NEW: for instructions on new Python version please click here!
Near-real-time, interactive, self-contained, non-proprietary implementation of the Cloude Decomposition [1] by Shane R Cloude for:
- rapid exploration of quad-polarized SAR data such as from nisar, uavsar, palsar, or radarsat2 and
- inspiring new PolSAR applications.
Sample data included.
This July 26 2002 presentation is a companion resource accompanying the article below [1].
"In this paper [1] we show how to develop the idea of orthogonality in radar polarimetry for enhanced target detection and land-use classification of POLSAR data. It is well known that every elliptical polarization has its orthogonal partner, uniquely defined as the antipodal point on the Poincaré sphere. Here we show that for scatterers, we can extend this idea so that every scattering matrix has a corresponding multi-dimensional orthogonal space [2]. We give a geometrical interpretation of this approach using a generalized Poincaré sphere representation. We then derive an algorithm for finding the peak signal in this ortho-space. We derive this optimum for both monostatic and bistatic radar systems, to illustrate how bistatic polarimetry offers great potential benefits in future POLSAR studies."
[1] "Generalized Poincaré Orthogonality: A New Approach to POLSAR Data Analysis" SR Cloude, A Richardson, https://arxiv.org/abs/2109.09093
[2] "Target Detection Using Rank-1 Polarimetric Processing" SR Cloude, IEEE Geoscience and Remote Sensing Letters (2020)
Inputs: A) fully-polarimetric SAR data in standard PolSARPro "T3 coherency matrix" format with config.txt file B) an RGB encoding such as the well-known Pauli encoding, used for selection of a target location C) image coordinates of user-selected target to be "cancelled". Output: optimised radar cross section (a greyscale image)
The interactive version supports exploring fully-polarimetric SAR data quickly, by selecting a target location using the mouse, then viewing the resulting optimised radar cross section promptly
- Tested on Ubuntu 20 LTS and MacOS. Windows is supported via WSL ( Windows Subsystem: Linux )
- Open your windows terminal (AKA "cmd.exe")
- If you didn't install WSL yet:
wsl --install
- Enter the wsl environment:
wsl
- Install "unzip" command (needed to open code) At the WSL prompt:
sudo apt install unzip
Please open your terminal and run the following commands:
(by the way, make sure to use the "copy" icon to the right of the code fragment, followed by pasting in your terminal, in order to avoid re-typing manually). All code fragments require pressing "RETURN" key afterwards to run the command.
curl -o cloude_decom.zip https://codeload.github.com/ashlinrichardson/cloude_decom/zip/refs/heads/master; unzip cloude_decom.zip; mv cloude_decom-master cloude_decom
cd cloude_decom
python3 cpp/compile.py
cd T3
cloude_view
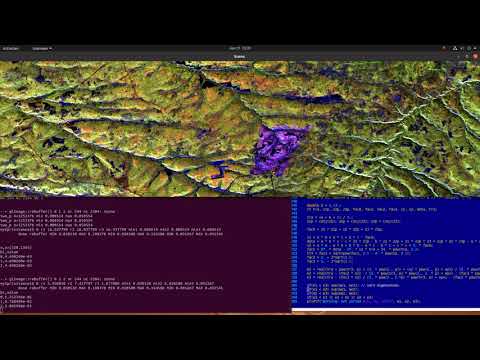
If all goes well, you should see an interactive visualization of the test data (as below).
Notes:
- compile.py installs, provided you enter your super-user password:
- required dependencies g++ and freeglut3-dev (on ubuntu). Mac users need to install xcode "command line" development tools
- binaries to /usr/bin/cloude_decom and /usr/bin/cloude_view
- Alternate downloading method, you could use this instead of step "1)" above if you're already connected to github by ssh:
git clone [email protected]:ashlinrichardson/cloude_decom.git
and then proceed to "2) enter the project folder". This approach would make sense if you're experienced with GitHub and considering making code revisions
- Another alternative downloading method (assuming you have wget installed)
wget https://github.com/ashlinrichardson/cloude_decom/archive/refs/heads/master.zip; unzip master.zip; mv cloude_decom-master cloude_decom
Assuming the mouse pointer is positioned somewhere over the image display:
- depressing the left button, restores the default visualization used (e.g. the pauli encoding)
- releasing the left button, runs the decomposition and displays the optimized radar cross section associated with the target area under the cursor, when the button was released
So it's necessary to engage and then release the left mouse button, to generate an output. Again the location where the mouse button is released, becomes the target area for processing
Depressing the right mouse botton, or the "esc" key on the keyboard, both close the program
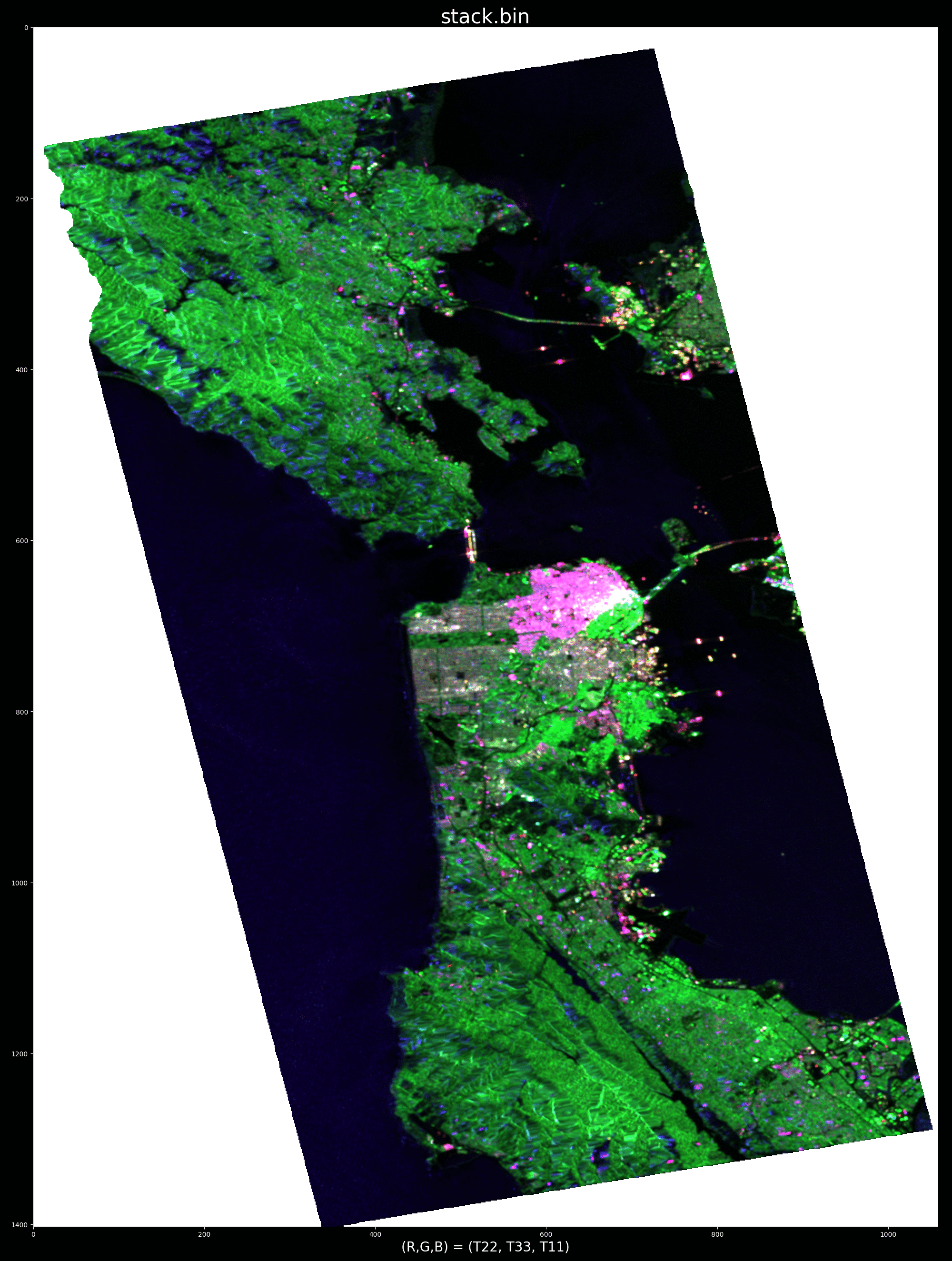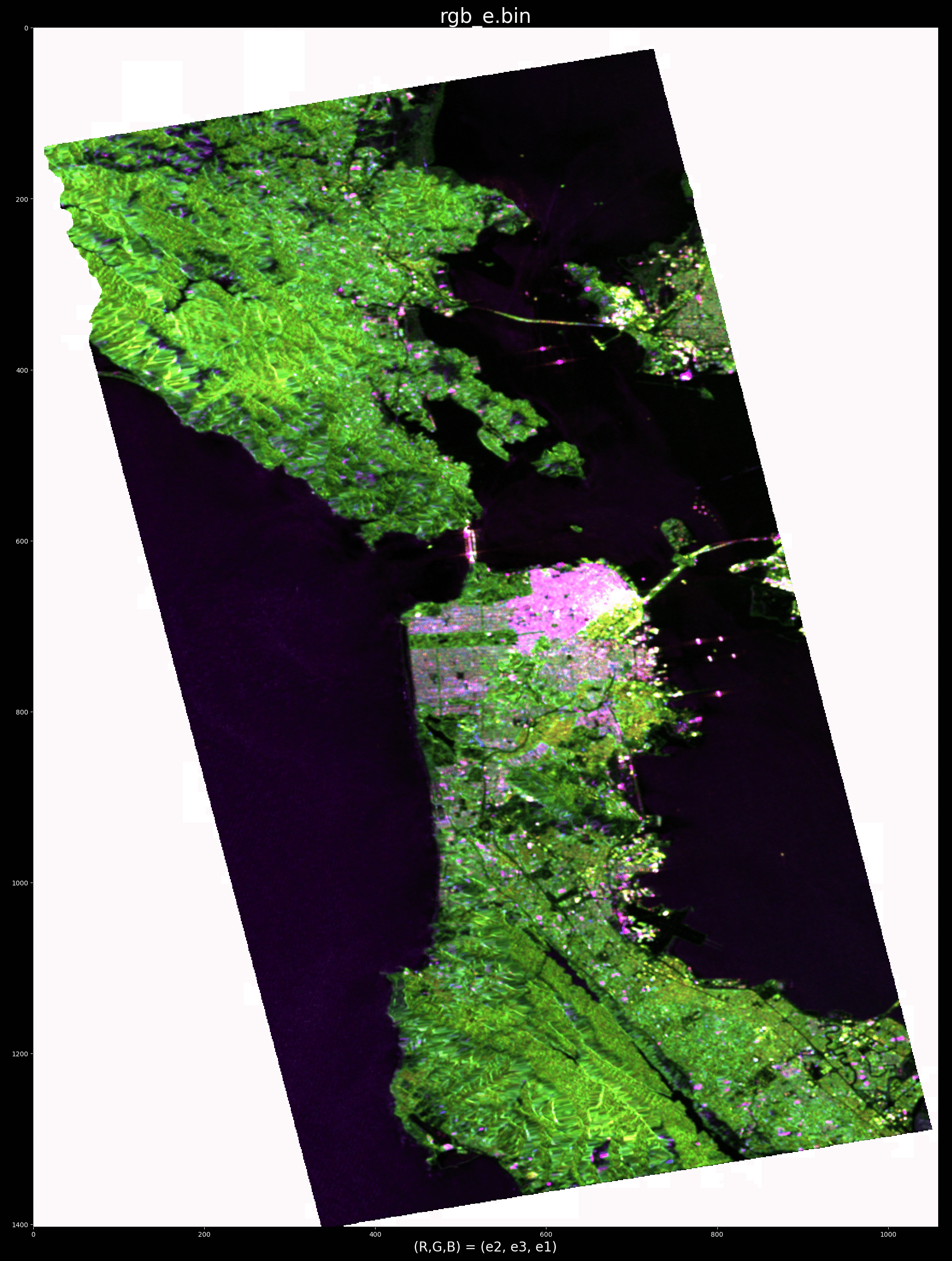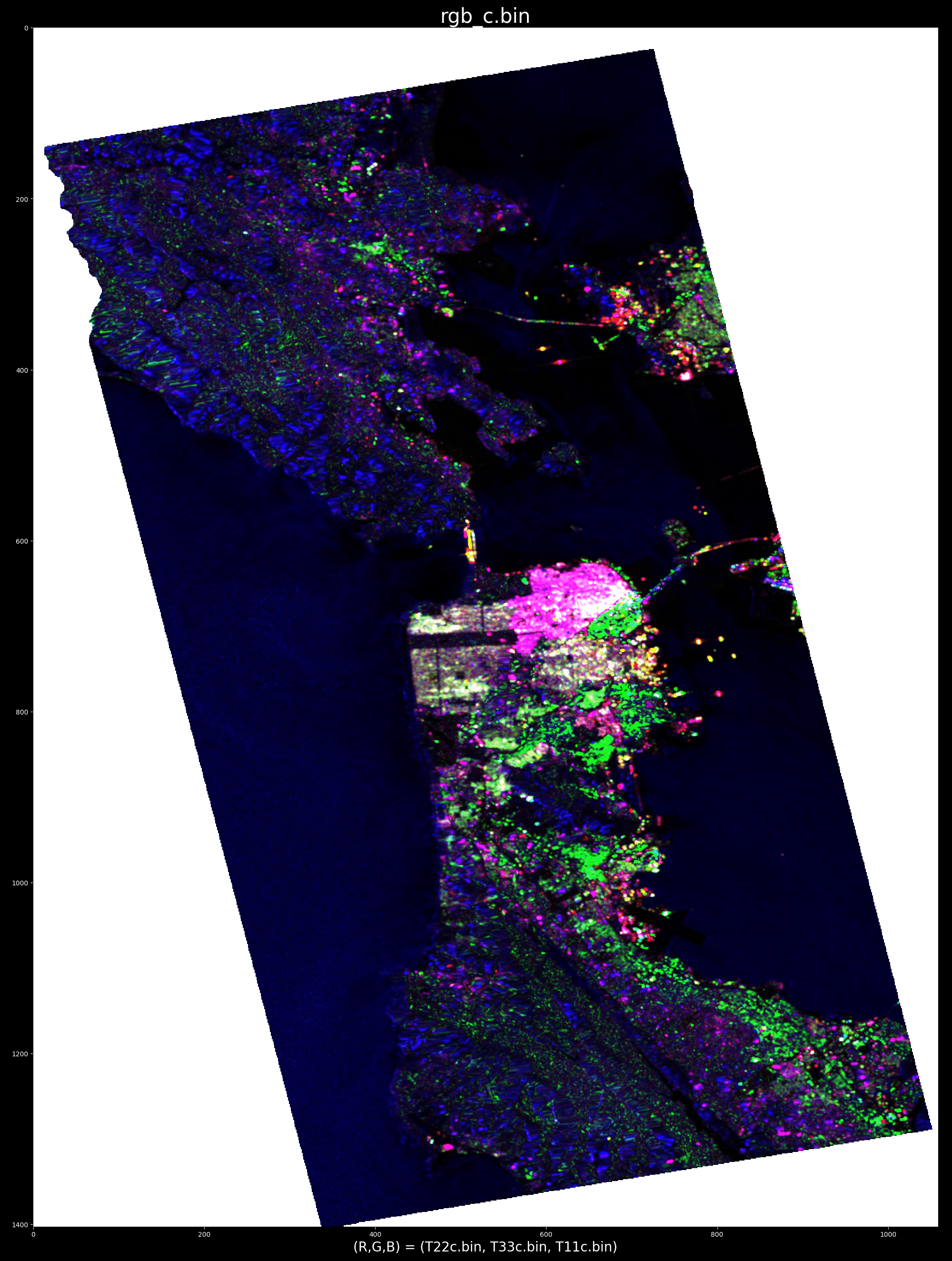
ALOS PALSAR data over SanFransisco, California in pauli encoding (r, g, b) = (T22, T33, T11):
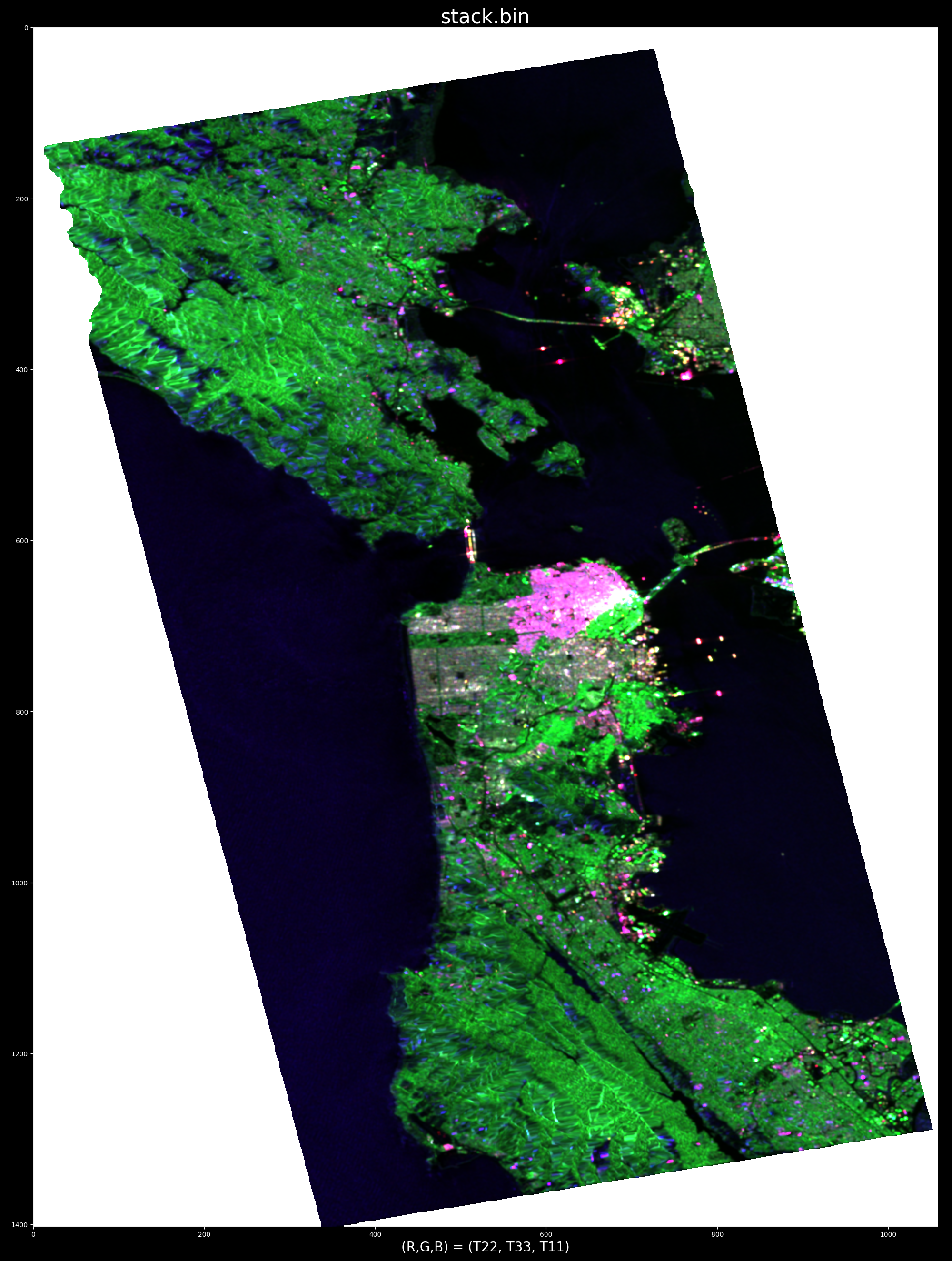
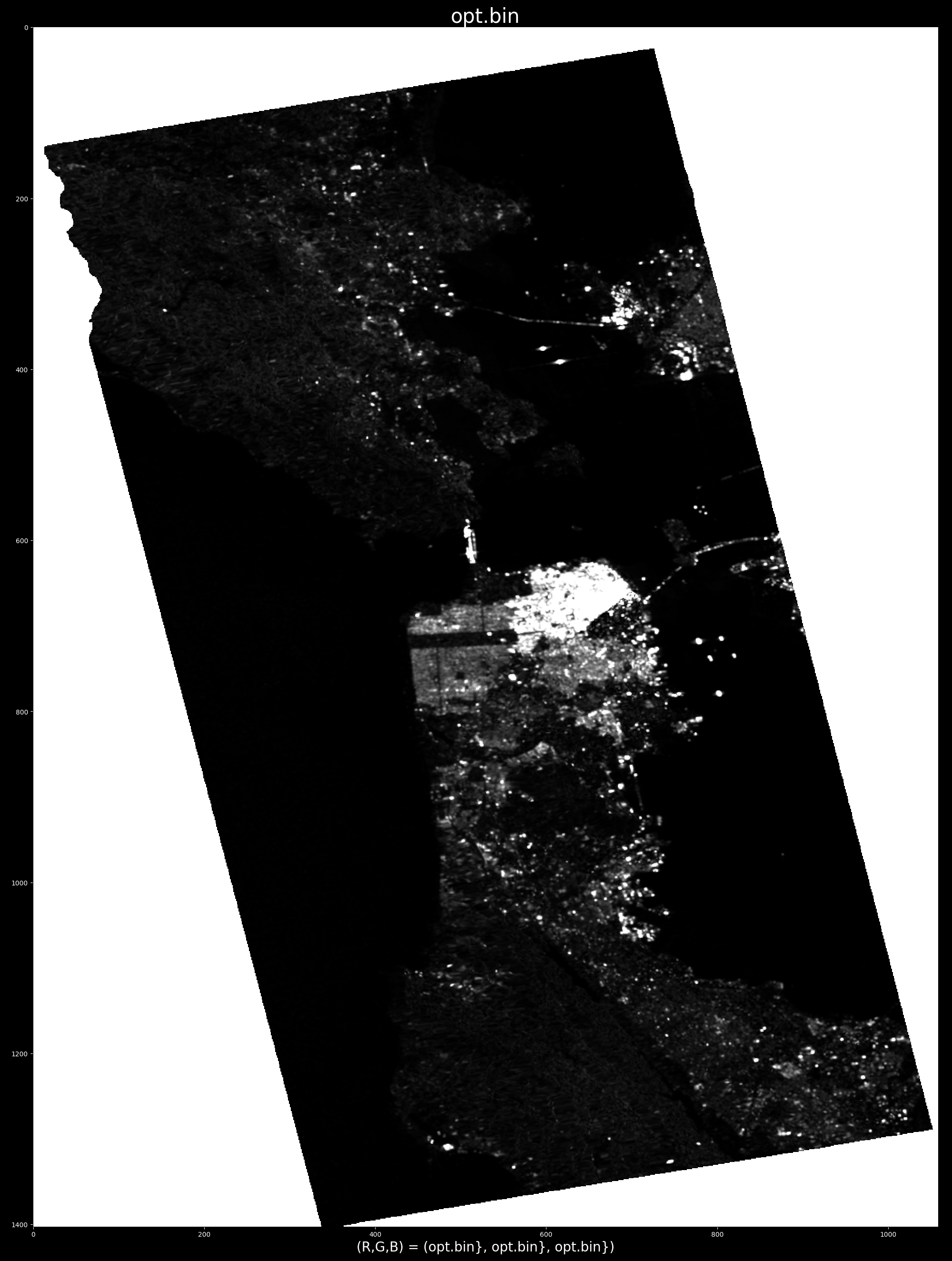
For example, triggering the processing by pointing at water, demonstrates (by "cancelling" water) the ability to pinpoint ships:
e.g. optimum radar cross section produced by selecting target at row/col index: (x,y)=863,739
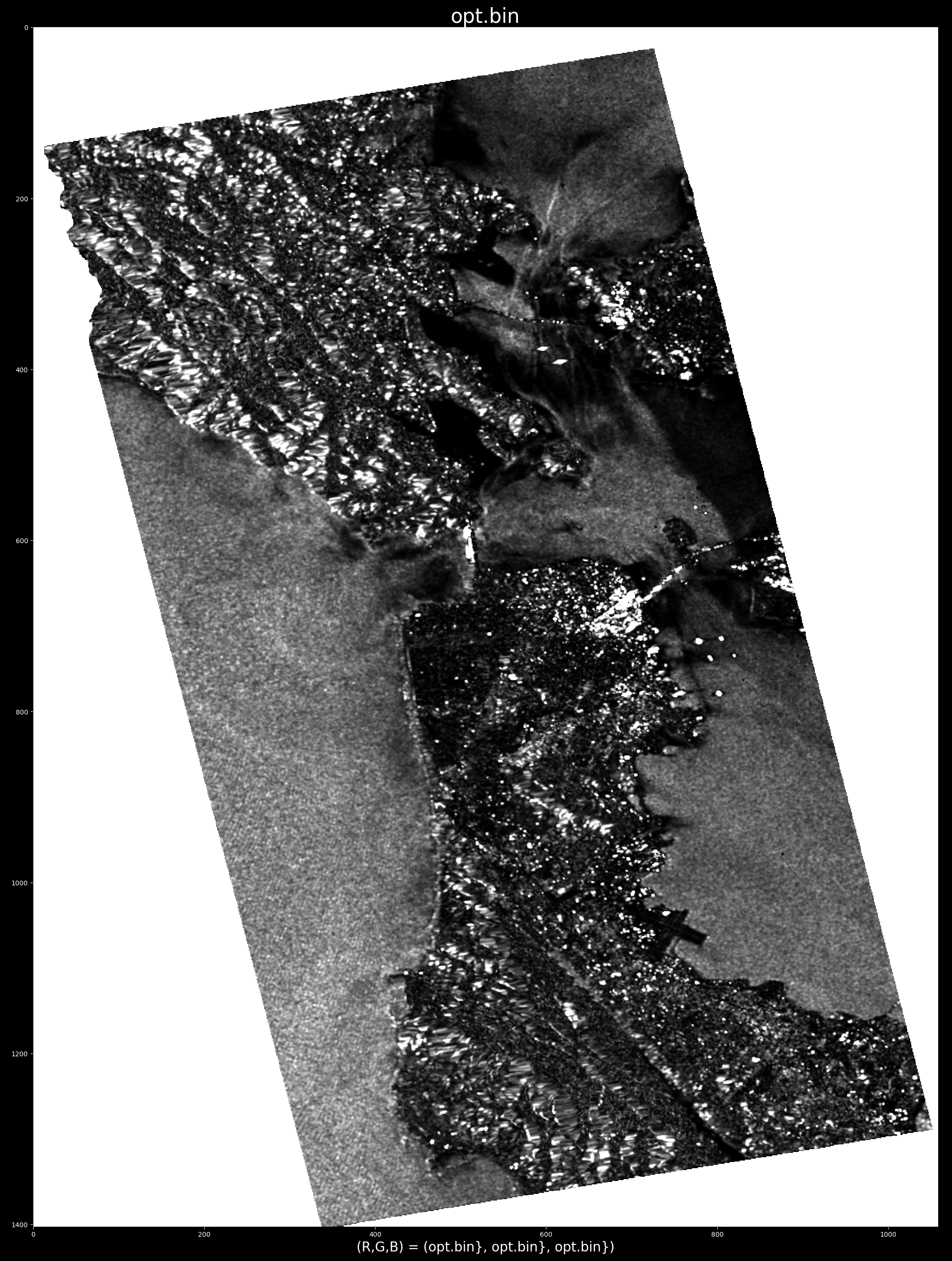
whereas cancelling an urban area highlights oceanographic and topographic features: e.g. optimum radar cross section produced by selecting target at point (x,y) = (688, 663)
At your terminal and from within the cloude_decom folder:
python3 cpp/uninstall.py
Default target window size is 3. To run with a different window size, e.g. 5:
cd T3
cloude_view stack.bin 5
If you already downloaded the code, proceed to step 3)
curl -o cloude_decom.zip https://codeload.github.com/ashlinrichardson/cloude_decom/zip/refs/heads/master; unzip cloude_decom.zip; mv cloude_decom-master cloude_decom
cd cloude_decom
python3 -m pip install numpy matplotlib
python3 py/cloude_decom.py T3
This gui works "the same" as the C++ gui:
- Depressing the mouse button will restore the "default" visualization e.g. (r,g,b) = (T22, T33, T11).
- Releasing the mouse button will run the decom ( and generate and display the product opt.bin).
Press on the keyboard, or use the "close window" button to exit.
Using the right mouse button draws an area target. Click the left mouse button once to close the polygon
Here is the sample command for running the decom using a "shapefile" target:
python3 py/cloude_decom.py T3 --shapefile=T3/shapefiles/water.shp
and e.g. cancelling a ship:
python3 py/cloude_decom.py T3 --shapefile=T3/shapefiles/ship.shp
Note that T11.bin must include georeferencing information in the T11.hdr file. For this option, the GUI is suppressed and outputs ( opt.bin, etc ) are made available in the T3 data folder.
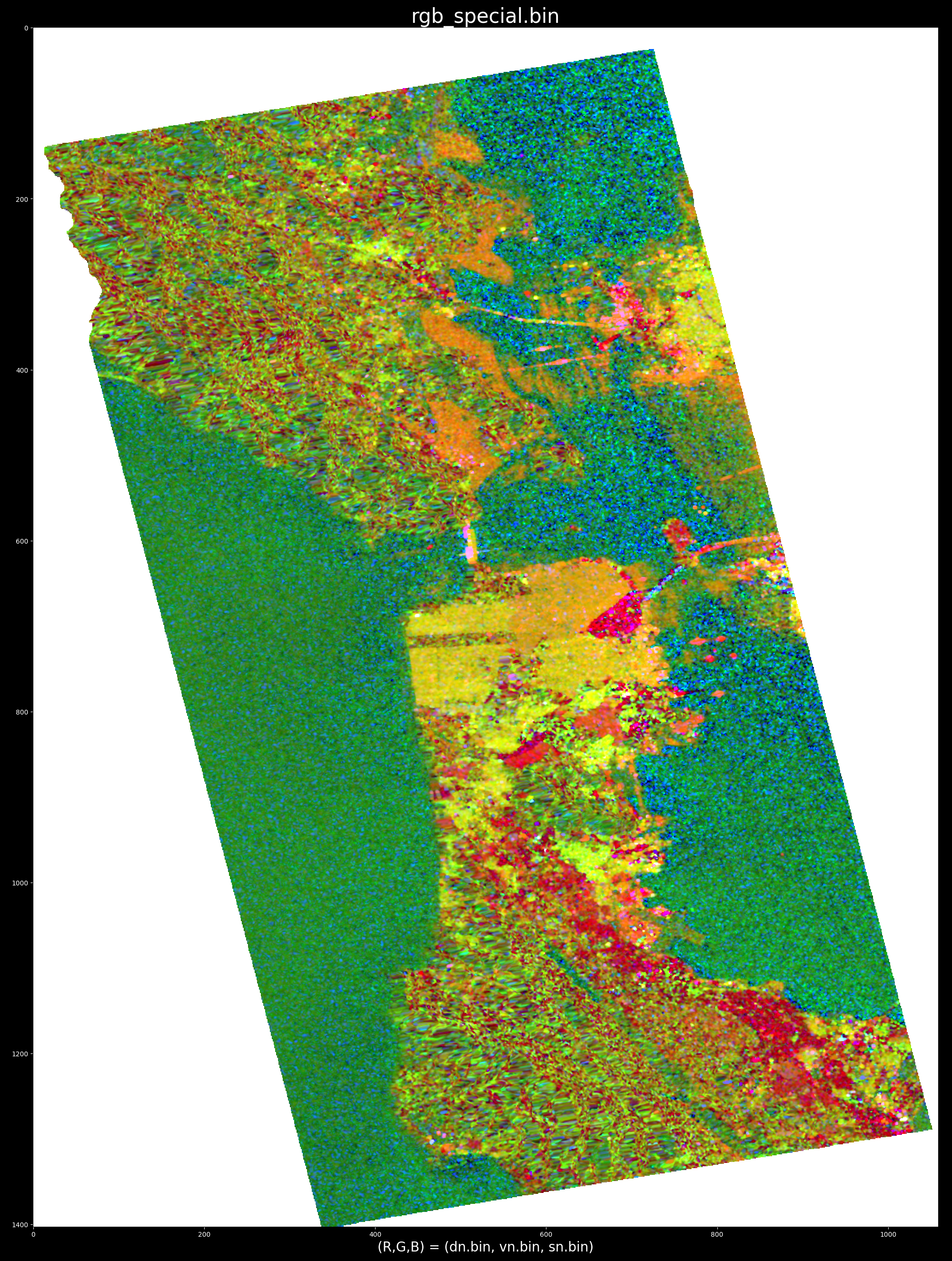
To run the python version at a specific target location (column, row index) examine the top of the python file for examples. For example, the flag --special_rgb can be used to display a special color-encoding:
Quite a few parameters are output by the program, such as the diagonal elements of the rank-1 T3 matrix:
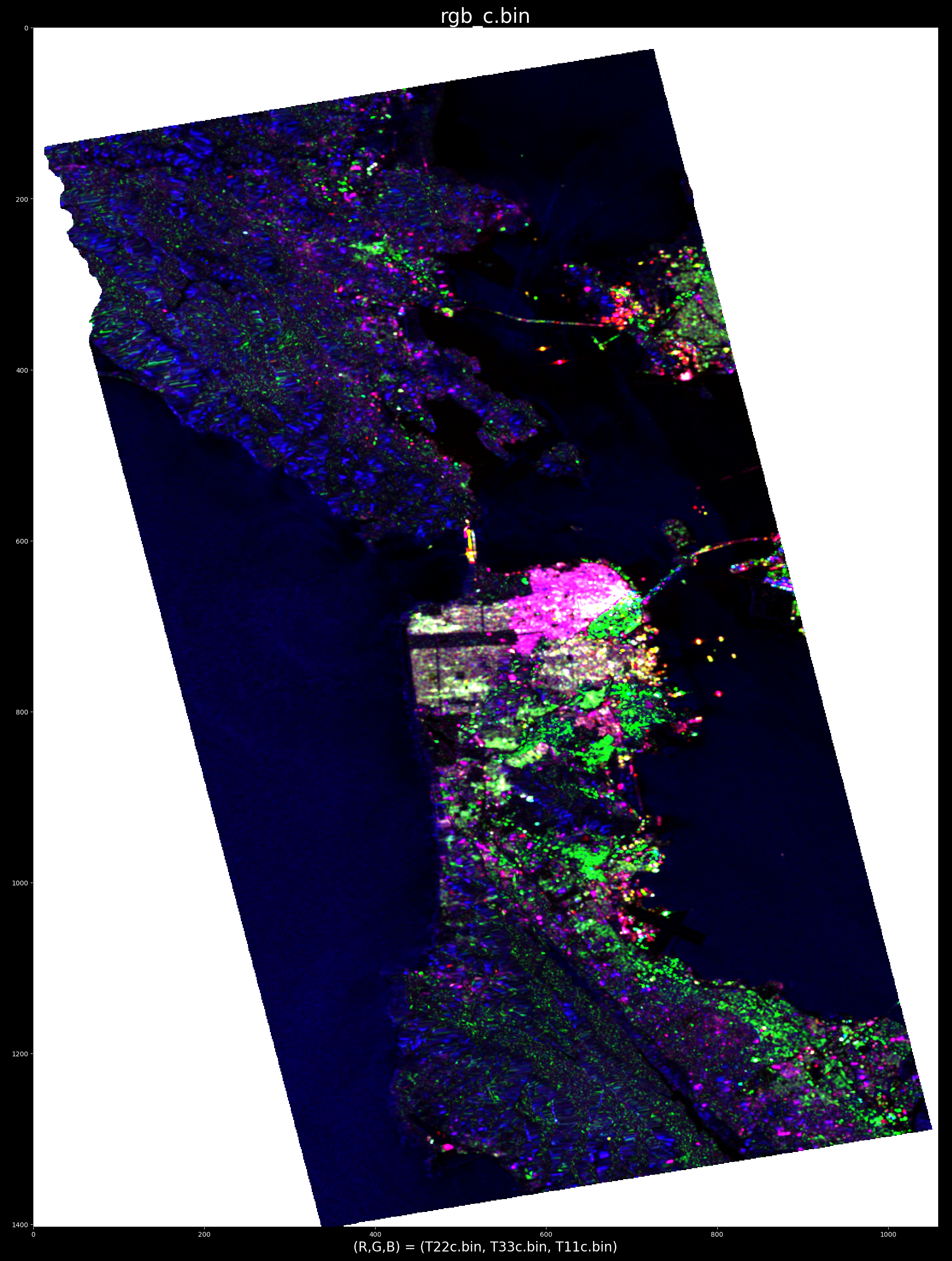
Comparing the pauli representation with the eigenvalues ( sorted ) and also with the rank-1 matrix elements:
- Make sure your dataset is in T3 matrix format ( PolSARPro standard ). ENVI header files ( one for each .bin file) are expected, as is the PolSARPro "config.txt" file.
Thanks to Eric Pottier and JAXA for providing ALOS-1 quad-pol data over San Fransisco, California. Please click here to see other sample data generously provided by Dr. Pottier.
Thanks to:
- Hao Chen, Canadian Forest Service
- Subhadip Dey, Indian Institute of Technology Bombay
- Shane Cloude, AELc
- Adithi Balaji (U.Vic)
for initial beta testing
At your convenience, please be welcome to:
- open an issue on this repository,
- submit a pull request, or
- provide feedback by email
- Radar --> Radiometric --> Calibrate (write complex output)
- Radar --> Geometric --> ALOS Deskewing
- Radar --> Polarimetric --> Faraday Rotation Correction
- Radar -> Polarimetrc --> Polarimetric Matrix Generation ( T3 matrix )
- Radar --> Radiometric --> Radiometric Terrain Flattening ( Small et al RTF )
- Radar --> Utilities --> Multilooking ( to approx square pixel )
- Radar -> Polarimetric --> Speckle filter (Boxcar filter, 7x7)
- Radar -> Geometric --> Terrain Correction --> Range Doppler Terrain Correction ( to put in geographic coordinates )