-
Notifications
You must be signed in to change notification settings - Fork 2
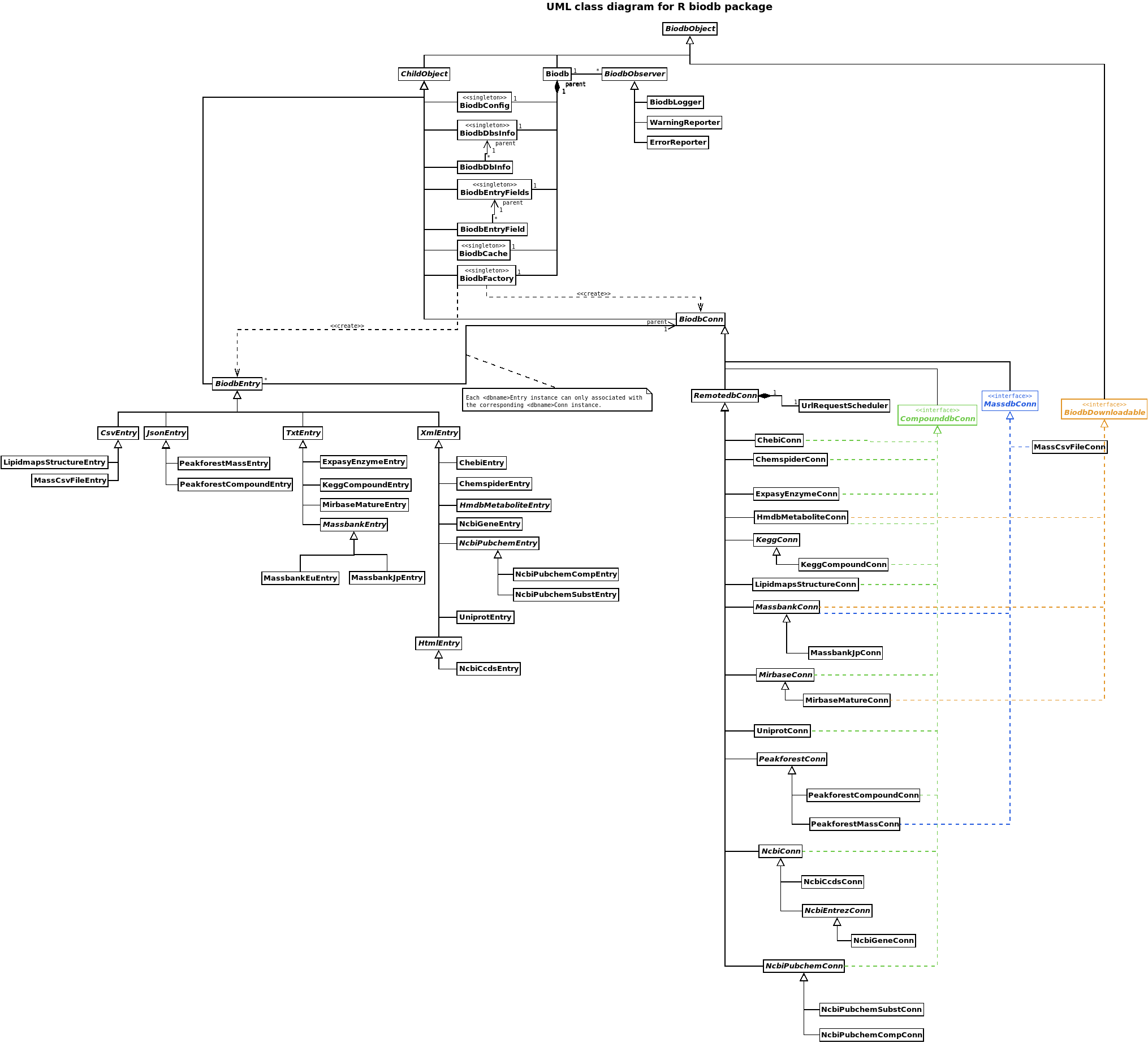
Architedture of biodb
This page has been generated automatically from a package vignette. Please do not edit, since your modifications will later be removed.
The architecture is organised around the following classes:
- The central
Biodbclass. - The singleton classes, attached to the
Biodbclass. - The connection classes
*Conn, responsible for connecting to databases. - The entry classes
*Entry, representing entries of the different databases. - The observer classes.
You will find below a UML class diagram, helpful to visualise all classes and their relationships.
All classes in biodb inherit directly on indirectly from the BiodbObject class, at the exception of UrlRequestScheduler and the observer classes.
The BiodbObject class contains mainly useful methods for:
- Sending messages.
- Assertions.
- Declaring methods as abstract in sub-classes.
- Declaring methods as deprecated in sub-classes.
The ChildObject class defines a parent field, allowing a class that inherits from it to be the child of another a class.
The class (Biodb) is the central point of the biodb package. To use the biodb package, an instance of this class Biodb has to be created. You can created as many instances as you want at the same time, though there would not be much purpose; all instances will be independent of each other.
From the instance of the class Biodb, you can access all singleton classes.
There are 5 singleton classes, each having each a distinct purpose:
-
BiodbConfigis meant for managing all configuration information. -
BiodbDbsInfostores database information. -
BiodbEntryFieldsstores entry fields information. -
BiodbFactoryis responsible for instantiating connection and entry classes. -
BiodbCachehandles the cache system.
The observer classes are used to transmit messages. You can create your own observer if you wish by creating a new class that inherits from BiodbObserver.
The observers are registered with the Biodb instance, either through the constructor or through the addObservers() method.
There are three concrete observer classes defined in the biodb package:
-
BiodbLoggerlogs messages either to standard error or to a file. -
WarningReporteremits a standard R warning if the message of this type. -
ErrorReporteremits a standard R error if the messages of this type.
Both WarningReporter and ErrorReporter are always defined in each instance of Biodb.
The connection classes handle the retrieval of information from databases. Their main purpose is the retrieval of and the search for entries.
BiodbConn class is the mother abstract class of all connection classes.
RemotedbConn class defines features specific to remote databases, such as an instance of the UrlRequestScheduler.
MassdbConn class defines methods for searching mass spectra databases.
CompunddbConn class defines methods for searching compound databases.
BiodbDownloadable interface defines methods for handling download of a database.
All those classes are abstract.
Generally, each connection concrete class inherits either directly from BiodbConn or from one or more of the other abstract classes. However, in order to factorize code, some intermediate abstract classes have been created: PeakforestConn, NcbiConn, etc.
The UrlRequestScheduler class is used inside the RemotedbConn class. For each instance of a concrete class that inherits from RemotedbConn, an instance of the UrlRequestScheduler class is defined. This instance is responsible for handling all URL requests, and especially to wait required time between two requests. Some databases define precisely what should be the frequency of the requests, and those specifications are defined in the instance of the UrlRequestScheduler class. Other databases do not define anything, however this does not mean one can send as many request as he wants; thus a default frequency of three requests per second is defined by default for a new instance of UrlRequestScheduler class.
The entry classes represent entries in the database. BiodbEntry is the mother abstract class of all entry classes. It handle the field values of the entries, and organize the parsing of entries from the string content returned by the database.
Different intermediate abstract classes have been created in order to ease parsing of the various type of contents returned by the different databases:
-
CsvEntryhandles CSV type content. The separator character can be chosen. -
HtmlEntryhandles HTML parsing through XPath expressions. -
JsonEntryhandles JSON parsing. -
TxtEntryhandles generic text parsing through regular expressions. -
XmlEntryhandles XML parsing through XPath expressions.
Here is a complete class diagram showing all classes and their relations.