forked from ESIPFed/bds-primer-best-practices
-
Notifications
You must be signed in to change notification settings - Fork 0
/
CONTEXT_UNDERSTANDABILITY.qmd
123 lines (64 loc) · 5.56 KB
/
CONTEXT_UNDERSTANDABILITY.qmd
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
# Provide Context and Understandability to Your Data
## Ecological Metadata Language (EML)
### What Is It?
EML is a community-developed metadata schema designed for ecological data, which encompasses biological data. EML is normally presented as [Extensible Markup Language (XML)](https://www.w3schools.com/xml/). An EML instance (XML document) holds metadata to describe one or more data objects. Data tables are the most common, but almost any data object can be accommodated.
### Why?
- Provide context to your data and improve reproducability of the data.
- Can capture linked data relationships within EML (dataset series)
- Standardized representation of information.
- EML was designed for ecological data, which encompasses biological data.
- It's taxonomic fields cover relationships (hierarchies), IDs, and authoritative material
### Key Information
- [EML Schema](https://eml.ecoinformatics.org/eml-schema)
- Mandatory for LTER, iLTER, OBIS, GBIF, Darwin Core Archive (DwC-A)
- Maintained, and github repo, managed by NCEAS
- Usually, what you would submit to a repository is a "data package" consisting of an EML document and one or more data objects.
### Top References
Tools or packages to help write EML:
- For data managers, coders:
- [EML-R package](https://cran.r-project.org/web/packages/EML/index.html)
- [Postgresql database with fields compatible with EML](https://github.com/lter/LTER-core-metabase)
- [R-code for generating EML from LTER-metabase (built on EML-R package)](https://github.com/BLE-LTER/MetaEgress)
- [EMLAssemblyline (built on EML-R package)](https://ediorg.github.io/EMLassemblyline/articles/overview.html)
- For scientists or those not inclined to write scripts
- [ezEML](https://ezeml.edirepository.org/)
## ISO 19115
### What Is It?
Content standard for describing geographic data sponsored by the International Standards Organization (ISO). At its most basic, it is written in narrative form with class diagrams. There are many implementations and extensions (e.g., <https://www.dcc.ac.uk/resources/metadata-standards/iso-19115>).
### Why?
- Provide context to your data (biological data is inherently 'geographic')
- Standardized representation of information
- **Mandated** by some US federal agencies, including NOAA, NASA, and USGS
- Can be used at different granularities, used to describe data packages or collections, as well as at a dataset level (?): content standard vs collection standard?
### What?
- Evolved from the need for to to harmonize the FGDC Content Standard for Digital Geospatial Metadata (CSDGM) with other formal and defacto standards that support the documentation of geospatial data and services.
- Many variations including 19115, 19115-1, 19115-2
- From [NCEI](https://www.ncei.noaa.gov/resources/metadata):
- ISO 19115 Geographic information -- Metadata: The ISO standard for documenting geospatial data.
- ISO 19115-2 Geographic information -- Metadata -- Part 2: Extensions for imagery and gridded data: An extension of ISO 19115 used to document information about imagery, gridded data, and remotely sensed data. The root of ISO 19115 metadata records will change from MD_Metadata to MI_Metadata when using ISO 19115-2.
- Usurped FGDC CSDGM - all users encouraged to migrate to ISO.
- Highly flexible for many uses compared FGDC CSDGM, but few required elements leaves room for incomplete metadata
### Top References
- [NOAA Workbook for ISO 19115-2](https://www.ncei.noaa.gov/sites/default/files/2020-04/ISO%2019115-2%20Workbook_Part%20II%20Extentions%20for%20imagery%20and%20Gridded%20Data.pdf)
- [How to Convert ISO to EML](https://nceas.github.io/arcticdatautils/reference/convert_iso_to_eml.html)
- [Work Flow Model](https://www.fgdc.gov/metadata/iso-implementation-model-workflow)
- [mdToolkit](https://www.mdtoolkit.org/home) - mdEditor is a writer for ISO 19115 metadata which uses mdJSON as an intermediary and mdTranslator allows translation to different metadata formats
## Minimum Information about any (x) Sequence (MIxS)
### Who?
This is a standard for molecular data, like DNA and RNA. It is used by molecular biologist and ecologists who generate, manage and archive these type of sequence data.
### What is it?
A set of checklists and packages for genomic sequence data.
### Why?
- Provide minimal standardized metadata about genetic sequence data
- Agreed upon and published by the Genome Standards Consortium
- Used by the INSDC (DDBJ, EMBL-EBI and NCBI)
### Key Information
- MIxS (pronounced MIX-ess) is a suite of checklists standards introduced the reporting of a breadth of environment-specific metadata variables to augment the genome-specific checklists.
- Enables mixing and matching of genome checklists and environmental-specific packages.
- [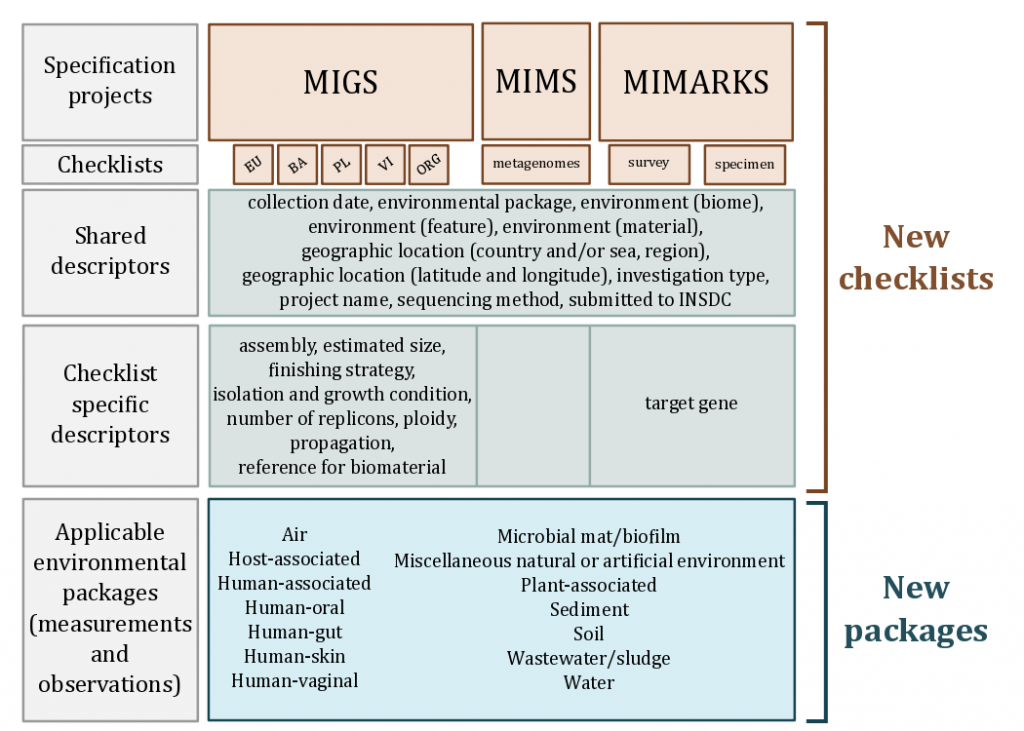](http://www.gensc.org/pages/standards-intro.html)
### Top References
- [MIxS Term Search Tool](http://www.gensc.org/pages/standards/search-terms.html)
- [Genomic Standards Consortium term list](https://genomicsstandardsconsortium.github.io/mixs/)
- [Minimum Information about Marker Gene Sequence (MIMARKS)](https://www.nature.com/articles/nbt.1823)
- [MIxS GitHub repo](https://github.com/GenomicsStandardsConsortium/mixs)
- [Minimum Information about Sequence Data from the Built Environment (MIxS-BE)](https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3869023/)