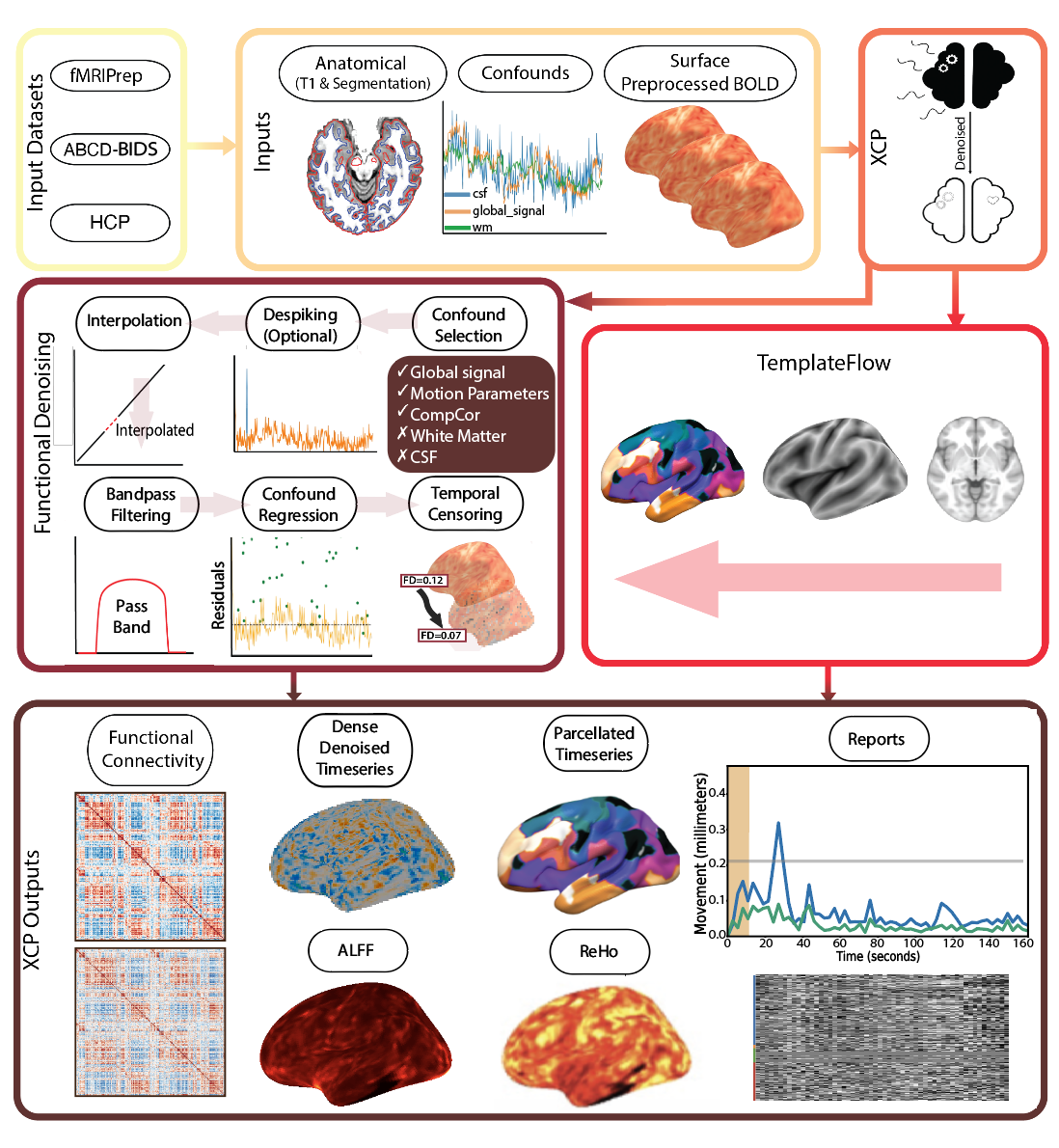
This fMRI post-processing and noise regression pipeline is developed by the Satterthwaite lab at the University of Pennslyvania (XCP; eXtensible Connectivity Pipeline) and Developmental Cognition and Neuroimaging lab at the University of Minnesota (-DCAN) for open-source software distribution.
XCP-D paves the final section of the reproducible and scalable route from the MRI scanner to functional connectivity data in the hands of neuroscientists. We developed XCP-D to extend the BIDS and NiPrep apparatus to the point where data is most commonly consumed and analyzed by neuroscientists studying functional connectivity. Thus, with the development of XCP-D, data can be automatically preprocessed and analyzed in BIDS format, using NiPrep-style containerized code, all the way from the scanner to functional connectivity matrices.
XCP-D picks up right where fMRIprep ends, directly consuming the outputs of fMRIPrep. XCP-D leverages the BIDS and NiPreps frameworks to automatically generate denoised BOLD images, parcellated time series, functional connectivity matrices, and quality assessment reports. XCP-D can also process outputs from: NiBabies, ABCD-BIDS, Minimally preprocessed HCP, and UK Biobank data.
Please note that XCP is only compatible with HCP-YA versions downloaded c.a. Feb 2023 at the moment.
See the documentation for more details.
XCP-D produces the following commonly-used outputs: matrices, parcellated time series, dense time series, and additional QC measures.
XCP-D is designed for resting-state or pseudo-resting-state functional connectivity analyses. XCP-D derivatives may be useful for seed-to-voxel and ROI-to-ROI functional connectivity analyses, as well as decomposition-based methods, such as ICA or NMF.
XCP-D is not designed as a general-purpose postprocessing pipeline. It is really only appropriate for certain analyses, and other postprocessing/analysis tools are better suited for many types of data/analysis.
XCP-D derivatives are not particularly useful for task-dependent functional connectivity analyses, such as psychophysiological interactions (PPIs) or beta series analyses. It is also not suitable for general task-based analyses, such as standard task GLMs, as we recommend including nuisance regressors in the GLM step, rather than denoising data prior to the GLM.
If you use XCP-D in your research, please use the boilerplate generated by the workflow. If you need an immediate citation, please cite the following paper:
Mehta, K., Salo, T., Madison, T. J., Adebimpe, A., Bassett, D. S., Bertolero, M., ... & Satterthwaite, T. D. (2024). XCP-D: A Robust Pipeline for the post-processing of fMRI data. Imaging Neuroscience, 2, 1-26. doi:10.1162/imag_a_00257.
Please also cite the Zenodo DOI for the version you're referencing.