Quantitative layer based analysis for renal magnetic resonance imaging.
The easiest way to install qlayersis via pip:
pip install qlayersor if you're a conda user:
conda install qlayers -c conda-forgeAlternatively, you can install qlayersfrom source in pips editable mode:
git clone https://github.com/alexdaniel654/qlayers.git
cd qlayers
pip install -e .For a more thorough example of how to use qlayers see the tutorials section of this reposetry/documentation, however if
you want to get started, the snippet of code below should get you going.
import nibabel as nib
from qlayers import QLayers
mask_img = nib.load("kidney_mask.nii.gz")
t2star_img = nib.load("t2star_map.nii.gz")
qlayers = QLayers(mask_img, pelvis_dist=10)
qlayers.add_map(t2star_img, "t2star")
df = qlayers.get_df(format="wide")
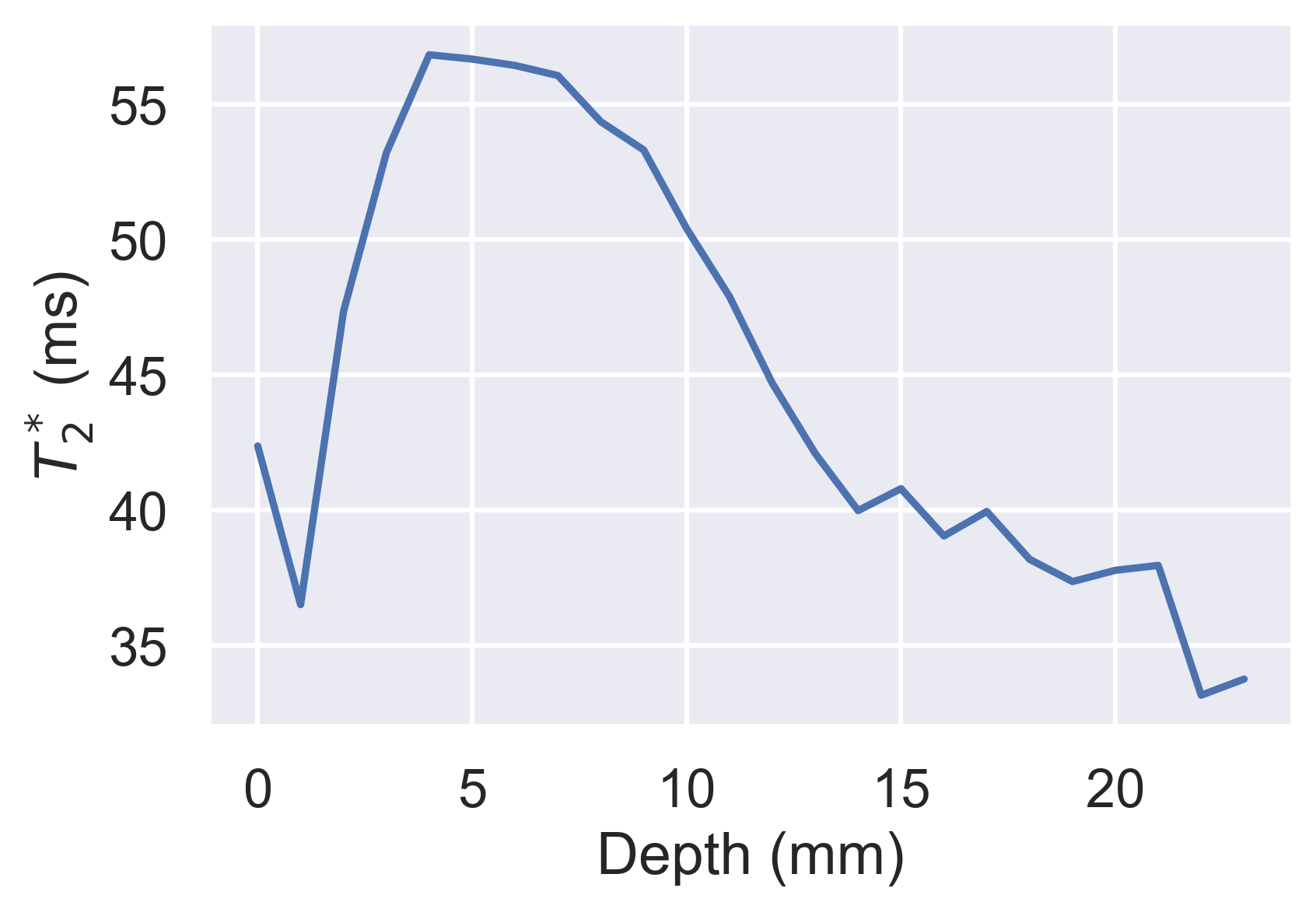
df.groupby("layer").median().loc[:, "t2star"].plot(
xlabel="Depth (mm)", ylabel="$T_2^*$ (ms)"
)The premise behind qlayers was first proposed by Pruijm et al and is based
on the idea to segment the kidney into layers based on each voxels distance from the surface of the kidney. The average of
a quantitative parameter can be calculated for each layer producing profiles of, for example, T2* with
depth. The outer and inner layers are analogous to the cortex and medulla respectively while the gradient of the profile is
representative of the cortico-medullary difference. qlayers extends this idea by allowing the user to define layers based on
a 3D mask and apply the layer to any quantitative parameter.
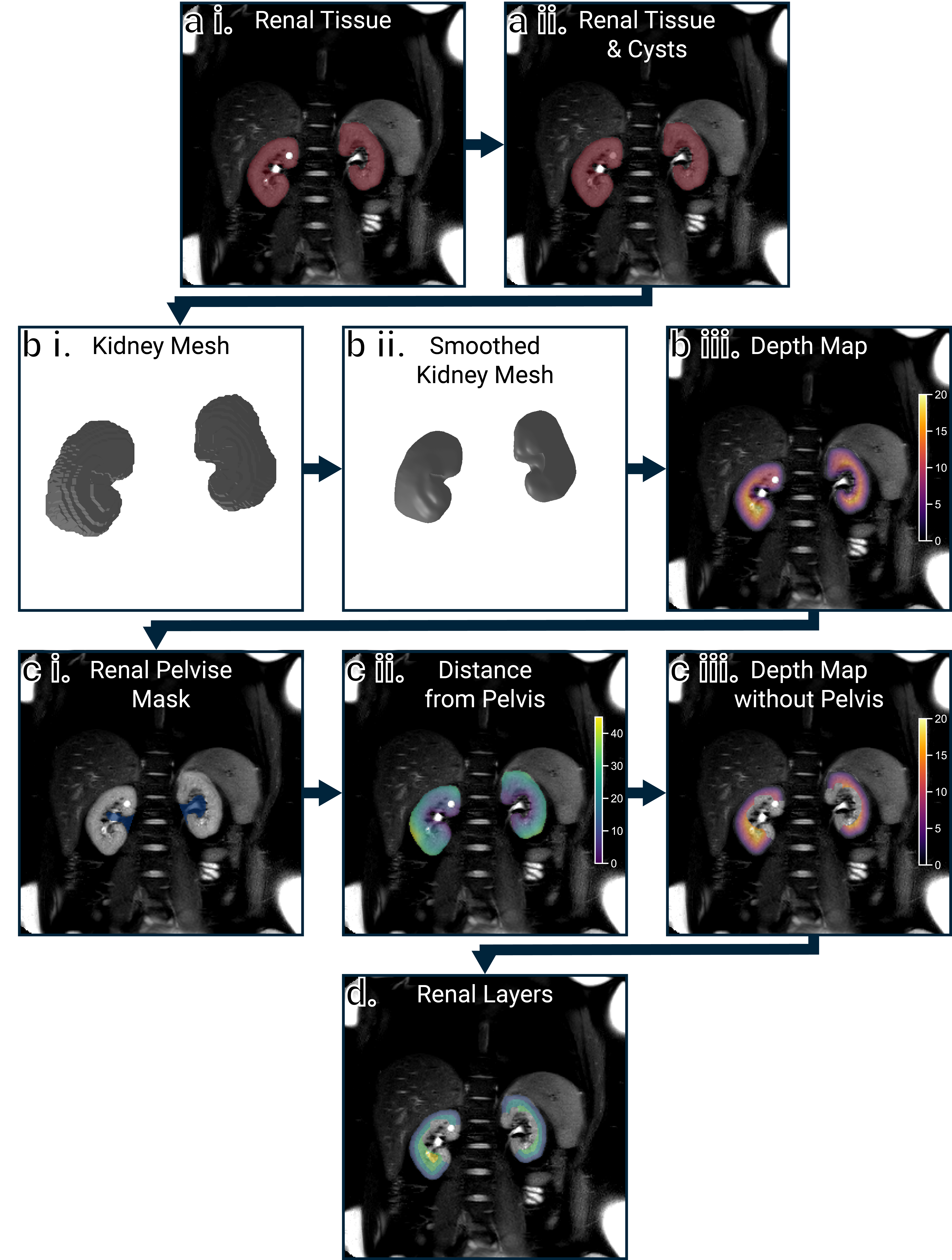
Layers are generated via the process outlined in the figure below.
a i. Shows the mask thats input to the QLayers class. This mask then has any holes smaller than fill_ml filled as these are
most likely cysts
and therefore not cortical surfaces, a ii. The mask is then converted from a voxel representation to a mesh surface
representation, b i, this mesh is then smoothed because anatomical scans of the kidneys often have a low through-plane
resolution, b ii. The distance from the centre of each voxel in the kidneys to the closest surface on the mesh is then
calculated, b iii. As the tissue adjacent to the renal pelvis is not representative of the medulla, this is automatically
excluded from the resulting depth maps. Fist the pelvis is automatically segmented, c i, and the distance from each voxel
in the kidneys to the pelvis calculated as above, c ii. Voxels closer than a specified threshold pelvis_dist are then
excluded from the depth maps, c iii. Finally, a layer image is generated by quantising the depth map to a desired layer
thickness, typically 1 mm although shown with 5 mm layers here for illustrative purposes, d.
If the space parameter of the QLayers object is set to layers, when a quantitative map is added to the QLayers
object, it is resampled to the same resolution and orientation as the layers. If the space parameter is set to map then
the layers are resampled to the resolution and orientation of the quantitative map. In both cases, Pandas DataFrames can
be generated with the quantitative value, depth and layer each voxel is in. These DataFrames can then be used for further
calculations such as generating profiles or linear regressions to explore the cortico-medullary difference. Some example
voxels are shown in the table below.
| Depth | Layer | T2* | R2* |
|---|---|---|---|
| 0 | 0 | 57 | 17.6 |
| 13.2 | 14 | 35.5 | 28.2 |
| 10.2 | 11 | 60.9 | 16.4 |
| 3.05 | 4 | 51.6 | 19.4 |
| 9.33 | 10 | 42.8 | 23.3 |
| 10.4 | 11 | 29.6 | 33.8 |
| 8.63 | 9 | 37.5 | 26.7 |
| 6.66 | 7 | 49.2 | 20.3 |
| 19.8 | 20 | 42.8 | 23.3 |
| 12.1 | 13 | 39.4 | 25.4 |
If you have used 3DQLayers in your research, please cite the following conference abstract:
Alternatively, if you want to cite a specific version of this software, each release has an individual DOI on Zenodo, the DOI for the latest release can be found here.
We welcome contributions to QLayers, a full contributing guide can be found here.