diff --git a/0.2/index.bs b/0.2/index.bs
new file mode 100644
index 00000000..6810c1d0
--- /dev/null
+++ b/0.2/index.bs
@@ -0,0 +1,650 @@
+
+Title: Next-generation file formats (NGFF)
+Shortname: ome-ngff
+Level: 1
+Status: w3c/CG-FINAL
+Group: ome
+URL: https://ngff.openmicroscopy.org/0.2/
+Repository: https://github.com/ome/ngff
+Issue Tracking: Forums https://forum.image.sc/tag/ome-ngff
+Logo: http://www.openmicroscopy.org/img/logos/ome-logomark.svg
+Local Boilerplate: header yes
+Local Boilerplate: copyright yes
+Boilerplate: style-darkmode off
+Markup Shorthands: markdown yes
+Editor: Josh Moore, Open Microscopy Environment (OME) https://www.openmicroscopy.org
+Editor: Sébastien Besson, Open Microscopy Environment (OME) https://www.openmicroscopy.org
+Abstract: This document contains next-generation file format (NGFF)
+Abstract: specifications for storing bioimaging data in the cloud.
+Abstract: All specifications are submitted to the https://image.sc community for review.
+Status Text: This is the 0.2 release of this specification. Migration scripts
+Status Text: will be provided between numbered versions. Data written with the latest version
+Status Text: (an "editor's draft") will not necessarily be supported.
+
+
+
+Introduction {#intro}
+=====================
+
+Bioimaging science is at a crossroads. Currently, the drive to acquire more,
+larger, preciser spatial measurements is unfortunately at odds with our ability
+to structure and share those measurements with others. During a global pandemic
+more than ever, we believe fervently that global, collaborative discovery as
+opposed to the post-publication, "data-on-request" mode of operation is the
+path forward. Bioimaging data should be shareable via open and commercial cloud
+resources without the need to download entire datasets.
+
+At the moment, that is not the norm. The plethora of data formats produced by
+imaging systems are ill-suited to remote sharing. Individual scientists
+typically lack the infrastructure they need to host these data themselves. When
+they acquire images from elsewhere, time-consuming translations and data
+cleaning are needed to interpret findings. Those same costs are multiplied when
+gathering data into online repositories where curator time can be the limiting
+factor before publication is possible. Without a common effort, each lab or
+resource is left building the tools they need and maintaining that
+infrastructure often without dedicated funding.
+
+This document defines a specification for bioimaging data to make it possible
+to enable the conversion of proprietary formats into a common, cloud-ready one.
+Such next-generation file formats layout data so that individual portions, or
+"chunks", of large data are reference-able eliminating the need to download
+entire datasets.
+
+
+Why "NGFF"? {#why-ngff}
+-------------------------------------------------------------------------------------------------
+
+A short description of what is needed for an imaging format is "a hierarchy
+of n-dimensional (dense) arrays with metadata". This combination of features
+is certainly provided by HDF5
+from the HDF Group, which a number of
+bioimaging formats do use. HDF5 and other larger binary structures, however,
+are ill-suited for storage in the cloud where accessing individual chunks
+of data by name rather than seeking through a large file is at the heart of
+parallelization.
+
+As a result, a number of formats have been developed more recently which provide
+the basic data structure of an HDF5 file, but do so in a more cloud-friendly way.
+In the [PyData](https://pydata.org/) community, the Zarr [[zarr]] format was developed
+for easily storing collections of [NumPy](https://numpy.org/) arrays. In the
+[ImageJ](https://imagej.net/) community, N5 [[n5]] was developed to work around
+the limitations of HDF5 ("N5" was originally short for "Not-HDF5").
+Both of these formats permit storing individual chunks of data either locally in
+separate files or in cloud-based object stores as separate keys.
+
+A [current effort](https://zarr-specs.readthedocs.io/en/core-protocol-v3.0-dev/protocol/core/v3.0.html)
+is underway to unify the two similar specifications to provide a single binary
+specification. The editor's draft will soon be entering a [request for comments (RFC)](https://github.com/zarr-developers/zarr-specs/issues/101) phase with the goal of having a first version early in 2021. As that
+process comes to an end, this document will be updated.
+
+OME-NGFF {#ome-ngff}
+--------------------
+
+The conventions and specifications defined in this document are designed to
+enable next-generation file formats to represent the same bioimaging data
+that can be represented in \[OME-TIFF](http://www.openmicroscopy.org/ome-files/)
+and beyond. However, the conventions will also be usable by HDF5 and other sufficiently advanced
+binary containers. Eventually, we hope, the moniker "next-generation" will no longer be
+applicable, and this will simply be the most efficient, common, and useful representation
+of bioimaging data, whether during acquisition or sharing in the cloud.
+
+Note: The following text makes use of OME-Zarr [[ome-zarr-py]], the current prototype implementation,
+for all examples.
+
+On-disk (or in-cloud) layout {#on-disk}
+=======================================
+
+An overview of the layout of an OME-Zarr fileset should make
+understanding the following metadata sections easier. The hierarchy
+is represented here as it would appear locally but could equally
+be stored on a web server to be accessed via HTTP or in object storage
+like S3 or GCS.
+
+Images {#image-layout}
+----------------------
+
+The following layout describes the expected Zarr hierarchy for images with
+multiple levels of resolutions and optionally associated labels.
+
+```
+. # Root folder, potentially in S3,
+│ # with a flat list of images by image ID.
+│
+├── 123.zarr # One image (id=123) converted to Zarr.
+│
+└── 456.zarr # Another image (id=456) converted to Zarr.
+ │
+ ├── .zgroup # Each image is a Zarr group, or a folder, of other groups and arrays.
+ ├── .zattrs # Group level attributes are stored in the .zattrs file and include
+ │ # "multiscales" and "omero" below)
+ │
+ ├── 0 # Each multiscale level is stored as a separate Zarr array,
+ │ ... # which is a folder containing chunk files which compose the array.
+ ├── n # The name of the array is arbitrary with the ordering defined by
+ │ │ # by the "multiscales" metadata, but is often a sequence starting at 0.
+ │ │
+ │ ├── .zarray # All image arrays are 5-dimensional
+ │ │ # with dimension order (t, c, z, y, x).
+ │ │
+ │ └─ t # Chunks are stored with the nested directory layout.
+ │ └─ c # All but the last chunk element are stored as directories.
+ │ └─ z # The terminal chunk is a file. Together the directory and file names
+ │ └─ y # provide the "chunk coordinate" (t, c, z, y, x), where the maximum coordinate
+ │ └─ x # will be `dimension_size / chunk_size`.
+ │
+ └── labels
+ │
+ ├── .zgroup # The labels group is a container which holds a list of labels to make the objects easily discoverable
+ │
+ ├── .zattrs # All labels will be listed in `.zattrs` e.g. `{ "labels": [ "original/0" ] }`
+ │ # Each dimension of the label `(t, c, z, y, x)` should be either the same as the
+ │ # corresponding dimension of the image, or `1` if that dimension of the label
+ │ # is irrelevant.
+ │
+ └── original # Intermediate folders are permitted but not necessary and currently contain no extra metadata.
+ │
+ └── 0 # Multiscale, labeled image. The name is unimportant but is registered in the "labels" group above.
+ ├── .zgroup # Zarr Group which is both a multiscaled image as well as a labeled image.
+ ├── .zattrs # Metadata of the related image and as well as display information under the "image-label" key.
+ │
+ ├── 0 # Each multiscale level is stored as a separate Zarr array, as above, but only integer values
+ │ ... # are supported.
+ └── n
+```
+
+High-content screening {#hcs-layout}
+------------------------------------
+
+The following specification defines the hierarchy for a high-content screening
+dataset. Three groups must be defined above the images:
+
+- the group above the images defines the well and MUST implement the
+ [well specification](#well-md). All images contained in a well are fields
+ of view of the same well
+- the group above the well defines a row of wells
+- the group above the well row defines an entire plate i.e. a two-dimensional
+ collection of wells organized in rows and columns. It MUST implement the
+ [plate specification](#plate-md)
+
+
+```
+. # Root folder, potentially in S3,
+│
+└── 5966.zarr # One plate (id=5966) converted to Zarr
+ ├── .zgroup
+ ├── .zattrs # Implements "plate" specification
+ ├── A # First row of the plate
+ │ ├── .zgroup
+ │ │
+ │ ├── 1 # First column of row A
+ │ │ ├── .zgroup
+ │ │ ├── .zattrs # Implements "well" specification
+ │ │ │
+ │ │ ├── 0 # First field of view of well A1
+ │ │ │ │
+ │ │ │ ├── .zgroup
+ │ │ │ ├── .zattrs # Implements "multiscales", "omero"
+ │ │ │ ├── 0
+ │ │ │ │ ... # Resolution levels
+ │ │ │ ├── n
+ │ │ │ └── labels # Labels (optional)
+ │ │ ├── ... # Fields of view
+ │ │ └── m
+ │ ├── ... # Columns
+ │ └── 12
+ ├── ... # Rows
+ └── H
+```
+
+Metadata {#metadata}
+====================
+
+The various `.zattrs` files throughout the above array hierarchy may contain metadata
+keys as specified below for discovering certain types of data, especially images.
+
+"multiscales" metadata {#multiscale-md}
+---------------------------------------
+
+Metadata about the multiple resolution representations of the image can be
+found under the "multiscales" key in the group-level metadata.
+The specification for the multiscale (i.e. "resolution") metadata is provided
+in [zarr-specs#50](https://github.com/zarr-developers/zarr-specs/issues/50).
+If only one multiscale is provided, use it. Otherwise, the user can choose by
+name, using the first multiscale as a fallback:
+
+```python
+datasets = []
+for named in multiscales:
+ if named["name"] == "3D":
+ datasets = [x["path"] for x in named["datasets"]]
+ break
+if not datasets:
+ # Use the first by default. Or perhaps choose based on chunk size.
+ datasets = [x["path"] for x in multiscales[0]["datasets"]]
+```
+
+The subresolutions in each multiscale are ordered from highest-resolution
+to lowest.
+
+"omero" metadata {#omero-md}
+----------------------------
+
+Information specific to the channels of an image and how to render it
+can be found under the "omero" key in the group-level metadata:
+
+```json
+"id": 1, # ID in OMERO
+"name": "example.tif", # Name as shown in the UI
+"version": "0.2", # Current version
+"channels": [ # Array matching the c dimension size
+ {
+ "active": true,
+ "coefficient": 1,
+ "color": "0000FF",
+ "family": "linear",
+ "inverted": false,
+ "label": "LaminB1",
+ "window": {
+ "end": 1500,
+ "max": 65535,
+ "min": 0,
+ "start": 0
+ }
+ }
+],
+"rdefs": {
+ "defaultT": 0, # First timepoint to show the user
+ "defaultZ": 118, # First Z section to show the user
+ "model": "color" # "color" or "greyscale"
+}
+```
+
+See https://docs.openmicroscopy.org/omero/5.6.1/developers/Web/WebGateway.html#imgdata
+for more information.
+
+"labels" metadata {#labels-md}
+------------------------------
+
+The special group "labels" found under an image Zarr contains the key `labels` containing
+the paths to label objects which can be found underneath the group:
+
+```json
+{
+ "labels": [
+ "orphaned/0"
+ ]
+}
+```
+
+Unlisted groups MAY be labels.
+
+"image-label" metadata {#label-md}
+----------------------------------
+
+Groups containing the `image-label` dictionary represent an image segmentation
+in which each unique pixel value represents a separate segmented object.
+`image-label` groups MUST also contain `multiscales` metadata and the two
+"datasets" series MUST have the same number of entries.
+
+The `colors` key defines a list of JSON objects describing the unique label
+values. Each entry in the list MUST contain the key "label-value" with the
+pixel value for that label. Additionally, the "rgba" key MAY be present, the
+value for which is an RGBA unsigned-int 4-tuple: `[uint8, uint8, uint8, uint8]`
+All `label-value`s must be unique. Clients who choose to not throw an error
+should ignore all except the _last_ entry.
+
+Some implementations may represent overlapping labels by using a specially assigned
+value, for example the highest integer available in the pixel range.
+
+The `properties` key defines a list of JSON objects which also describes the unique
+label values. Each entry in the list MUST contain the key "label-value" with the
+pixel value for that label. Additionally, an arbitrary number of key-value pairs
+MAY be present for each label value denoting associated metadata. Not all label
+values must share the same key-value pairs within the properties list.
+
+The `source` key is an optional dictionary which contains information on the
+image the label is associated with. If included it MAY include a key `image`
+whose value is the relative path to a Zarr image group. The default value is
+"../../" since most labels are stored under a subgroup named "labels/" (see
+above).
+
+
+```json
+"image-label":
+ {
+ "version": "0.2",
+ "colors": [
+ {
+ "label-value": 1,
+ "rgba": [255, 255, 255, 0]
+ },
+ {
+ "label-value": 4,
+ "rgba": [0, 255, 255, 128]
+ },
+ ...
+ ],
+ "properties": [
+ {
+ "label-value": 1,
+ "area (pixels)": 1200,
+ "class": "foo"
+
+ },
+ {
+ "label-value": 4,
+ "area (pixels)": 1650
+ },
+ ...
+ ]
+ },
+ "source": {
+ "image": "../../"
+ }
+]
+```
+
+"plate" metadata {#plate-md}
+----------------------------
+
+For high-content screening datasets, the plate layout can be found under the
+custom attributes of the plate group under the `plate` key.
+
+
+ - acquisitions
+ - An optional list of JSON objects defining the acquisitions for a given
+ plate. Each acquisition object MUST contain an `id` key providing an
+ unique identifier within the context of the plate to which fields of
+ view can refer to. It SHOULD contain a `name` key identifying the name
+ of the acquisition. It SHOULD contain a `maximumfieldcount` key
+ indicating the maximum number of fields of view for the acquisition. It
+ MAY contain a `description` key providing a description for the
+ acquisition. It MAY contain a `startime` and/or `endtime` key specifying
+ the start and/or end timestamp of the acquisition using an epoch
+ string.
+ - columns
+ - A list of JSON objects defining the columns of the plate. Each column
+ object defines the properties of the column at the index of the object
+ in the list. If not empty, it MUST contain a `name` key specifying the
+ column name.
+ - field_count
+ - An integer defining the maximum number of fields per view across all
+ wells.
+ - name
+ - A string defining the name of the plate.
+ - rows
+ - A list of JSON objects defining the rows of the plate. Each row object
+ defines the properties of the row at the index of the object in the
+ list. If not empty, it MUST contain a `name` key specifying the row
+ name.
+ - version
+ - A string defining the version of the specification.
+ - wells
+ - A list of JSON objects defining the wells of the plate. Each well object
+ MUST contain a `path` key identifying the path to the well subgroup.
+
+
+For example the following JSON object defines a plate with two acquisition and
+6 wells (2 rows and 3 columns), containing up 2 fields of view per acquistion.
+
+```json
+"plate": {
+ "acquisitions": [
+ {
+ "id": 1,
+ "maximumfieldcount": 2,
+ "name": "Meas_01(2012-07-31_10-41-12)",
+ "starttime": 1343731272000
+ },
+ {
+ "id": 2,
+ "maximumfieldcount": 2,
+ "name": "Meas_02(201207-31_11-56-41)",
+ "starttime": 1343735801000
+ }
+ ],
+ "columns": [
+ {
+ "name": "1"
+ },
+ {
+ "name": "2"
+ },
+ {
+ "name": "3"
+ }
+ ],
+ "field_count": 4,
+ "name": "test",
+ "rows": [
+ {
+ "name": "A"
+ },
+ {
+ "name": "B"
+ }
+ ],
+ "version": "0.2",
+ "wells": [
+ {
+ "path": "2020-10-10/A/1"
+ },
+ {
+ "path": "2020-10-10/A/2"
+ },
+ {
+ "path": "2020-10-10/A/3"
+ },
+ {
+ "path": "2020-10-10/B/1"
+ },
+ {
+ "path": "2020-10-10/B/2"
+ },
+ {
+ "path": "2020-10-10/B/3"
+ }
+ ]
+ }
+```
+
+"well" metadata {#well-md}
+--------------------------
+
+For high-content screening datasets, the metadata about all fields of views
+under a given well can be found under the "well" key in the attributes of the
+well group.
+
+
+ - images
+ - A list of JSON objects defining the fields of views for a given well.
+ Each object MUST contain a `path` key identifying the path to the
+ field of view. If multiple acquisitions were performed in the plate, it
+ SHOULD contain an `acquisition` key identifying the id of the
+ acquisition which must match one of acquisition JSON objects defined in
+ the plate metadata.
+ - version
+ - A string defining the version of the specification.
+
+
+For example the following JSON object defines a well with four fields of
+views. The first two fields of view were part of the first acquisition while
+the last two fields of view were part of the second acquisition.
+
+```json
+"well": {
+ "images": [
+ {
+ "acquisition": 1,
+ "path": "0"
+ },
+ {
+ "acquisition": 1,
+ "path": "1"
+ },
+ {
+ "acquisition": 2,
+ "path": "2"
+ },
+ {
+ "acquisition": 2,
+ "path": "3"
+ }
+ ],
+ "version": "0.2"
+ }
+```
+
+Implementations {#implementations}
+==================================
+
+Projects which support reading and/or writing OME-NGFF data include:
+
+
+
+ - [omero-ms-zarr](https://github.com/ome/omero-ms-zarr)
+ - A microservice for OMERO.server that converts images stored in OMERO to OME Zarr files on the fly, served via a web API.
+
+ - [idr-zarr-tools](https://github.com/IDR/idr-zarr-tools)
+ - A full workflow demonstrating the conversion of IDR images to OME Zarr images on S3.
+
+ - [OMERO CLI Zarr plugin](https://github.com/ome/omero-cli-zarr)
+ - An OMERO CLI plugin that converts images stored in OMERO.server into a local Zarr file.
+
+ - [ome-zarr-py](https://github.com/ome/ome-zarr-py)
+ - A napari plugin for reading ome-zarr files.
+
+ - [bioformats2raw](https://github.com/glencoesoftware/bioformats2raw)
+ - A performant, Bio-Formats image file format converter.
+
+ - [vizarr](https://github.com/hms-dbmi/vizarr/)
+ - A minimal, purely client-side program for viewing Zarr-based images with Viv & ImJoy.
+
+
+
+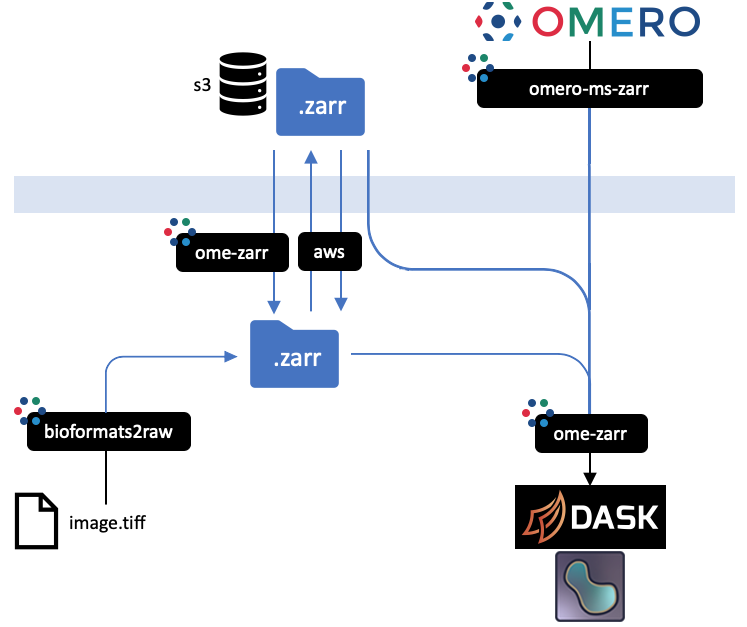 +
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
+Citing {#citing}
+================
+
+[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.2)
+J. Moore, *et al*. Editors. Open Microscopy Environment Consortium, 20 November 2020.
+This edition of the specification is [https://ngff.openmicroscopy.org/0.2/](https://ngff.openmicroscopy.org/0.2/]).
+The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
+[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
+
+Version History {#history}
+==========================
+
+
+
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
+Citing {#citing}
+================
+
+[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.2)
+J. Moore, *et al*. Editors. Open Microscopy Environment Consortium, 20 November 2020.
+This edition of the specification is [https://ngff.openmicroscopy.org/0.2/](https://ngff.openmicroscopy.org/0.2/]).
+The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
+[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
+
+Version History {#history}
+==========================
+
+
+
+
+ | Revision |
+ Date |
+ Description |
+
+
+
+ | 0.2.0 |
+ 2021-03-29 |
+ Change chunk dimension separator to "/" |
+
+
+ | 0.1.4 |
+ 2020-11-26 |
+ Add HCS specification |
+
+
+ | 0.1.3 |
+ 2020-09-14 |
+ Add labels specification |
+
+
+ | 0.1.2 |
+ 2020-05-07 |
+ Add description of "omero" metadata |
+
+
+ | 0.1.1 |
+ 2020-05-06 |
+ Add info on the ordering of resolutions |
+
+
+ | 0.1.0 |
+ 2020-04-20 |
+ First version for internal demo |
+
+
+
+
+
+{
+ "blogNov2020": {
+ "href": "https://blog.openmicroscopy.org/file-formats/community/2020/11/04/zarr-data/",
+ "title": "Public OME-Zarr data (Nov. 2020)",
+ "authors": [
+ "OME Team"
+ ],
+ "status": "Informational",
+ "publisher": "OME",
+ "id": "blogNov2020",
+ "date": "04 November 2020"
+ },
+ "imagesc26952": {
+ "href": "https://forum.image.sc/t/ome-s-position-regarding-file-formats/26952",
+ "title": "OME’s position regarding file formats",
+ "authors": [
+ "OME Team"
+ ],
+ "status": "Informational",
+ "publisher": "OME",
+ "id": "imagesc26952",
+ "date": "19 June 2020"
+ },
+ "n5": {
+ "id": "n5",
+ "href": "https://github.com/saalfeldlab/n5/issues/62",
+ "title": "N5---a scalable Java API for hierarchies of chunked n-dimensional tensors and structured meta-data",
+ "status": "Informational",
+ "authors": [
+ "John A. Bogovic",
+ "Igor Pisarev",
+ "Philipp Hanslovsky",
+ "Neil Thistlethwaite",
+ "Stephan Saalfeld"
+ ],
+ "date": "2020"
+ },
+ "ome-zarr-py": {
+ "id": "ome-zarr-py",
+ "href": "https://doi.org/10.5281/zenodo.4113931",
+ "title": "ome-zarr-py: Experimental implementation of next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.",
+ "status": "Informational",
+ "publisher": "Zenodo",
+ "authors": [
+ "OME",
+ "et al"
+ ],
+ "date": "06 October 2020"
+ },
+ "zarr": {
+ "id": "zarr",
+ "href": "https://doi.org/10.5281/zenodo.4069231",
+ "title": "Zarr: An implementation of chunked, compressed, N-dimensional arrays for Python.",
+ "status": "Informational",
+ "publisher": "Zenodo",
+ "authors": [
+ "Alistair Miles",
+ "et al"
+ ],
+ "date": "06 October 2020"
+ }
+}
+
 +
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
+Citing {#citing}
+================
+
+[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.2)
+J. Moore, *et al*. Editors. Open Microscopy Environment Consortium, 20 November 2020.
+This edition of the specification is [https://ngff.openmicroscopy.org/0.2/](https://ngff.openmicroscopy.org/0.2/]).
+The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
+[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
+
+Version History {#history}
+==========================
+
+
+
+All implementations prevent an equivalent representation of a dataset which can be downloaded or uploaded freely. An interactive
+version of this diagram is available from the [OME2020 Workshop](https://downloads.openmicroscopy.org/presentations/2020/Dundee/Workshops/NGFF/zarr_diagram/).
+Mouseover the blackboxes representing the implementations above to get a quick tip on how to use them.
+
+Note: If you would like to see your project listed, please open an issue or PR on the [ome/ngff](https://github.com/ome/ngff) repository.
+
+Citing {#citing}
+================
+
+[Next-generation file format (NGFF) specifications for storing bioimaging data in the cloud.](https://ngff.openmicroscopy.org/0.2)
+J. Moore, *et al*. Editors. Open Microscopy Environment Consortium, 20 November 2020.
+This edition of the specification is [https://ngff.openmicroscopy.org/0.2/](https://ngff.openmicroscopy.org/0.2/]).
+The latest edition is available at [https://ngff.openmicroscopy.org/latest/](https://ngff.openmicroscopy.org/latest/).
+[(doi:10.5281/zenodo.4282107)](https://doi.org/10.5281/zenodo.4282107)
+
+Version History {#history}
+==========================
+
+ +
+
+
+