-
Notifications
You must be signed in to change notification settings - Fork 97
Home
Documentation has been moved to http://docs.opencb.org/display/opencga
OpenCGA is an open-source project that aims to provide a Big Data storage engine and analysis framework for genomic scale data analysis of hundreds of terabytes or even petabytes. For users, its main features will include uploading and downloading files to a repository, storing their information in a generic way (non-dependant of the original file-format) and retrieving this information efficiently. For developers, it will be a platform for supporting the most used bioinformatics file formats and accelerating the development of visualization and analysis applications.
There already exist some platforms with the same objectives as OpenCGA. However, they are focused on representing strictly the information from the files they stored. OpenCGA, on the other side, not only includes this information, but its generic databases include extra fields of interest and allow to combine data from different studies seamlessly.
Plain access to the files stored in the system is simply not fast enough for giving a real-time, interactive user experience. For this reason, we are exploring and using the most modern advanced technologies from different fields:
- Different NoSQL databases for storage. Users can choose which database fits bets its current infrastructure and data size
- Apache Hadoop for big data processing and storage
- High-performance Computing (HPC) for computation-intensive analysis
- HTML5 and RESTful web services for information retrieval and data visualization
If you want to see some projects using OpenCGA successfully, please visit the Use Cases section.
The image below show a global view of the infrastructure used by OpenCGA. When a file is uploaded to the system, it is stored in:
- A filesystem for archiving purposes. This filesystem could be UNIX-based or Hadoop-based.
- A database for interactive queries. We plan to support MongoDB and HBase databases.
This dual schema will always allow to download the original files from the archiving file system, and to use the databases to retrieve information much faster than reading the files.
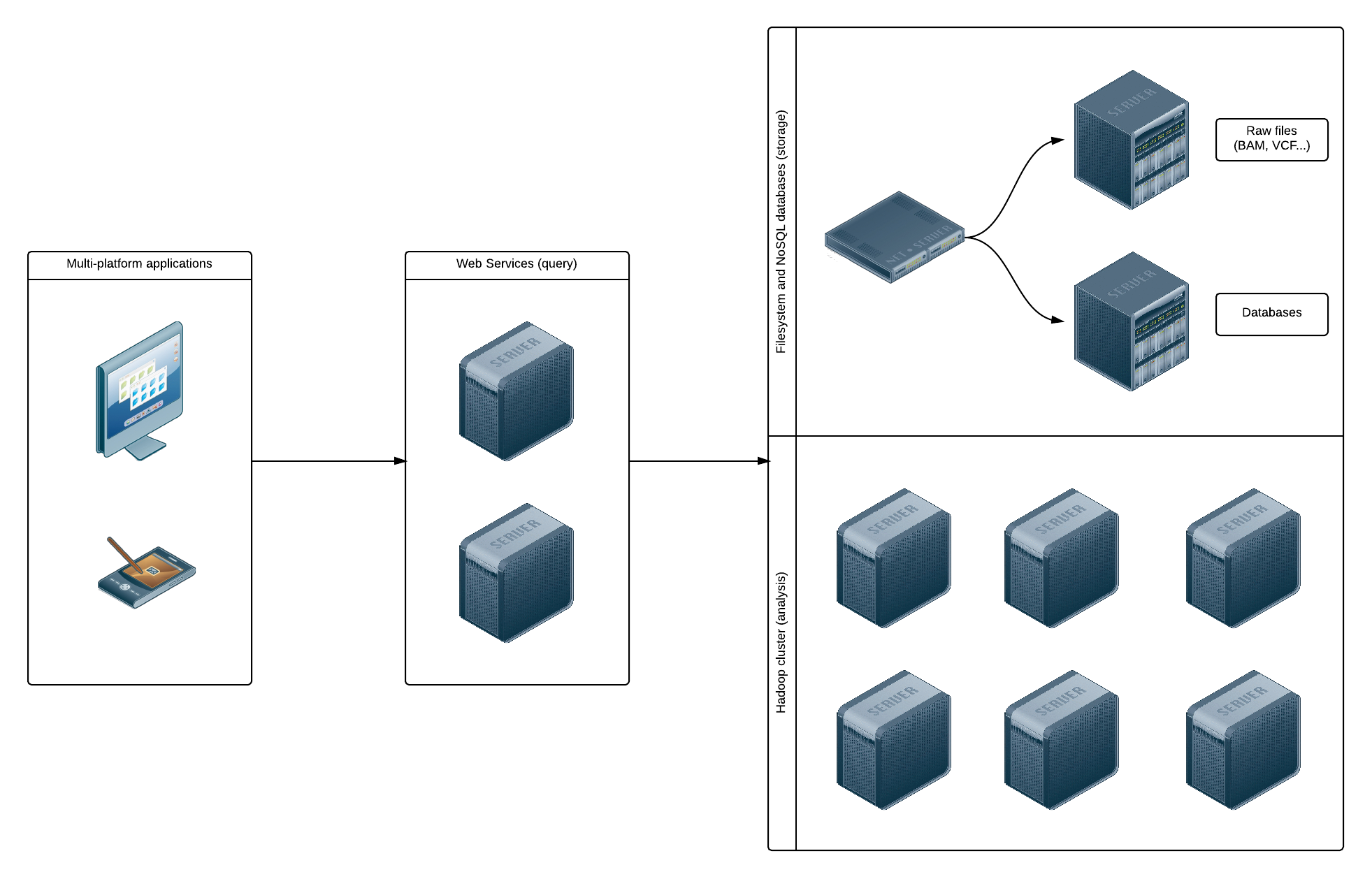
Apache Hadoop could be considered the de facto standard for Big Data analysis. OpenCGA will allow several ways of accessing and analyzing a file using Hadoop. First, by storing a file in an HDFS filesystem, it is be possible to read it directly and run Map/Reduce jobs. If its data is also saved to a HBase database, real-time queries could also be executed.
It is important to note that the storage and analysis subsystems are not completely separated: Hadoop uses the same machines for data storage and computation in order to reduce network traffic and improve efficiency.
Users do not have direct access to the storage and analysis subsystems, but they access data using a web services API. This provides both an homogeneous way of presenting the information and a security layer that can be easily implemented and improved.
At this section you can find some useful links and information for researchers and software developers who are planning to deploy and/or integrating OpenCGA services with their software applications and tools. These are working documents:
- Data models: Describes data models for representing Variant and alignment data.
- Architecture: Describes the technologies and architecture of OpenCGA and some other implementation details.
- Storage implementation: Describes how the data models are mapped to the different database backends (Mongo and HBase).
- Releases and Roadmap: Do you want to know what's coming next?
- Download and install: Please have a look at the README file in the repository.
##Getting Involved OpenCGA is an open-source project and as such, we welcome everyone who wants to contribute in any way they can :) Helping with the development, documentation or testing and bug reporting are always useful! If you want to get involved in some way, please join our mailing list [email protected].
OpenCGA is an open source project and it is freely available.
General
- Home
- Architecture
- Data Models
- RESTful Web Services
- Configuration
- Download and Installation
- Tutorials
OpenCGA Catalog
OpenCGA Storage
About