Clinical and epidemiological data tend to explain most of the variation of health-related traits, and its joint modeling with Omics data is crucial to increase the algorithm’s predictive ability. However, the nature of non-Omics data, such as heterogeneity, lack of standardization, high complexity, and loose links to Omics data types, make it hard to use both Omics and non-Omics data for ML analyses.
OmicsMLRepo project aims to build the first large-scale, platform-independent, curated, ML-ready data repository for diverse Omics and associated non-Omics data, starting from two Bioconductor data packages - curatedMetagenomicData containing human microbiome data and cBioPortalData package on cancer genomics data.
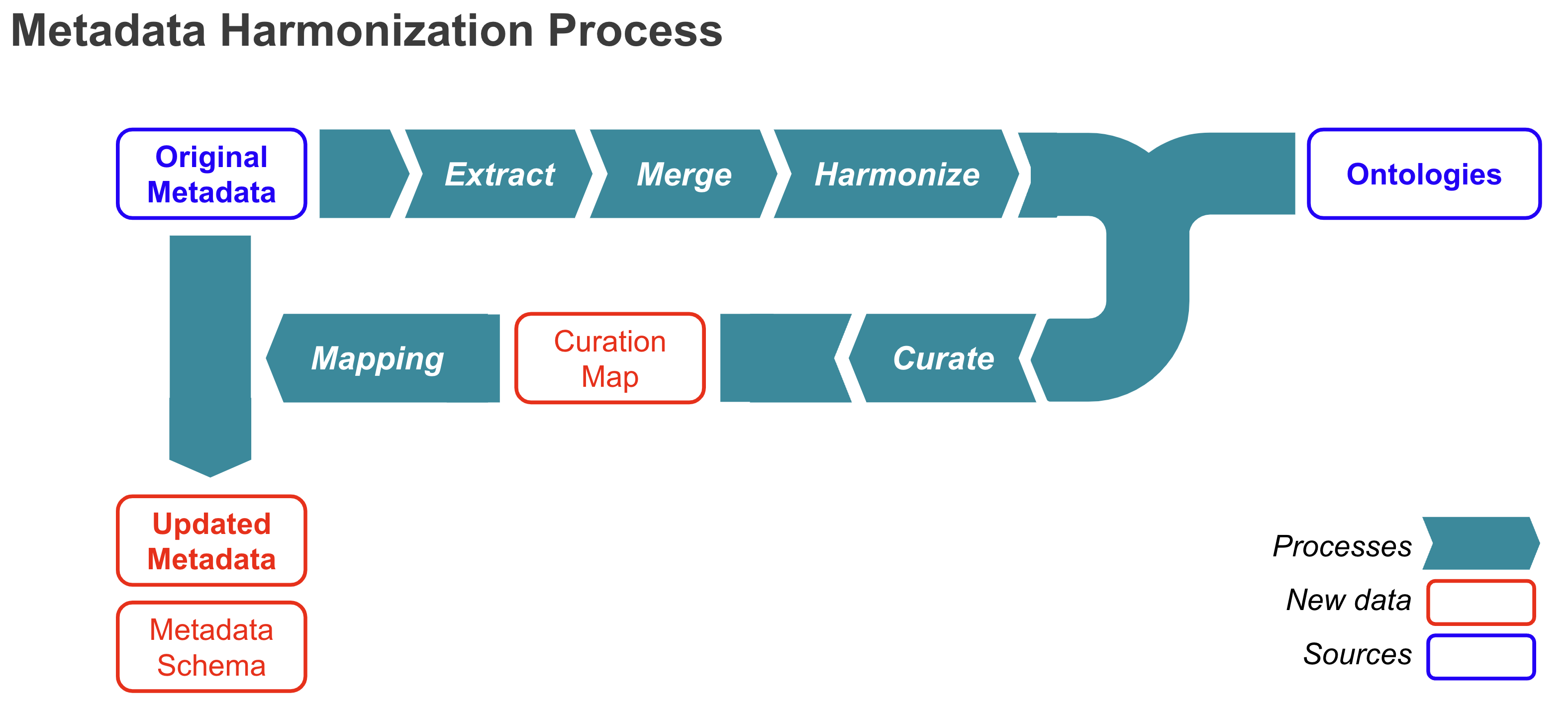
This repository, OmicsMLRepoData, documents the hamonization/curation processes and the artifacts generated throughout. We are also developing a software package, OmicsMLRepoR, allowing users to leverage ontology in metadata search.
In summary, the OmicsMLRepo project simplifies the process of cross-study, multi-faceted data analyses through metadata harmonization and standardization, making Omics data more AI/ML-ready.
You can access the harmonized version of metadata using the
OmicsMLRepoR::getMetadata function:
if (!require("devtools"))
install.packages("devtools")
devtools::install_github("shbrief/OmicsMLRepoR")
library(OmicsMLRepoR)
cmd <- getMetadata("cMD")
cbio <- getMetadata("cBioPortal")