-
Couldn't load subscription status.
- Fork 179
reproduce OutbreakTools example using ggtree
Guangchuang Yu edited this page Jul 24, 2019
·
9 revisions
This example is presented in vignette of OutbreakTools.
library(OutbreakTools)
data(FluH1N1pdm2009)
attach(FluH1N1pdm2009)
x <- new("obkData", individuals = individuals, dna = FluH1N1pdm2009$dna,
dna.individualID = samples$individualID, dna.date = samples$date,
trees = FluH1N1pdm2009$trees)
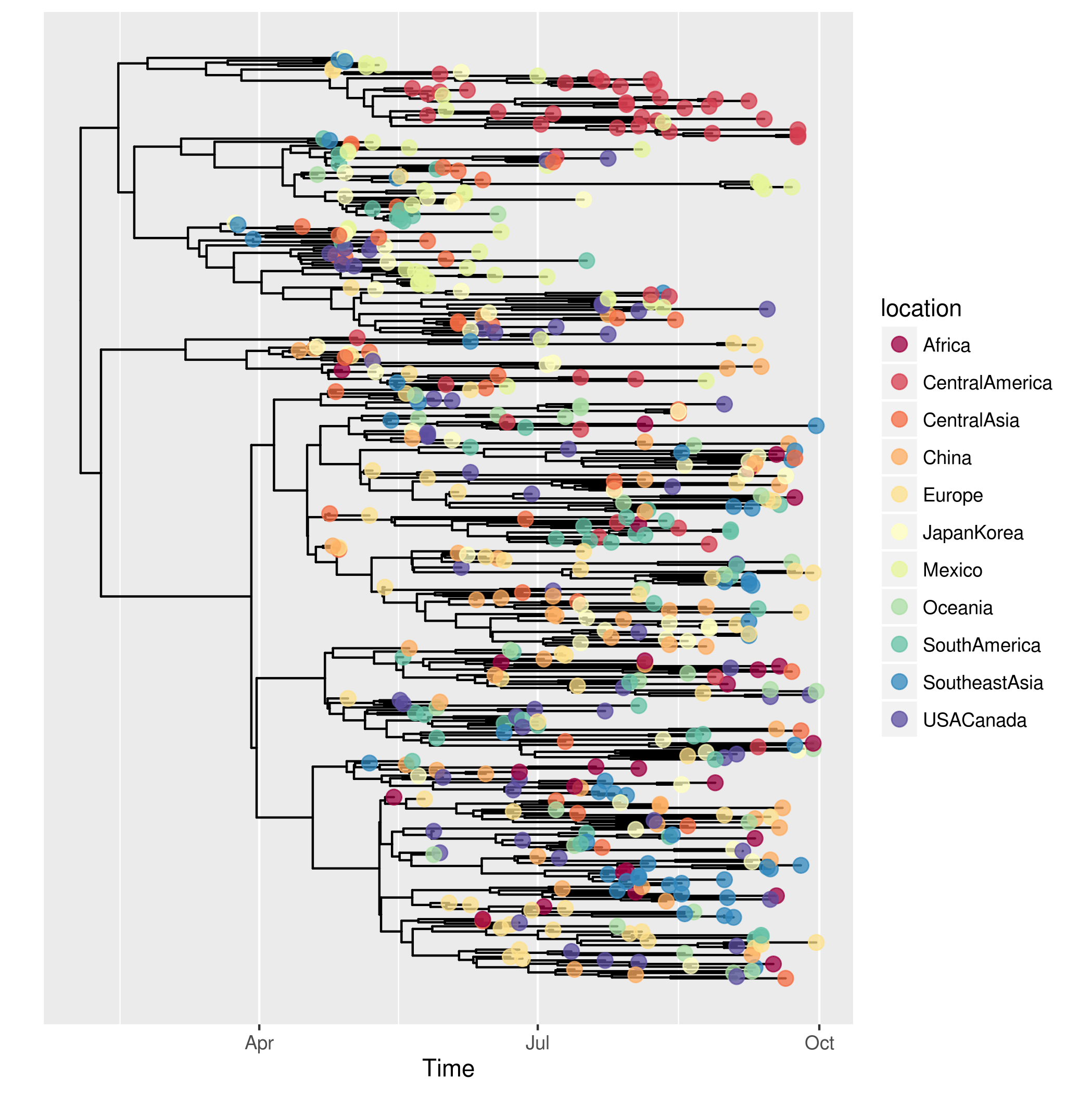
p <- plotggphy(x, ladderize = TRUE, branch.unit = "year",
tip.color = "location", tip.size = 3, tip.alpha = 0.75)In this figure, it use Date for x-axis and the tree was annotated with Location. In an epidemic, time and location that of the virus sampled is important information and plotggphy is designed for viewing such information.
In ggtree, it's very easy to reproduce such figure by adding a layer of location information.
library(ggtree)
draw_OutbreakTools_ggtree <- function(x) {
tree <- x@trees[[1]]
g <- ggtree(tree, right=T, mrsd="2009-09-30", as.Date=TRUE) + theme_tree2()
g <- g + theme(panel.grid.major = element_line(color="grey"),
panel.grid.major.y=element_blank())
## extract location info
meta.df <- x@dna@meta
meta.df <- data.frame(taxa=rownames(meta.df), meta.df)
loc <- x@individuals
loc <- data.frame(individualID=rownames(loc), loc)
meta_loc <- merge(meta.df, loc, by="individualID")
meta_loc <- meta_loc[,-1]
## attach additional information to tree view via %<+% operator
## that was created in ggtree
g <- g %<+% meta_loc
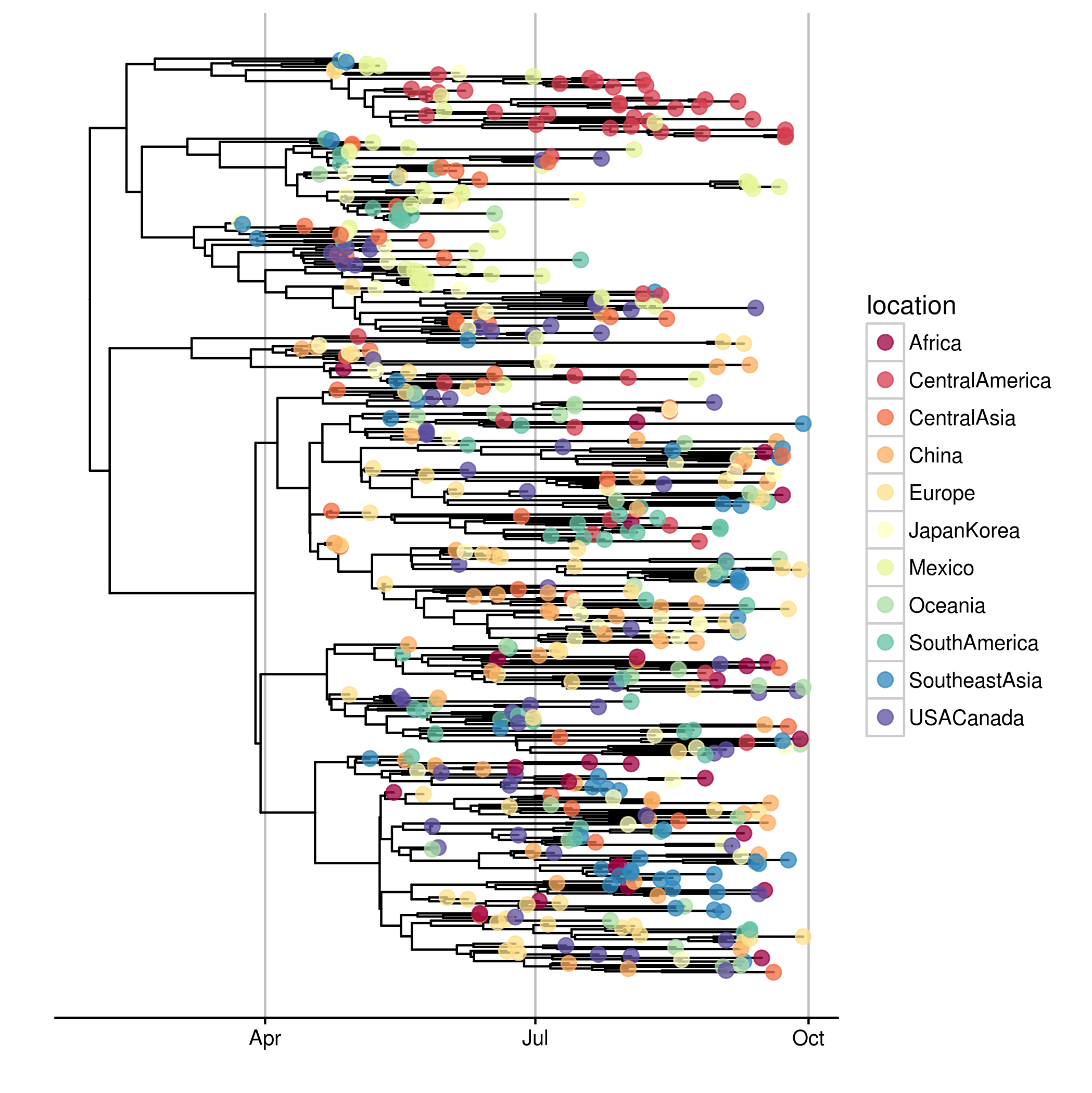
g + geom_tippoint(aes(color=location), size=3, alpha=.75) +
theme(legend.position="right") +
scale_color_brewer("location", palette="Spectral")
}
draw_OutbreakTools_ggtree(x)require(microbenchmark)
bm <- microbenchmark(
OutbreakTools = plotggphy(x, ladderize = TRUE, branch.unit = "year",
tip.color = "location", tip.size = 3, tip.alpha = 0.75),
ggtree = draw_OutbreakTools_ggtree(x),
times=100L
)
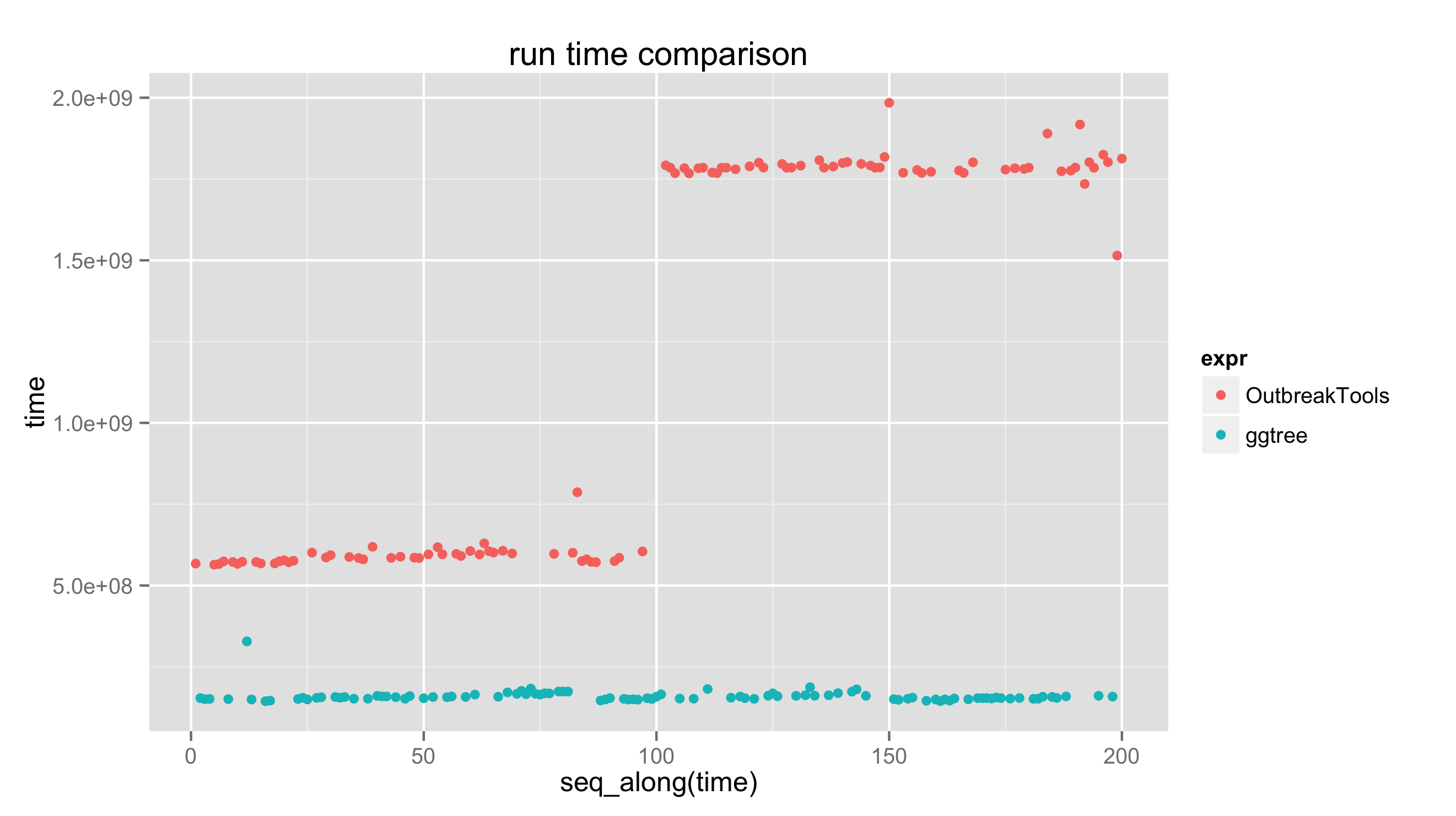
print(bm)
## Unit: milliseconds
## expr min lq mean median uq max neval
## OutbreakTools 564.0195 585.7710 1225.8299 1767.713 1784.8457 1984.3352 100
## ggtree 144.2246 151.7099 159.3318 155.200 160.9121 328.2016 100
qplot(y=time, data=bm, color=expr) + ggtitle("run time comparison")