-
Notifications
You must be signed in to change notification settings - Fork 3
Stats
One of our main objectives one developping SCALPEL-Analysis was to build ready-to-use, out of the box descriptive statistics over the CohortFlow. This allows to assess quickly the Flowchart building, depict correlations ans biases and especially check the quality of the data.
In this tutorial, we will show how use existing stats to perform basic statistical analysis. We will also list all the available stats functions. Finally, we show how to add new functions to the library while using decorators to automatise boiler plat code.
%matplotlib inline
%load_ext autoreload
%autoreload 2
%config InlineBackend.figure_format = 'retina'
import pandas as pd
pd.set_option('display.max_rows', 500)SCALPEL-Analysis is not yet available through PIP or Conda channels to be installed. However, as is the case for any Python library, it is pretty straight forward to add and use.
Before, proceeding you will need to download SCALPEL-Analysis as a zip from here. Put the zip wherever you judge suitable for you. Please make sure that you meet all the requirements explicited here.
I have downloaded the zip file, and put it under the path /home/sebiat/builds/cnam-model-001-interactions/dist/scalpel.zip.
There is two ways of doing it:
- Permanently add a directory to PYTHONPATH environmment variable. This will allow you to add it once and for all.
- Add it through
sysimport as shown below.
import sys
project_path = '/home/sebiat/builds/cnam-model-001-interactions/dist/scalpel.zip'
sys.path.append(project_path)As stated in the first comment cell, you will need to import the Metadata JSON. There is a straightforward way of doing it that will save the pain of a lot of boiler plate code. The following cell shows how.
from scalpel.core.cohort_collection import CohortCollection
import pytz
AGE_REFERENCE_DATE = pytz.datetime.datetime(2015, 1, 1, tzinfo=pytz.UTC)
cc = CohortCollection.from_json("metadata_fall_2020_01_27_16_46_39.json")
cc.add_subjects_information('omit_all', reference_date=AGE_REFERENCE_DATE)
cc.cohorts_names2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
2020-03-09 15:55:42 WARNING : Some patients and their events might be ommited
{'Cardiac',
'HTA',
'IPP',
'Opioids',
'acts',
'control_drugs_exposures',
'control_drugs_purchases',
'diagnoses',
'drug_purchases',
'epileptics',
'exposures',
'extract_patients',
'filter_patients',
'follow_up',
'fractures',
'interactions',
'liberal_acts',
'prescriptions',
'prescriptions_exposures'}
Right, we officially have a CohortCollection with a number of of Cohorts. But wait, what it is a CohortCollection? No rush, here is the definition from the SCALPEL paper, section 2.5:
The
CohortCollectionabstraction is a collection ofCohorts on which operations can be jointly performed. TheCohortCollectionhas metadata that keeps information about eachCohort, such as the successive operations performed on it, the Parquet files they are stored in and a git commit hash of the code producing the extraction from theSource.
CohortCollection can be seen in the same way a list or dict in Python is seen. It is a bag full of Cohorts that allows to iterate over, but also allows to apply operations to all the Cohorts at once. Finally, it allows to do specific operations for the CohortCollection such as finding the basic subjects Cohort which is the largest Cohort in the CohortCollection. We have a dedicated tutorial for CohortCollection.
What is usely called a Flowchart in epidemiology, is referenced in our library as CohortFlow. This naming is partly explained by the behaviour of the abstraction. It can be seen as an initial Cohort flowing through the steps of the chart.
Here is the definition as is in the SCALPEL3 paper:
The
CohortFlowabstraction is an orderedCohortCollection, where eachCohortis included in the previous one. It is meant to track the stages leading to a finalCohort, where each intermediateCohortis stored along with textual information about the filtering rules used to go from each stage to the next one.
Remember that each Cohort stores a description in human-readable format.
Technically, a CohortFlow is just an ordered collection of Cohorts. A user just pass a simple list of Cohort to initiate a CohortFlow object. The CohortFlow object than stores in two Python properties all the needed information:
-
ordered_cohorts: just a copy of the initial passed list. -
steps: are the realCohortsthat appear in epidemiological studies. Eachstep[n]being theCohortintersection betweenordered_cohorts[n]andstep[n-1].
# Import the necessary Python objects
from scalpel.core.cohort_flow import CohortFlow
# create the CohortFlow
cf = CohortFlow([
cc.get('extract_patients'),
cc.get('exposures'),
cc.get('filter_patients'),
cc.get('fractures')
])The above cohort flow can be translated to the following human readable steps:
- Get all the subjects available.
- Keep only exposed subjects.
- Keep only filtered subjects. In this particular case, subjects not exposed in the first year of the study.
- Keep only fractured subjects.
# Explore the ordered_cohorts
for cohort in cf.ordered_cohorts:
print(cohort.describe())This a subject cohort, no event needed. Subjects are from operation extract_patients.
Events are exposures. Events contain only subjects with event exposures.
This a subject cohort, no event needed. Subjects are from operation filter_patients.
Events are fractures. Events contain only subjects with event fractures.
All stat functions that are available have the exact same signature
def plot_xxxx(figure: Figure, cohort: Cohort, kwargs**: Dict) -> FigureSo all the functions that are made available follow the exact same pattern. This has numerous advantages:\
- Allow to use closures easily to decorate the functions and avoid boilerplate code.
- Allow to iterate over the functions without knowing them by name. They are just callable where you need to pass two mandatory parameters.
- You can also pass optional paramaters, if you want more control over the behaviour, but this will force you to know which method you are dealing with.
Below is the example of a function defined in stats.patients module in SCALPEL-Analysis.
@register
@title("Gender distribution")
@xlabel("Gender")
@ylabel("Subjects count")
def distribution_by_gender(figure, cohort):
df = agg(cohort.subjects, frozenset(["gender"]), "count")
ax = figure.gca()
ax = sns.barplot(x=df["gender"].values, y=df["count(1)"].values, ax=ax)
ax.set_xticklabels(["Male", "Female"])
return figureThe plot function allows to get the distribution of the subjects based on their gender. Notice the use of the decorators and an agg method. We will have a deeper look at why these are helpful when discussing the developpement of new stat methods.
The stats module and the CohortFlow are the perfect combination. Together, they leverage the user to get insights in the studied data and spot eventual biases. One major advantage is that all the stats that exist can be easily used on all the steps of the cohort flow. Look at the example below.
from scalpel.stats.patients import distribution_by_gender
import matplotlib.pyplot as pltfor cohort in cf.steps:
figure = plt.figure(figsize=(8, 4.5))
distribution_by_gender(cohort=cohort, figure=figure)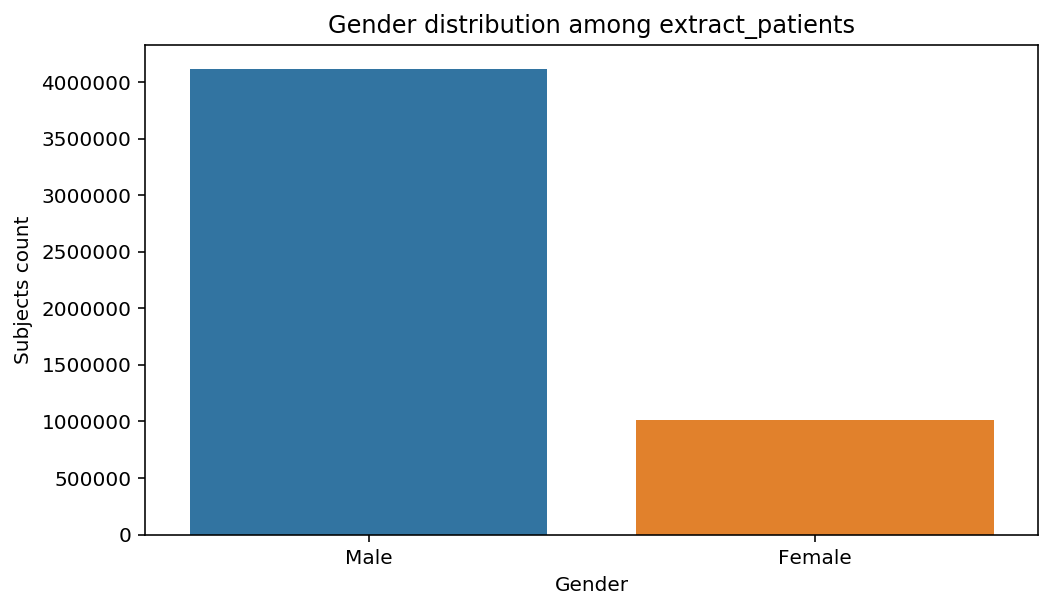
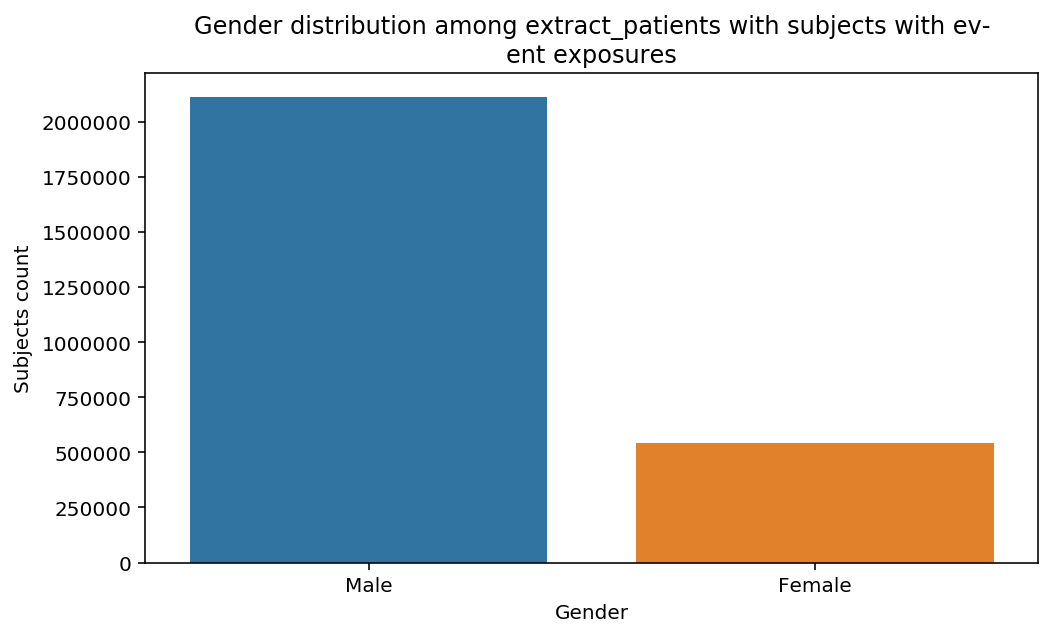
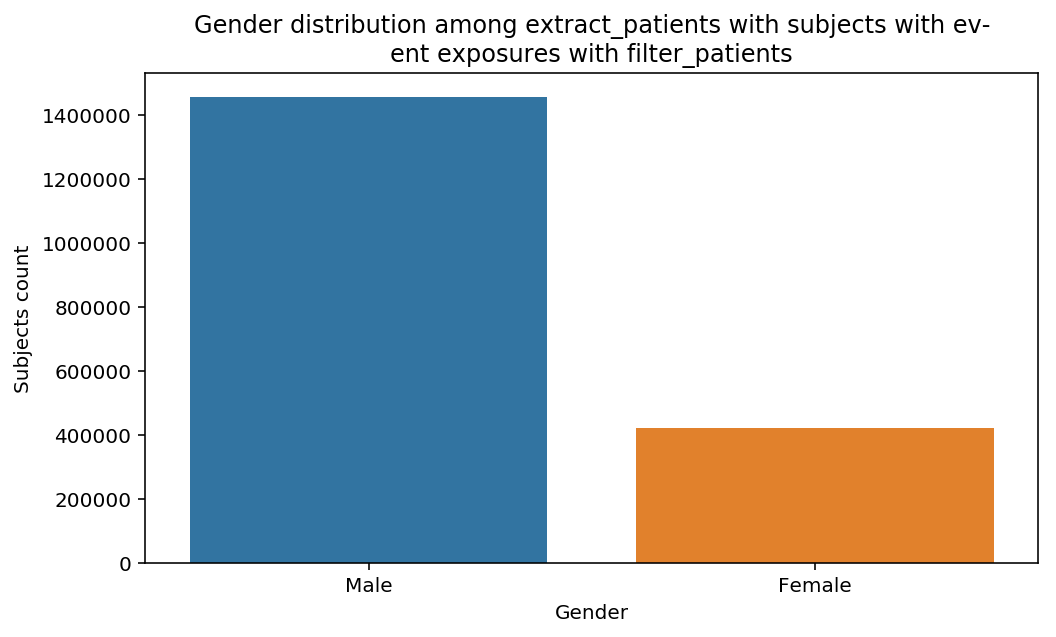
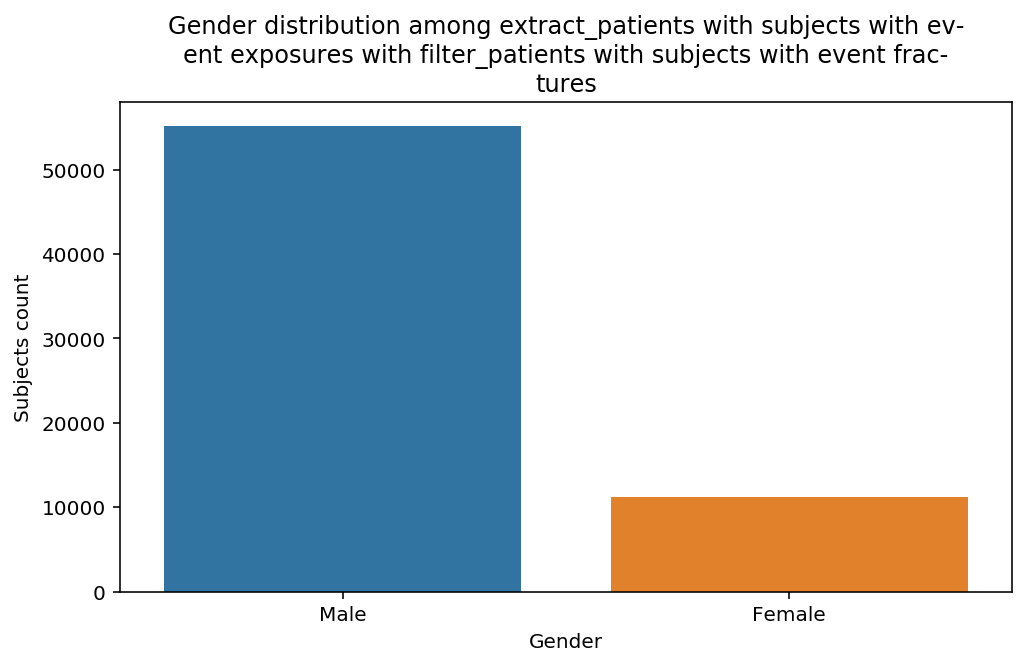
The graphs show a bias toward Male subjects, especially in the last step. Notice that the title of the of the graph adapted automatically to the decscription of the Cohort. This when advantage of the title decorator used above.
One following some stat through the CohortFlow one might wonder if is it possible to get not the absolute number, but a percentage to follow. Here how is it possible.
from scalpel.stats.decorators import percentage_yfor cohort in cf.steps:
figure = plt.figure(figsize=(8, 4.5))
total = cohort.subjects.count()
percentage_y(total=total, y_limit=90)(distribution_by_gender)(cohort=cohort, figure=figure)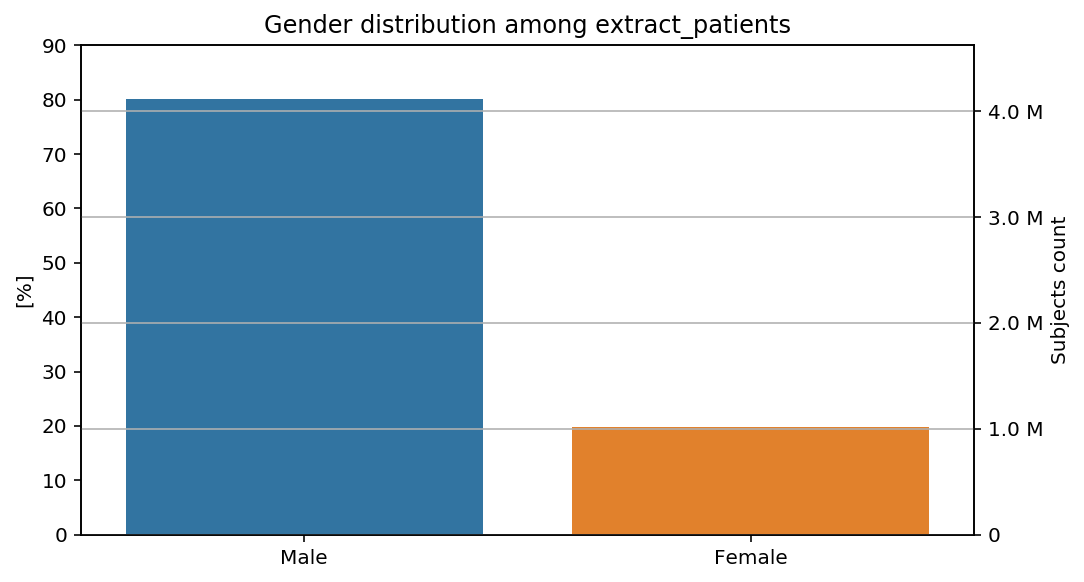
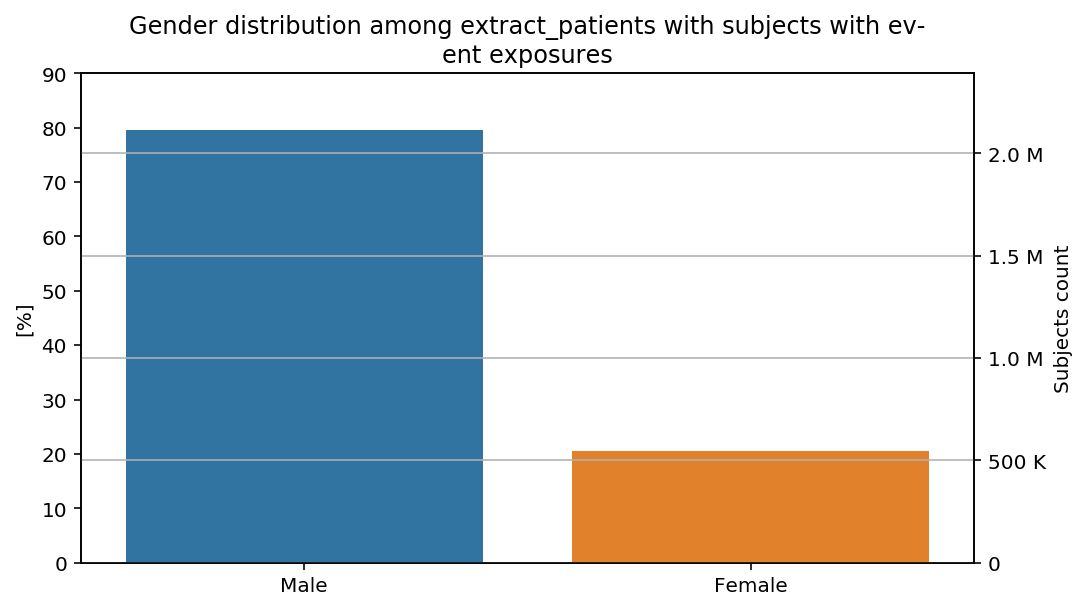
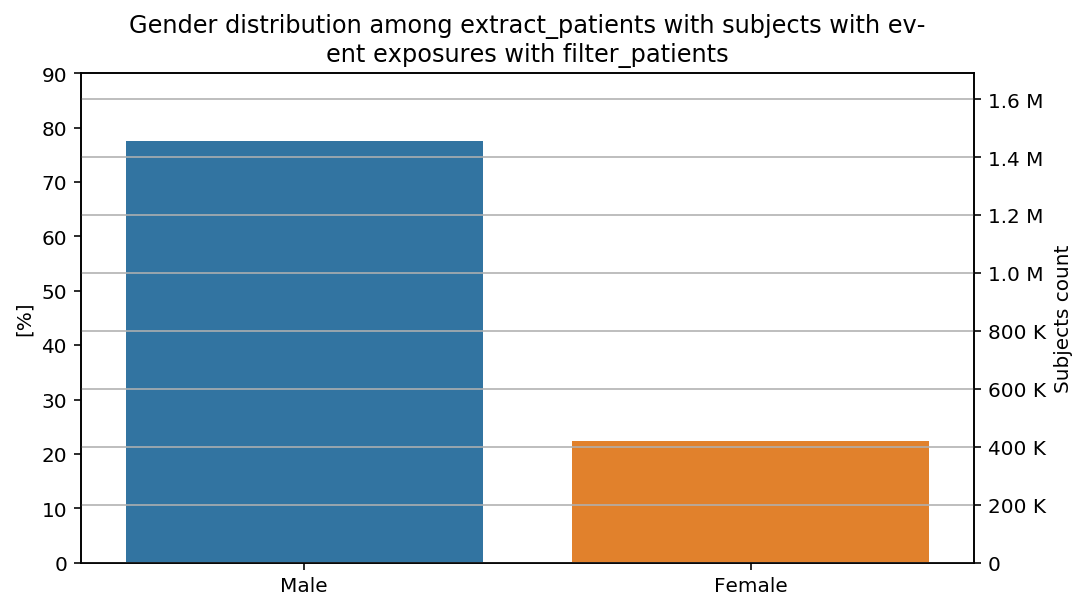
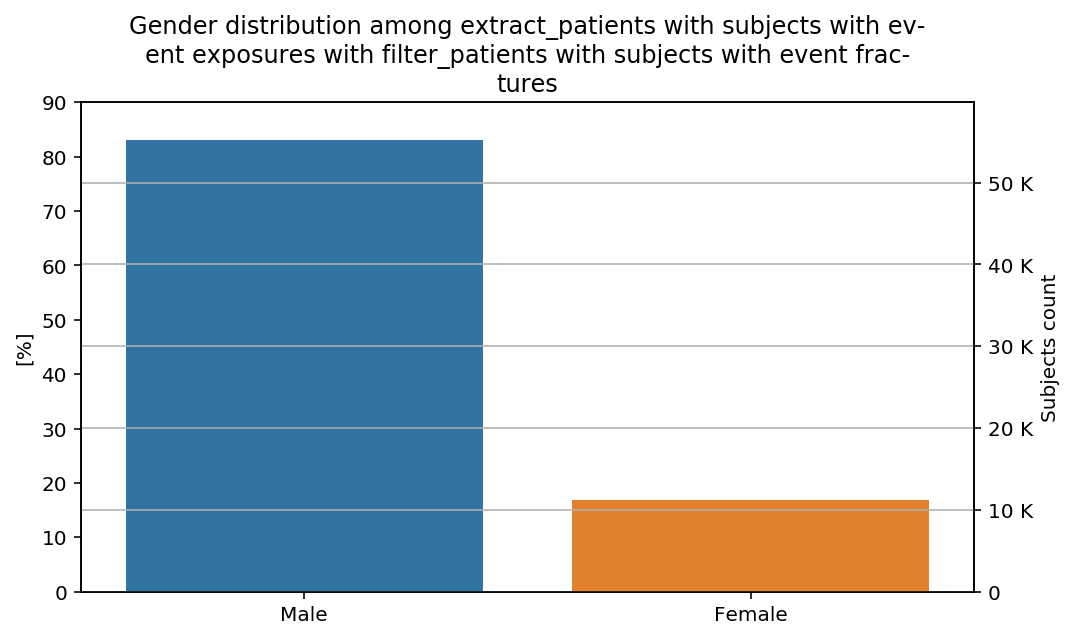
The percentages show our previous remarks: the percentage of male subjects is slightly higher in the last step than in the previous steps of the CohortFlow.
As stated above, the rigid structure allows the user to invoke plot functions without specifically naming them. This for instance can be very helpful to launch all available stats at once.
Every script in the stats module has a registry variable that can be imported. registry is just a python list that stores all of the available functions in the script. To add a function to registry just call the @register decorator above your plot function. Below an example of how to use it.
from scalpel.stats.event_distribution import registryfor plot in registry:
for cohort in cf.prepend_cohort(cc.get('fractures')).steps:
figure = plt.figure(figsize=(8, 4.5))
plot(cohort=cohort, figure=figure)/usr/local/lib/anaconda3/lib/python3.6/site-packages/matplotlib/pyplot.py:537: RuntimeWarning: More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).
max_open_warning, RuntimeWarning)
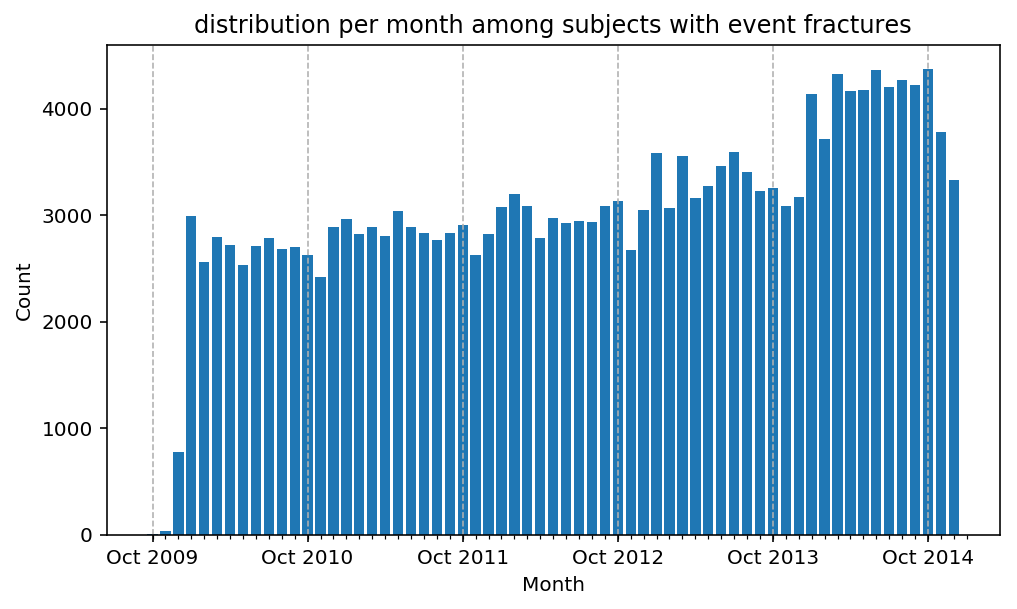
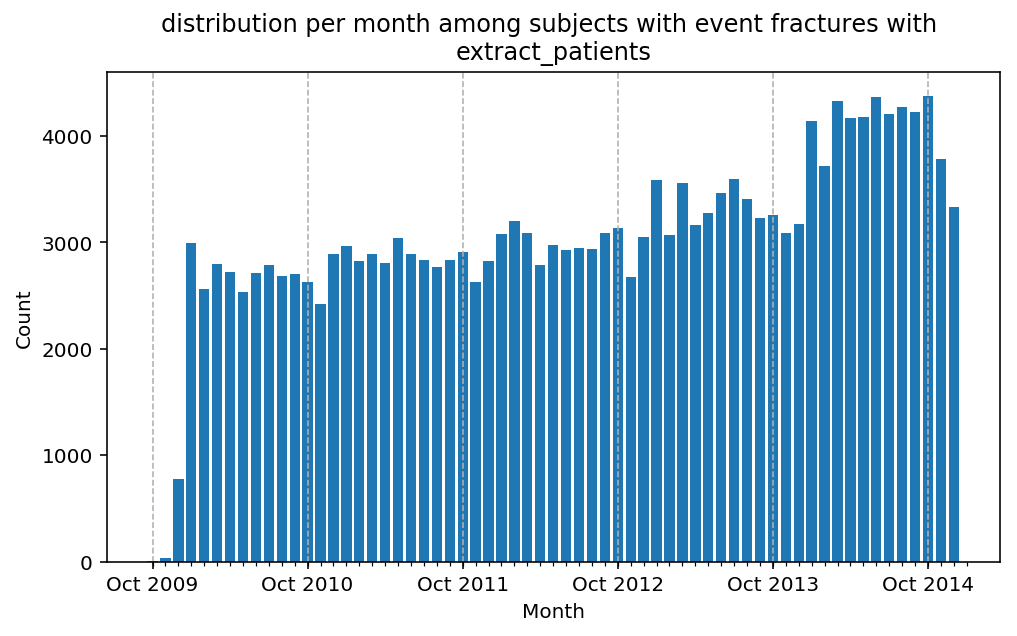
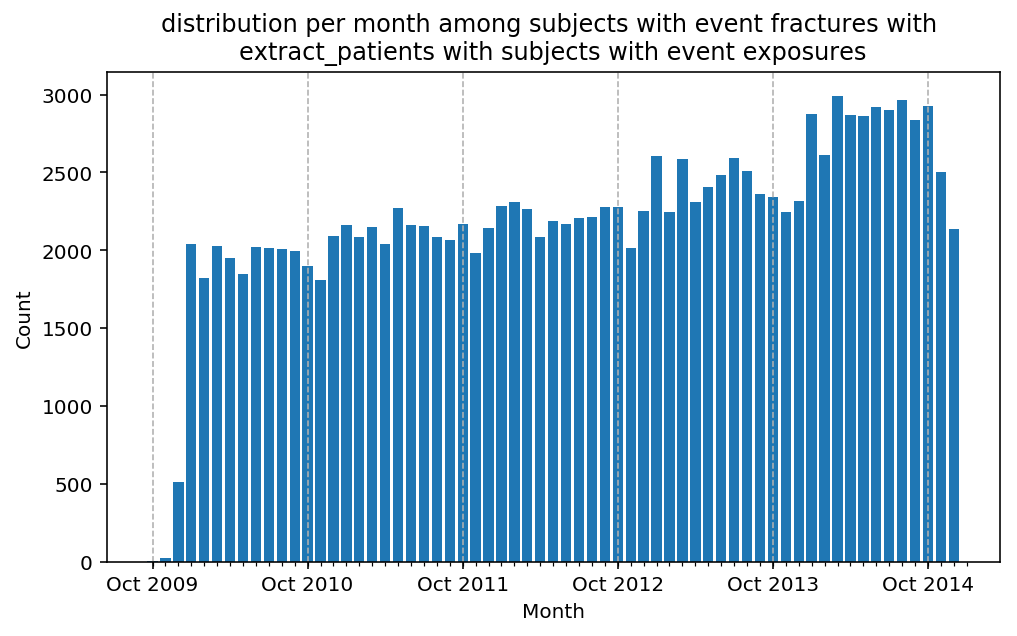
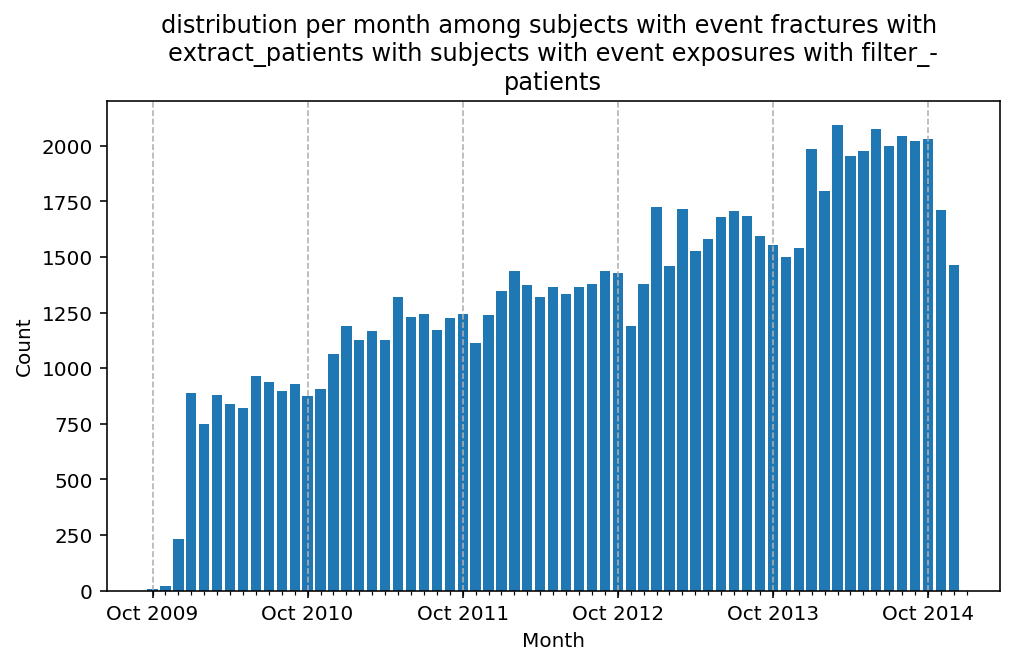
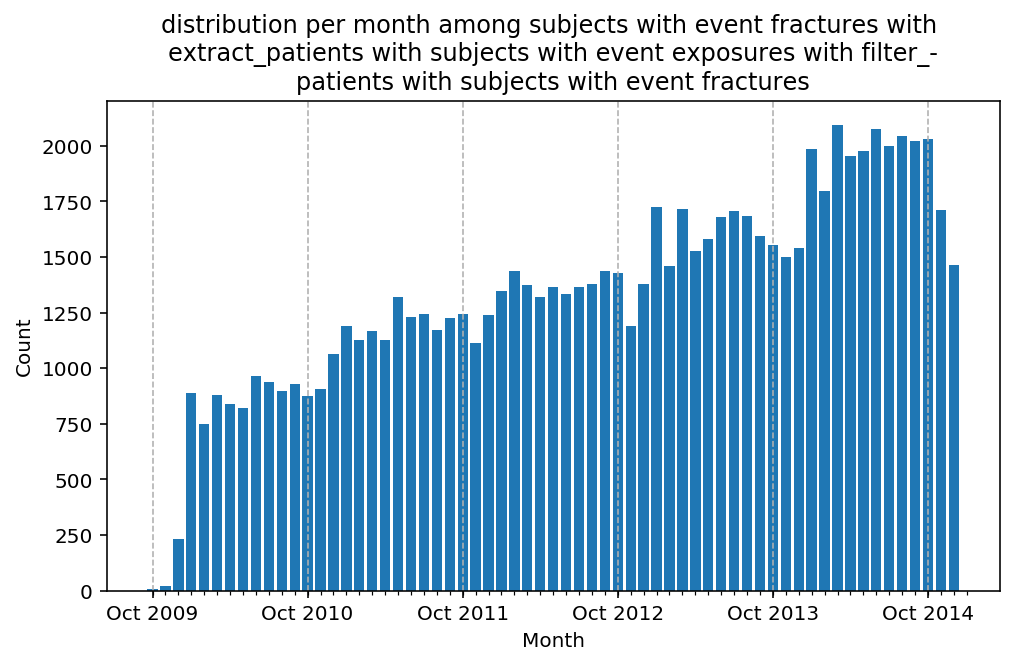
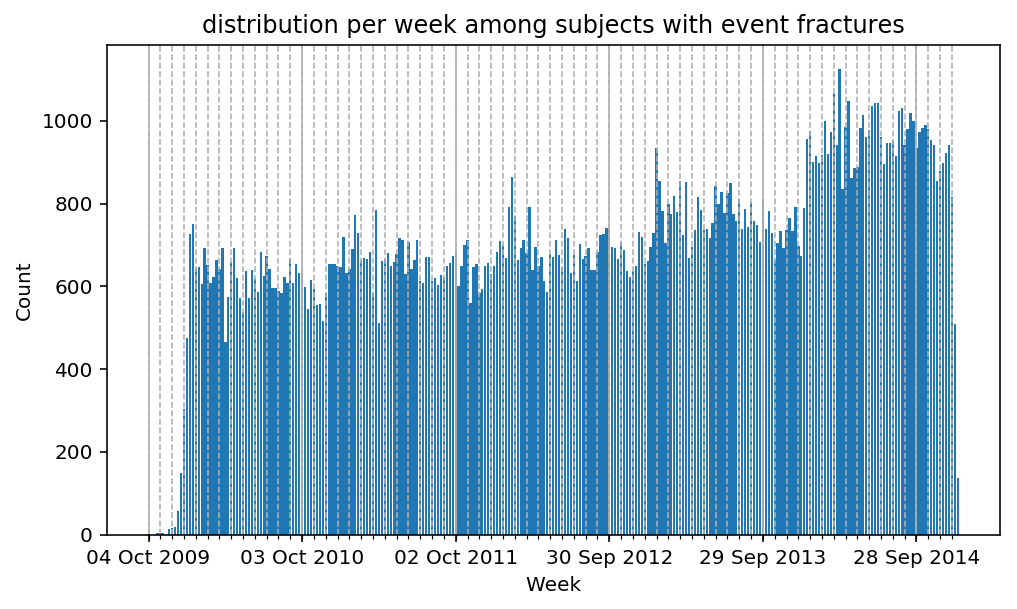
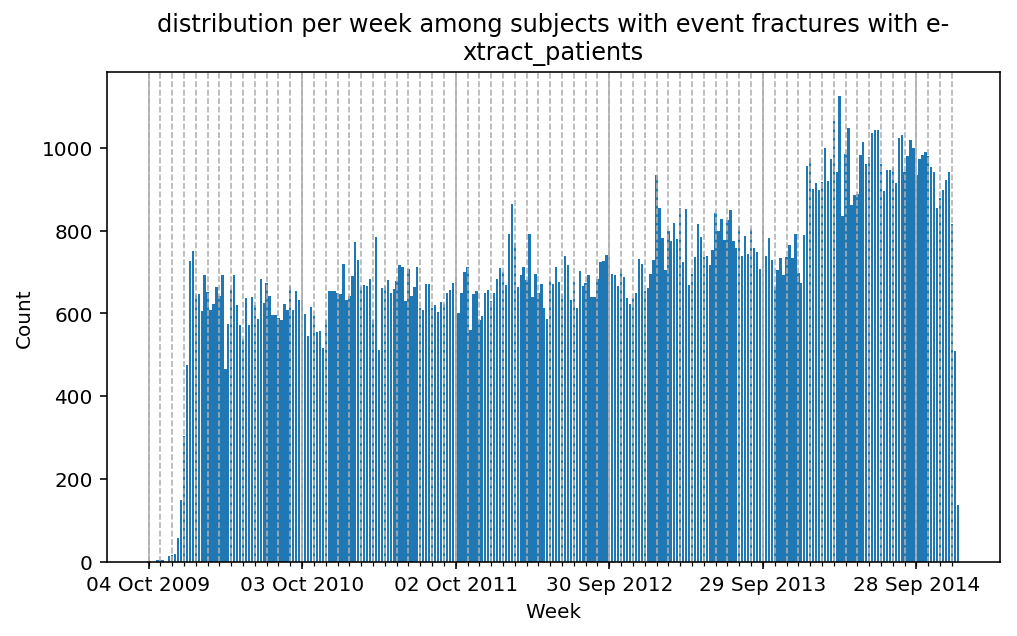
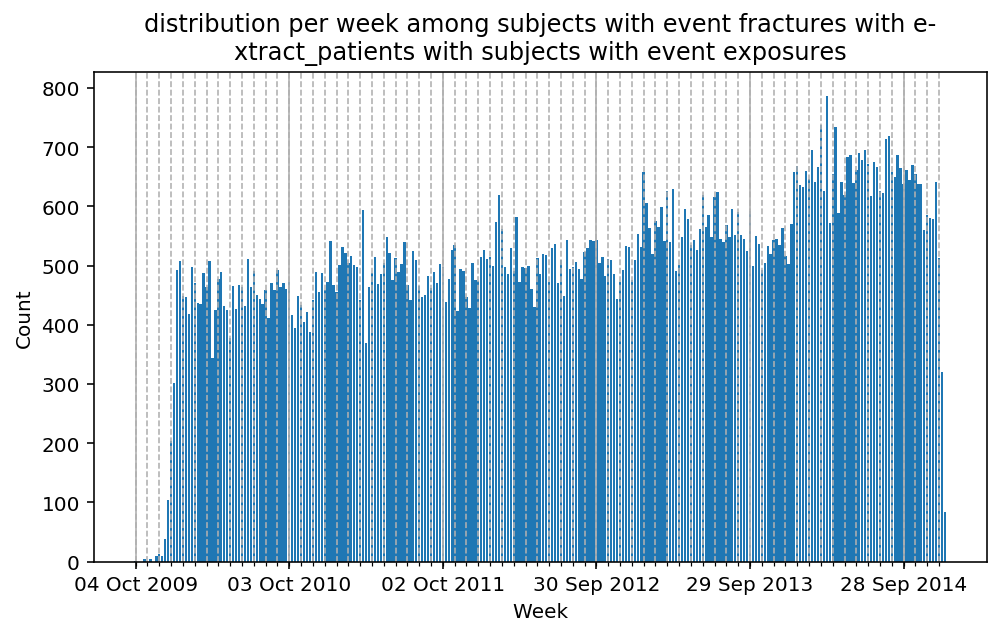
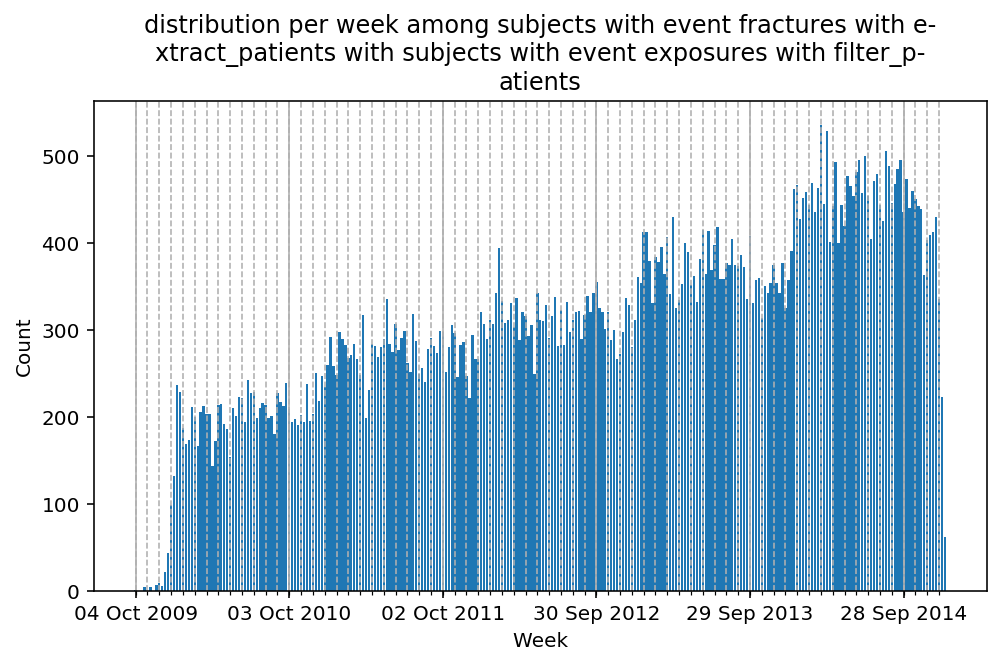
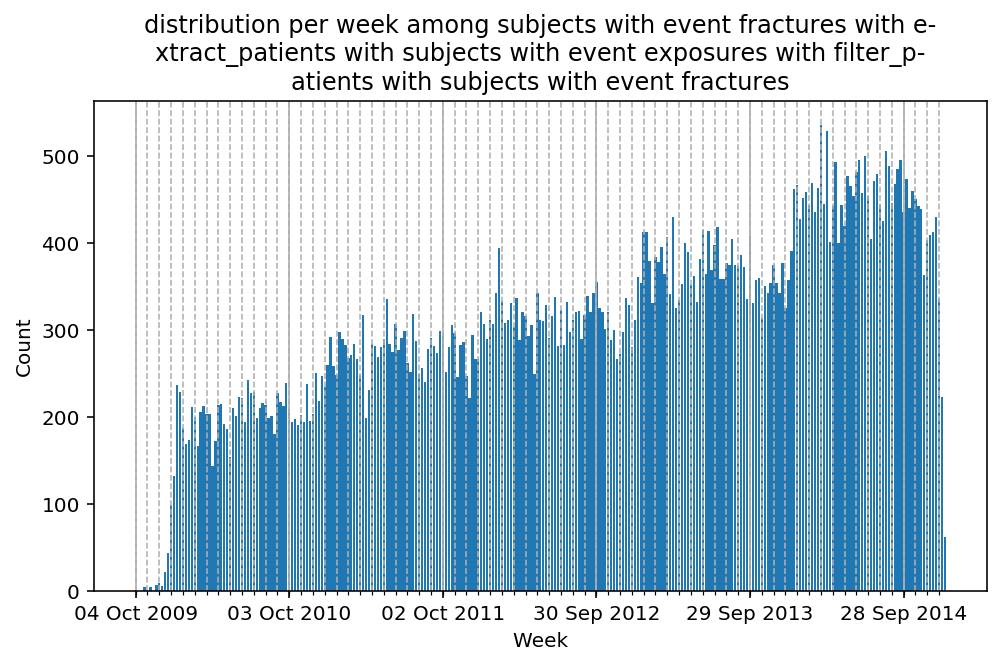
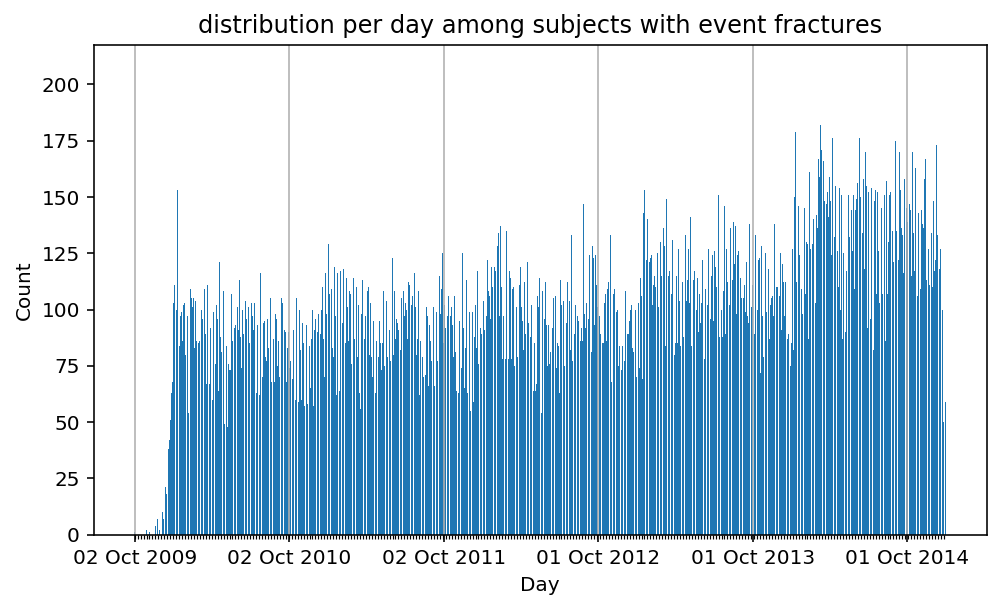
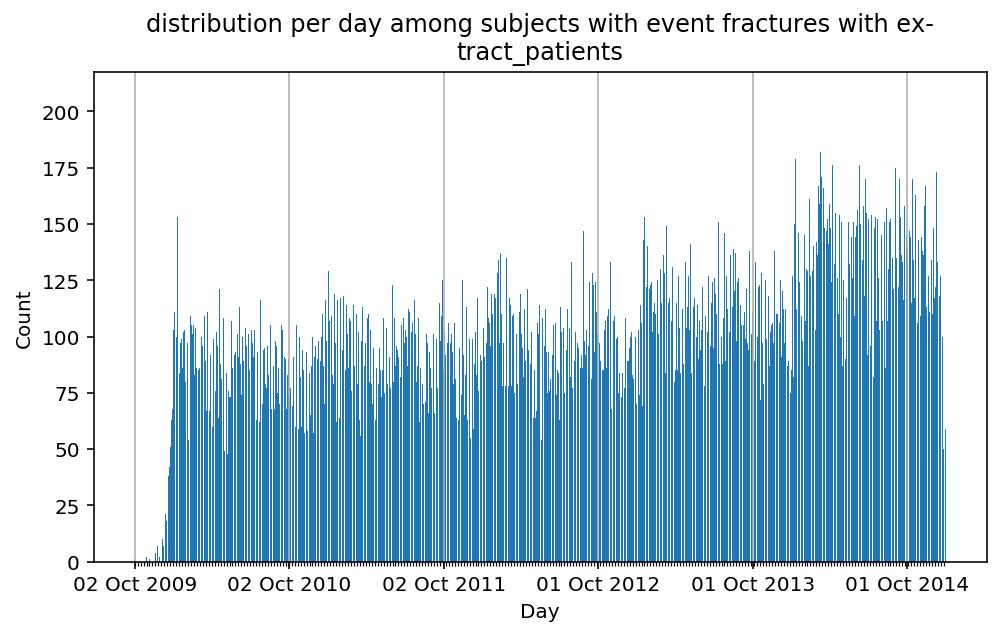
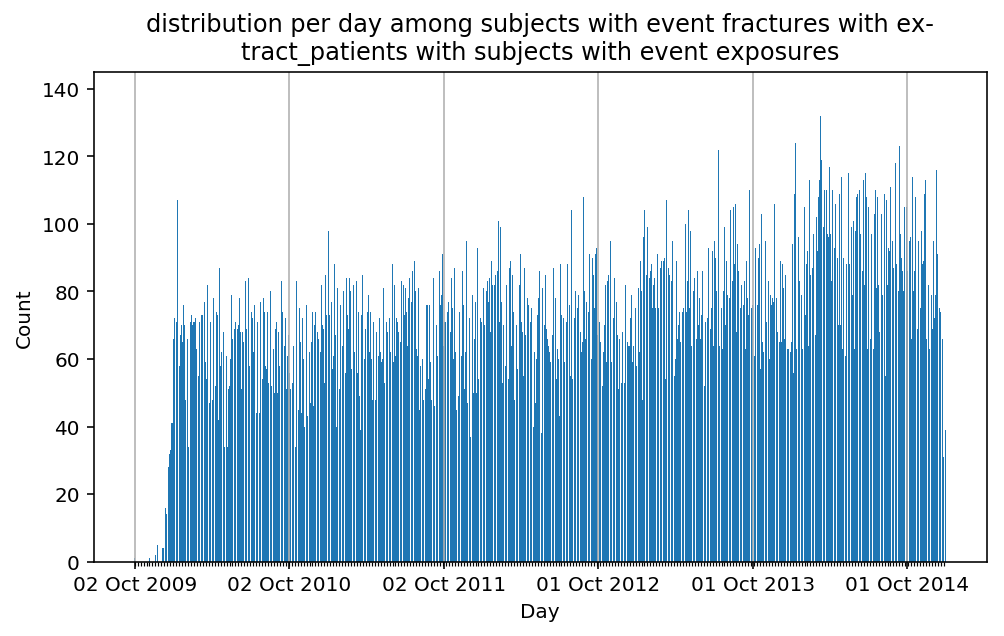
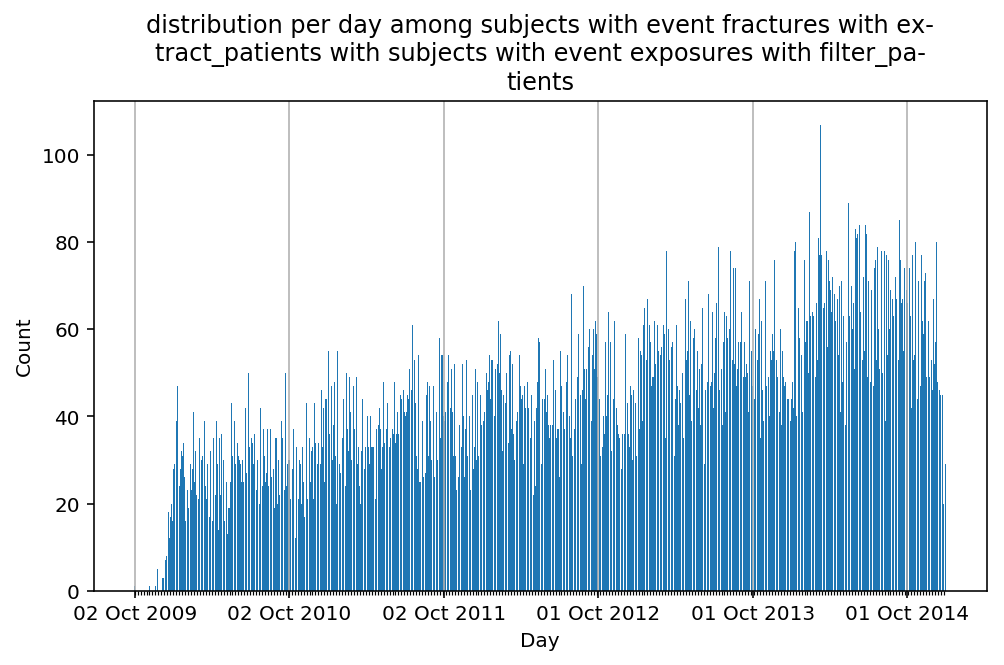
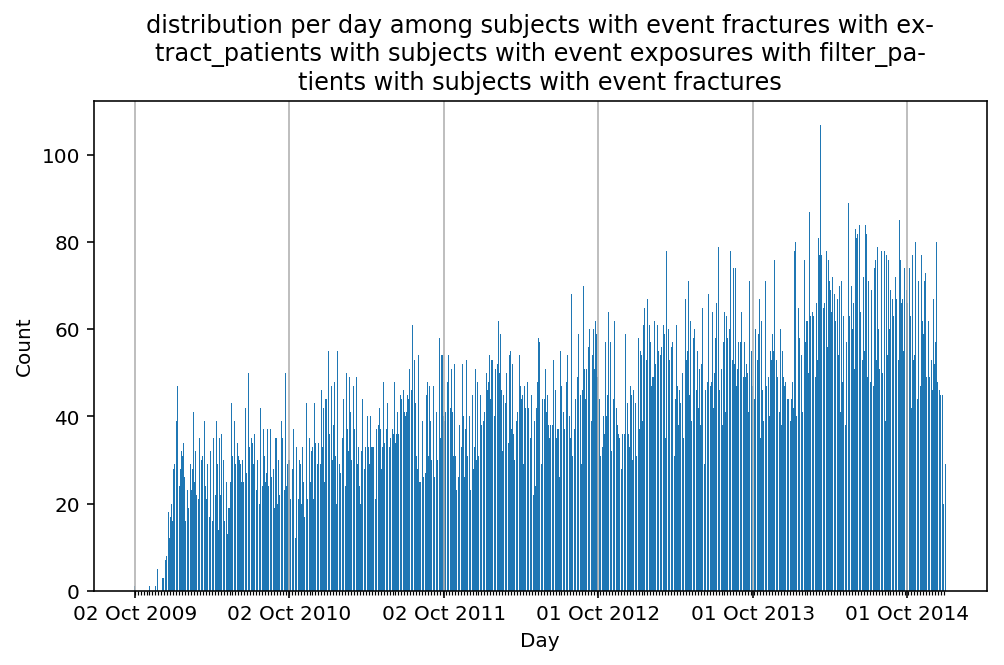
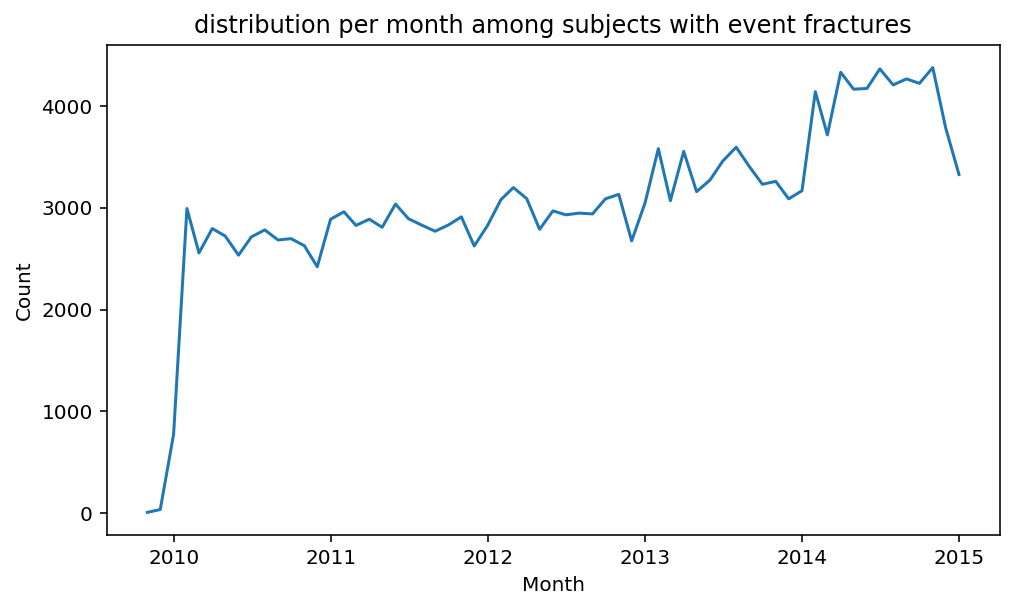
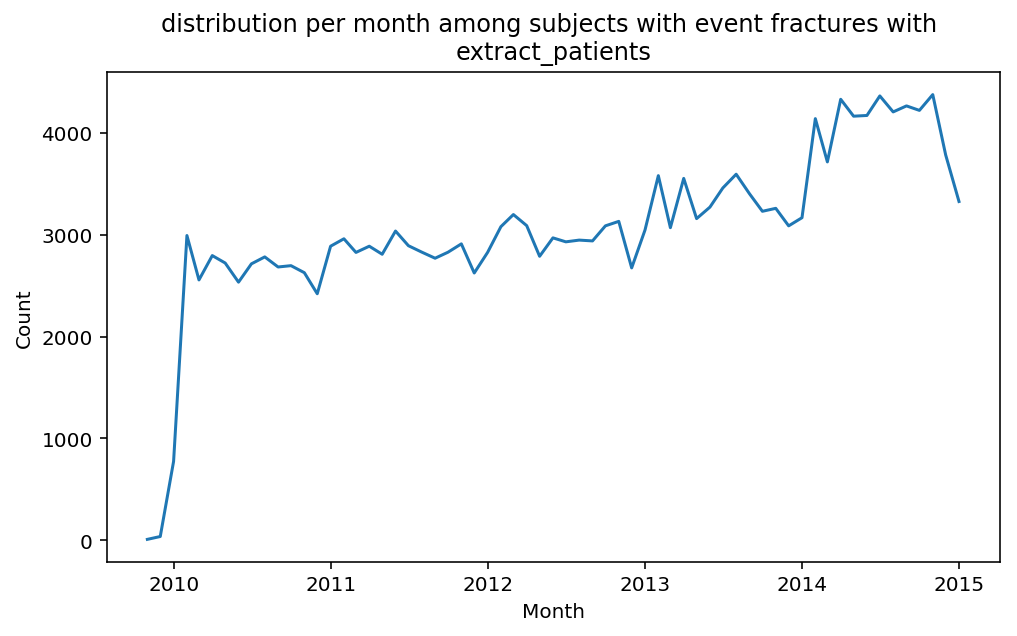
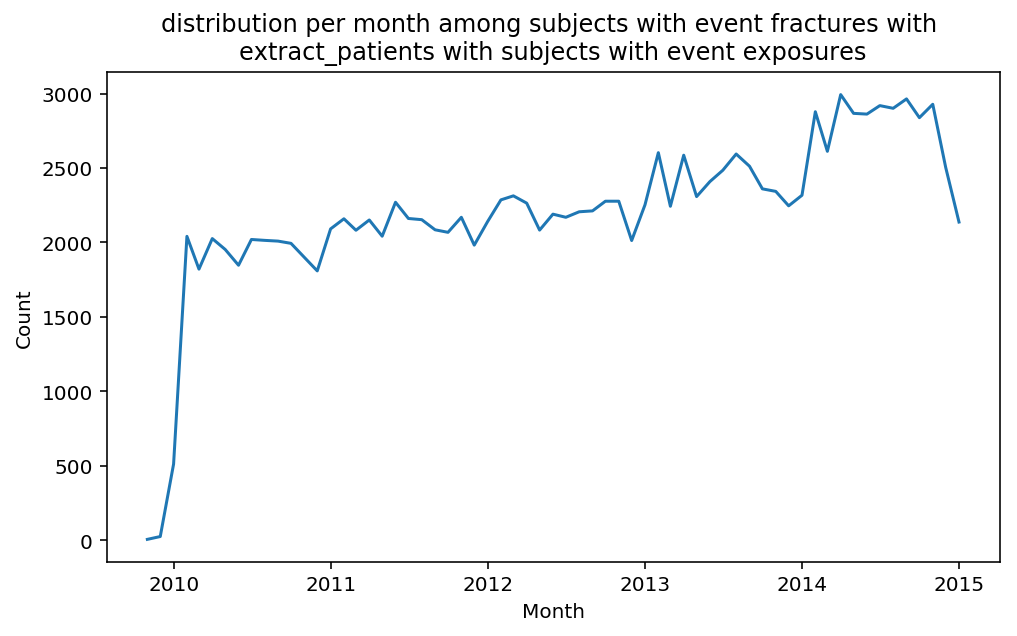
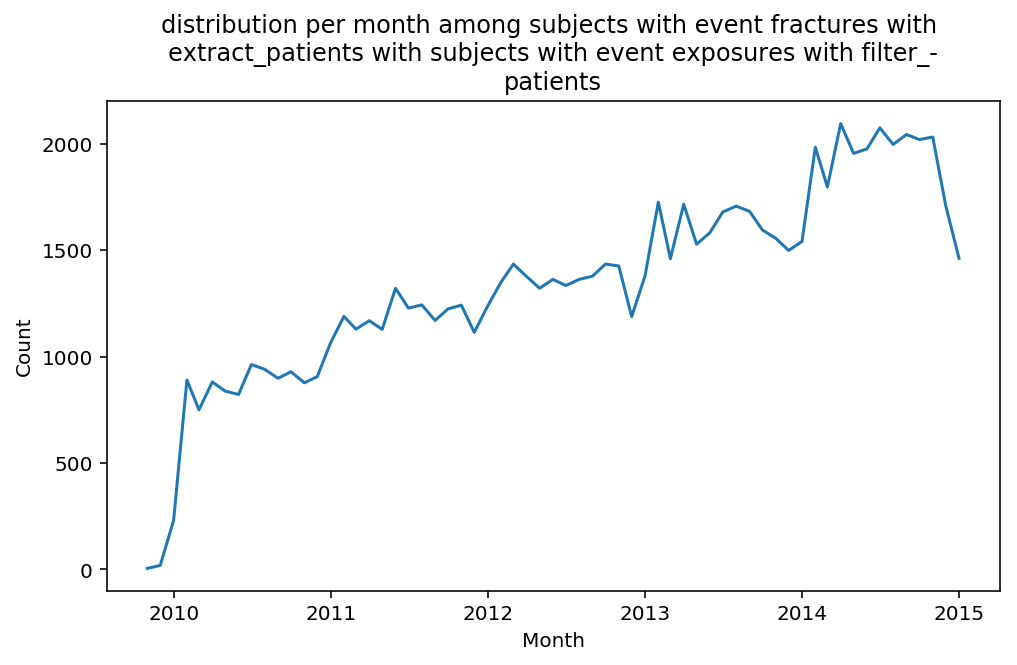
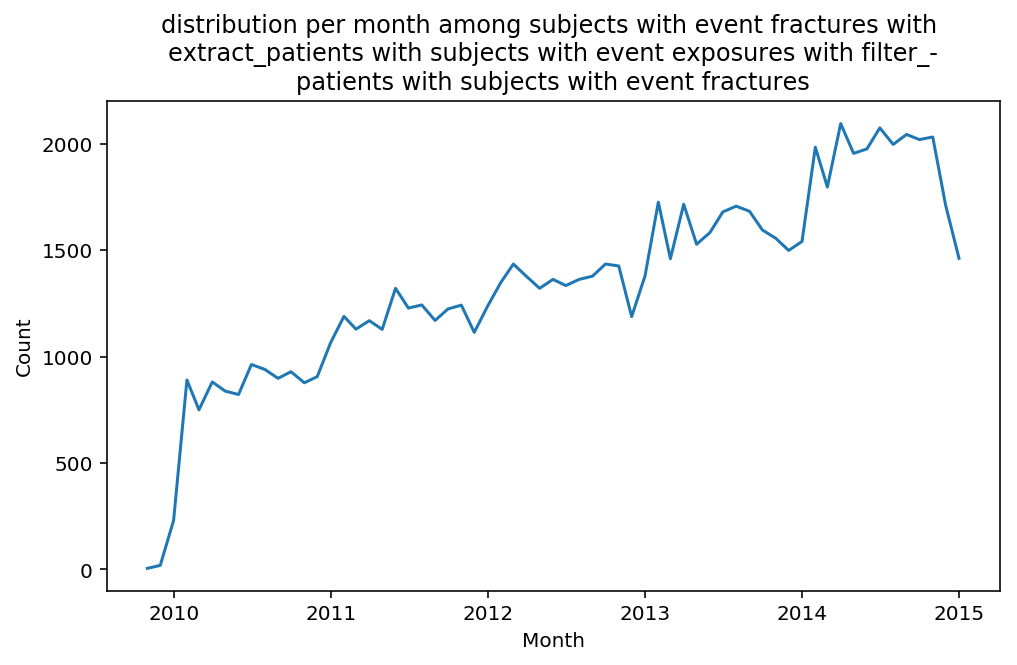
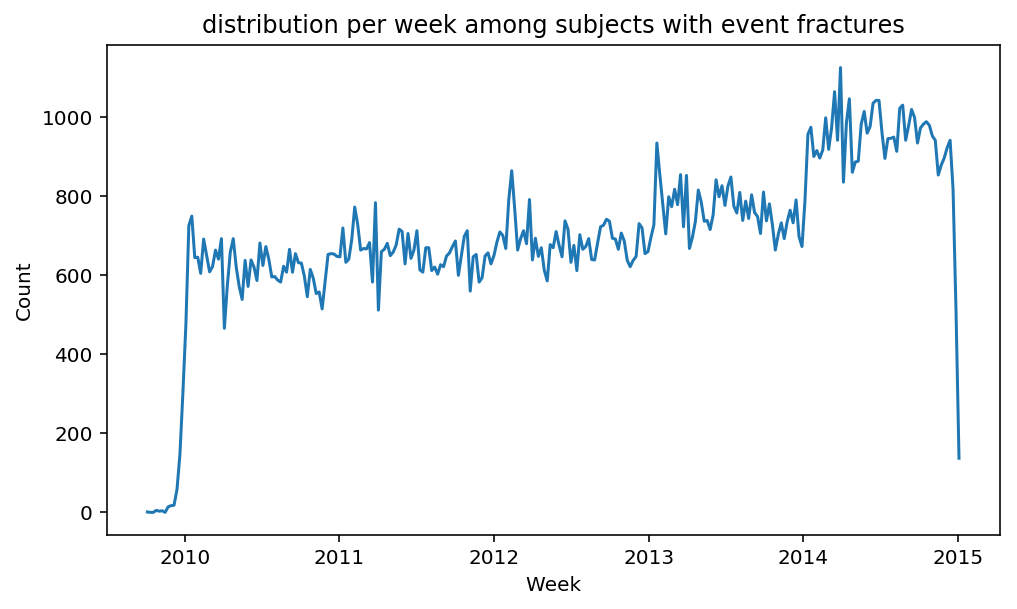
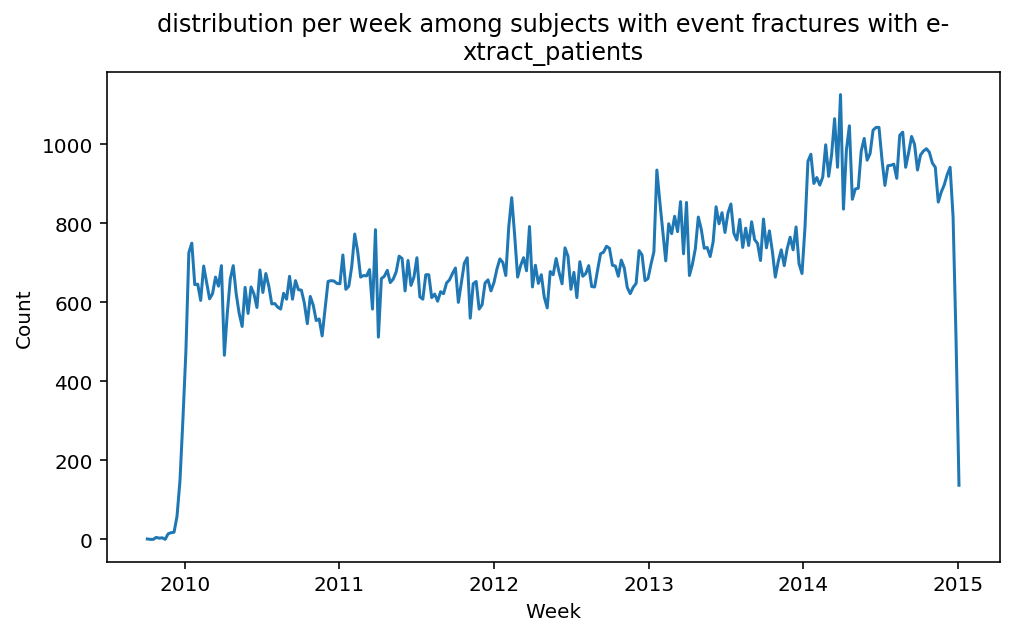
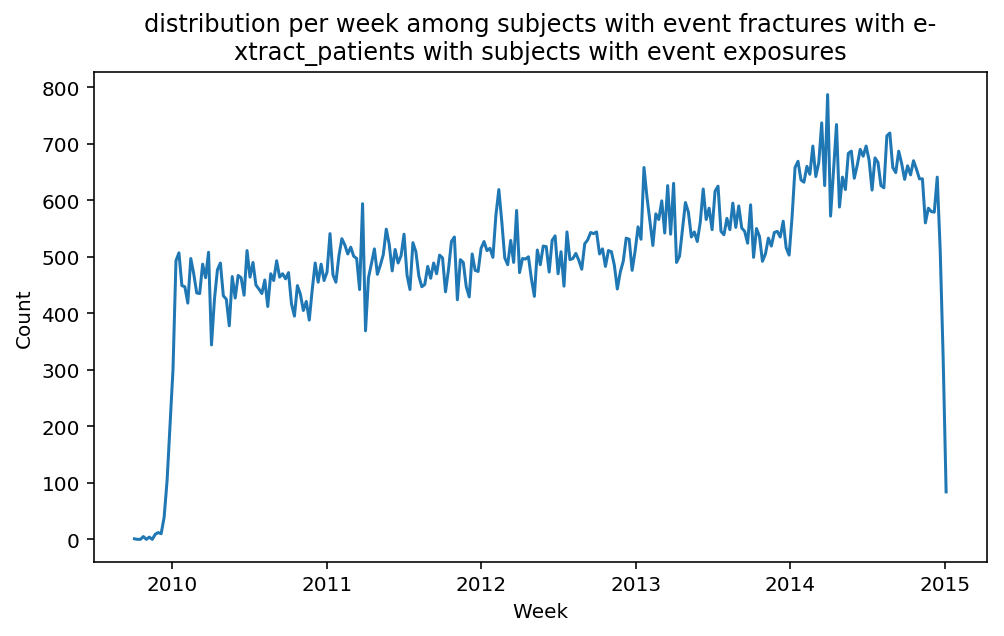
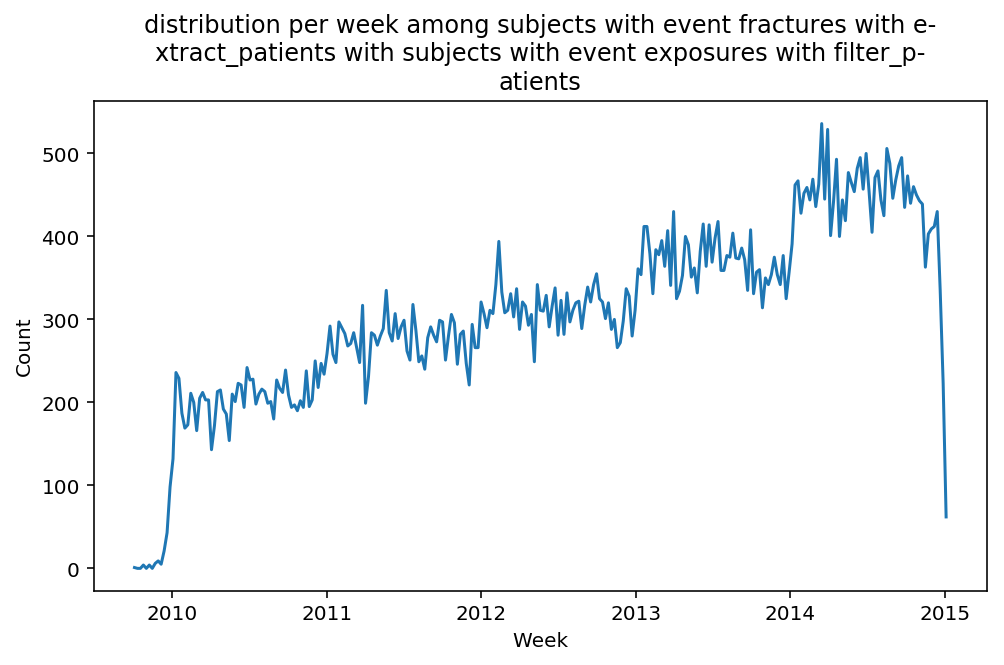
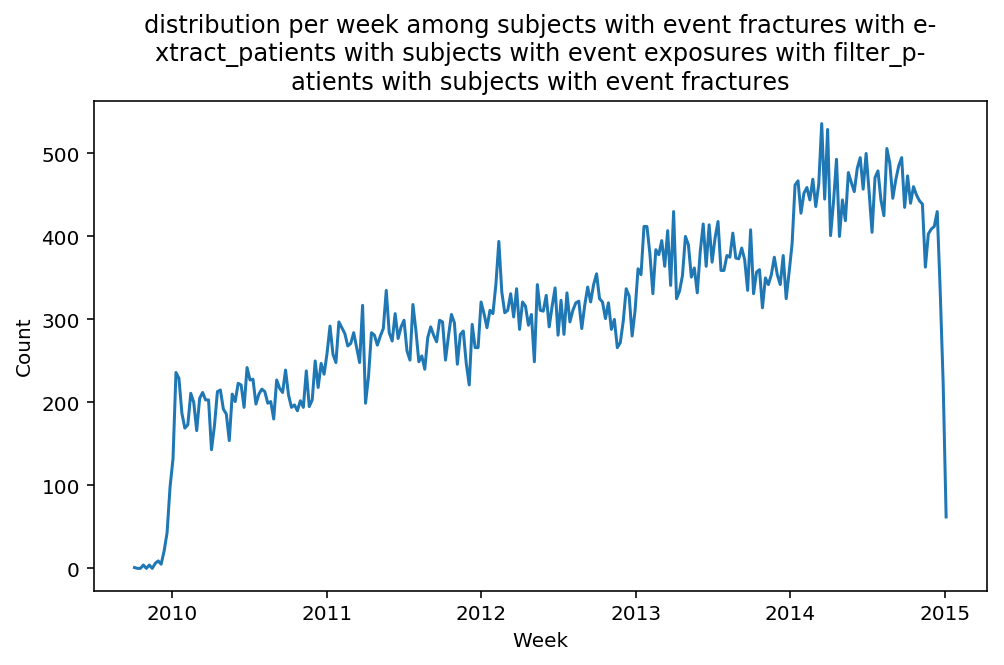
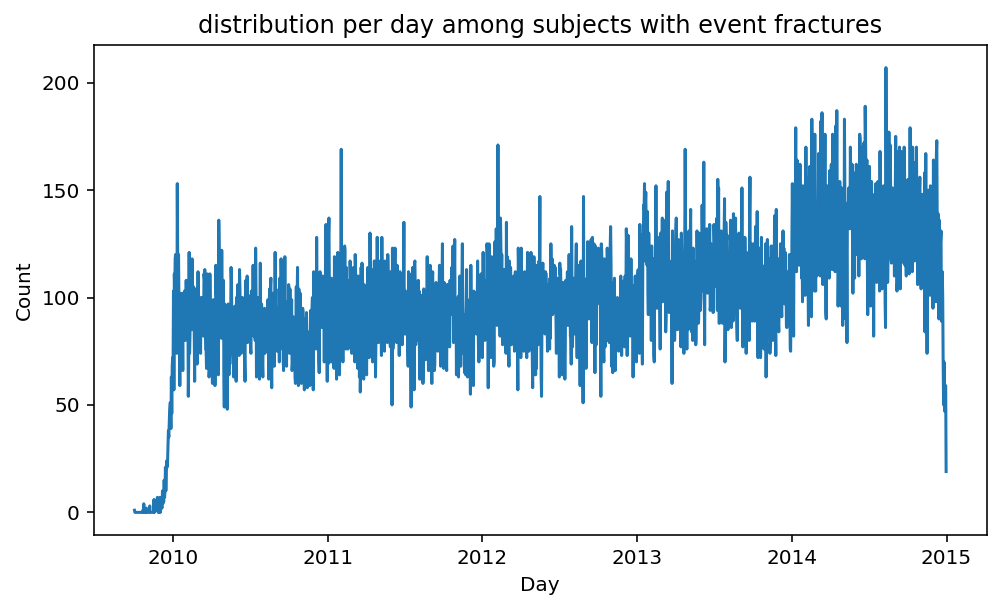
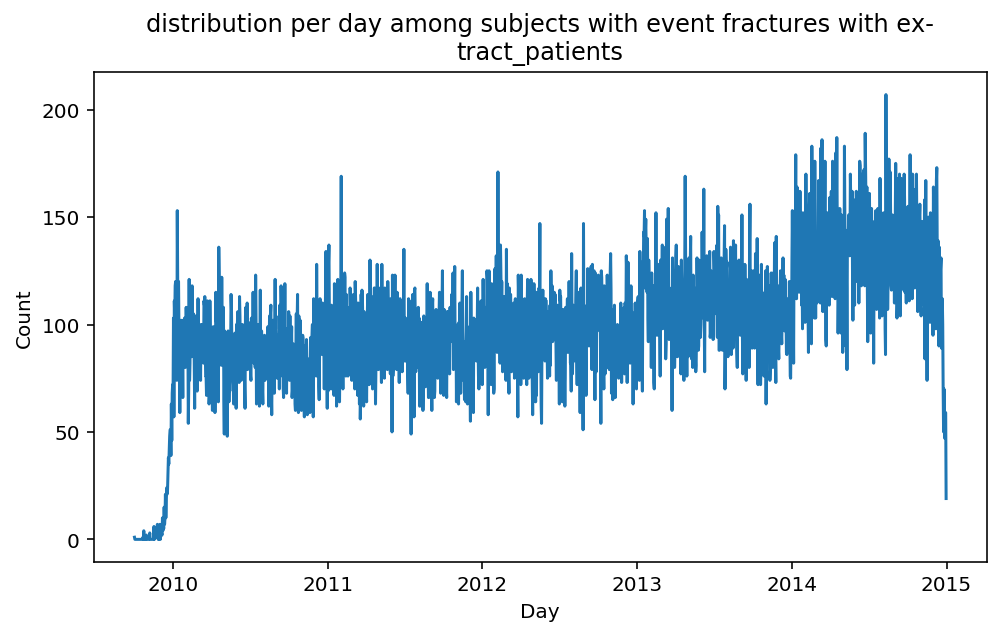
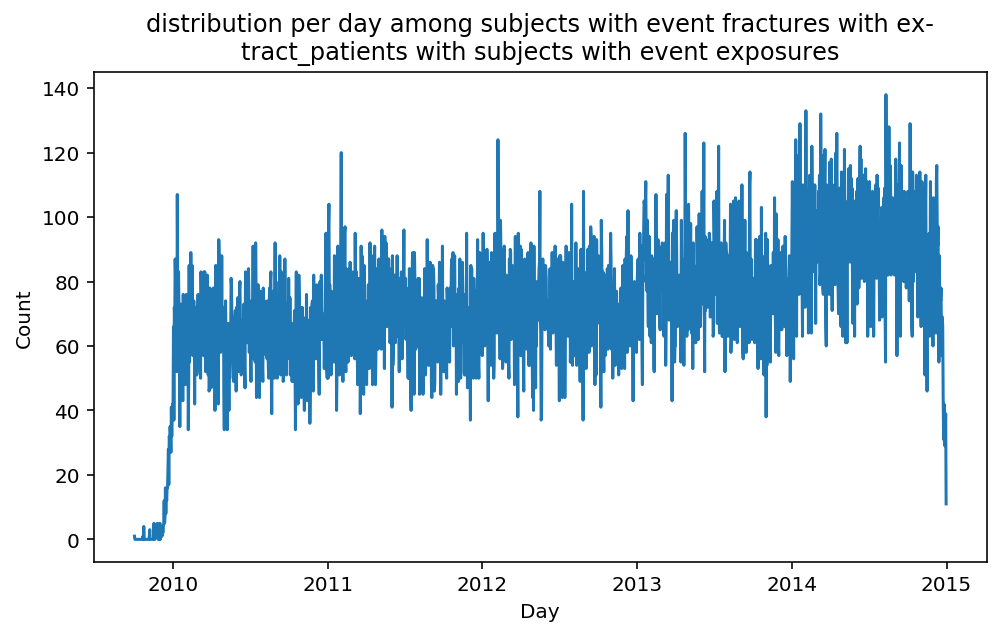
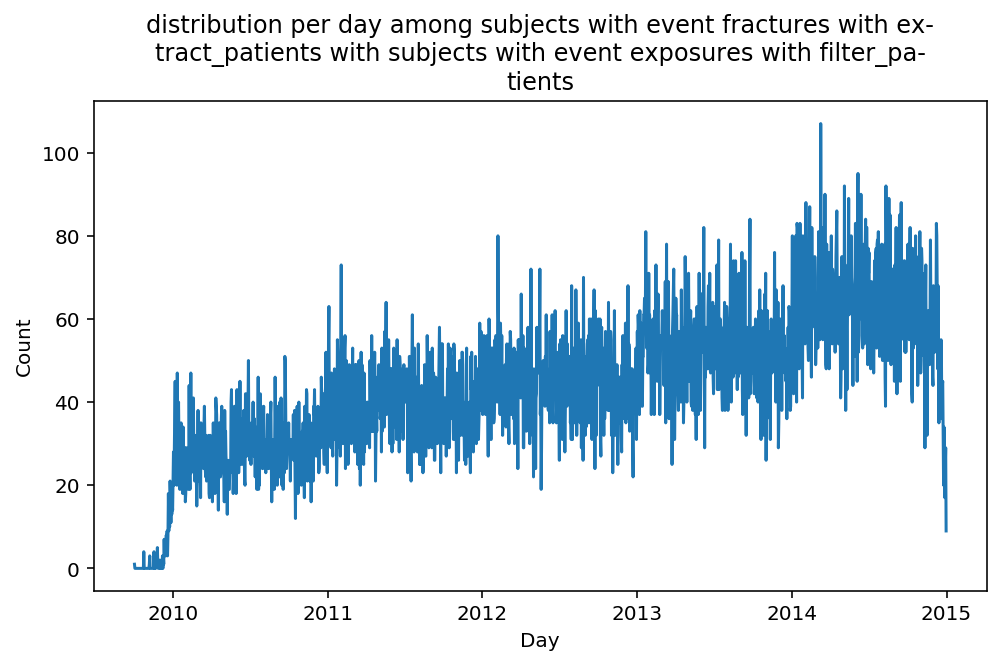
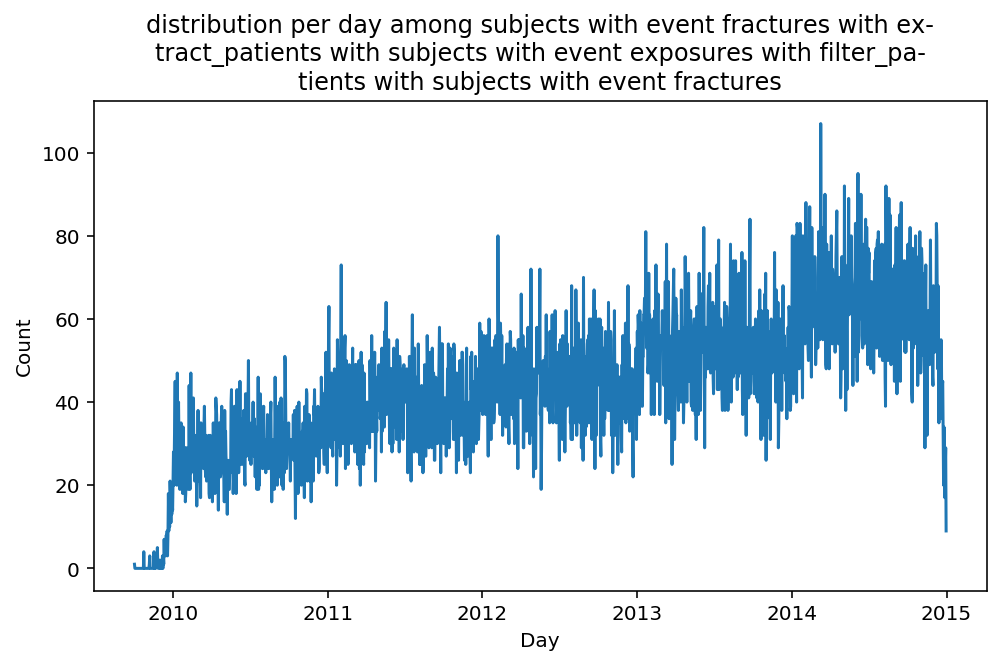
Besides have the same signature, developping a new stat can made easier in SCALPEL-Analysis.
Matplotlib is notorious for tha amount of boilerplate code it engenders. One solution is to use decorators to avoid writing down the title, axis names etc. Since the Cohort maintains a description of the underlyning data, decorators take advantage of this to generate automatically the titles. Here is an example. Previous show h
from scalpel.stats.decorators import title, xlabel, ylabel #import decorators
from scalpel.stats import agg
registry = []
@register
@title("Gender distribution")
@xlabel("Gender")
@ylabel("Subjects count")
def distribution_by_gender(figure, cohort):
df = agg(cohort.subjects, frozenset(["gender"]), "count")
ax = figure.gca()
ax = sns.barplot(x=df["gender"].values, y=df["count(1)"].values, ax=ax)
ax.set_xticklabels(["Male", "Female"])
return figureIn general, stats begin with an aggregation. That agregation is then used to produce the results of the statistics module. In our SCALPEL-Analysis, we tought that is might be a good idea to cache the results of aggregation to avoid hefety computations. In order to use it, one has just to import the cached method and use it to peform the aggregation. The example below shows how.
from scalpel.stats.decorators import title, xlabel, ylabel
from scalpel.stats import agg # import the agg function
registry = []
@register
@title("Gender distribution")
@xlabel("Gender")
@ylabel("Subjects count")
def distribution_by_gender(figure, cohort):
df = agg(cohort.subjects, frozenset(["gender"]), "count") # use it to perform the aggregation
ax = figure.gca()
ax = sns.barplot(x=df["gender"].values, y=df["count(1)"].values, ax=ax)
ax.set_xticklabels(["Male", "Female"])
return figure| Module name | Function name | Description | Conditions |
|---|---|---|---|
| scalpel.stats.patients | distribution_by_gender | Plots the distribution per gender. | Cohort must contain demographics. |
| scalpel.stats.patients | distribution_by_age_bucket | Plots distribution by age bucket. | Cohort must contain demographics. |
| scalpel.stats.patients | distribution_by_gender_age_bucket | Plots distribution by age bucket and gender. | Cohort must contain demographics. |
| scalpel.stats.event_distribution | plot_events_per_month_as_bars | Plots distribution of events start date per month as bars. | Cohort must be event cohort. |
| scalpel.stats.event_distribution | plot_events_per_week_as_bars | Plots distribution of events start date per week as bars. | Cohort must be event cohort. |
| scalpel.stats.event_distribution | plot_events_per_day_as_bars | Plots distribution of events start date per day as bars. | Cohort must be event cohort. |
| scalpel.stats.event_distribution | plot_events_per_month_as_timeseries | Plots distribution of events start date per month as timeseries. | Cohort must be event cohort. |
| scalpel.stats.event_distribution | plot_events_per_week_as_timeseries | Plots distribution of events start date per week as timeseries. | Cohort must be event cohort. |
| scalpel.stats.event_distribution | plot_events_per_day_as_timeseries | Plots distribution of events start date per day as timeseries. | Cohort must be event cohort. |
| scalpel.stats.event_distribution | plot_events_per_day_as_timeseries | Plots distribution of events start date per day as timeseries. | Cohort must be event cohort. |
| scalpel.stats.event_patient_distribution | plot_unique_event_distribution_per_patient | Plots distribution of number unique events per subject. | Cohort must be event cohort. |
| scalpel.stats.event_patient_distribution | plot_patient_distribution_per_unique_event | Plots distribution of number unique subjects per events type. | Cohort must be event cohort. |
| scalpel.stats.event_patient_distribution | plot_patient_distribution_per_unique_event | Plots distribution of number unique subjects per events type. | Cohort must be event cohort. |
| scalpel.stats.duration_distribution | plot_duration_distribution_per_day_as_line | Plots distribution of duration of Events using a day unit as line. | Cohort must be duration cohort. |
| scalpel.stats.duration_distribution | plot_duration_distribution_per_month_as_bar | Plots distribution of duration of Events using a month unit as line. | Cohort must be duration cohort. |
| scalpel.stats.duration_distribution | plot_mean_duration_per_value | Plots mean duration per Event type. | Cohort must be duration cohort. |
| scalpel.stats.co_occurence_stats | plot_co_events | Plots how many Events co-occur in matrice fashion. Axis are defined by Event Value. | Cohort must be duration cohort. |
SCALPEL3