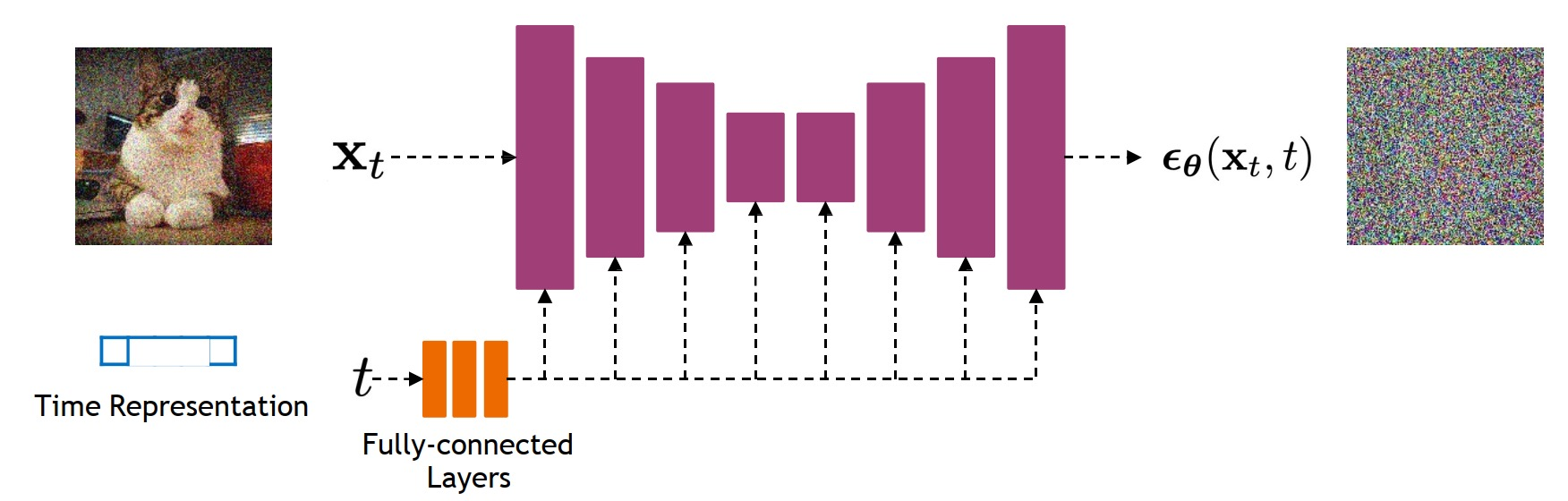
MedSegDiff is a comprehensive PyTorch implementation of the MedSegDiff paper, presenting the first Diffusion Probabilistic Model (DPM) designed specifically for general medical image segmentation tasks. This repository aims to provide researchers and practitioners with a clear, step-by-step codebase and documentation to facilitate understanding and application of MedSegDiff across various medical imaging modalities.
| Source Code | Website |
|---|---|
| github.com/deepmancer/medseg-diffusion | deepmancer.github.io/medseg-diffusion |
- General Medical Image Segmentation: Tailored to handle diverse medical imaging tasks, including segmentation of brain tumors, optic cups, and thyroid nodules.
- Dynamic Conditional Encoding: Implements step-wise adaptive conditions to enhance regional attention during diffusion.
- Feature Frequency Parser (FF-Parser): Leverages Fourier domain operations to reduce high-frequency noise, improving segmentation quality.
- From-Scratch Implementation: Offers a clean and well-documented PyTorch codebase for easy learning and experimentation.
- Community-Friendly: Welcomes contributions, issues, and discussions to foster community engagement.
- MedSegDiff: Medical Image Segmentation with Diffusion Probabilistic Models 🚀
- 🌟 Key Features
- 📖 Table of Contents
- 🔍 Overview
- 🛠️ Methodology
- 🔧 Dynamic Conditional Encoding
- ⏳ Time Encoding Block
- 🏗️ Encoder & Decoder Blocks
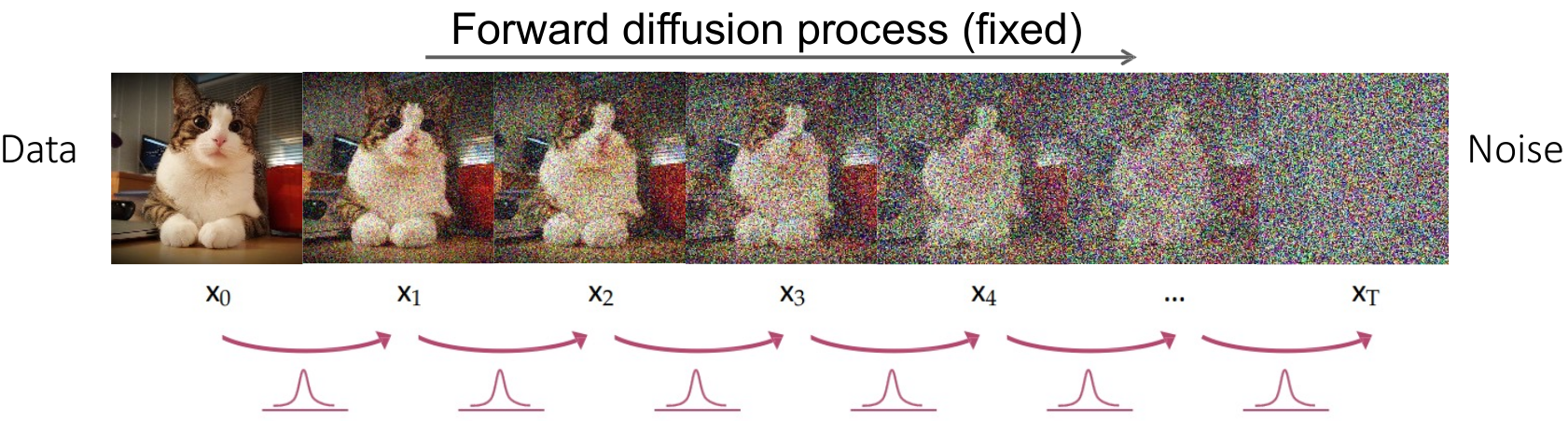
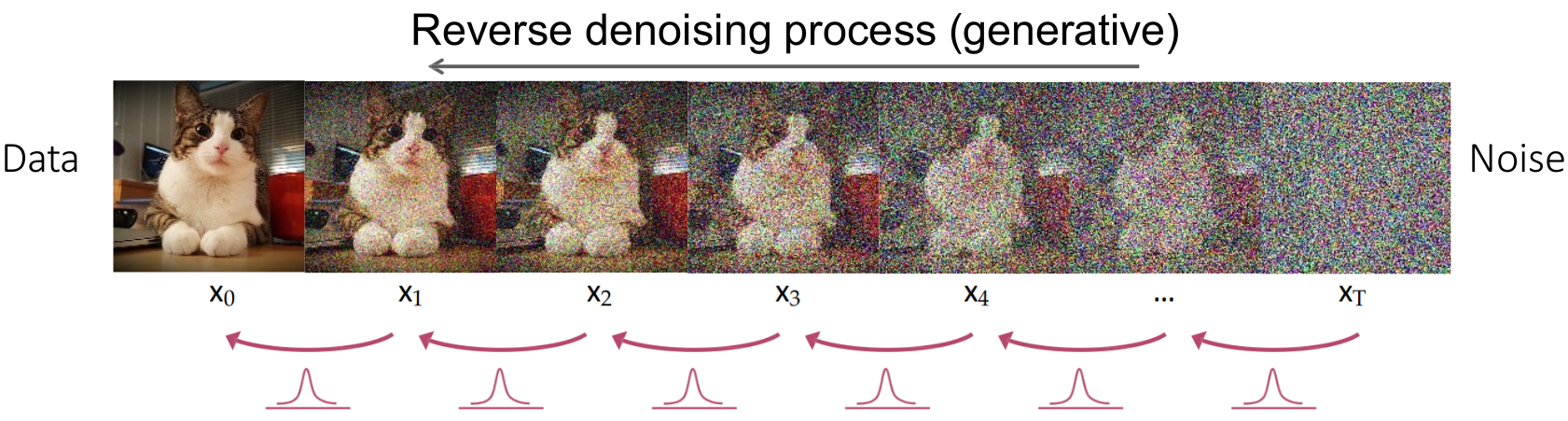
- 🔄 Diffusion Process (Forward & Reverse)
- 🟢 Forward Diffusion
- 🔴 Reverse Diffusion
- 🎯 Results
- 🚀 Installation & Usage
- 📁 Repository Structure
- 📝 License
- 🙏 Acknowledgments
- 🌟 Support the Project
- 📚 Citations
MedSegDiff addresses a fundamental challenge in medical imaging: achieving accurate and robust segmentation across various imaging modalities. Building upon the principles of Diffusion Probabilistic Models (DPMs), MedSegDiff introduces innovative techniques like dynamic conditional encoding and the Feature Frequency Parser (FF-Parser) to enhance the model's ability to focus on critical regions, reduce high-frequency noise, and achieve state-of-the-art segmentation results.
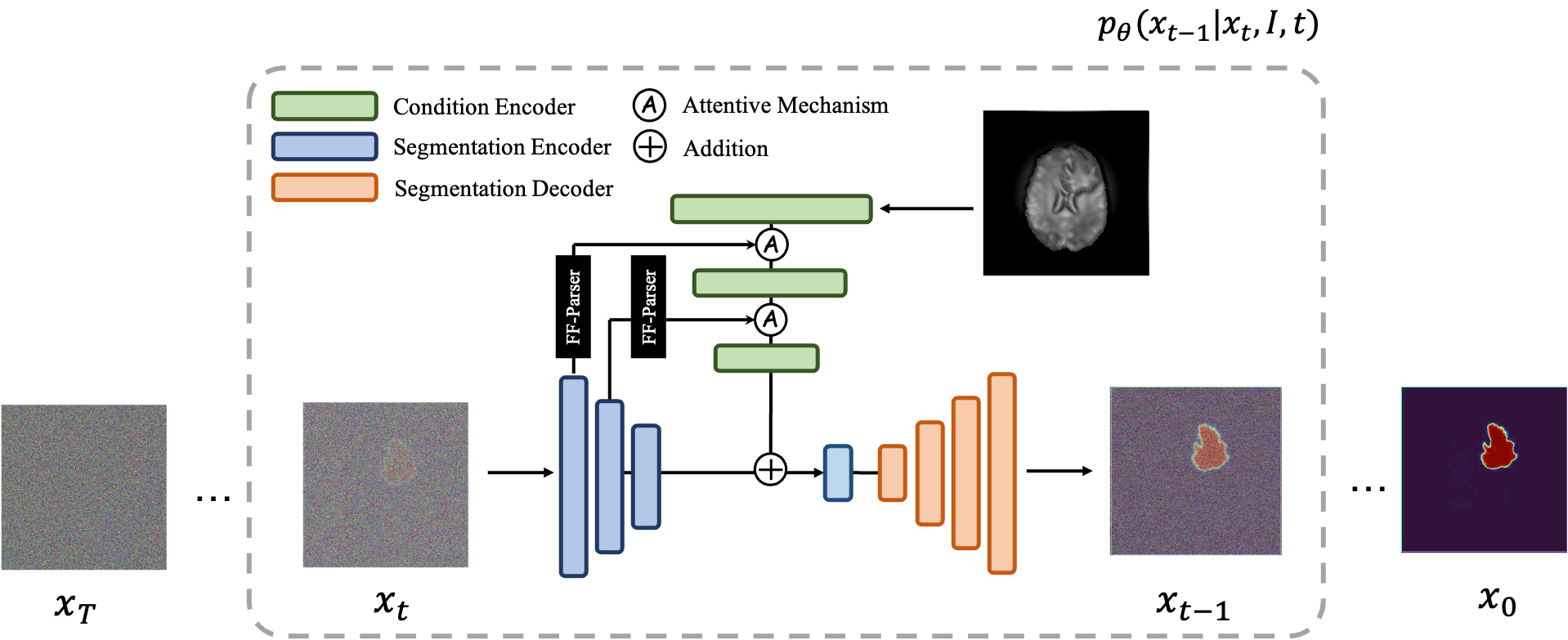
An overview of the MedSegDiff architecture. The time step encoding component is omitted for clarity.
Formally, at each diffusion step, the model estimates:
Here:
-
$\mathbf{E_t^I}$ : Conditional feature embedding from the input image. -
$\mathbf{E_t^x}$ : Feature embedding of the evolving segmentation mask. -
$D$ : A U-Net decoder guiding reconstruction.
The training objective:
This loss encourages the model to accurately predict the noise added at each step, ultimately guiding the segmentation toward a clean, high-quality mask.
MedSegDiff employs a U-Net-based architecture enriched with diffusion steps, dynamic conditional encoding, and Fourier-based noise reduction. The key idea is to iteratively refine a noisy segmentation map into a clean, accurate mask using reverse diffusion steps guided by learned conditioning from the original image.
-
Feature Frequency Parser (FF-Parser): The segmentation map first passes through the FF-Parser, which utilizes Fourier transforms to filter out high-frequency noise components, thereby refining the feature representation.
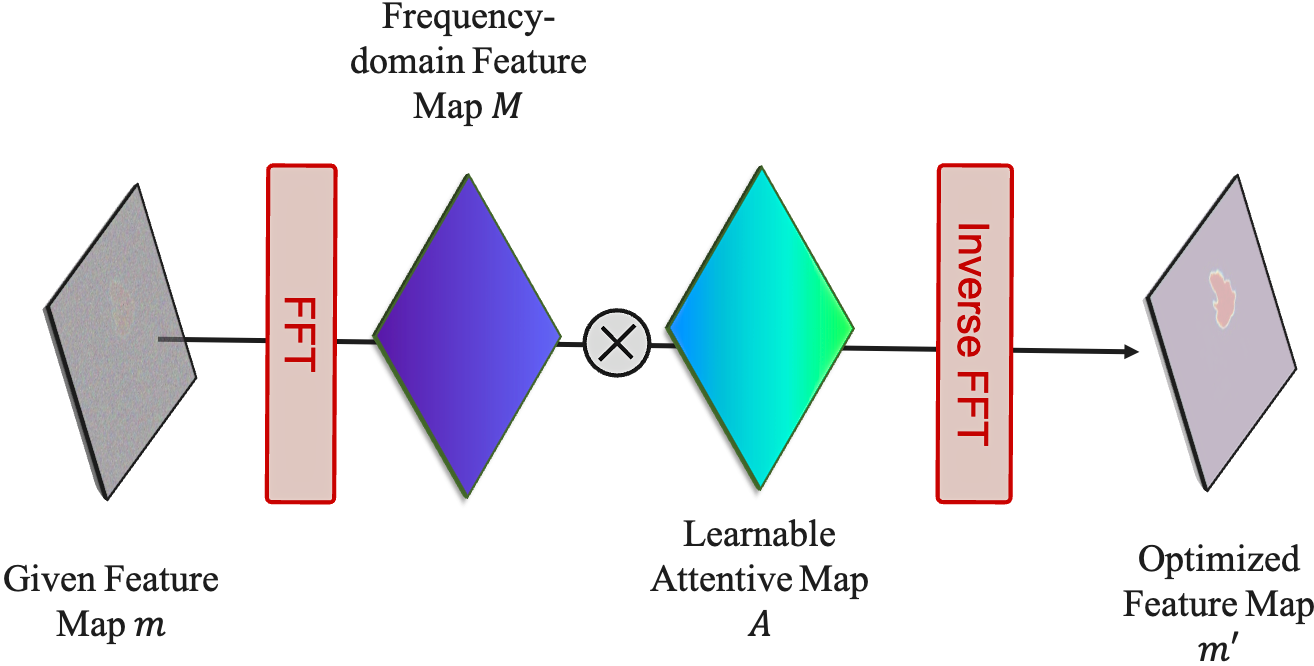
The FF-Parser integrates FFT-based denoising before feature fusion. -
Attentive Fusion: The denoised feature map is then fused with the image embeddings through an attentive mechanism, enhancing regional attention and improving segmentation precision.
-
Iterative Refinement: This combined feature undergoes further refinement, culminating in a bottleneck phase that integrates with encoder features.
-
Bottleneck Integration: The refined features merge with the encoder outputs, resulting in the final segmentation mask.
- Sinusoidal Embeddings: Timestep embeddings are computed using sinusoidal functions, capturing temporal information of the diffusion process.
- Integration into Residual Blocks: These time features are injected into the model's residual blocks, providing temporal context at each diffusion step.
- Initial Convolutions: Separate initial convolutional layers process the input image and the segmentation mask.
- Residual Blocks: The backbone consists of ResNet-like blocks with convolutional layers, GroupNorm, and activation functions.
- Attention Mechanisms: Multi-head attention modules are incorporated to enhance spatial focus on critical regions.
In the forward diffusion process, Gaussian noise is progressively added to the segmentation mask over a series of timesteps, degrading it into pure noise.
-
Noise Addition: Starting from the original segmentation mask
$\text{mask}_0$ , Gaussian noise is added iteratively at each timestep$t$ , controlled by a variance schedule$\beta_t$ . -
Progressive Degradation: This process produces a sequence of increasingly noisy masks
$\text{mask}_0, \text{mask}_1, \dots, \text{mask}_T$ . -
Convergence to Noise: As
$T \to \infty$ , the mask becomes indistinguishable from pure Gaussian noise.
The reverse diffusion process aims to reconstruct the original segmentation mask from the noisy data by iteratively denoising.
-
Noise Prediction: A U-Net is trained to predict the noise added at each timestep, learning a mapping
$\epsilon_\theta(\text{mask}_t, t)$ . -
Stepwise Denoising: Starting from
$\text{mask}_T$ , the model refines the mask by subtracting the predicted noise at each timestep, moving backward from$t = T$ to$t = 0$ . -
Final Reconstruction: After
$T$ steps, the output$\text{mask}_0$ approximates the original segmentation mask.
MedSegDiff demonstrates superior performance across various medical image segmentation tasks, outperforming state-of-the-art methods by a significant margin.
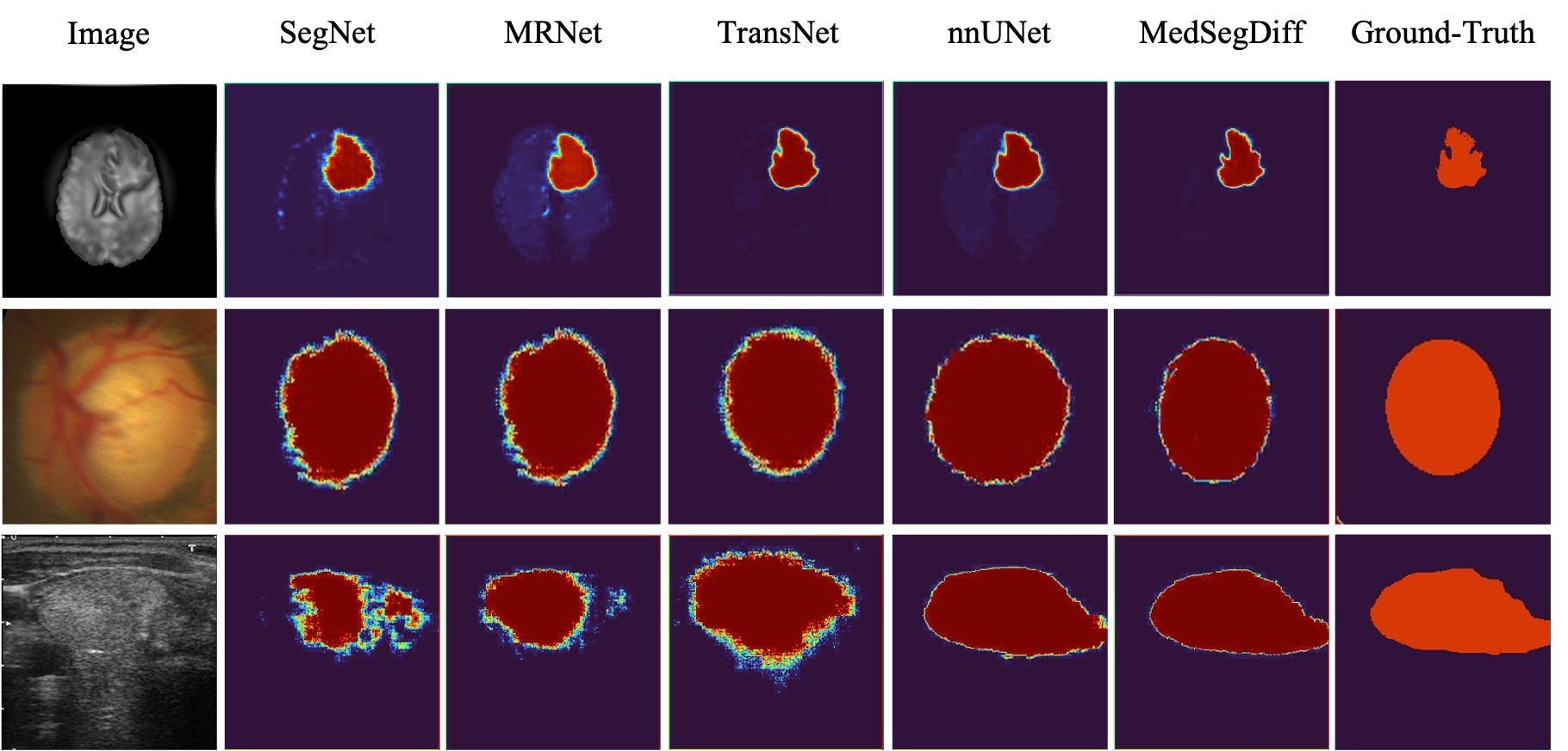
Visual comparisons with other segmentation methods.
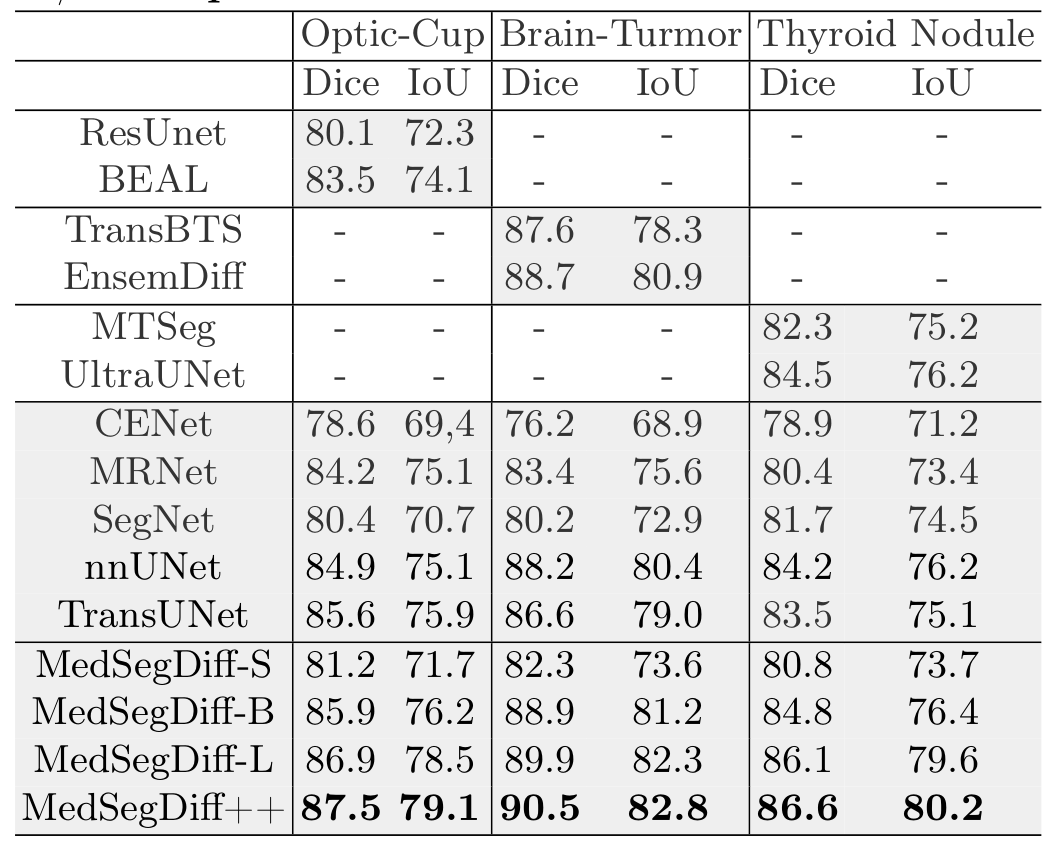
Quantitative results comparing MedSegDiff with state-of-the-art methods. Best results are highlighted in bold.
- Python 3.8 or higher
- PyTorch
- Other dependencies as specified in
requirements.txt
Clone the repository and install the required packages:
git clone https://github.com/deepmancer/medseg-diffusion.git
cd medseg-diffusion
pip install -r requirements.txt- Explore
MedSegDiff.ipynbfor a comprehensive, step-by-step notebook demonstration. - Adjust hyperparameters and diffusion steps as needed within the notebook.
- To use your own datasets, modify the data loading sections accordingly.
This project is licensed under the MIT License. See the LICENSE file for details.
We extend our gratitude to the authors of the MedSegDiff paper and other referenced works for their valuable research and insights that inspired this implementation.
If you find MedSegDiff valuable for your research or projects, please consider starring ⭐ this repository on GitHub. Your support helps others discover this work!
If you utilize this repository, please consider citing the following works:
@article{Wu2022MedSegDiffMI,
title = {MedSegDiff: Medical Image Segmentation with Diffusion Probabilistic Model},
author = {Junde Wu and Huihui Fang and Yu Zhang and Yehui Yang and Yanwu Xu},
journal = {ArXiv},
year = {2022},
volume = {abs/2211.00611}
}@inproceedings{Hoogeboom2023simpleDE,
title = {simple diffusion: End-to-end diffusion for high resolution images},
author = {Emiel Hoogeboom and Jonathan Heek and Tim Salimans},
year = {2023}
}