-
Notifications
You must be signed in to change notification settings - Fork 4
Configuration
Metadata-Manager is designed to receive a configuration XML file with the needed parameters to configure all its steps: downloading, transforming, cleaning, mapping, enriching, checking, flattening and loading. An XSD schema file is designed to validate any configuration XML file given as input for the tool.
The schema includes a root node where general settings and a list of sources are stored. Sources represent NGS data providers which provide those genomic data and experimental metadata divided in datasets (examples are ENCODE, TCGA, GDC, Roadmap Epigenomics). Each source contains a list of datasets. After processing, each dataset represents a GDM dataset where:
- every sample has a region data file;
- every sample has a metadata file;
- every sample share the same region data schema.
The configuration XSD is organized in a tree structure which starts from the root node. At this level a list of sources is contained. Each source can feature multiple datasets.
Below we show the details of XSDs for these three levels, in addition to the XSD of the parameter element, which contains basic information to be read by the single steps of the procedure.
Note that we allow for enabling/disabling each step of the process at different granularities. E.g. the XML element downloader_enabled is present at the level of the entire repositories (i.e., for all sources), at the level of a single source (i.e., for all the datasets from that origin), and at the level of a single dataset (i.e., the smallest organizational unit in this project).
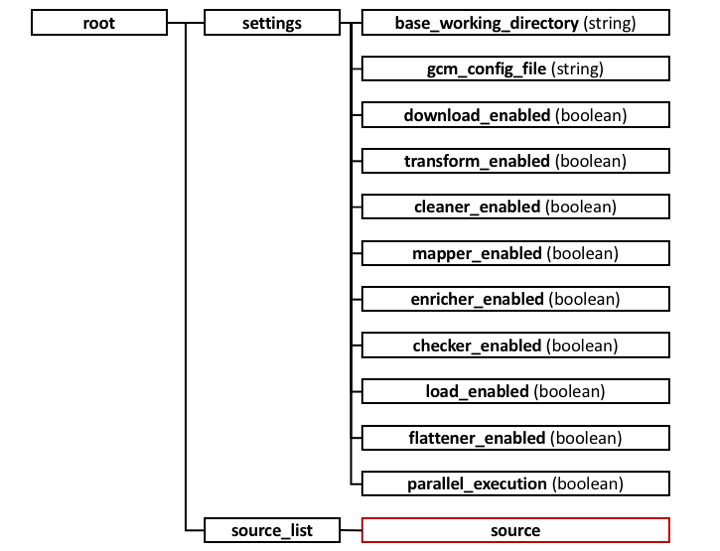
- root: contains general settings and a list for sources to import.
-
settings:
- base_working_directory: folder used to save downloaded, transformed, cleaned and flattened files.
- download_enabled, ..., load_enabled: flags used to enable separate steps of the overall process.
- parallel_execution: flag to enable execution with single thread processing or multi-thread processing.
- source_list: collection of sources to be imported.

- source: represents an NGS databank, contains basic information for the process.
- name: identification for the source.
- url: address of the source.
- source_working_directory: sub directory where the source’s files will be processed.
- downloader, transformer, loader: indicate the specific classes to be used to perform respectively downloading, transforming and loading.
- download_enabled, ..., load_enabled: flags used to enable separate steps of the overall process for a specific source.
- parameter_list: collection of parameters regarding different steps of the process.
- dataset_list: collection of datasets to import from the source.
Example parameters used at the source level are used to:
- know which user in gmql is going to be imported
- define the metadata name separation characters
- compose the URLs to download list of files from sources
- filter useless/wrong files from such lists (e.g. Useless: files from Encode with no biological replicates, Wrong: files from Encode with assembly different from the one specified)
- specify the extensions of produced files
- specify location of Rule Base for Cleaning step
- specify location of Mappings Base for Mapping step
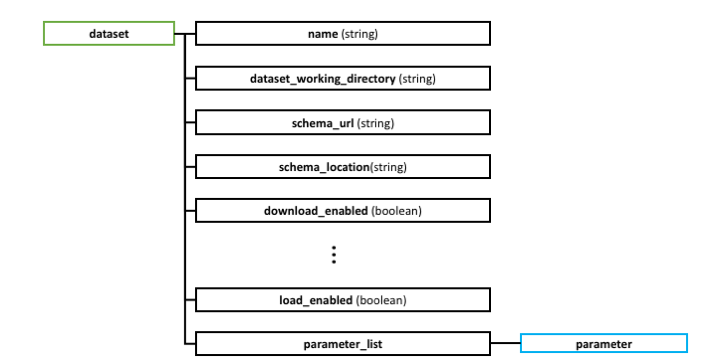
- dataset: represents a set of samples that share the same region data schema and the same types of experimental or clinical metadata.
- name: identifier for the dataset.
- dataset_working_directory: sub-folder where the download and transformation of this dataset is performed.
- schema_url: address where the schema file can be found.
- schema_location: indicates whether the schema is located in FTP, HTTP or LOCAL destination.
- download_enabled, ..., load_enabled: flags used to enable separate steps of the overall process for a specific dataset.
- parameter_list: list of dataset specific parameters for downloading, transforming or loading this dataset.
Example parameters used at the dataset level are used to:
- specify loading name and description for the dataset
- specify the source URL to retrieve the files interesting for the specific dataset
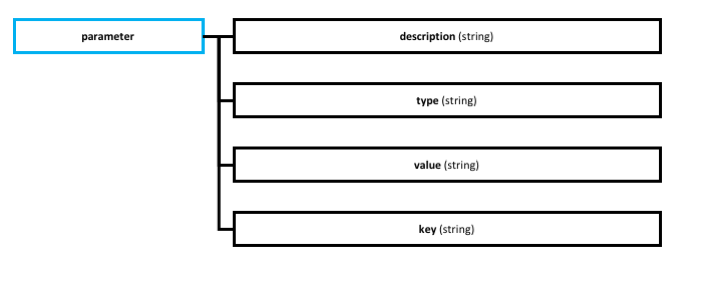
- parameter: defines specific information for a source or a dataset, this information is useful for downloading, transforming or loading procedures.
- key: is the name for the parameter, its identifier.
- value: parameter information.
- description: explains what the parameter is used for.
- type: optional tag for the parameter.
Note that newer modules can be added to configurations and different parameters could emerge in the future as addition of other sources and datasets to the project is ongoing.
Usage
Supporting Tools