This repository has been archived by the owner on Feb 2, 2022. It is now read-only.
-
Notifications
You must be signed in to change notification settings - Fork 99
Home
Aditya Panchal edited this page Aug 14, 2015
·
4 revisions
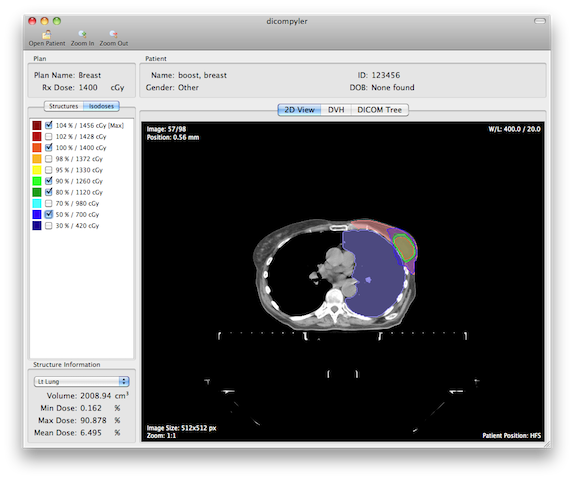 dicompyler is an extensible open source radiation therapy research platform based on the DICOM standard. It also functions as a cross-platform DICOM RT viewer.
dicompyler is an extensible open source radiation therapy research platform based on the DICOM standard. It also functions as a cross-platform DICOM RT viewer.dicompyler is written in Python and is built on a number of technologies including: pydicom, wxPython, PIL, and matplotlib and runs on Windows, Mac OS X and Linux.
Take a tour of dicompyler by checking out some screenshots or download your own copy today.


Google has deprecated downloads via Google Code, downloads are available through Google Drive:
Version 0.4.2: Windows | Mac | Source | Test Data
- New Features:
- Added Quick DICOM Import plugin
- Plugins can now access the 2D View drawing canvas
- Plugins can now be enabled/disabled
- DICOM data can now be loaded via command line argument or dragging and dropping a folder on the dicompyler icon on the Windows version
- Added persistence of the window size and position
- Added Quick DICOM Import plugin
- Many bug fixes
For additional changes, please see the release notes.
- Technology from dicompyler has been implemented as part of ASTRO/ROI's National Radiation Oncology Registry (NROR). Please see poster 2764 presented at the 2013 ASTRO Annual Meeting.
Please see the News archive for older news.
- Import CT/MR/PET Images, DICOM RT structure set, RT dose and RT plan files
- Extensible plugin system with included plugins:
- 2D image viewer with dose and structure overlay
- Dose volume histogram viewer with the ability to analyze DVH parameters
- DICOM data tree viewer
- Patient anonymizer
For upcoming features, see the project roadmap.
- 2D image viewer with dose and structure overlay
- Windows 2000/XP/Vista/7 (32-bit)
- Windows XP/Vista/7 (64-bit) - coming soon
- Mac OS X 10.5 - 10.9 (Intel 32-bit)
- Linux - via a package from PyPI or a Debian package (courtesy of debian-med)
If you are interested in building from source, please check out the build instructions.
- As a starting point, please read the FAQ as it answers the most commonly asked questions about dicompyler.
- If you need to cite dicompyler as a reference in your publication, please use the following citation:
-
A Panchal and R Keyes. "SU-GG-T-260: dicompyler: An Open Source Radiation Therapy Research Platform with a Plugin Architecture" Med. Phys. 37, 3245 , 2010.
-
A Panchal and R Keyes. "SU-GG-T-260: dicompyler: An Open Source Radiation Therapy Research Platform with a Plugin Architecture" Med. Phys. 37, 3245 , 2010.
- You can view the original poster (SU-GG-T-260) which was presented at the AAPM 2010 Annual Meeting. The reference in Medical Physics can be accessed via http://dx.doi.org/10.1118/1.3468652


