000..f1ec4db5
--- /dev/null
+++ b/0.5/schemas/strict_label.schema
@@ -0,0 +1,21 @@
+{
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/strict_label.schema",
+ "allOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/label.schema"
+ },
+ {
+ "properties": {
+ "ome": {
+ "properties": {
+ "image-label": {
+ "required": [
+ "colors"
+ ]
+ }
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/0.5/schemas/strict_plate.schema b/0.5/schemas/strict_plate.schema
new file mode 100644
index 00000000..ce678ce8
--- /dev/null
+++ b/0.5/schemas/strict_plate.schema
@@ -0,0 +1,31 @@
+{
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/strict_plate.schema",
+ "allOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/plate.schema"
+ },
+ {
+ "properties": {
+ "ome": {
+ "properties": {
+ "plate": {
+ "properties": {
+ "acquisitions": {
+ "items": {
+ "required": [
+ "name",
+ "maximumfieldcount"
+ ]
+ }
+ }
+ },
+ "required": [
+ "name"
+ ]
+ }
+ }
+ }
+ }
+ }
+ ]
+}
diff --git a/0.5/schemas/strict_well.schema b/0.5/schemas/strict_well.schema
new file mode 100644
index 00000000..43cb58cb
--- /dev/null
+++ b/0.5/schemas/strict_well.schema
@@ -0,0 +1,8 @@
+{
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/strict_well.schema",
+ "allOf": [
+ {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/well.schema"
+ }
+ ]
+}
diff --git a/0.5/schemas/well.schema b/0.5/schemas/well.schema
new file mode 100644
index 00000000..447f51df
--- /dev/null
+++ b/0.5/schemas/well.schema
@@ -0,0 +1,56 @@
+{
+ "$schema": "https://json-schema.org/draft/2020-12/schema",
+ "$id": "https://ngff.openmicroscopy.org/0.5/schemas/well.schema",
+ "title": "OME-Zarr well schema",
+ "description": "JSON from OME-Zarr zarr.json",
+ "type": "object",
+ "properties": {
+ "ome": {
+ "description": "The versioned OME-Zarr Metadata namespace",
+ "type": "object",
+ "properties": {
+ "well": {
+ "type": "object",
+ "properties": {
+ "images": {
+ "description": "The fields of view for this well",
+ "type": "array",
+ "items": {
+ "type": "object",
+ "properties": {
+ "acquisition": {
+ "description": "A unique identifier within the context of the plate",
+ "type": "integer"
+ },
+ "path": {
+ "description": "The path for this field of view subgroup",
+ "type": "string",
+ "pattern": "^[A-Za-z0-9]+$"
+ }
+ },
+ "required": [
+ "path"
+ ]
+ },
+ "minItems": 1,
+ "uniqueItems": true
+ }
+ },
+ "required": [
+ "images"
+ ]
+ },
+ "version": {
+ "$ref": "https://ngff.openmicroscopy.org/0.5/schemas/_version.schema"
+ }
+ },
+ "required": [
+ "well",
+ "version"
+ ]
+ }
+ },
+ "required": [
+ "ome"
+ ]
+}
diff --git a/0.5/tests/image_suite.json b/0.5/tests/image_suite.json
new file mode 100644
index 00000000..df359ce4
--- /dev/null
+++ b/0.5/tests/image_suite.json
@@ -0,0 +1,1040 @@
+{
+ "description": "TBD",
+ "schema": {
+ "id": "schemas/image.schema"
+ },
+ "tests": [
+ {
+ "formerly": "valid/mismatch_axes_units.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [0.13, 0.13],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid/untyped_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "angle"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid/invalid_axis_units.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micron"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [0.13, 0.13],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid/missing_name.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "datasets": [
+ {
+ "path": "path/to/0",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [1, 1]
+ }
+ ]
+ }
+ ],
+ "type": "gaussian",
+ "metadata": {
+ "method": "skimage.transform.pyramid_gaussian",
+ "version": "0.16.1",
+ "args": ["true", "false"],
+ "kwargs": {
+ "multichannel": true
+ }
+ },
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid/custom_type_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "angle",
+ "type": "custom"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "invalid/duplicate_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_space_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_transformation_type.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "translation"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_scale.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "translation": [1, 1],
+ "type": "translation"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/too_many_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "angle",
+ "type": "custom"
+ },
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space"
+ },
+ {
+ "name": "y",
+ "type": "space"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 1, 1, 1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_channels_color.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ],
+ "omero": {
+ "channels": [
+ {
+ "active": true,
+ "coefficient": 1.0,
+ "color": 255,
+ "family": "linear",
+ "label": "1234",
+ "window": {
+ "end": 1765.0,
+ "max": 2555.0,
+ "min": 5.0,
+ "start": 0.0
+ }
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_axes_name.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "type": "space",
+ "unit": "micron"
+ },
+ {
+ "type": "space",
+ "unit": "micron"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [0.13, 0.13],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_axes_count.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/one_space_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_path.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": 0,
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_multiscales_transformations.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ],
+ "coordinateTransformations": [
+ {
+ "scale": ["invalid"],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_transformations.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0"
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/no_datasets.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": []
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_datasets.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_axis_type.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "invalid",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/duplicate_scale.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ },
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/no_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/too_many_space_axes.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "X",
+ "type": "space"
+ },
+ {
+ "name": "z",
+ "type": "space"
+ },
+ {
+ "name": "y",
+ "type": "space"
+ },
+ {
+ "name": "x",
+ "type": "space"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/no_multiscales.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": []
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/invalid_channels_window.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ],
+ "omero": {
+ "channels": [
+ {
+ "active": true,
+ "coefficient": 1.0,
+ "color": "ff0000",
+ "family": "linear",
+ "label": "1234",
+ "window": {
+ "end": "100",
+ "max": 2555.0,
+ "min": 5.0,
+ "start": 0.0
+ }
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/empty_transformations.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": []
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "invalid/missing_path.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": false
+ }
+ ]
+}
diff --git a/0.5/tests/label_suite.json b/0.5/tests/label_suite.json
new file mode 100644
index 00000000..66a2d35d
--- /dev/null
+++ b/0.5/tests/label_suite.json
@@ -0,0 +1,158 @@
+{
+ "description": "Tests for the image-label JSON schema",
+ "schema": {
+ "id": "schemas/label.schema"
+ },
+ "tests": [
+ {
+ "formerly": "image-label/minimal",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": [
+ {
+ "label-value": 1,
+ "rgba": [0, 0, 0, 0]
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "image-label/minimal_properties",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": [
+ {
+ "label-value": 1,
+ "rgba": [0, 0, 0, 0]
+ }
+ ],
+ "properties": [
+ {
+ "label-value": 1
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "image-label/empty_colors",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": []
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "image-label/empty_properties",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "properties": []
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "image-label/colors_no_label_value",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": [
+ {
+ "rgba": [0, 0, 0, 0]
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "image-label/properties_no_label_value",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "properties": [
+ {
+ "value": "foo"
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "image-label/colors_rgba_length",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": [
+ {
+ "label-value": 1,
+ "rgba": [0, 0, 0]
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "image-label/colors_rgba_type",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": [
+ {
+ "label-value": 1,
+ "rgba": [0, 0, 0, 500]
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "image-label/colors_duplicate",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {
+ "colors": [
+ {
+ "label-value": 1,
+ "rgba": [0, 0, 0, 0]
+ },
+ {
+ "label-value": 1,
+ "rgba": [0, 0, 0, 0]
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ }
+ ]
+}
diff --git a/0.5/tests/plate_suite.json b/0.5/tests/plate_suite.json
new file mode 100644
index 00000000..3db70de7
--- /dev/null
+++ b/0.5/tests/plate_suite.json
@@ -0,0 +1,883 @@
+{
+ "description": "Tests for the plate JSON schema",
+ "schema": {
+ "id": "schemas/plate.schema"
+ },
+ "tests": [
+ {
+ "formerly": "plate/minimal_no_acquisitions",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "plate/minimal_acquisitions",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "plate/missing_rows",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/empty_rows",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/duplicate_rows",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ },
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_columns",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/empty_columns",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/duplicate_columns",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ },
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_wells",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/empty_wells",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": {}
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/duplicate_rows",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ },
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ },
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_column_name",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "concentration": 10
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_row_name",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "concentration": 10
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_well_path",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_well_rowIndex",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_well_columnIndex",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/well_1group",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A1",
+ "rowIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/well_3groups",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "plate/A/1",
+ "rowIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/non_alphanumeric_column",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A-1"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A-1/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/non_alphanumeric_row",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "A1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/A1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "plate/missing_acquisition_id",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "maximumfieldcount": 1
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/non_integer_acquisition_id",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": "0"
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/negative_acquisition_id",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": -1
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/non_integer_acquisition_maximumfieldcount",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "maximumfieldcount": "0"
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/acquisition_zero_maximumfieldcount",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "maximumfieldcount": 0
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/acquisition_noninteger_starttime",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "starttime": "2022-05-13T13:48:06+00:00"
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/acquisition_negative_starttime",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "starttime": -1
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/acquisition_noninteger_endtime",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "endtime": "2022-05-13T13:48:06+00:00"
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/negative_endtime",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "endtime": -1
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/zero_field_count",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "field_count": 0,
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ }
+ ]
+}
diff --git a/0.5/tests/strict_image_suite.json b/0.5/tests/strict_image_suite.json
new file mode 100644
index 00000000..b52fd2d3
--- /dev/null
+++ b/0.5/tests/strict_image_suite.json
@@ -0,0 +1,341 @@
+{
+ "description": "TBD",
+ "schema": {
+ "id": "schemas/strict_image.schema"
+ },
+ "tests": [
+ {
+ "formerly": "valid_strict/multiscales_example.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "name": "example",
+ "axes": [
+ {
+ "name": "t",
+ "type": "time",
+ "unit": "millisecond"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [1.0, 1.0, 0.5, 0.5, 0.5]
+ }
+ ]
+ },
+ {
+ "path": "1",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [1.0, 1.0, 1.0, 1.0, 1.0]
+ }
+ ]
+ },
+ {
+ "path": "2",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [1.0, 1.0, 2.0, 2.0, 2.0]
+ }
+ ]
+ }
+ ],
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [0.1, 1.0, 1.0, 1.0, 1.0]
+ }
+ ],
+ "type": "gaussian",
+ "metadata": {
+ "description": "the fields in metadata depend on the downscaling implementation. Here, the parameters passed to the skimage function are given",
+ "method": "skimage.transform.pyramid_gaussian",
+ "version": "0.16.1",
+ "args": "[true]",
+ "kwargs": {
+ "multichannel": true
+ }
+ }
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid_strict/multiscales_transformations.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ],
+ "coordinateTransformations": [
+ {
+ "scale": [10, 10],
+ "type": "scale"
+ }
+ ],
+ "name": "image_with_coordinateTransformations",
+ "type": "foo",
+ "metadata": {
+ "key": "value"
+ }
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid_strict/image_metadata.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "@id": "top",
+ "@type": "ngff:Image",
+ "multiscales": [
+ {
+ "@id": "inner",
+ "name": "example",
+ "datasets": [
+ {
+ "path": "path/to/0",
+ "coordinateTransformations": [
+ {
+ "type": "scale",
+ "scale": [1, 1]
+ }
+ ]
+ }
+ ],
+ "type": "gaussian",
+ "metadata": {
+ "method": "skimage.transform.pyramid_gaussian",
+ "version": "0.16.1",
+ "args": ["true", "false"],
+ "kwargs": {
+ "multichannel": true
+ }
+ },
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ]
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid_strict/image.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1],
+ "type": "scale"
+ }
+ ]
+ }
+ ],
+ "name": "simple_image",
+ "type": "foo",
+ "metadata": {
+ "key": "value"
+ }
+ }
+ ]
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "valid_strict/image_omero.json",
+ "description": "TBD",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "multiscales": [
+ {
+ "axes": [
+ {
+ "name": "t",
+ "type": "time"
+ },
+ {
+ "name": "c",
+ "type": "channel"
+ },
+ {
+ "name": "z",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "y",
+ "type": "space",
+ "unit": "micrometer"
+ },
+ {
+ "name": "x",
+ "type": "space",
+ "unit": "micrometer"
+ }
+ ],
+ "datasets": [
+ {
+ "path": "0",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 0.5, 0.13, 0.13],
+ "type": "scale"
+ },
+ {
+ "translation": [0, 9, 0.5, 25.74, 21.58],
+ "type": "translation"
+ }
+ ]
+ },
+ {
+ "path": "1",
+ "coordinateTransformations": [
+ {
+ "scale": [1, 1, 1, 0.26, 0.26],
+ "type": "scale"
+ }
+ ]
+ }
+ ],
+ "name": "image_with_omero_metadata",
+ "type": "foo",
+ "metadata": {
+ "key": "value"
+ }
+ }
+ ],
+ "omero": {
+ "channels": [
+ {
+ "active": true,
+ "coefficient": 1.0,
+ "color": "00FF00",
+ "family": "linear",
+ "inverted": false,
+ "label": "FITC",
+ "window": {
+ "end": 813.0,
+ "max": 870.0,
+ "min": 102.0,
+ "start": 82.0
+ }
+ },
+ {
+ "active": true,
+ "coefficient": 1.0,
+ "color": "FF0000",
+ "family": "linear",
+ "inverted": false,
+ "label": "RD-TR-PE",
+ "window": {
+ "end": 815.0,
+ "max": 441.0,
+ "min": 129.0,
+ "start": 78.0
+ }
+ }
+ ],
+ "id": 1,
+ "rdefs": {
+ "defaultT": 0,
+ "defaultZ": 2,
+ "model": "color"
+ },
+ "version": "0.5-dev"
+ }
+ }
+ },
+ "valid": true
+ }
+ ]
+}
diff --git a/0.5/tests/strict_label_suite.json b/0.5/tests/strict_label_suite.json
new file mode 100644
index 00000000..1b3535c2
--- /dev/null
+++ b/0.5/tests/strict_label_suite.json
@@ -0,0 +1,18 @@
+{
+ "description": "Tests for the strict image-label JSON schema",
+ "schema": {
+ "id": "schemas/strict_label.schema"
+ },
+ "tests": [
+ {
+ "formerly": "image-label/no_colors",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "image-label": {}
+ }
+ },
+ "valid": false
+ }
+ ]
+}
diff --git a/0.5/tests/strict_plate_suite.json b/0.5/tests/strict_plate_suite.json
new file mode 100644
index 00000000..7cea0747
--- /dev/null
+++ b/0.5/tests/strict_plate_suite.json
@@ -0,0 +1,171 @@
+{
+ "description": "Tests for the strict plate JSON schema",
+ "schema": {
+ "id": "schemas/strict_plate.schema"
+ },
+ "tests": [
+ {
+ "formerly": "plate/strict_no_acquisitions",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "name": "test plate",
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "plate/missing_name",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/strict_acquisitions",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "name": "0",
+ "maximumfieldcount": 1
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "name": "test plate",
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "plate/missing_acquisition_name",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "maximumfieldcount": 1
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "name": "test plate",
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ },
+ {
+ "formerly": "plate/missing_acquisition_maximumfieldcount",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "plate": {
+ "acquisitions": [
+ {
+ "id": 0,
+ "name": "0"
+ }
+ ],
+ "columns": [
+ {
+ "name": "A"
+ }
+ ],
+ "name": "test plate",
+ "rows": [
+ {
+ "name": "1"
+ }
+ ],
+ "wells": [
+ {
+ "path": "A/1",
+ "rowIndex": 0,
+ "columnIndex": 0
+ }
+ ]
+ }
+ }
+ },
+ "valid": false
+ }
+ ]
+}
diff --git a/0.5/tests/strict_well_suite.json b/0.5/tests/strict_well_suite.json
new file mode 100644
index 00000000..f56b6c24
--- /dev/null
+++ b/0.5/tests/strict_well_suite.json
@@ -0,0 +1,41 @@
+{
+ "description": "Tests for the strict well JSON schema",
+ "schema": {
+ "id": "schemas/strict_well.schema"
+ },
+ "tests": [
+ {
+ "formerly": "well/strict_no_acquisitions",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "well": {
+ "images": [
+ {
+ "path": "0"
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ },
+ {
+ "formerly": "plate/strict_acquisitions",
+ "data": {
+ "ome": {
+ "version": "0.5",
+ "well": {
+ "images": [
+ {
+ "acquisition": 0,
+ "path": "0"
+ }
+ ]
+ }
+ }
+ },
+ "valid": true
+ }
+ ]
+}
diff --git a/latest/tests/test_validation.py b/0.5/tests/test_validation.py
similarity index 83%
rename from latest/tests/test_validation.py
rename to 0.5/tests/test_validation.py
index 2b862785..b9c7c929 100644
--- a/latest/tests/test_validation.py
+++ b/0.5/tests/test_validation.py
@@ -16,9 +16,16 @@
schema = json.load(f)
schema_store[schema["$id"]] = schema
+GENERIC_SCHEMA = schema_store[
+ "https://ngff.openmicroscopy.org/0.5/schemas/ome_zarr.schema"
+]
+
+print(schema_store)
+
+
@dataclass
class Suite:
- schema: dict
+ schema: dict
data: dict
valid: bool = True
@@ -29,6 +36,10 @@ def validate(self, validator) -> None:
else:
validator.validate(self.data)
+ def maybe_validate(self, validator) -> None:
+ if self.valid:
+ validator.validate(self.data)
+
def pytest_generate_tests(metafunc):
"""
@@ -72,8 +83,11 @@ def pytest_generate_tests(metafunc):
for filename in glob.glob(f"{example_folder}/*.json"):
with open(filename) as f:
# Strip comments
- data = ''.join(line for line in f if not line.lstrip().startswith('//'))
+ data = "".join(
+ line for line in f if not line.lstrip().startswith("//")
+ )
data = json.loads(data)
+ data = data["attributes"] # Only validate the attributes object
ids.append("example_" + str(filename).split("/")[-1][0:-5])
suites.append(Suite(schema, data, True)) # Assume true
@@ -91,6 +105,12 @@ def test_run(suite):
suite.validate(validator)
+def test_generic_run(suite):
+ resolver = RefResolver.from_schema(GENERIC_SCHEMA, store=schema_store)
+ validator = Validator(GENERIC_SCHEMA, resolver=resolver)
+ suite.maybe_validate(validator)
+
+
def test_example_configs():
"""
Test that all example folders have a config file
diff --git a/latest/tests/well_suite.json b/0.5/tests/well_suite.json
similarity index 67%
rename from latest/tests/well_suite.json
rename to 0.5/tests/well_suite.json
index 0f752551..7fbf8ca2 100644
--- a/latest/tests/well_suite.json
+++ b/0.5/tests/well_suite.json
@@ -7,12 +7,15 @@
{
"formerly": "well/minimal_no_acquisition",
"data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ]
+ "ome": {
+ "version": "0.5",
+ "well": {
+ "images": [
+ {
+ "path": "0"
+ }
+ ]
+ }
}
},
"valid": true
@@ -20,13 +23,16 @@
{
"formerly": "well/minimal_acquisitions",
"data": {
- "well": {
- "images": [
- {
- "acquisition": 1,
- "path": "0"
- }
- ]
+ "ome": {
+ "version": "0.5",
+ "well": {
+ "images": [
+ {
+ "acquisition": 1,
+ "path": "0"
+ }
+ ]
+ }
}
},
"valid": true
@@ -56,20 +62,6 @@
},
"valid": false
},
- {
- "formerly": "well/invalid_version",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ],
- "version": "foo"
- }
- },
- "valid": false
- },
{
"formerly": "well/non_integer_acquisition_id",
"data": {
diff --git a/latest/tox.ini b/0.5/tox.ini
similarity index 100%
rename from latest/tox.ini
rename to 0.5/tox.ini
diff --git a/about/index.md b/about/index.md
index 855ecd60..8009f7b6 100644
--- a/about/index.md
+++ b/about/index.md
@@ -19,11 +19,11 @@ factor before publication is possible. Without a common effort, each lab or
resource is left building the tools they need and maintaining that
infrastructure often without dedicated funding.
-This document defines a specification for bioimaging data to make it possible
-to enable the conversion of proprietary formats into a common, cloud-ready one.
-Such next-generation file formats layout data so that individual portions, or
-"chunks", of large data are reference-able eliminating the need to download
-entire datasets.
+This document defines OME-Zarr, a specification for bioimaging data to make it
+possible to enable the conversion of proprietary formats into a common,
+cloud-ready one. Such a next-generation file format layouts data so that
+individual portions, or "chunks", of large data are reference-able eliminating
+the need to download entire datasets.
Why "NGFF"?
@@ -47,13 +47,14 @@ the limitations of HDF5 ("N5" was originally short for "Not-HDF5").
Both of these formats permit storing individual chunks of data either locally in
separate files or in cloud-based object stores as separate keys.
-An [updated Zarr version (v3)](https://zarr-specs.readthedocs.io/)
-is underway to unify the two similar specifications to provide a single binary
+An [updated Zarr version (v3)](https://zarr-specs.readthedocs.io/en/latest/v3/core/v3.0.html)
+has been accepted to unify the two similar specifications to provide a single binary
specification. See this [blog post](https://zarr.dev/blog/zep1-update/) for more information.
+The file format is called "OME-Zarr". The term "NGFF" still refers to the community and [specification process](../rfc/index.md).
-In addition to the next-generation file format (NGFF) [specifications](../specifications/index.md),
+In addition to the [OME-Zarr specification](../specifications/index.md),
the pages listed below are intended to provide an overview of external resources available
-for working with NGFF data.
+for working with OME-Zarr data.
The following pages are intended to provide an overview of the available resources in the NGFF space:
@@ -74,3 +75,4 @@ Additionally, notes and recordings of the past NGFF community calls are availabl
|6 (0.4/axes)| 2022-01-27| Constantin, Will, Seb| [image.sc](https://forum.image.sc/t/next-call-on-next-gen-bioimaging-data-tools-2022-01-27/60885)| [hackmd](https://hackmd.io/QfiBKHIoTZ-CJSp3q0Wykg)|
|7 (0.5/tables & transforms)| 2022-10-05| Kevin & John| [image.sc](https://forum.image.sc/t/ome-ngff-community-call-transforms-and-tables/71792)| [hackmd](https://hackmd.io/TyfrLiCqRteL0Xfc8HRiOA) |
|8 (Metadata) | 2023-03-15 | Wouter-Michiel | [image.sc](https://forum.image.sc/t/community-call-metadata-in-ome-ngff/77570/10) | [hackmd](https://hackmd.io/BqnK9Wm4QpGYAhYOoaFBQQ) |
+|9 (Collections and labels) | 2024-04-03 | Norman | [image.sc](https://forum.image.sc/t/ome-ngff-community-call-labels-and-other-collections/93815) | [hackmd](https://hackmd.io/So61knrQR0iLftd2LGAVjA) |
diff --git a/conf.py b/conf.py
index f18fb755..5f64deaa 100644
--- a/conf.py
+++ b/conf.py
@@ -6,9 +6,9 @@
# -- Project information -----------------------------------------------------
# https://www.sphinx-doc.org/en/master/usage/configuration.html#project-information
-project = 'NGFF'
-copyright = '2023, NGFF Community'
-author = 'NGFF Community'
+project = "NGFF"
+copyright = "2024, NGFF Community"
+author = "NGFF Community"
# -- General configuration ---------------------------------------------------
# https://www.sphinx-doc.org/en/master/usage/configuration.html#general-configuration
@@ -18,44 +18,53 @@
myst_heading_anchors = 5
myst_enable_extensions = ["deflist"]
-templates_path = ['_templates']
-exclude_patterns = ['_build', 'Thumbs.db', '.DS_Store', '.git', '.pytest_cache', '**/.pytest_cache', '**/.tox', 'README.md', 'LICENSE.md', 'CONTRIBUTING.md']
-
+templates_path = ["_templates"]
+exclude_patterns = [
+ "_build",
+ "Thumbs.db",
+ ".DS_Store",
+ ".git",
+ ".pytest_cache",
+ "**/.pytest_cache",
+ "**/.tox",
+ "README.md",
+ "LICENSE.md",
+ "CONTRIBUTING.md",
+]
# -- Options for HTML output -------------------------------------------------
# https://www.sphinx-doc.org/en/master/usage/configuration.html#options-for-html-output
-html_theme = 'sphinx_book_theme'
+html_theme = "sphinx_book_theme"
-html_static_path = ['_static']
+html_static_path = ["_static"]
html_css_files = [
- 'https://cdn.datatables.net/v/dt/dt-1.11.5/datatables.min.css',
+ "https://cdn.datatables.net/v/dt/dt-1.11.5/datatables.min.css",
]
html_js_files = [
- 'https://cdn.datatables.net/v/dt/dt-1.11.5/datatables.min.js',
- 'main.js',
+ "https://cdn.datatables.net/v/dt/dt-1.11.5/datatables.min.js",
+ "main.js",
]
html_extra_path = [
- '_bikeshed',
+ "_bikeshed",
]
# ####################################
# Run bikeshed build
# ####################################
-def bikeshed():
+def bikeshed():
import glob
import os
import shutil
import subprocess
for index_file in ["latest/index.bs"] + glob.glob("[0-9]*/index.bs"):
-
output_file = index_file.replace("bs", "html")
output_dir = os.path.dirname(output_file)
target_dir = os.path.join("_bikeshed", output_dir)
@@ -71,11 +80,14 @@ def bikeshed():
run_bikeshed = False
if run_bikeshed:
- subprocess.check_call(f"bikeshed spec {index_file} {output_file}", shell=True)
+ subprocess.check_call(
+ f"bikeshed spec {index_file} {output_file}", shell=True
+ )
if os.path.exists(target_dir):
shutil.rmtree(target_dir)
shutil.copytree(output_dir, target_dir)
+
bikeshed()
del bikeshed
diff --git a/data/index.md b/data/index.md
index be0f179e..f2591ea0 100644
--- a/data/index.md
+++ b/data/index.md
@@ -15,7 +15,9 @@ Data Resources
| [Sanger](https://www.sanger.ac.uk/project/ome-zarr/) | Sanger, UK | 10 | 1 TB |
| [SpatialData](https://github.com/scverse/spatialdata-notebooks/tree/main/datasets) | EMBL-HD | 10 | 25 GB |
| [SSBD](https://ssbd.riken.jp/ssbd-ome-ngff-samples) | SSBD | 12 | 196 GB |
-| [webKnossos](https://zarr.webknossos.org) | scalableminds GmbH | 69 | 70 TB |
+| [webKnossos](https://zarr.webknossos.org) | scalable minds | 69 | 70 TB |
+
+OME-Zarr 0.5 data has been collected during [the OME 2024 NGFF challenge](https://ome.github.io/ome2024-ngff-challenge/).
[dandi2]: https://identifiers.org/DANDI:000108
[dandi3]: https://github.com/dandisets/000108
diff --git a/index.rst b/index.rst
index ecbb8bab..85db7d2b 100644
--- a/index.rst
+++ b/index.rst
@@ -6,13 +6,13 @@
Next-generation file formats (NGFF)
===================================
-OME-NGFF is an imaging format specification being developed by the bioimaging community to
+NGFF is an initiative by the bioimaging community to develop imaging format specifications to
address issues of scalability and interoperability.
Please see the :doc:`about/index` section for an introduction.
-The OME-NGFF specification is detailed under :doc:`specifications/index`.
-Various Image viewers and other software for working with NGFF data
+The OME-Zarr specification is detailed under :doc:`specifications/index`.
+Various Image viewers and other software for working with OME-Zarr data
are listed on the :doc:`tools/index` page.
-Sample NGFF datasets provided by the community can be found under :doc:`data/index`.
+Sample OME-Zarr datasets provided by the community can be found under :doc:`data/index`.
.. toctree::
:maxdepth: 1
diff --git a/latest b/latest
new file mode 120000
index 00000000..ea2303bc
--- /dev/null
+++ b/latest
@@ -0,0 +1 @@
+0.5
\ No newline at end of file
diff --git a/latest/examples/bf2raw/image.json b/latest/examples/bf2raw/image.json
deleted file mode 100644
index c5eadb8a..00000000
--- a/latest/examples/bf2raw/image.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "bioformats2raw.layout" : 3
-}
\ No newline at end of file
diff --git a/latest/examples/bf2raw/plate.json b/latest/examples/bf2raw/plate.json
deleted file mode 100644
index bd98a16e..00000000
--- a/latest/examples/bf2raw/plate.json
+++ /dev/null
@@ -1,22 +0,0 @@
-{
- "bioformats2raw.layout" : 3,
- "plate" : {
- "columns" : [ {
- "name" : "1"
- } ],
- "name" : "Plate Name 0",
- "wells" : [ {
- "path" : "A/1",
- "rowIndex" : 0,
- "columnIndex" : 0
- } ],
- "field_count" : 1,
- "rows" : [ {
- "name" : "A"
- } ],
- "acquisitions" : [ {
- "id" : 0
- } ],
- "version" : "0.4"
- }
-}
diff --git a/latest/examples/label_strict/colors_properties.json b/latest/examples/label_strict/colors_properties.json
deleted file mode 100644
index c96820e1..00000000
--- a/latest/examples/label_strict/colors_properties.json
+++ /dev/null
@@ -1,31 +0,0 @@
-{
- "image-label": {
- "version": "0.5-dev",
- "colors": [
- {
- "label-value": 0,
- "rgba": [0, 0, 128, 128]
- },
- {
- "label-value": 1,
- "rgba": [0, 128, 0, 128]
- }
- ],
- "properties": [
- {
- "label-value": 0,
- "area (pixels)": 1200,
- "class": "intercellular space"
- },
- {
- "label-value": 1,
- "area (pixels)": 1650,
- "class": "cell",
- "cell type": "neuron"
- }
- ],
- "source": {
- "image": "../../"
- }
- }
-}
diff --git a/latest/examples/multiscales_strict/multiscales_example.json b/latest/examples/multiscales_strict/multiscales_example.json
deleted file mode 100644
index 73e5286c..00000000
--- a/latest/examples/multiscales_strict/multiscales_example.json
+++ /dev/null
@@ -1,54 +0,0 @@
-{
- "multiscales": [
- {
- "version": "0.5-dev",
- "name": "example",
- "axes": [
- {"name": "t", "type": "time", "unit": "millisecond"},
- {"name": "c", "type": "channel"},
- {"name": "z", "type": "space", "unit": "micrometer"},
- {"name": "y", "type": "space", "unit": "micrometer"},
- {"name": "x", "type": "space", "unit": "micrometer"}
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [{
- // the voxel size for the first scale level (0.5 micrometer)
- "type": "scale",
- "scale": [1.0, 1.0, 0.5, 0.5, 0.5]
- }]
- },
- {
- "path": "1",
- "coordinateTransformations": [{
- // the voxel size for the second scale level (downscaled by a factor of 2 -> 1 micrometer)
- "type": "scale",
- "scale": [1.0, 1.0, 1.0, 1.0, 1.0]
- }]
- },
- {
- "path": "2",
- "coordinateTransformations": [{
- // the voxel size for the third scale level (downscaled by a factor of 4 -> 2 micrometer)
- "type": "scale",
- "scale": [1.0, 1.0, 2.0, 2.0, 2.0]
- }]
- }
- ],
- "coordinateTransformations": [{
- // the time unit (0.1 milliseconds), which is the same for each scale level
- "type": "scale",
- "scale": [0.1, 1.0, 1.0, 1.0, 1.0]
- }],
- "type": "gaussian",
- "metadata": {
- "description": "the fields in metadata depend on the downscaling implementation. Here, the parameters passed to the skimage function are given",
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": "[true]",
- "kwargs": {"multichannel": true}
- }
- }
- ]
-}
\ No newline at end of file
diff --git a/latest/examples/multiscales_strict/multiscales_transformations.json b/latest/examples/multiscales_strict/multiscales_transformations.json
deleted file mode 100644
index 80dbedab..00000000
--- a/latest/examples/multiscales_strict/multiscales_transformations.json
+++ /dev/null
@@ -1,47 +0,0 @@
-{
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "scale": [
- 10,
- 10
- ],
- "type": "scale"
- }
- ],
- "version": "0.5-dev",
- "name": "image_with_coordinateTransformations",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ]
-}
\ No newline at end of file
diff --git a/latest/examples/ome/series-2.json b/latest/examples/ome/series-2.json
deleted file mode 100644
index be7e9ed6..00000000
--- a/latest/examples/ome/series-2.json
+++ /dev/null
@@ -1,3 +0,0 @@
-{
- "series" : [ "0", "1" ]
-}
diff --git a/latest/examples/plate_strict/plate_2wells.json b/latest/examples/plate_strict/plate_2wells.json
deleted file mode 100644
index 3578798f..00000000
--- a/latest/examples/plate_strict/plate_2wells.json
+++ /dev/null
@@ -1,91 +0,0 @@
-{
- "plate": {
- "acquisitions": [
- {
- "id": 1,
- "maximumfieldcount": 1,
- "name": "single acquisition",
- "starttime": 1343731272000
- }
- ],
- "columns": [
- {
- "name": "1"
- },
- {
- "name": "2"
- },
- {
- "name": "3"
- },
- {
- "name": "4"
- },
- {
- "name": "5"
- },
- {
- "name": "6"
- },
- {
- "name": "7"
- },
- {
- "name": "8"
- },
- {
- "name": "9"
- },
- {
- "name": "10"
- },
- {
- "name": "11"
- },
- {
- "name": "12"
- }
- ],
- "field_count": 1,
- "name": "sparse test",
- "rows": [
- {
- "name": "A"
- },
- {
- "name": "B"
- },
- {
- "name": "C"
- },
- {
- "name": "D"
- },
- {
- "name": "E"
- },
- {
- "name": "F"
- },
- {
- "name": "G"
- },
- {
- "name": "H"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "C/5",
- "rowIndex": 2,
- "columnIndex": 4
- },
- {
- "path": "D/7",
- "rowIndex": 3,
- "columnIndex": 6
- }
- ]
- }
-}
diff --git a/latest/examples/plate_strict/plate_6wells.json b/latest/examples/plate_strict/plate_6wells.json
deleted file mode 100644
index d2f06389..00000000
--- a/latest/examples/plate_strict/plate_6wells.json
+++ /dev/null
@@ -1,72 +0,0 @@
-{
- "plate": {
- "acquisitions": [
- {
- "id": 1,
- "maximumfieldcount": 2,
- "name": "Meas_01(2012-07-31_10-41-12)",
- "starttime": 1343731272000
- },
- {
- "id": 2,
- "maximumfieldcount": 2,
- "name": "Meas_02(201207-31_11-56-41)",
- "starttime": 1343735801000
- }
- ],
- "columns": [
- {
- "name": "1"
- },
- {
- "name": "2"
- },
- {
- "name": "3"
- }
- ],
- "field_count": 4,
- "name": "test",
- "rows": [
- {
- "name": "A"
- },
- {
- "name": "B"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- },
- {
- "path": "A/2",
- "rowIndex": 0,
- "columnIndex": 1
- },
- {
- "path": "A/3",
- "rowIndex": 0,
- "columnIndex": 2
- },
- {
- "path": "B/1",
- "rowIndex": 1,
- "columnIndex": 0
- },
- {
- "path": "B/2",
- "rowIndex": 1,
- "columnIndex": 1
- },
- {
- "path": "B/3",
- "rowIndex": 1,
- "columnIndex": 2
- }
- ]
- }
-}
diff --git a/latest/examples/well_strict/well_2fields.json b/latest/examples/well_strict/well_2fields.json
deleted file mode 100644
index 6f43a911..00000000
--- a/latest/examples/well_strict/well_2fields.json
+++ /dev/null
@@ -1,15 +0,0 @@
-{
- "well": {
- "images": [
- {
- "acquisition": 0,
- "path": "0"
- },
- {
- "acquisition": 3,
- "path": "1"
- }
- ],
- "version": "0.5-dev"
- }
-}
\ No newline at end of file
diff --git a/latest/examples/well_strict/well_4fields.json b/latest/examples/well_strict/well_4fields.json
deleted file mode 100644
index c2285d53..00000000
--- a/latest/examples/well_strict/well_4fields.json
+++ /dev/null
@@ -1,23 +0,0 @@
-{
- "well": {
- "images": [
- {
- "acquisition": 1,
- "path": "0"
- },
- {
- "acquisition": 1,
- "path": "1"
- },
- {
- "acquisition": 2,
- "path": "2"
- },
- {
- "acquisition": 2,
- "path": "3"
- }
- ],
- "version": "0.5-dev"
- }
-}
\ No newline at end of file
diff --git a/latest/schemas/bf2raw.schema b/latest/schemas/bf2raw.schema
deleted file mode 100644
index 834aee24..00000000
--- a/latest/schemas/bf2raw.schema
+++ /dev/null
@@ -1,14 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/bf2raw.schema",
- "title": "NGFF container produced by bioformats2raw",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "bioformats2raw.layout": {
- "description": "The top-level identifier metadata added by bioformats2raw",
- "type": "number",
- "enum": [3]
- }
- }
-}
diff --git a/latest/schemas/ome.schema b/latest/schemas/ome.schema
deleted file mode 100644
index bd600a2a..00000000
--- a/latest/schemas/ome.schema
+++ /dev/null
@@ -1,17 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/ome.schema",
- "title": "NGFF group produced by bioformats2raw to contain OME metadata",
- "description": "JSON from OME-NGFF OME/.zattrs linked to an OME-XML file",
- "type": "object",
- "properties": {
- "series": {
- "description": "An array of the same length and the same order as the images defined in the OME-XML",
- "type": "array",
- "items": {
- "type": "string"
- },
- "minContains": 1
- }
- }
-}
diff --git a/latest/schemas/plate.schema b/latest/schemas/plate.schema
deleted file mode 100644
index 1b7ed532..00000000
--- a/latest/schemas/plate.schema
+++ /dev/null
@@ -1,140 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/plate.schema",
- "title": "OME-NGFF plate schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "plate": {
- "type": "object",
- "properties": {
- "acquisitions": {
- "description": "The acquisitions for this plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "id": {
- "description": "A unique identifier within the context of the plate",
- "type": "integer",
- "minimum": 0
- },
- "maximumfieldcount": {
- "description": "The maximum number of fields of view for the acquisition",
- "type": "integer",
- "exclusiveMinimum": 0
- },
- "name": {
- "description": "The name of the acquisition",
- "type": "string"
- },
- "description": {
- "description": "The description of the acquisition",
- "type": "string"
- },
- "starttime": {
- "description": "The start timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
- "type": "integer",
- "minimum": 0
- },
- "endtime": {
- "description": "The end timestamp of the acquisition, expressed as epoch time i.e. the number seconds since the Epoch",
- "type": "integer",
- "minimum": 0
- }
- },
- "required": [
- "id"
- ]
- }
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.5-dev"
- ]
- },
- "field_count": {
- "description": "The maximum number of fields per view across all wells",
- "type": "integer",
- "exclusiveMinimum": 0
- },
- "name": {
- "description": "The name of the plate",
- "type": "string"
- },
- "columns": {
- "description": "The columns of the plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "description": "The column name",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "rows": {
- "description": "The rows of the plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "name": {
- "description": "The row name",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "name"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "wells": {
- "description": "The wells of the plate",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "path": {
- "description": "The path to the well subgroup",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+/[A-Za-z0-9]+$"
- },
- "rowIndex": {
- "description": "The index of the well in the rows list",
- "type": "integer",
- "minimum": 0
- },
- "columnIndex": {
- "description": "The index of the well in the columns list",
- "type": "integer",
- "minimum": 0
- }
- },
- "required": [
- "path", "rowIndex", "columnIndex"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- }
- },
- "required": [
- "columns", "rows", "wells", "version"
- ]
- }
- }
-}
diff --git a/latest/schemas/strict_image.schema b/latest/schemas/strict_image.schema
deleted file mode 100644
index bcecc003..00000000
--- a/latest/schemas/strict_image.schema
+++ /dev/null
@@ -1,19 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/strict_image.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/latest/schemas/image.schema"
- },
- {
- "properties": {
- "multiscales": {
- "items": {
- "required": [
- "version", "metadata", "type", "name"
- ]
- }
- }
- }
- }
- ]
-}
\ No newline at end of file
diff --git a/latest/schemas/strict_label.schema b/latest/schemas/strict_label.schema
deleted file mode 100644
index c52dd51b..00000000
--- a/latest/schemas/strict_label.schema
+++ /dev/null
@@ -1,18 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/latest/schemas/strict_label.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/latest/schemas/label.schema"
- },
- {
- "properties": {
- "image-label": {
- "required": [
- "version",
- "colors"
- ]
- }
- }
- }
- ]
-}
diff --git a/latest/schemas/strict_plate.schema b/latest/schemas/strict_plate.schema
deleted file mode 100644
index 5a88ab72..00000000
--- a/latest/schemas/strict_plate.schema
+++ /dev/null
@@ -1,28 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/strict_plate.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.4/schemas/plate.schema"
- },
- {
- "properties": {
- "plate": {
- "properties": {
- "acquisitions": {
- "items": {
- "required": [
- "name",
- "maximumfieldcount"
- ]
- }
- }
- },
- "required": [
- "name",
- "version"
- ]
- }
- }
- }
- ]
-}
diff --git a/latest/schemas/strict_well.schema b/latest/schemas/strict_well.schema
deleted file mode 100644
index 1e200294..00000000
--- a/latest/schemas/strict_well.schema
+++ /dev/null
@@ -1,17 +0,0 @@
-{
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/strict_well.schema",
- "allOf": [
- {
- "$ref": "https://ngff.openmicroscopy.org/0.4/schemas/well.schema"
- },
- {
- "properties": {
- "well": {
- "required": [
- "version"
- ]
- }
- }
- }
- ]
-}
diff --git a/latest/schemas/well.schema b/latest/schemas/well.schema
deleted file mode 100644
index 891abd05..00000000
--- a/latest/schemas/well.schema
+++ /dev/null
@@ -1,47 +0,0 @@
-{
- "$schema": "https://json-schema.org/draft/2020-12/schema",
- "$id": "https://ngff.openmicroscopy.org/0.4/schemas/well.schema",
- "title": "OME-NGFF well schema",
- "description": "JSON from OME-NGFF .zattrs",
- "type": "object",
- "properties": {
- "well": {
- "type": "object",
- "properties": {
- "images": {
- "description": "The fields of view for this well",
- "type": "array",
- "items": {
- "type": "object",
- "properties": {
- "acquisition": {
- "description": "A unique identifier within the context of the plate",
- "type": "integer"
- },
- "path": {
- "description": "The path for this field of view subgroup",
- "type": "string",
- "pattern": "^[A-Za-z0-9]+$"
- }
- },
- "required": [
- "path"
- ]
- },
- "minItems": 1,
- "uniqueItems": true
- },
- "version": {
- "description": "The version of the specification",
- "type": "string",
- "enum": [
- "0.5-dev"
- ]
- }
- },
- "required": [
- "images"
- ]
- }
- }
-}
diff --git a/latest/tests/image_suite.json b/latest/tests/image_suite.json
deleted file mode 100644
index bec52214..00000000
--- a/latest/tests/image_suite.json
+++ /dev/null
@@ -1,1185 +0,0 @@
-{
- "description": "TBD",
- "schema": {
- "id": "schemas/image.schema"
- },
- "tests": [
- {
- "formerly": "valid/mismatch_axes_units.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 0.13,
- 0.13
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/untyped_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "angle"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "invalid/missing_version.json",
- "description": "TBD",
- "data": {
- "@type": "ngff:Image",
- "multiscales": [
- {
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1,
- 1
- ]
- }
- ]
- }
- ],
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ]
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "valid/invalid_axis_units.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micron"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 0.13,
- 0.13
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/missing_name.json",
- "description": "TBD",
- "data": {
- "@type": "ngff:Image",
- "multiscales": [
- {
- "version": "0.5-dev",
- "datasets": [
- {
- "path": "path/to/0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1,
- 1
- ]
- }
- ]
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ]
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid/custom_type_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "invalid/duplicate_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_space_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_transformation_type.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "translation"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_scale.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "translation": [
- 1,
- 1
- ],
- "type": "translation"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/too_many_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "angle",
- "type": "custom"
- },
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1,
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_channels_color.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": 255,
- "family": "linear",
- "label": "1234",
- "window": {
- "end": 1765.0,
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_axes_name.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "type": "space",
- "unit": "micron"
- },
- {
- "type": "space",
- "unit": "micron"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 0.13,
- 0.13
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_axes_count.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/one_space_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_path.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": 0,
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_multiscales_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "scale": [
- "invalid"
- ],
- "type": "scale"
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0"
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/no_datasets.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_datasets.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_version.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.3"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_version.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ]
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_axis_type.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "invalid",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/duplicate_scale.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- },
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/no_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/too_many_space_axes.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "X",
- "type": "space"
- },
- {
- "name": "z",
- "type": "space"
- },
- {
- "name": "y",
- "type": "space"
- },
- {
- "name": "x",
- "type": "space"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/no_multiscales.json",
- "description": "TBD",
- "data": {
- "@type": "ngff:Image",
- "multiscales": []
- },
- "valid": false
- },
- {
- "formerly": "invalid/invalid_channels_window.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": "ff0000",
- "family": "linear",
- "label": "1234",
- "window": {
- "end": "100",
- "max": 2555.0,
- "min": 5.0,
- "start": 0.0
- }
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "invalid/empty_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": []
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- },
- {
- "formerly": "invalid/missing_path.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev"
- }
- ]
- },
- "valid": false
- }
- ]
-}
diff --git a/latest/tests/label_suite.json b/latest/tests/label_suite.json
deleted file mode 100644
index 634225c8..00000000
--- a/latest/tests/label_suite.json
+++ /dev/null
@@ -1,131 +0,0 @@
-{
- "description": "Tests for the image-label JSON schema",
- "schema": {
- "id": "schemas/label.schema"
- },
- "tests": [
- {
- "formerly": "image-label/minimal",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "image-label/minimal_properties",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ],
- "properties": [
- {
- "label-value": 1
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "image-label/empty_colors",
- "data": {
- "image-label": {
- "colors": []
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/empty_properties",
- "data": {
- "image-label": {
- "properties": []
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_no_label_value",
- "data": {
- "image-label": {
- "colors": [
- {
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/properties_no_label_value",
- "data": {
- "image-label": {
- "properties": [
- {
- "value": "foo"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_rgba_length",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_rgba_type",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 500]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/colors_duplicate",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- },
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/latest/tests/plate_suite.json b/latest/tests/plate_suite.json
deleted file mode 100644
index 876d7c45..00000000
--- a/latest/tests/plate_suite.json
+++ /dev/null
@@ -1,874 +0,0 @@
-{
- "description": "Tests for the plate JSON schema",
- "schema": {
- "id": "schemas/plate.schema"
- },
- "tests": [
- {
- "formerly": "plate/minimal_no_acquisitions",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/minimal_acquisitions",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_rows",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/empty_rows",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/duplicate_rows",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- },
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_columns",
- "data": {
- "version": "0.5-dev",
- "plate": {
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/empty_columns",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/duplicate_columns",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- },
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_wells",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/empty_wells",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": {}
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/duplicate_rows",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- },
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- },
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_column_name",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "concentration": 10
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_row_name",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "concentration": 10
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_well_path",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_well_rowIndex",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_well_columnIndex",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/well_1group",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A1",
- "rowIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/well_3groups",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "plate/A/1",
- "rowIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/invalid_version",
- "data": {
- "plate": {
- "version": "foo",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_version",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_alphanumeric_column",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A-1"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A-1/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_alphanumeric_row",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "A1"
- }
- ],
- "wells": [
- {
- "path": "A/A1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_acquisition_id",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "maximumfieldcount": 1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_integer_acquisition_id",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": "0"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/negative_acquisition_id",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": -1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/non_integer_acquisition_maximumfieldcount",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0,
- "maximumfieldcount": "0"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_zero_maximumfieldcount",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0,
- "maximumfieldcount": 0
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_noninteger_starttime",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0,
- "starttime": "2022-05-13T13:48:06+00:00"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_negative_starttime",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0,
- "starttime": -1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/acquisition_noninteger_endtime",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0,
- "endtime": "2022-05-13T13:48:06+00:00"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/negative_endtime",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "acquisitions": [
- {
- "id": 0,
- "endtime": -1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/zero_field_count",
- "data": {
- "plate": {
- "version": "0.5-dev",
- "columns": [
- {
- "name": "A"
- }
- ],
- "field_count": 0,
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/latest/tests/strict_image_suite.json b/latest/tests/strict_image_suite.json
deleted file mode 100644
index 7bff90ca..00000000
--- a/latest/tests/strict_image_suite.json
+++ /dev/null
@@ -1,388 +0,0 @@
-{
- "description": "TBD",
- "schema": {
- "id": "schemas/strict_image.schema"
- },
- "tests": [
- {
- "formerly": "valid_strict/multiscales_example.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "version": "0.5-dev",
- "name": "example",
- "axes": [
- {
- "name": "t",
- "type": "time",
- "unit": "millisecond"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1.0,
- 1.0,
- 0.5,
- 0.5,
- 0.5
- ]
- }
- ]
- },
- {
- "path": "1",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1.0,
- 1.0,
- 1.0,
- 1.0,
- 1.0
- ]
- }
- ]
- },
- {
- "path": "2",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1.0,
- 1.0,
- 2.0,
- 2.0,
- 2.0
- ]
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 0.1,
- 1.0,
- 1.0,
- 1.0,
- 1.0
- ]
- }
- ],
- "type": "gaussian",
- "metadata": {
- "description": "the fields in metadata depend on the downscaling implementation. Here, the parameters passed to the skimage function are given",
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": "[true]",
- "kwargs": {
- "multichannel": true
- }
- }
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/multiscales_transformations.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "coordinateTransformations": [
- {
- "scale": [
- 10,
- 10
- ],
- "type": "scale"
- }
- ],
- "version": "0.5-dev",
- "name": "image_with_coordinateTransformations",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/image_metadata.json",
- "description": "TBD",
- "data": {
- "@id": "top",
- "@type": "ngff:Image",
- "multiscales": [
- {
- "@id": "inner",
- "version": "0.5-dev",
- "name": "example",
- "datasets": [
- {
- "path": "path/to/0",
- "coordinateTransformations": [
- {
- "type": "scale",
- "scale": [
- 1,
- 1
- ]
- }
- ]
- }
- ],
- "type": "gaussian",
- "metadata": {
- "method": "skimage.transform.pyramid_gaussian",
- "version": "0.16.1",
- "args": [
- "true",
- "false"
- ],
- "kwargs": {
- "multichannel": true
- }
- },
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ]
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/image.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev",
- "name": "simple_image",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ]
- },
- "valid": true
- },
- {
- "formerly": "valid_strict/image_omero.json",
- "description": "TBD",
- "data": {
- "multiscales": [
- {
- "axes": [
- {
- "name": "t",
- "type": "time"
- },
- {
- "name": "c",
- "type": "channel"
- },
- {
- "name": "z",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "y",
- "type": "space",
- "unit": "micrometer"
- },
- {
- "name": "x",
- "type": "space",
- "unit": "micrometer"
- }
- ],
- "datasets": [
- {
- "path": "0",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 0.5,
- 0.13,
- 0.13
- ],
- "type": "scale"
- },
- {
- "translation": [
- 0,
- 9,
- 0.5,
- 25.74,
- 21.58
- ],
- "type": "translation"
- }
- ]
- },
- {
- "path": "1",
- "coordinateTransformations": [
- {
- "scale": [
- 1,
- 1,
- 1,
- 0.26,
- 0.26
- ],
- "type": "scale"
- }
- ]
- }
- ],
- "version": "0.5-dev",
- "name": "image_with_omero_metadata",
- "type": "foo",
- "metadata": {
- "key": "value"
- }
- }
- ],
- "omero": {
- "channels": [
- {
- "active": true,
- "coefficient": 1.0,
- "color": "00FF00",
- "family": "linear",
- "inverted": false,
- "label": "FITC",
- "window": {
- "end": 813.0,
- "max": 870.0,
- "min": 102.0,
- "start": 82.0
- }
- },
- {
- "active": true,
- "coefficient": 1.0,
- "color": "FF0000",
- "family": "linear",
- "inverted": false,
- "label": "RD-TR-PE",
- "window": {
- "end": 815.0,
- "max": 441.0,
- "min": 129.0,
- "start": 78.0
- }
- }
- ],
- "id": 1,
- "rdefs": {
- "defaultT": 0,
- "defaultZ": 2,
- "model": "color"
- },
- "version": "0.5-dev"
- }
- },
- "valid": true
- }
- ]
-}
diff --git a/latest/tests/strict_label_suite.json b/latest/tests/strict_label_suite.json
deleted file mode 100644
index 9fe0cda2..00000000
--- a/latest/tests/strict_label_suite.json
+++ /dev/null
@@ -1,31 +0,0 @@
-{
- "description": "Tests for the strict image-label JSON schema",
- "schema": {
- "id": "schemas/strict_label.schema"
- },
- "tests": [
- {
- "formerly": "image-label/no_version",
- "data": {
- "image-label": {
- "colors": [
- {
- "label-value": 1,
- "rgba": [0, 0, 0, 0]
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "image-label/no_colors",
- "data": {
- "image-label": {
- "version": "0.5-dev"
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/latest/tests/strict_plate_suite.json b/latest/tests/strict_plate_suite.json
deleted file mode 100644
index 931cf7e7..00000000
--- a/latest/tests/strict_plate_suite.json
+++ /dev/null
@@ -1,187 +0,0 @@
-{
- "description": "Tests for the strict plate JSON schema",
- "schema": {
- "id": "schemas/strict_plate.schema"
- },
- "tests": [
- {
- "formerly": "plate/strict_no_acquisitions",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_name",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_version",
- "data": {
- "plate": {
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/strict_acquisitions",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "name": "0",
- "maximumfieldcount": 1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_acquisition_name",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "maximumfieldcount": 1
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/missing_acquisition_maximumfieldcount",
- "data": {
- "plate": {
- "acquisitions": [
- {
- "id": 0,
- "name": "0"
- }
- ],
- "columns": [
- {
- "name": "A"
- }
- ],
- "name": "test plate",
- "rows": [
- {
- "name": "1"
- }
- ],
- "version": "0.5-dev",
- "wells": [
- {
- "path": "A/1",
- "rowIndex": 0,
- "columnIndex": 0
- }
- ]
- }
- },
- "valid": false
- }
- ]
-}
diff --git a/latest/tests/strict_well_suite.json b/latest/tests/strict_well_suite.json
deleted file mode 100644
index edfb2936..00000000
--- a/latest/tests/strict_well_suite.json
+++ /dev/null
@@ -1,50 +0,0 @@
-{
- "description": "Tests for the strict well JSON schema",
- "schema": {
- "id": "schemas/strict_well.schema"
- },
- "tests": [
- {
- "formerly": "well/strict_no_acquisitions",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ],
- "version": "0.5-dev"
- }
- },
- "valid": true
- },
- {
- "formerly": "plate/missing_version",
- "data": {
- "well": {
- "images": [
- {
- "path": "0"
- }
- ]
- }
- },
- "valid": false
- },
- {
- "formerly": "plate/strict_acquisitions",
- "data": {
- "well": {
- "images": [
- {
- "acquisition": 0,
- "path": "0"
- }
- ],
- "version": "0.5-dev"
- }
- },
- "valid": true
- }
- ]
-}
diff --git a/requirements.txt b/requirements.txt
index 9cc69f43..9ffd7c09 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,3 +1,4 @@
bikeshed<4.2
myst-parser
sphinx-book-theme
+testresources
\ No newline at end of file
diff --git a/specifications/index.md b/specifications/index.md
index a40e0708..b33772b6 100644
--- a/specifications/index.md
+++ b/specifications/index.md
@@ -1,9 +1,7 @@
Specifications
==============
-The current released version of the OME-NGFF specification is 0.4.
-
-The latest version is still undergoing changes and is not yet released.
+The current released version of the OME-Zarr specification is 0.5.
Version history:
@@ -12,5 +10,6 @@ Version history:
0.2 March 2021
0.3 August 2021
0.4 February 2022
+ 0.5 November 2024
latest
diff --git a/tools/index.md b/tools/index.md
index 1a4a62ec..59eed830 100644
--- a/tools/index.md
+++ b/tools/index.md
@@ -1,6 +1,6 @@
# Tools
-A list of tools and libraries with OME-Zarr support. These are developed by various members of the OME-NGFF community. If you think your tool/library should be listed here, please [open a pull request](https://github.com/ome/ngff).
+A list of tools and libraries with OME-Zarr support. These are developed by various members of the NGFF community. If you think your tool/library should be listed here, please [open a pull request](https://github.com/ome/ngff).
In addition to this collection, an evaluation of selected tools/libraries is available on .
 +
+
+
+  +
+
+
+  -
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+See [Tools](https://ngff.openmicroscopy.org/tools/index.html).
Citing {#citing}
================
[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.4)
J. Moore, *et al*. Open Microscopy Environment Consortium, 8 February 2022.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.4/](https://ngff.openmicroscopy.org/0.4/]).
+This edition of the specification is [https://ngff.openmicroscopy.org/0.5/](https://ngff.openmicroscopy.org/0.5/]).
The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
@@ -623,6 +590,11 @@ Version History {#history}
-
-All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
-version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
-Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
-
-Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+See [Tools](https://ngff.openmicroscopy.org/tools/index.html).
Citing {#citing}
================
[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.4)
J. Moore, *et al*. Open Microscopy Environment Consortium, 8 February 2022.
-This edition of the specification is [https://ngff.openmicroscopy.org/0.4/](https://ngff.openmicroscopy.org/0.4/]).
+This edition of the specification is [https://ngff.openmicroscopy.org/0.5/](https://ngff.openmicroscopy.org/0.5/]).
The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
@@ -623,6 +590,11 @@ Version History {#history}