- Documentation: https://dcmri.org
- Source code: https://github.com/dcmri/dcmri
Note: dcmri is under construction. At this stage, the API may still change and features may be deprecated without warning.
Install the latest version of dcmri:
pip install dcmriimport dcmri as dc
# Generate some test data
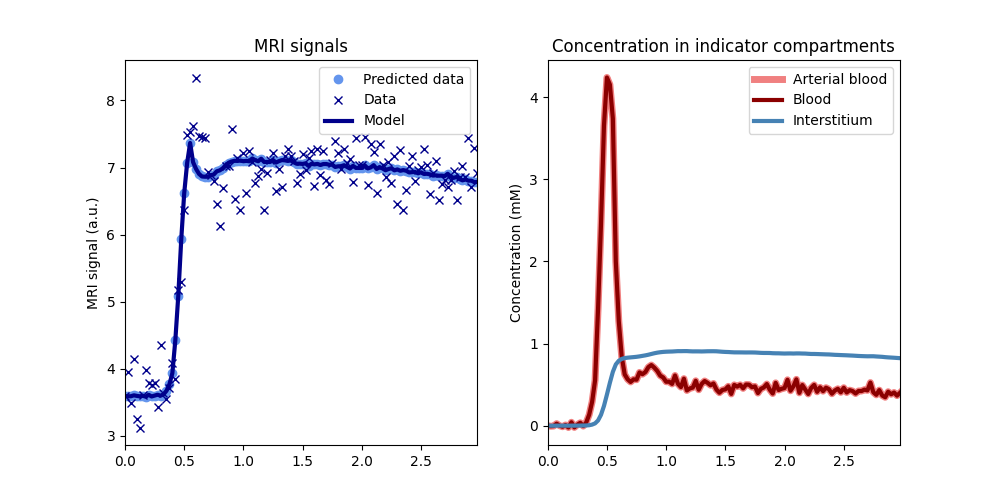
time, aif, roi, _ = dc.fake_tissue(CNR=50)
# Construct a tissue
tissue = dc.Tissue(aif=aif, t=time)
# Train the tissue on the data
tissue.train(time, roi)
# Check the fit to the data
tissue.plot(time, roi)# Print the fitted parameters
tissue.print_params(round_to=3)--------------------------------
Free parameters with their stdev
--------------------------------
Blood volume (vb): 0.018 (0.002) mL/cm3
Interstitial volume (vi): 0.174 (0.004) mL/cm3
Permeability-surface area product (PS): 0.002 (0.0) mL/sec/cm3
----------------------------
Fixed and derived parameters
----------------------------
Plasma volume (vp): 0.01 mL/cm3
Interstitial mean transit time (Ti): 74.614 sec# Generate some test data
n = 128
time, signal, aif, _ = dc.fake_brain(n)
# Construct an array of tissues
image = dc.TissueArray((n,n),
aif = aif, t = time,
kinetics = '2CU', verbose = 1)
# Train the tissue array on the data
image.train(time, signal)
# Plot the parameter maps
image.plot(time, signal)Released under the Apache 2.0 license:
Copyright (C) 2023-2024 dcmri developers