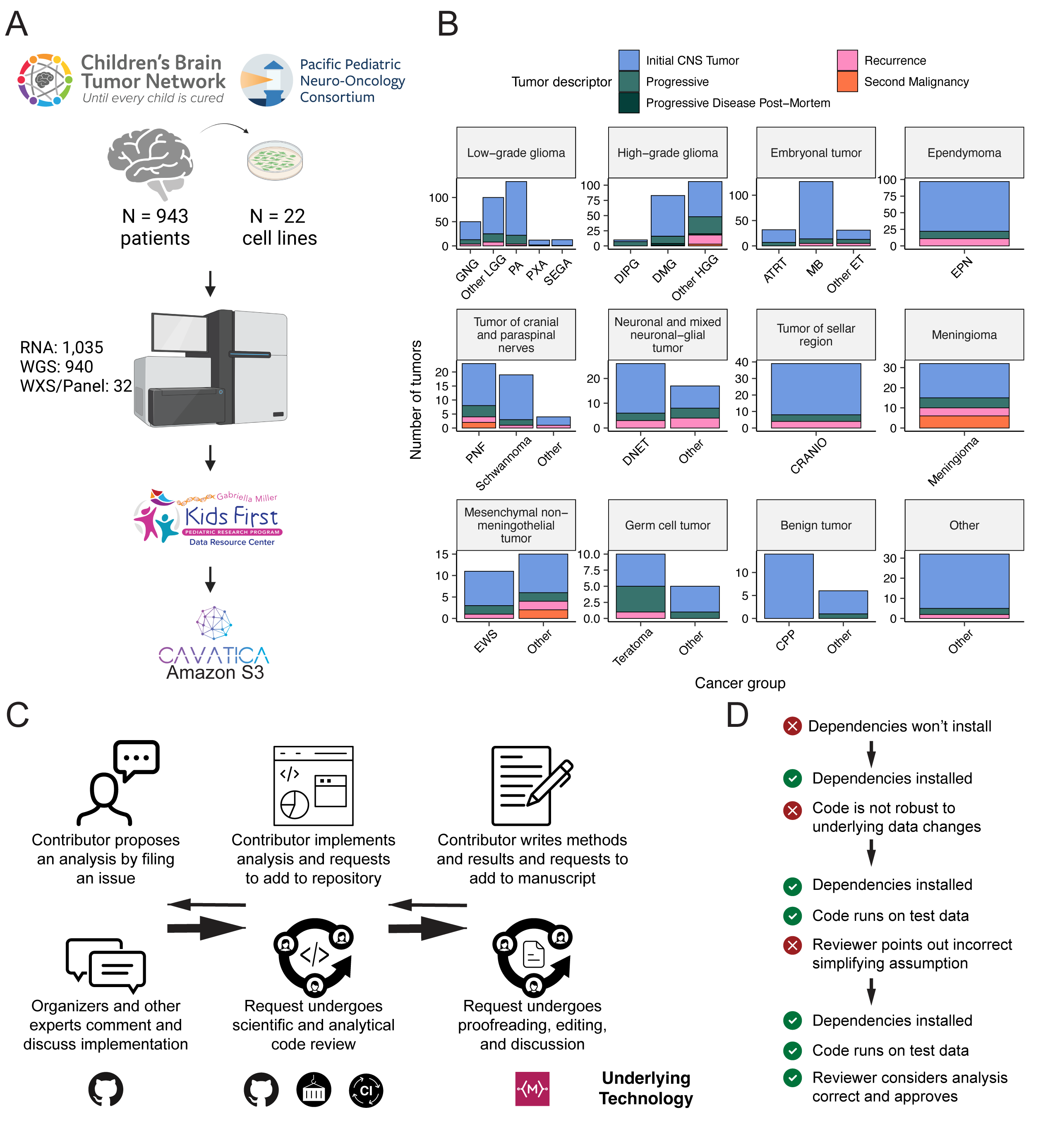
We previously performed whole genome sequencing (WGS), whole exome sequencing (WXS), and RNA sequencing (RNA-Seq) on matched tumor/normal tissues and selected cell lines [@doi:10.1093/neuonc/noz192] from 943 patients from the Pediatric Brain Tumor Atlas (PBTA), consisting of 911 patients from the CBTN [@doi:10.1016/j.neo.2022.100846] and 32 patients from PNOC [@doi:10.1002/ijc.32258; @doi:10.1158/1078-0432.CCR-22-0803] (Figure {@fig:Fig1}A) across various histologies phrases of therapy (Figure {@fig:Fig1}B). We harnessed and extended the benchmarking efforts of the Gabriella Miller Kids First Data Resource Center to develop robust and reproducible data analysis workflows within the CAVATICA platform for comprehensive somatic analyses (Figure {@fig:S1}) and STAR Methods) of the PBTA.
A key innovative feature of OpenPBTA is its open contribution framework used for analytical code and manuscript writing. We created a public Github analysis repository (https://github.com/AlexsLemonade/OpenPBTA-analysis) to hold all analysis code downstream of Kids First workflows and a GitHub manuscript repository (https://github.com/AlexsLemonade/OpenPBTA-manuscript) with Manubot [@doi:10.1371/journal.pcbi.1007128] integration to enable real-time manuscript creation. As all analyses and manuscript writing were conducted in public repositories, any researcher in the world could contribute to OpenPBTA following the process outlined in Figure {@fig:Fig1}C. First, a potential contributor proposed an analysis by filing an issue in the GitHub analysis repository. Next, project organizers or other contributors with expertise provided feedback about the proposed analysis (Figure {@fig:Fig1}C). The contributor formally requested to include their analytical code and results – written in their own copy (fork) of repository – in the OpenPBTA analysis repository by filing a GitHub pull request (PR). All PRs underwent peer review to ensure scientific accuracy, maintainability, and readability of code and documentation (Figure {@fig:Fig1}C-D).
Beyond peer review, we implemented additional checks to ensure consistent results for all collaborators over time (Figure {@fig:Fig1}D). To provide a consistent software development environment, we created a monolithic image with all OpenPBTA dependencies using Docker® [@https://dl.acm.org/doi/10.5555/2600239.2600241] and the Rocker project [@arxiv:1710.03675]. We used the continuous integration (CI) service CircleCI® to run analytical code in PRs on a test dataset before formal code review, allowing us to detect code bugs or sensitivity to data release changes.
We followed a similar process in our Manubot-powered [@doi:10.1371/journal.pcbi.1007128] repository for proposed manuscript additions (Figure {@fig:Fig1}C); peer reviewers ensured clarity and scientific accuracy, and Manubot performed spell-checking.
Since 2000, neuro-oncology experts and the WHO have collaborated to iteratively redefine central nervous system (CNS) tumor classifications [@pubmed:11895036; @doi:10.1007/s00401-007-0243-4].
In 2016 [@doi:10.1007/s00401-016-1545-1], molecular subtypes driven by genetic alterations were integrated into these classifications.
Since CBTN specimen collection began in 2011, most tumors lacked molecular subtype information when tissue was collected.
Moreover, PBTA does not yet feature methylation arrays which are increasingly used to inform molecular subtyping and cancer diagnosis.
Therefore, we created analysis modules to systematically consider key genomic features of tumors described by the WHO in 2016 or Ryall and colleagues [@doi:10.1016/j.ccell.2020.03.011].
Coupled with clinician and pathologist review, we generated high-confidence research-grade integrated diagnoses for 60% (644/1074) of tumors (Table S1) without methylation data, a major innovation of this project.
We then aligned OpenPBTA specimen diagnoses with WHO classifications (e.g., tumors formerly ascribed primitive neuro-ectodermal tumor [PNET] diagnoses), discovered rarer tumor entities (e.g., H3-mutant ependymoma, meningioma with YAP1::FAM118B fusion), as well as identified and corrected data entry errors (e.g., an embryonal tumor with multilayer rosettes (ETMR) incorrectly entered as a medulloblastoma) and histologically mis-identified specimens (e.g., Ewing sarcoma sample labeled as a craniopharyngioma).
Uniquely, we used transcriptomic classification to subtype 122 medulloblastomas into SHH, WNT, Group 3, or Group 4 with MedulloClassifier [@doi:10.1371/journal.pcbi.1008263] and MM2S [@doi:10.1186/s13029-016-0053-y], with 95% (41/43) and 91% (39/43) accuracy, respectively.
In total, we subtyped low-grade gliomas (LGGs) (N = 290), HGGs (N = 141), embryonal tumors (N = 126), ependymomas (N = 33), tumors of sellar region (N = 27), mesenchymal non-meningothelial tumors (N = 11), glialneuronal tumors (N = 10), and chordomas (N = 6), where Ns represent unique tumors (Table {@tbl:Table1}). For detailed methods, see STAR Methods and Figure {@fig:S1}.
| Broad histology group | OpenPBTA molecular subtype | Patients | Tumors |
|---|---|---|---|
| Chordoma | CHDM, conventional | 2 | 2 |
| Chordoma | CHDM, poorly differentiated | 2 | 4 |
| Embryonal tumor | CNS Embryonal, NOS | 13 | 13 |
| Embryonal tumor | CNS HGNET-MN1 | 1 | 1 |
| Embryonal tumor | CNS NB-FOXR2 | 2 | 3 |
| Embryonal tumor | ETMR, C19MC-altered | 5 | 5 |
| Embryonal tumor | ETMR, NOS | 1 | 1 |
| Embryonal tumor | MB, Group3 | 14 | 14 |
| Embryonal tumor | MB, Group4 | 48 | 49 |
| Embryonal tumor | MB, SHH | 24 | 30 |
| Embryonal tumor | MB, WNT | 10 | 10 |
| Ependymoma | EPN, H3 K28 | 1 | 1 |
| Ependymoma | EPN, ST RELA | 25 | 28 |
| Ependymoma | EPN, ST YAP1 | 3 | 4 |
| High-grade glioma | DMG, H3 K28 | 18 | 24 |
| High-grade glioma | DMG, H3 K28, TP53 activated | 10 | 13 |
| High-grade glioma | DMG, H3 K28, TP53 loss | 30 | 40 |
| High-grade glioma | HGG, H3 G35 | 3 | 3 |
| High-grade glioma | HGG, H3 G35, TP53 loss | 1 | 1 |
| High-grade glioma | HGG, H3 wildtype | 26 | 31 |
| High-grade glioma | HGG, H3 wildtype, TP53 activated | 5 | 5 |
| High-grade glioma | HGG, H3 wildtype, TP53 loss | 14 | 21 |
| High-grade glioma | HGG, IDH, TP53 activated | 1 | 2 |
| High-grade glioma | HGG, IDH, TP53 loss | 1 | 1 |
| Low-grade glioma | GNG, BRAF V600E | 13 | 13 |
| Low-grade glioma | GNG, BRAF V600E, CDKN2A/B | 1 | 1 |
| Low-grade glioma | GNG, FGFR | 1 | 1 |
| Low-grade glioma | GNG, H3 | 1 | 1 |
| Low-grade glioma | GNG, IDH | 1 | 2 |
| Low-grade glioma | GNG, KIAA1549-BRAF | 5 | 5 |
| Low-grade glioma | GNG, MYB/MYBL1 | 1 | 1 |
| Low-grade glioma | GNG, NF1-germline | 1 | 1 |
| Low-grade glioma | GNG, NF1-somatic, BRAF V600E | 1 | 1 |
| Low-grade glioma | GNG, other MAPK | 4 | 4 |
| Low-grade glioma | GNG, other MAPK, IDH | 1 | 1 |
| Low-grade glioma | GNG, RTK | 2 | 3 |
| Low-grade glioma | GNG, wildtype | 14 | 14 |
| Low-grade glioma | LGG, BRAF V600E | 25 | 27 |
| Low-grade glioma | LGG, BRAF V600E, CDKN2A/B | 5 | 5 |
| Low-grade glioma | LGG, FGFR | 8 | 8 |
| Low-grade glioma | LGG, IDH | 3 | 3 |
| Low-grade glioma | LGG, KIAA1549-BRAF | 106 | 113 |
| Low-grade glioma | LGG, KIAA1549-BRAF, NF1-germline | 1 | 1 |
| Low-grade glioma | LGG, KIAA1549-BRAF, other MAPK | 1 | 1 |
| Low-grade glioma | LGG, MYB/MYBL1 | 2 | 2 |
| Low-grade glioma | LGG, NF1-germline | 6 | 6 |
| Low-grade glioma | LGG, NF1-germline, CDKN2A/B | 1 | 1 |
| Low-grade glioma | LGG, NF1-germline, FGFR | 1 | 2 |
| Low-grade glioma | LGG, NF1-somatic | 2 | 2 |
| Low-grade glioma | LGG, NF1-somatic, FGFR | 1 | 1 |
| Low-grade glioma | LGG, NF1-somatic, NF1-germline, CDKN2A/B | 1 | 1 |
| Low-grade glioma | LGG, other MAPK | 11 | 12 |
| Low-grade glioma | LGG, RTK | 8 | 10 |
| Low-grade glioma | LGG, RTK, CDKN2A/B | 1 | 1 |
| Low-grade glioma | LGG, wildtype | 33 | 34 |
| Low-grade glioma | SEGA, RTK | 1 | 1 |
| Low-grade glioma | SEGA, wildtype | 10 | 11 |
| Mesenchymal non-meningothelial tumor | EWS | 9 | 11 |
| Neuronal and mixed neuronal-glial tumor | CNC | 2 | 2 |
| Neuronal and mixed neuronal-glial tumor | EVN | 1 | 1 |
| Neuronal and mixed neuronal-glial tumor | GNT, BRAF V600E | 1 | 1 |
| Neuronal and mixed neuronal-glial tumor | GNT, KIAA1549-BRAF | 1 | 2 |
| Neuronal and mixed neuronal-glial tumor | GNT, other MAPK | 1 | 1 |
| Neuronal and mixed neuronal-glial tumor | GNT, other MAPK, FGFR | 1 | 1 |
| Neuronal and mixed neuronal-glial tumor | GNT, RTK | 1 | 2 |
| Tumor of sellar region | CRANIO, ADAM | 27 | 27 |
| Total | 577 | 644 |
Table: Molecular subtypes generated through the OpenPBTA project. Broad tumor histologies, molecular subtypes generated, and number of patients and tumors subtyped within OpenPBTA. {#tbl:Table1}
We performed a comprehensive genomic analysis of somatic SNVs, CNVs, SVs, and fusions across all 1,074 PBTA tumors (N = 1,019 RNA-Seq, N = 918 WGS, N = 32 WXS/Panel) and 22 cell lines (N = 16 RNA-Seq, N = 22 WGS), from 943 patients, 833 with paired normal specimens (N = 801 WGS, N = 32 WXS/Panel). Tumor purity across PBTA samples was high (median 76%), though we observed some cancer groups with lower purity, including SEGA, PXA, and teratoma (Figure {@fig:S3}A). Unless otherwise noted, each analysis was performed for diagnostic tumors using one tumor per patient.
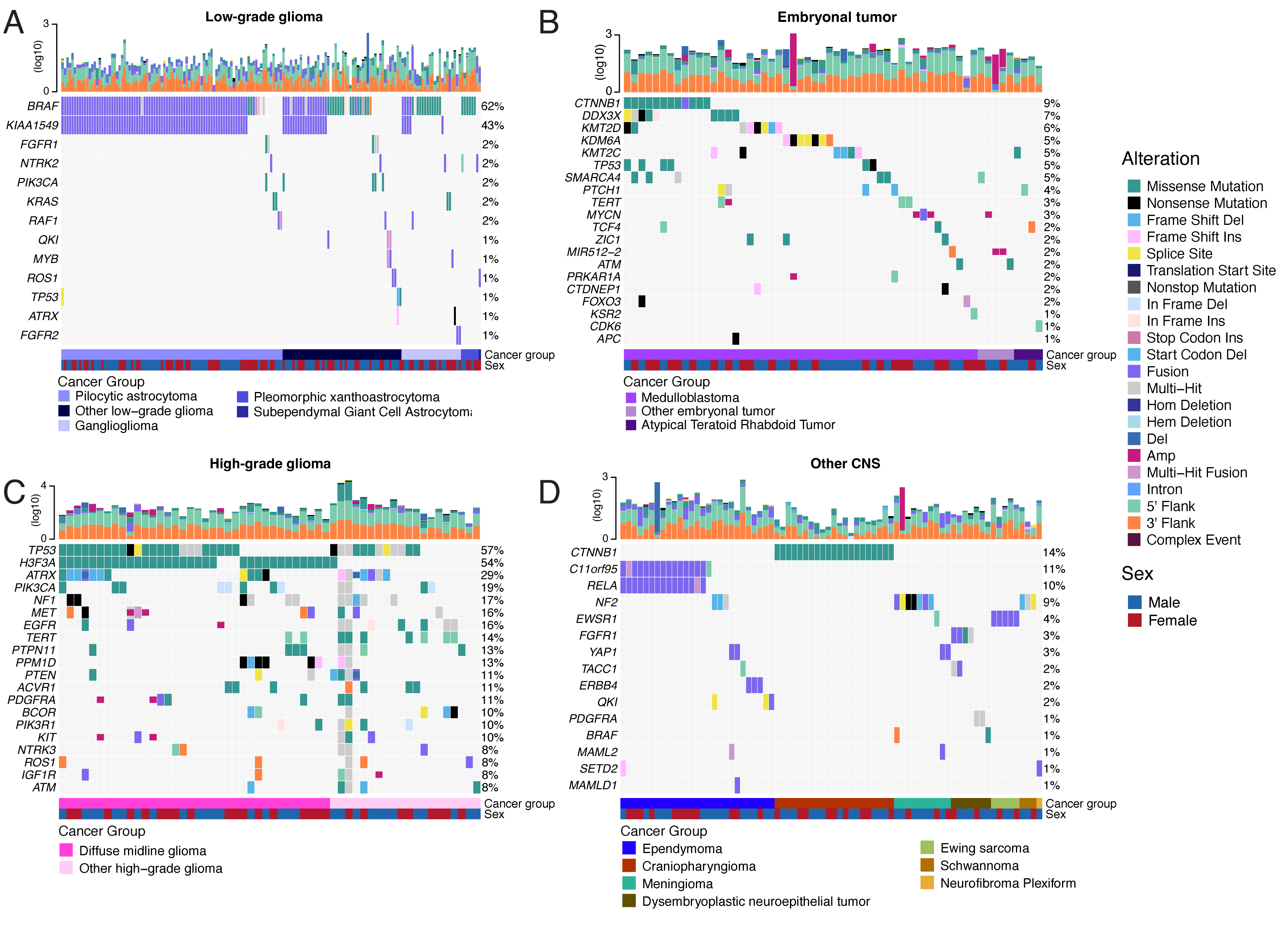
SNV consensus calling (Figure {@fig:S1} and Figure {@fig:S2}A-G) revealed, as expected, lower tumor mutation burden (TMB) (Figure {@fig:S2}H) in pediatric tumors compared to adult brain tumors from The Cancer Genome Atlas (TCGA) (Figure {@fig:S2}I), with hypermutant (> 10 Mut/Mb) and ultra-hypermutant (> 100 Mut/Mb) tumors [@doi:10.1016/j.cell.2017.09.048] only found within HGGs and embryonal tumors. Figure {@fig:Fig2} and Figure {@fig:S3}B depict oncoprints recapitulating known histology-specific driver genes in primary tumors across OpenPBTA histologies, and Table S2 summarizes all detected alterations across cancer groups.
As expected, most (62%, 140/226) LGGs harbored a somatic alteration in BRAF, with canonical BRAF::KIAA1549 fusions as the major oncogenic driver [@doi:10.1186/s40478-020-00902-z] (Figure {@fig:Fig2}A). We observed additional mutations in FGFR1 (2%), PIK3CA (2%), KRAS (2%), TP53 (1%), and ATRX (1%) and fusions in NTRK2 (2%), RAF1 (2%), MYB (1%), QKI (1%), ROS1 (1%), and FGFR2 (1%), concordant with previous studies reporting near-universal upregulation of the RAS/MAPK pathway in LGGs [@doi:10.1186/s40478-020-00902-z; @doi:10.1016/j.ccell.2020.03.011]. Indeed, gene set variant analysis (GSVA) revealed significant upregulation (ANOVA Bonferroni-corrected p < 0.01) of the KRAS signaling pathway in LGGs (Figure {@fig:Fig5}B).
Most (N = 95) embryonal tumors were medulloblastomas from four characterized molecular subtypes (WNT, SHH, Group3, and Group 4; see Molecular Subtyping of CNS Tumors), as identified by subtype-specific canonical mutations (Figure {@fig:Fig2}B). We detected canonical SMARCB1/SMARCA4 deletions or inactivating mutations in atypical teratoid rhabdoid tumors (ATRTs; Table S2) and C19MC amplification in ETMRs (displayed within "Other embryonal tumors" in Figure {@fig:Fig2}B) [@doi:10.1007/s00401-020-02182-2; @doi:10.1093/neuonc/noab178; @doi:10.1186/s40478-020-00984-9; @doi:10.1038/nature22973].
Across HGGs, TP53 (57%, 36/63) and H3F3A (54%, 34/63) were both most mutated and co-occurring genes (Figure {@fig:Fig2}A and C), followed by frequent mutations in ATRX (29%, 18/63) which is commonly mutated in gliomas [@doi:10.1080/14728222.2018.1487953]. We observed recurrent amplifications and fusions in EGFR, MET, PDGFRA, and KIT, highlighting that these tumors leverage multiple oncogenic mechanisms to activate tyrosine kinases, as previously reported [@doi:10.1002/ijc.32258; @doi:10.1016/j.ccell.2017.08.017; @doi:10.1186/s40478-020-00905-w]. GSVA showed upregulation (ANOVA Bonferroni-corrected p < 0.01) of DNA repair, G2M checkpoint, and MYC pathways as well as downregulation of the TP53 pathway (Figure {@fig:Fig5}B). The two ultra-hypermutated tumors (> 100 Mutations/Mb) were from patients with mismatch repair deficiency syndrome [@doi:10.1093/neuonc/noz192].
We observed that 25% (15/60) of ependymomas were C11orf95::RELA (now, ZFTA::RELA) fusion-positive [@doi:10.1038/nature13109] and 68% (21/31) of craniopharyngiomas contained CTNNB1 mutations (Figure {@fig:Fig2}D). We observed somatic mutations or fusions in NF2 in 41% (7/17) of meningiomas, 5% (3/60) of ependymomas, and 25% (3/12) of schwannomas, as well as rare fusions in ERBB4, YAP1, and/or QKI in 10% (6/60) of ependymomas. DNETs harbored alterations in MAPK/PI3K pathway genes, as was previously reported [@doi:10.1093/jnen/nlz101], including FGFR1 (21%, 4/19), PDGFRA (10%, 2/19), and BRAF (5%, 1/19).
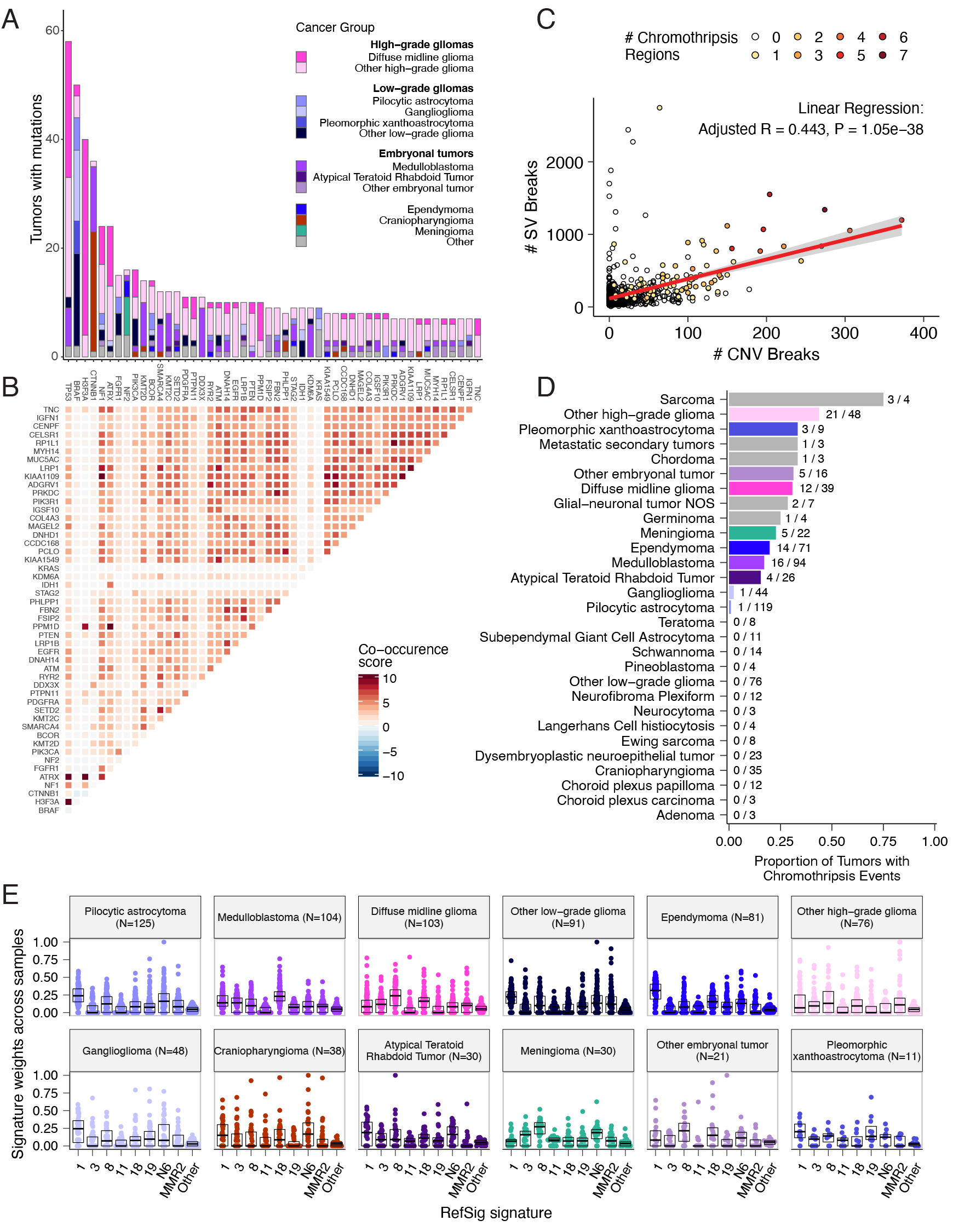
We analyzed mutational co-occurrence across the OpenPBTA, using a single tumor from each patient (N = 668) with WGS. The top 50 mutated genes (see STAR Methods for details) in primary tumors are shown in Figure {@fig:Fig3} by tumor type (A, bar plots), with co-occurrence scores illustrated in the heatmap (B). As expected, TP53 was the most frequently mutated gene across the OpenPBTA (8.7%, 58/668), significantly co-occurring with H3F3A (OR = 30.05, 95% CI: 14.5 - 62.3, q = 2.34e-16), ATRX (OR = 23.3, 95% CI: 9.6 - 56.3, q = 8.72e-9), NF1 (OR = 8.26, 95% CI: 3.5 - 19.4, q = 7.40e-5), and EGFR (OR = 17.5, 95% CI: 4.8 - 63.9, q = 2e-4), with all of these driven by HGGs and consistent with previous reports [@doi:10.1016/j.ccell.2017.08.017; @doi:10.1093/neuonc/noaa251; @doi:10.1038/ng.2938].
In embryonal tumors, CTNNB1 mutations significantly co-occurred with TP53 mutations (OR = 43.6 95% CI: 7.1 - 265.8, q = 1.52e-3) as well as with DDX3X mutations (OR = 21.4, 95% CI: 4.7 - 97.9, q = 4.15e-3), events driven by medulloblastomas as previously reported [@doi:10.1038/nrc3410; @doi:10.1200/JCO.2010.31.1670]. FGFR1 and PIK3CA mutations significantly co-occurred in LGGs (OR = 77.25, 95% CI: 10.0 - 596.8, q = 3.12e-3), consistent with previous findings [@doi:10.1200/JCO.2010.31.1670; @doi:10.1186/s40478-020-01027-z]. Of HGG tumors with TP53 or PPM1D mutations, 53/55 (96.3%) had mutations in only one of these genes (OR = 0.17, 95% CI: 0.04 - 0.89, q = 0.056), recapitulating previous observations that these mutations are usually mutually exclusive in HGGs [@https://doi.org/10.1038/ng.2938].
CNV and SV analyses revealed that HGG, DMG, and medulloblastoma tumors had the most unstable genomes, while craniopharyngiomas and schwannomas generally lacked somatic CNV (Figure {@fig:S3}C). These CNV patterns largely aligned with our TMB estimates (Figure {@fig:S2}H). SV and CNV breakpoint densities were significantly correlated (linear regression p = 1.05e-38; Figure {@fig:Fig3}C), and as expected, the number of chromothripsis regions called increased with breakpoint density (Figure {@fig:S3}D-E). We identified chromothripsis events in 31% (N = 12/39) of DMGs and in 44% (N = 21/48) of other HGGs (Figure {@fig:Fig3}D), and found evidence of chromothripsis in over 15% of sarcomas, PXAs, metastatic secondary tumors, chordomas, glial-neuronal tumors, germinomas, meningiomas, ependymomas, medulloblastomas, ATRTs, and other embryonal tumors.
We assessed the contributions of eight adult CNS-specific mutational signatures from the RefSig database [@doi:10.1038/s43018-020-0027-5] across tumors (Figure {@fig:Fig3}E and Figure {@fig:S4}A). Signature 1, which reflects normal spontaneous deamination of 5-methylcytosine, predominated in stage 0 and/or 1 tumors characterized by low TMBs (Figure {@fig:S2}H) such as pilocytic astrocytomas, gangliogliomas, other LGGs, and craniopharyngiomas (Figure {@fig:S4}A). Signature 1 weights were generally higher in tumors sampled at diagnosis (pre-treatment) compared to tumors from later phases of therapy (Figure {@fig:S4}B). This trend may have emerged from therapy-induced mutations that produced additional signatures (e.g., temozolomide treatment has been suggested to drive Signature 11 [@doi:10.1053/j.gastro.2014.07.052]), subclonal expansion, and/or acquisition of additional driver mutations during tumor progression, leading to detection of additional signatures. We observed the CNS-specific signature N6 in nearly all tumors. Signature 18 drivers (TP53, APC, NOTCH1; found at https://signal.mutationalsignatures.com/explore/referenceCancerSignature/31/drivers) are also canonical medulloblastoma drivers, and indeed, Signature 18 had the highest signature weight in medulloblastomas. Finally, signatures 3, 8, 18, and MMR2 were prevalent in HGGs, including DMGs.
Most RNA-Seq samples in the PBTA were prepared with ribosomal RNA depletion followed by stranded sequencing (N = 977), while remaining samples were prepared with poly-A selection (N = 58). Since batch correction was not feasible (see Limitations of the Study and Figure {@fig:S7}A), the following transcriptomic analyses considered only stranded samples.
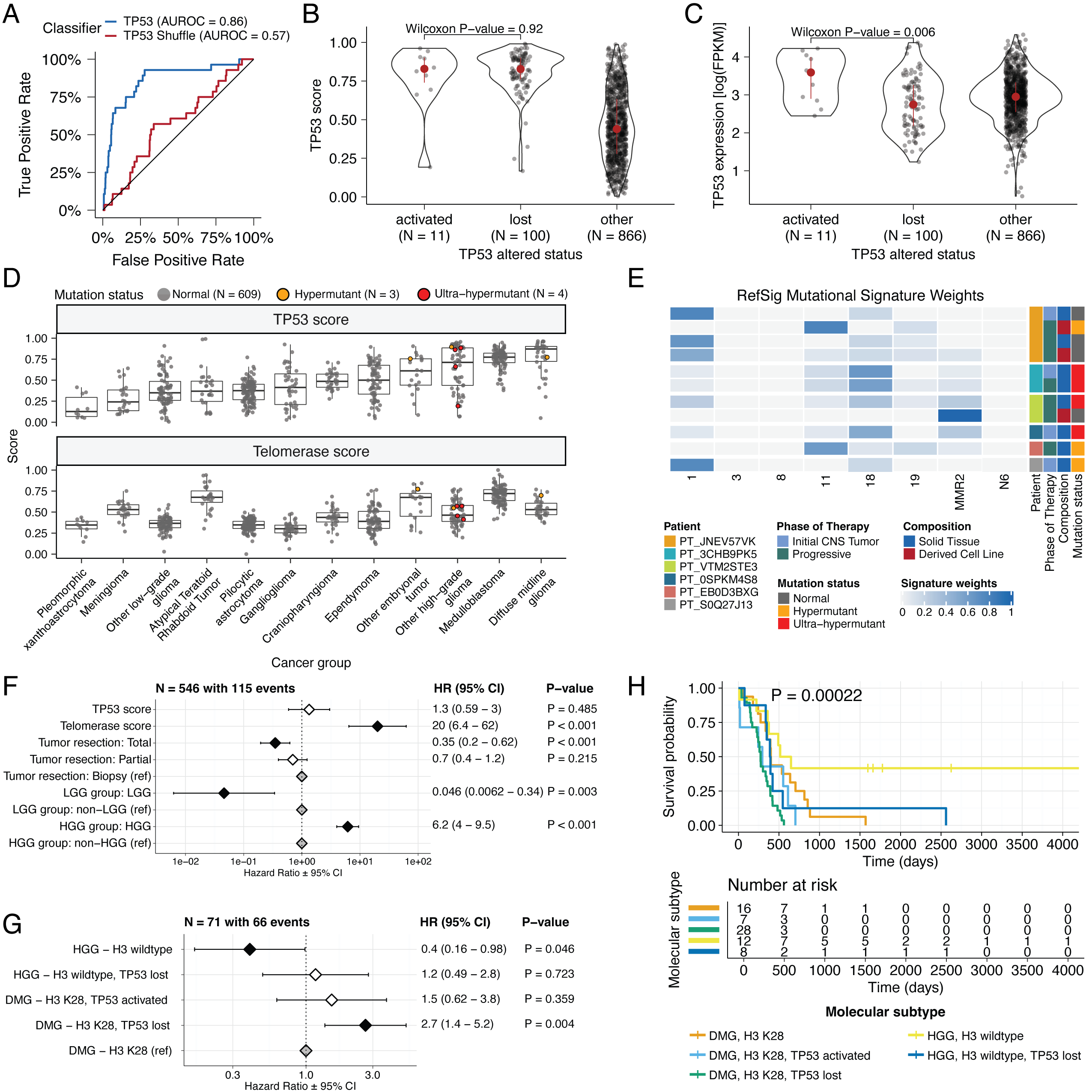
We applied a TCGA-trained classifier [@doi:10.1016/j.celrep.2018.03.076] to calculate a TP53 score, a proxy for TP53 gene or pathway dysregulation, and subsequently infer tumor TP53 inactivation status. We identified "true positive" TP53 alterations from high-confidence SNVs, CNVs, SVs, and fusions in TP53, annotating tumors as "activated" if they harbored one of p.R273C or p.R248W gain-of-function mutations [@doi:10.1038/ng0593-42], or "lost" if 1) the patient had a Li Fraumeni Syndrome (LFS) predisposition diagnosis, 2) the tumor harbored a known hotspot mutation, or 3) the tumor contained two hits (e.g. both SNV and CNV), suggesting both alleles were affected. If the TP53 mutation did not reside within the DNA-binding domain or no alterations in TP53 were detected, we annotated the tumor as "other," indicating an unknown TP53 alteration status. The classifier achieved a high accuracy (AUROC = 0.86) for rRNA-depleted, stranded tumors, but it did not perform as well on the poly-A tumors in this cohort (AUROC = 0.62; Figure {@fig:S5}A).
We observed that "activated" and "lost" tumors had similar TP53 scores (Figure {@fig:Fig4}B, Wilcoxon p = 0.92), contrasting our expectation that "lost" tumors would have higher TP53 scores. This difference suggests that classifier scores > 0.5 may actually represent an oncogenic, or altered, TP53 phenotype rather than solely TP53 inactivation, as interpreted previously [@doi:10.1016/j.celrep.2018.03.076]. However, "activated" tumors showed higher TP53 expression compared to those with TP53 "loss" mutations (Wilcoxon p = 0.006, Figure {@fig:Fig4}C). DMGs, medulloblastomas, HGGs, DNETs, ependymomas, and craniopharyngiomas, all known to harbor TP53 mutations, had the highest median TP53 scores (Figure {@fig:Fig4}D). By contrast, gangliogliomas, LGGs, meningiomas, and schwannomas had the lowest median scores.
We hypothesized that tumors (N = 10) from patients with LFS (N = 8) would have higher TP53 scores, which we indeed observed for 8/10 tumors (Table S3).
Although two tumors had low TP53 scores (BS_DEHJF4C7 at 0.09 and BS_ZD5HN296 at 0.28), pathology reports confirmed that both patients were diagnosed with LFS and harbored a TP53 pathogenic germline variant.
These two LFS tumors also had low tumor purity (16% and 37%, respectively), suggesting that accurate classification may require a certain level of tumor content.
We suggest this classifier could be generally applied to infer TP53 function in the absence of a predicted oncogenic TP53 alteration or DNA sequencing.
We used gene expression data to predict telomerase activity using EXpression-based Telomerase ENzymatic activity Detection (EXTEND) [@doi:10.1038/s41467-020-20474-9] as a surrogate measure of malignant potential [@doi:10.1038/s41467-020-20474-9; @doi:10.1093/carcin/bgp268], where higher EXTEND scores indicate higher telomerase activity.
Aggressive tumors such as DMGs, other HGGs, and MB had high EXTEND scores (Figure {@fig:Fig4}D), and low-grade lesions such as schwannomas, GNGs, DNETs, and other LGGs had among the lowest scores (Table S3), supporting previous reports that aggressive tumor phenotypes have higher telomerase activity [@doi:10.1007/s13277-016-5045-7; @doi:10.1038/labinvest.3700710; @doi:10.1007/s12032-016-0736-x; @doi:10.1111/j.1750-3639.2010.00372.x].
While EXTEND scores were not significantly higher in tumors with TERT promoter (TERTp) mutations (N = 6; Wilcoxon p-value = 0.1196), scores were significantly correlated with TERC (R = 0.619, p < 0.01) and TERT (R = 0.491, p < 0.01) log2 FPKM expression values (Figure {@fig:S5}B-C).
Since catalytically-active telomerase requires full-length TERT, TERC, and certain accessory proteins [@pubmed:9751630], we expect that EXTEND scores may not be exclusively correlated with TERT alterations and expression.
We investigated the mutational signature profiles of hypermutant (TMB > 10 Mut/Mb; N = 3) and ultra-hypermutant (TMB > 100 Mut/Mb; N = 4) tumors and/or derived cell lines from six patients in OpenPBTA (Figure {@fig:Fig4}E).
Five tumors were HGGs and one was a brain metastasis of a MYCN non-amplified neuroblastoma tumor.
Signature 11, which is associated with exposure to temozolomide plus MGMT promoter and/or mismatch repair deficiency [@doi:10.1038/s41588-019-0525-5], was indeed present in tumors with previous exposure to the drug (Table {@tbl:Table2}).
We detected the MMR2 signature in tumors of four patients (PT_0SPKM4S8, PT_3CHB9PK5, PT_JNEV57VK, and PT_VTM2STE3) diagnosed with either constitutional mismatch repair deficiency (CMMRD) or Lynch syndrome (Table {@tbl:Table2}), genetic predisposition syndromes caused by a variant in a mismatch repair gene such as PMS2, MLH1, MSH2, MSH6, or others [@doi:10.1136/jmedgenet-2020-107627].
Three of these patients harbored pathogenic germline variants in one of the aforementioned genes.
While we did not detect a known pathogenic variant in the germline of PT_VTM2STE3, this patient's pathology report contained a self-reported PMS2 variant, and we indeed found 19 intronic variants of unknown significance (VUS) in their PMS2.
This is not surprising since an estimated 49% of germline PMS2 variants in patients with CMMRD and/or Lynch syndrome are VUS [@doi:10.1136/jmedgenet-2020-107627].
Interestingly, while the cell line derived from patient PT_VTM2STE3's tumor at progression was not hypermutated (TMB = 5.7 Mut/Mb), it only contained the MMR2 signature, suggesting selective pressure to maintain a mismatch repair (MMR) phenotype in vitro.
Only one of the two cell lines derived from patient PT_JNEV57VK's progressive tumor was hypermutated (TMB = 35.9 Mut/Mb).
The hypermutated cell line was strongly weighted towards signature 11, while the non-hypermutated cell line showed several lesser signature weights (1, 11, 18, 19, MMR2; Table S2).
This mutational process plasticity highlights the importance of careful genomic characterization and model selection for preclinical studies.
Signature 18, which has been associated with high genomic instability and can induce a hypermutator phenotype [@doi:10.1038/s43018-020-0027-5], was uniformly represented among hypermutant solid tumors. Additionally, all hypermutant HGG tumors or cell lines had dysfunctional TP53 (Table {@tbl:Table2}), consistent with previous findings that tumors with high genomic instability signatures require TP53 dysregulation [@doi:10.1038/s43018-020-0027-5]. With one exception, hypermutant and ultra-hypermutant tumors had high TP53 scores (> 0.5) and telomerase activity. Interestingly, none of the hypermutant tumors showed evidence of signature 3 (present in homologous recombination deficient tumors), signature 8 (arises from double nucleotide substitutions/unknown etiology), or signature N6 (a universal CNS tumor signature). The mutual exclusivity of signatures 3 and MMR2 corroborates previous suggestions that tumors do not generally feature both deficient homologous repair and mismatch repair [@doi:10.1016/j.celrep.2018.03.076].
| Kids First Participant ID | Kids First Biospecimen ID | CBTN ID | Phase of therapy | Composition | Therapy post-biopsy | Cancer predisposition | Pathogenic germline variant | TMB | OpenPBTA molecular subtype |
|---|---|---|---|---|---|---|---|---|---|
| PT_0SPKM4S8 | BS_VW4XN9Y7 | 7316-2640 | Initial CNS Tumor | Solid Tissue | Radiation, Temozolomide, CCNU | None documented | NM_000535.7(PMS2):c.137G>T (p.Ser46Ile) (LP) | 187.4 | HGG, H3 wildtype, TP53 activated |
| PT_3CHB9PK5 | BS_20TBZG09 | 7316-515 | Initial CNS Tumor | Solid Tissue | Radiation, Temozolomide, Irinotecan, Bevacizumab | CMMRD | NM_000179.3(MSH6):c.3439-2A>G (LP) | 307 | HGG, H3 wildtype, TP53 loss |
| PT_3CHB9PK5 | BS_8AY2GM4G | 7316-2085 | Progressive | Solid Tissue | Radiation, Temozolomide, Irinotecan, Bevacizumab | CMMRD | NM_000179.3(MSH6):c.3439-2A>G (LP) | 321.6 | HGG, H3 wildtype, TP53 loss |
| PT_EB0D3BXG | BS_F0GNWEJJ | 7316-3311 | Progressive | Solid Tissue | Radiation, Nivolumab | None documented | None detected | 26.3 | Metastatic NBL, MYCN non-amplified |
| PT_JNEV57VK | BS_85Q5P8GF | 7316-2594 | Initial CNS Tumor | Solid Tissue | Radiation, Temozolomide | Lynch Syndrome | NM_000251.3(MSH2):c.1906G>C (p.Ala636Pro) (P) | 4.7 | DMG, H3 K28, TP53 loss |
| PT_JNEV57VK | BS_HM5GFJN8 | 7316-3058 | Progressive | Derived Cell Line | Radiation, Temozolomide, Nivolumab | Lynch Syndrome | NM_000251.3(MSH2):c.1906G>C (p.Ala636Pro) (P) | 35.9 | DMG, H3 K28, TP53 loss |
| PT_JNEV57VK | BS_QWM9BPDY | 7316-3058 | Progressive | Derived Cell Line | Radiation, Temozolomide, Nivolumab | Lynch Syndrome | NM_000251.3(MSH2):c.1906G>C (p.Ala636Pro) (P) | 7.4 | DMG, H3 K28, TP53 loss |
| PT_JNEV57VK | BS_P0QJ1QAH | 7316-3058 | Progressive | Solid Tissue | Radiation, Temozolomide, Nivolumab | Lynch Syndrome | NM_000251.3(MSH2):c.1906G>C (p.Ala636Pro) (P) | 6.3 | DMG, H3 K28, TP53 activated |
| PT_S0Q27J13 | BS_P3PF53V8 | 7316-2307 | Initial CNS Tumor | Solid Tissue | Radiation, Temozolomide, Irinotecan | None documented | None detected | 15.5 | HGG, H3 wildtype, TP53 activated |
| PT_VTM2STE3 | BS_ERFMPQN3 | 7316-2189 | Progressive | Derived Cell Line | Unknown | Lynch Syndrome | None detected | 5.7 | HGG, H3 wildtype, TP53 loss |
| PT_VTM2STE3 | BS_02YBZSBY | 7316-2189 | Progressive | Solid Tissue | Unknown | Lynch Syndrome | None detected | 274.5 | HGG, H3 wildtype, TP53 activated |
Table: Patients with hypermutant tumors. Patients with at least one hypermutant or ultra-hypermutant tumor or cell line. Pathogenic (P) or likely pathogenic (LP) germline variants, coding region TMB, phase of therapy, therapeutic interventions, cancer predisposition (CMMRD = Constitutional mismatch repair deficiency), and molecular subtypes are included. {#tbl:Table2}
Next, we asked whether transcriptomic classification of TP53 dysregulation and/or telomerase activity recapitulate these oncogenic biomarkers' known prognostic influence.
We identified several expected trends, including a significant overall survival benefit following full tumor resection (HR = 0.35, 95% CI = 0.2 - 0.62, p < 0.001) or if the tumor was an LGG (HR = 0.046, 95% CI = 0.0062 - 0.34, p = 0.003), and a significant risk if the tumor was an HGG (HR = 6.2, 95% CI = 4.0 - 9.5, p < 0.001) (Figure {@fig:Fig4}F; STAR Methods).
High telomerase scores were associated with poor prognosis across brain tumor histologies (HR = 20, 95% CI = 6.4 - 62, p < 0.001), demonstrating that EXTEND scores calculated from RNA-Seq are an effective rapid surrogate measure for telomerase activity.
Higher TP53 scores were associated with significant survival risks (Table S4) within DMGs (HR = 6436, 95% CI = 2.67 - 1.55e7, p = 0.03) and ependymomas (HR = 2003, 95% CI = 9.9 - 4.05e5, p = 0.005).
Given this result, we next assessed whether different HGG molecular subtypes carry different survival risks if stratified by TP53 status.
We found that DMG H3 K28 tumors with TP53 loss had significantly worse prognosis (HR = 2.8, CI = 1.4-5.6, p = 0.003) than those with wildtype TP53 (Figure {@fig:Fig4}G and Figure {@fig:Fig4}H), recapitulating results from two recent restrospective analyses of DIPG tumors [@doi:10.1158/1078-0432.CCR-22-0803; @doi:10.1007/s11060-021-03890-9].
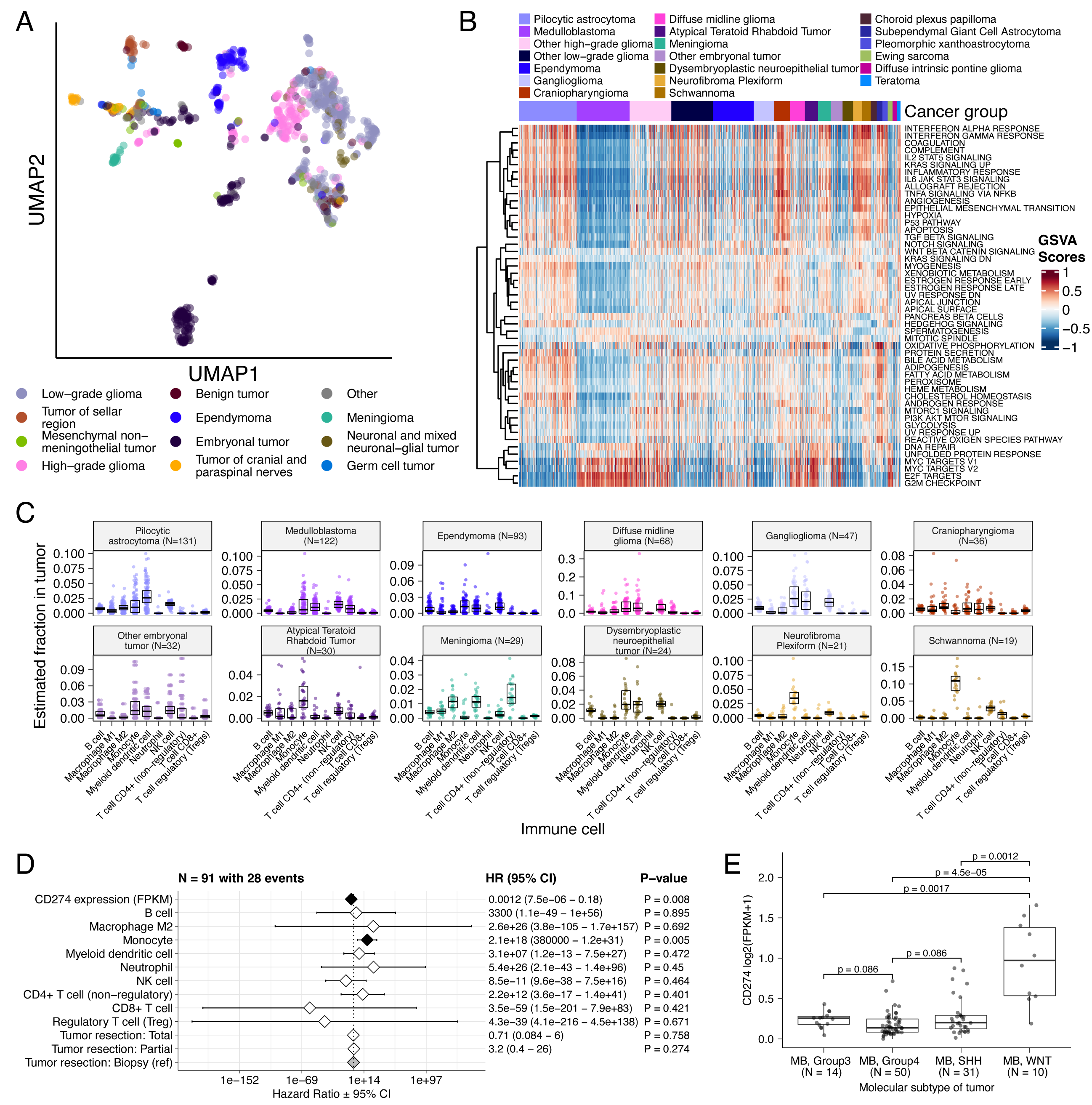
UMAP visualization of gene expression variation across brain tumors (Figure {@fig:Fig5}A) showed expected histological clustering of brain tumors. We further observed that, except for three outliers, C11orf95::RELA (ZFTA::RELA) fusion-positive ependymomas fell within distinct clusters (Figure {@fig:S6}A). Medulloblastoma (MB) tumors clustered by molecular subtype, with WNT and SHH in distinct clusters and Groups 3 and 4 showing some expected overlap (Figure {@fig:S6}B). Notably, two MB tumors annotated as SHH did not cluster with the other MB tumors and one clustered with Group 3/4 tumors, suggesting potential subtype misclassification or different underlying biology of these two tumors. BRAF-driven LGGs (Figure {@fig:S6}C) fell into three separate clusters, suggesting additional shared biology within each cluster. Histone H3 G35-mutant HGGs generally clustered together and away from K28-mutant tumors (Figure {@fig:S6}D). Interestingly, although H3 K28-mutant and H3 wildtype tumors have different biological drivers [@doi:10.1126/science.1232245], they did not form distinct clusters. This pattern suggests these subtypes may be driven by common transcriptional programs, have other much stronger biological drivers than their known distinct epigenetic drivers, or we lack power to detect transcriptional differences.
We performed GSVA for Hallmark cancer gene sets (Figure {@fig:Fig5}B) and quantified immune cell fractions using quanTIseq (Figure {@fig:Fig5}C and Figure {@fig:S6}E), results from which recapitulated previously-described tumor biology. For example, HGG, DMG, MB, and ATRT tumors are known to upregulate MYC [@doi:10.3390/genes8040107] which in turn activates E2F and S phase [@pubmed:11511364]. Indeed, we detected significant (Bonferroni-corrected p < 0.05) upregulation of MYC and E2F targets, as well as G2M (cell cycle phase following S phase) in MBs, ATRTs, and HGGs compared to several other cancer groups. In contrast, LGGs showed significant downregulation (Bonferroni-corrected p < 0.05, multiple cancer group comparisons) of these pathways. Schwannomas and neurofibromas, which have an inflammatory immune microenvironment of T and B lymphocytes and tumor-associated macrophages (TAMs), are driven by upregulation of cytokines such as IFN$\gamma$, IL-1, and IL-6, and TNF$\alpha$ [@doi:10.1093/noajnl/vdaa023]. GSVA revealed significant upregulation of these cytokines in hallmark pathways (Bonferroni-corrected p < 0.05, multiple cancer group comparisons) (Figure {@fig:Fig5}B), and monocytes dominated these tumors' immune cell repertoire (Figure {@fig:Fig5}C). We also observed significant upregulation of pro-inflammatory cytokines IFN$\alpha$ and IFN$\gamma$ in both LGGs and craniopharyngiomas when compared to either medulloblastoma or ependymomas (Bonferroni-corrected p < 0.05) (Figure {@fig:Fig5}B). Together, these results support previous proteogenomic findings that aggressive medulloblastomas and ependymomas have lower immune infiltration compared to BRAF-driven LGGs and craniopharyngiomas [@doi:10.1016/j.cell.2020.10.044].
Although CD8+ T-cell infiltration across all cancer groups was minimal (Figure {@fig:Fig5}C), we observed signal in specific cancer molecular subtypes (Groups 3 and 4 medulloblastoma) as well as outlier tumors (BRAF-driven LGG, BRAF-driven and wildtype ganglioglioma, and CNS embryonal NOS; Figure {@fig:S6}E) Surprisingly, the classically immunologically-cold HGGs and DMGs [@doi:10.1186/s40478-018-0553-x; @doi:10.1093/brain/awab155] contained higher overall fractions of immune cells, primarily monocytes, dendritic cells, and NK cells (Figure {@fig:Fig5}C). Thus, quanTIseq might have actually captured microglia within these immune cell fractions.
While we did not detect notable prognostic effects of immune cell infiltration on overall survival in HGGs or DMGs, we found that high levels of macrophage M1 and monocytes were associated with poorer overall survival (monocyte HR = 2.1e18, 95% CI = 3.80e5 - 1.2e31, p = 0.005, multivariate Cox) in medulloblastomas (Figure {@fig:Fig5}D). We further reproduced previous findings (Figure {@fig:Fig5}E) that medulloblastomas typically have low expression of CD274 (PD-L1) [@doi:10.18632/oncotarget.24951]. We also found that higher expression of CD274 was significantly associated with improved overall prognosis for medulloblastoma tumors, although marginal (HR = 0.0012, 95% CI = 7.5e−06 - 0.18, p = 0.008, multivariate Cox) (Figure {@fig:Fig5}D). This result may be explained by the higher expression of CD274 observed in WNT subtype tumors by us and others [@doi:10.1080/2162402X.2018.1462430], as this diagnosis carries the best prognosis of all medulloblastoma subgroups (Figure {@fig:Fig5}E).
We additionally explored the ratio of CD8+ to CD4+ T cells across tumor subtypes. This ratio has been associated with better immunotherapy response and prognosis following PD-L1 inhibition in non-small cell lung cancer or adoptive T cell therapy in multiple stage III or IV cancers [@doi:10.1136/jitc-2021-004012; @doi:10.4236/jct.2013.48164]. While adamantinomatous craniopharyngiomas and Group 3 and Group 4 medulloblastomas had the highest ratios (Figure {@fig:S6}F), very few tumors had ratios greater than 1, highlighting an urgent need to identify novel therapeutics for pediatric brain tumors with poor prognosis.
Finally, we explored the potential influence of tumor purity by repeating selected transcriptomic analyses restricted to only samples with high tumor purity (see STAR Methods). Results from these analyses were broadly consistent (Figure {@fig:S7}D-I) with results derived from all stranded RNA-Seq samples.