-
Notifications
You must be signed in to change notification settings - Fork 28
Merkle_PCB2016
bernhardsteiert edited this page Aug 8, 2016
·
2 revisions
Reference: Merkle et al. Identification of cell type-specific differences in erythropoietin receptor signaling in primary erythroid and lung cancer cells. PLoS Computational Biology 12(8): e1005049, 2016.
Folder: /Examples/Merkle_JAK2STAT5_PCB2016.
Facts: The model contains 1127 data points, 238 free parameters and 59 experimental conditions.
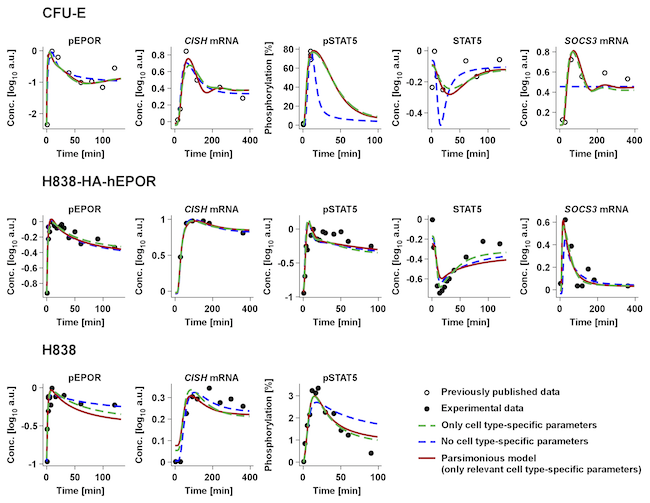
Figure: Shows only parts of the data.
- Installation and system requirements
- Setting up models
- First steps
- Advanced events and pre-equilibration
- Computation of integration-based prediction bands
- How is the architecture of the code and the most important commands?
- What are the most important fields of the global variable ar?
- What are the most important functions?
- Optimization algorithms available in the d2d-framework
- Objective function, likelhood and chi-square in the d2d framework
- How to set up priors?
- How to set up steady state constraints?
- How do I restart the solver upon a step input?
- How to deal with integrator tolerances?
- How to implement a bolus injection?
- How to implement washing and an injection?
- How to implement a moment ODE model?
- How to run PLE calculations on a Cluster?