-
Notifications
You must be signed in to change notification settings - Fork 3
Plotting in python matplotlib
Hi everyone, to help with everyone's plotting for the model I thought I'd include a couple of guides on plotting in python, specifically using matplotlib.
Just to put it somewhere useful, here is a link to the matplotlib website where you can find more examples and documentation: https://matplotlib.org/
The plot function basically takes a set of x,y coordinates and joins the dots together, nothing particularly exciting but it works!
A couple of things that are worth mentioning now is that the length (number) of the x values must equal the length (number) of the y values
# Create dummy data
x = np.linspace(0, np.pi * 3, 100)
y = np.sin(x)
# Plot with the basic line function
plt.plot(x, y, color='r', alpha=0.3)
# Create graph labels
plt.xlabel('x')
plt.ylabel('sin(x)')
plt.title('A riveting sine wave')
# Set the max and minimum values shown on the x and y axis
plt.ylim([- 1.5, 1.5])
plt.xlim([0, 10])
plt.show()
plt.clf()
Example of a plotting a confidence interval
# Create dummy data
x = np.linspace(0, np.pi * 3, 100)
y = np.sin(x)
# Create a dummy confidence interval
y_upper_CI = np.add(y, 0.2)
y_lower_CI = np.add(y, - 0.2)
# Plot with the basic line function
plt.plot(x, y, color='r', alpha=0.3, label='Estimate')
# Plot the 95% C.I.
plt.fill_between(x, y_lower_CI, y_upper_CI, color='r', alpha=0.2, label='95% C.I.')
# Create graph labels
plt.xlabel('x')
plt.ylabel('sin(x)')
plt.legend()
plt.title('A riveting sine wave')
# Set the max and minimum values shown on the x and y axis
plt.ylim([- 1.5, 1.5])
plt.xlim([0, 10])
plt.show()
plt.clf()
Simple bar plots are easy to do
# Bar charts
# Create dummy data
data = [10, 20, 30, 40, 50]
plt.bar(np.arange(len(data)), # create the x values to plot the bars on
data, # give the 'y' values to plot on the bars
color='lightseelblue', # choose a colour
)
plt.xticks(np.arange(len(data)), ['a', 'b', 'c', 'd', 'e'])
plt.ylabel('Dummy y')
plt.title('Some title')
plt.show()
Stacked bar chars are essentially the same as normal bar charts but with a small change, using the bottom option to stack one bar on top of another
# Create dummy data
total_DALYs = 100000
total_YLD = 40000
total_YLL = 60000
plt.bar([0], [total_DALYs], color=['lightsteelblue'], label='DALYs')
plt.bar([1], [total_YLD], color=['lightsalmon'], label='YLD')
plt.bar([1], [total_YLL], color=['darksalmon'], label='YLL', bottom=[total_YLD])
plt.xticks([0, 1], ['DALYs', 'Breakdown'])
plt.legend()
plt.title('Average number of DALYs and the YLL, YLD composition')
plt.show()
# Square plots
import squarify # These plots require an additional package
# create dummy data
dummy_data = [1004, 720, 366, 360, 80]
district_names = ["D1", "D2", "D3", "D4", "D5"]
colours = ['r', 'b', 'g', 'c', 'y']
# create a square plot
squarify.plot(dummy_data, # the data values which need to be sorted into squares
label=district_names, # state the labels, ordered the same way as the data
color=colours, # state the colours used in the plot, these are ordered in the same way as the data
pad=2 # create some space between the squares
)
plt.axis("off")
plt.title('Districts causing the most cases')
plt.show()
# Plotting a map
# First thing needed is to get the shape file ready by geopandas
zimbabwe = geopandas.read_file(zimShapeFile)
# Create dummy case numbers for each district
max_n_cases = 100000
dummy_cases = np.random.randint(10, high=max_n_cases, size=len(zimbabwe), dtype=int)
# Store the number of cases in the geopandas df to be plotted later
zimbabwe['Cases'] = dummy_cases
######### ------- very important to note that the data you are storing on to the Zimbabwe file needs to be ordered by district
######### so say you have data like (d_1, 10), (d_2, 20),..., (d_60, 60). When you are adding this onto the geopandas dataframe it must
######### be ordered in ascending district order
# Create an axis which we can plot on (necessary)
fig, ax = plt.subplots()
# use df.plot to produce the graph
zimbabwe.plot(ax=ax, column='Cases', cmap='Greens', edgecolor='k', legend=True, vmax=max_n_cases)
plt.title('Covid cases in Zimbabwe')
# Turn off the x and y axis to make it look neater
plt.axis('off')
plt.show()
# Lolipop plots
# You can create a lolipop plot fairly easily by using the stem function from matplotlib
# create data
x = range(1, 41)
y = np.log(x)
# stem function, linefmt changes the line format (dotted, solid dashed etc...) and colour, markerfmt does the same for
# the marker
plt.stem(x, y, linefmt='r-.', markerfmt='g*')
plt.xlabel('x')
plt.ylabel('Log(x)')
plt.title('Lolipop plots (absolute eyesore of a colour scheme)')
plt.show()
Unfortunately I can't seem to find a simple way to do the same thing where the points attach to a y-axis so I've made a function to do this:
def add_lolipop_stalks_on_y_axis(x_values, y_values, colour):
"""
x_values: the x coordinates
y_values: the y coordinates
colour: what colour you want the line to be
"""
for idx, y_val in enumerate(y_values):
plt.hlines(y=y_val, xmin=min(x_values), xmax=x_values[idx], color=colour)
x = np.arange(0, 20)
y = x**2
plt.scatter(x, y, color='r')
plt.xlabel('x')
plt.ylabel('x squared')
add_lolipop_stalks_on_y_axis(x, y, 'r')
plt.show()
# Pie chart
# Create dummy data
dummy_cfr = [0.97, 0.03]
# Create the pie chart
plt.pie(dummy_cfr, # tell it what data to plot
explode=[0, 0.3], # 'pop' out one of the pie segments
labels=['Non-fatal', 'Fatal'], # Create labels for each segment
colors=['lightsteelblue', 'lightsalmon'], # Give color each segment
autopct='%.1f' # Format the auto-percent labelling, .1 refers to the number of decimal places
)
plt.title('Percent of Covid-19 cases that are fatal')
plt.show()
These look very complicated, but it's actually not as hard to make them as you would think. Really all that happens in these plots is we plot a bunch of lines with the same x values on top of one another (offsetting the y values each time to make sure we aren't plotting things on top of one-another). With a few extra specified parameters we can change how the figures look (in this case making them black) to make the recognisable style of the plots
# Create new Figure with black background
fig = plt.figure(figsize=(8, 8), facecolor='black')
# Add a subplot with no frame
ax = plt.subplot(111, frameon=False)
# Format data from model run
onedrive_file_path = "/Users/robbiework/Library/CloudStorage/OneDrive-UniversityCollegeLondon/data/output/ICCS/" \
"extended_submission/ver_3 (multiDist)/50_perc/beta_0.3/output/"
# Generate epidemic curves from different runs
# Create a list to store the number of cases in
lines = []
# Iterate over each output file
for file in os.listdir(onedrive_file_path):
# load the data
data = pd.read_csv(onedrive_file_path + file, delimiter='\t')
# drop the column that is normally there
data = data.drop('Unnamed: 10', axis=1)
# Calculate the total number of cases in all districts at each point in time
data = data.groupby('time').sum()
# Calculate the total new cases (both symptomatic and asymptomatic)
data['new_cases'] = data['metric_new_cases_asympt'] + data['metric_new_cases_sympt']
# Store this in lines
lines.append(list(data['new_cases']))
# Generate line plots, iterate over number of runs
for i in range(len(os.listdir(onedrive_file_path))):
# We need something to push the lines up on the plot, otherwise each line will be plotted on top of one another and
# it will look messy, create this y-axis offset (this is just made up and will be individual to the graph, but works
# for this graph at least
y_offset = i * max([max(j) for j in lines]) / len(os.listdir(onedrive_file_path)) * 2
# Plot the line
ax.plot(np.arange(len(lines[i])), np.add(y_offset, lines[i]), color="w", lw=2)
plt.title('Joy division where okay, never really got into them,\n but at least we have cool graphs', color='w')
plt.show()
plt.clf()
Subplots in matplotlib can be really annoying, the most simple way to plot them it to use the plt.subplots(n_rows, n_columns, plot_number) route but this can lead to strange bunching up of the plots and general messiness. But if you're after a quick plot then this is the way to go. Let's plot a few (bad) examples:
x = np.linspace(0, 20, 1000)
sin_x = np.sin(x)
cos_x = np.cos(x)
tan_x = np.tan(x)
plt.subplot(1, 3, 1)
plt.plot(x, sin_x, color='r')
plt.xlabel('x')
plt.ylabel('sin(x)')
plt.title('The sine function')
plt.subplot(1, 3, 2)
plt.plot(x, cos_x, color='g')
plt.xlabel('x')
plt.ylabel('cos(x)')
plt.title('The cosine function')
plt.subplot(1, 3, 3)
plt.plot(x, tan_x, color='b')
plt.xlabel('x')
plt.ylabel('tan(x)')
plt.title('The tangent function')
plt.show()
So although this was a fairly quick to make plot, it doesn't look very nice. After we make the figure we can adjust things in the subplot to improve the way it looks however:
x = np.linspace(0, 20, 1000)
sin_x = np.sin(x)
cos_x = np.cos(x)
tan_x = np.tan(x)
plt.subplot(1, 3, 1)
plt.plot(x, sin_x, color='r')
plt.xlabel('x')
plt.ylabel('sin(x)')
plt.title('The sine function')
plt.subplot(1, 3, 2)
plt.plot(x, cos_x, color='g')
plt.xlabel('x')
plt.ylabel('cos(x)')
plt.title('The cosine function')
plt.subplot(1, 3, 3)
plt.plot(x, tan_x, color='b')
plt.xlabel('x')
plt.ylabel('tan(x)')
plt.title('The tangent function')
plt.subplots_adjust(wspace=1)
plt.show()
This is a little bit better, but it's still ugly and we can do better. One issue is that the figure size is small, which is a problem when we are trying to fit multiple subplots on the same figure. We can improve the look of these plots with a few extra steps as shown in the next section!
In the next level of subplot plotting we are going to firstly create a figure and specify a figure size, this stops subplots being crammed together in a small space. We are then going to create a bunch of axis (thing of these as the actual subplots) and plot onto the axis. For some absolutely horrible reason the functions used to change things on subplots changes when you specify each axis.
number_rows = 1
number_columns = 3
x = np.linspace(0, 20, 1000)
sin_x = np.sin(x)
cos_x = np.cos(x)
tan_x = np.tan(x)
fig = plt.figure(figsize=(18, 6))
(ax11, ax12, ax13) = fig.subplots(number_rows, number_columns)
ax11.plot(x, sin_x, color='r')
ax11.set_xlabel('x')
ax11.set_ylabel('sin(x)')
ax11.set_title('The sine function')
ax12.plot(x, cos_x, color='g')
ax12.set_xlabel('x')
ax12.set_ylabel('cos(x)')
ax12.set_title('The cosine function')
ax13.plot(x, tan_x, color='b')
ax13.set_xlabel('x')
ax13.set_ylabel('tan(x)')
ax13.set_title('The tangent function')
plt.show()
We can also gain access to additional labelling tools which are useful for giving meaning to multiple plots
number_rows = 1
number_columns = 3
x = np.linspace(0, 20, 1000)
sin_x = np.sin(x)
cos_x = np.cos(x)
tan_x = np.tan(x)
fig = plt.figure(figsize=(18, 6))
(ax11, ax12, ax13) = fig.subplots(number_rows, number_columns)
ax11.plot(x, sin_x, color='r')
ax11.set_xlabel('x')
ax11.set_ylabel('sin(x)')
ax11.set_title('The sine function')
ax12.plot(x, cos_x, color='g')
ax12.set_xlabel('x')
ax12.set_ylabel('cos(x)')
ax12.set_title('The cosine function')
ax13.plot(x, tan_x, color='b')
ax13.set_xlabel('x')
ax13.set_ylabel('tan(x)')
ax13.set_title('The tangent function')
fig.suptitle('The SohCahToa business')
plt.show()
Say for example, you have information from two model scenarios (e.g. green scenario, red scenario) and you want plot multiple output from these scenarios. These outputs will have either come from the green or red scenario so it would be nice if we can clearly group the scenarios on the same graph.
We can do this using the fanciest form of subplotting I know how to use, where we first make the figure object, then attach subfigures to the figure, and then subplots to the subfigures... It's a bit of a headache, but doing this allows you to control things in a lot of detail and label things nicely.
Below we plot green and red squares first as columns and then as rows. The only thing that changes in the following code sections is how we define the shape of the subfigures and subplots
Plotting shared scenarios in a shared column:
# Create the figure, specifying the size and background colour
fig = plt.figure(figsize=(8, 8), constrained_layout=True, facecolor='black')
# Create two subfigures each of which have some meaning behind or reason to group them together
(subfig1, subfig2) = fig.subfigures(1, 2, facecolor='black')
# On subfig1, add two green axis (subplots)
(ax11, ax12) = subfig1.subplots(2, 1)
ax11.set_facecolor('g')
ax12.set_facecolor('g')
# Add a title over these two plots which describes the information relevant to both plots
subfig1.suptitle('Usefully grouped green subplots on subfig1', color='g')
# Add a y and x label over both plots which describes the information relevant to both plots
subfig1.supylabel('Something on shared on the y-axis, subfig1', color='g')
subfig1.supxlabel('Something on shared on the x-axis, subfig1', color='g')
# On subfig1, add two red axis (subplots)
(ax21, ax22) = subfig2.subplots(2, 1)
ax21.set_facecolor('r')
ax22.set_facecolor('r')
# Add a title over these two plots which describes the information relevant to both plots
subfig2.suptitle('Usefully grouped red subplots on subfig2', color='r')
# Add a ylabel over both plots which describes the information relevant to both plots
subfig2.supylabel('Something on shared on the y-axis, subfig2', color='r')
subfig2.supxlabel('Something on shared on the x-axis, subfig2', color='r')
# add a title to describe what we are seeing in the figure overall
fig.suptitle('A plot showing the grouping of subplot objects by color', color='w')
fig.supylabel('Overall figure ylabel', color='w')
fig.supxlabel('Overall figure xlabel', color='w')
plt.show()
Plotting shared scenarios in a shared row:
# Create the figure, specifying the size and background colour
fig = plt.figure(figsize=(8, 8), constrained_layout=True, facecolor='black')
# Create two subfigures each of which have some meaning behind or reason to group them together
(subfig1, subfig2) = fig.subfigures(2, 1, facecolor='black')
# On subfig1, add two green axis (subplots)
(ax11, ax12) = subfig1.subplots(1, 2)
ax11.set_facecolor('g')
ax12.set_facecolor('g')
# Add a title over these two plots which describes the information relevant to both plots
subfig1.suptitle('Usefully grouped green subplots on subfig1', color='g')
# Add a y and x label over both plots which describes the information relevant to both plots
subfig1.supylabel('Something on shared on the y-axis, subfig1', color='g')
subfig1.supxlabel('Something on shared on the x-axis, subfig1', color='g')
# On subfig1, add two red axis (subplots)
(ax21, ax22) = subfig2.subplots(1, 2)
ax21.set_facecolor('r')
ax22.set_facecolor('r')
# Add a title over these two plots which describes the information relevant to both plots
subfig2.suptitle('Usefully grouped red subplots on subfig2', color='r')
# Add a ylabel over both plots which describes the information relevant to both plots
subfig2.supylabel('Something on shared on the y-axis, subfig2', color='r')
subfig2.supxlabel('Something on shared on the x-axis, subfig2', color='r')
# add a title to describe what we are seeing in the figure overall
fig.suptitle('A plot showing the grouping of subplot objects by color', color='w')
fig.supylabel('Overall figure ylabel', color='w')
fig.supxlabel('Overall figure xlabel', color='w')
plt.show()
Now let's put this into a bit of context with some model output from the ICCS submission runs, let's plot the number of cases and deaths in each of the 5% runs, and then do the same for the 50% runs.
import pandas as pd
from matplotlib import pyplot as plt
import numpy as np
import os
# Fanciest subplot options, including subfigures (aka a group of subplots with the possibility to share labels),
# subplots, and joy division plots
# Get the data from the ICCS runs and format it:
# create a shorthand reference to where we stored the output files (onedrive)
v3_50_file_path = "/Users/robbiework/Library/CloudStorage/OneDrive-UniversityCollegeLondon/data/output/ICCS/" \
"extended_submission/ver_3 (multiDist)/50_perc/beta_0.3/output/"
v3_5_file_path = "/Users/robbiework/Library/CloudStorage/OneDrive-UniversityCollegeLondon/data/output/ICCS/" \
"extended_submission/ver_3 (multiDist)/5_perc/beta_0.3/output/"
# Generate epidemic curves from different runs
# Create a list to store the number of cases in the 50 percent runs
v3_50_perc_cases_lines = []
v3_50_perc_deaths_lines = []
# Iterate over each output file
for file in os.listdir(v3_50_file_path):
# load the data
data = pd.read_csv(v3_50_file_path + file, delimiter='\t')
# drop the column that is normally there
data = data.drop('Unnamed: 10', axis=1)
# Calculate the total number of cases in all districts at each point in time
data = data.groupby('time').sum()
# Calculate the total new cases (both symptomatic and asymptomatic)
data['new_cases'] = data['metric_new_cases_asympt'] + data['metric_new_cases_sympt']
# Store this in lines
v3_50_perc_cases_lines.append(list(data['new_cases']))
# Death lines
v3_50_perc_deaths_lines.append(list(data['metric_new_deaths']))
# Create a list to store the number of cases in the 5 percent runs
v3_5_perc_cases_lines = []
v3_5_perc_deaths_lines = []
# Iterate over each output file
for file in os.listdir(v3_5_file_path):
# load the data
data = pd.read_csv(v3_5_file_path + file, delimiter='\t')
# drop the column that is normally there
data = data.drop('Unnamed: 10', axis=1)
# Calculate the total number of cases in all districts at each point in time
data = data.groupby('time').sum()
# Calculate the total new cases (both symptomatic and asymptomatic)
data['new_cases'] = data['metric_new_cases_asympt'] + data['metric_new_cases_sympt']
# Store this in lines
v3_5_perc_cases_lines.append(list(data['new_cases']))
# Death lines
v3_5_perc_deaths_lines.append(list(data['metric_new_deaths']))
# Create the figure, specifying the size and background colour
fig = plt.figure(figsize=(12, 8), constrained_layout=True, facecolor='black')
# Create two subfigures which will store results from each sample size
(subfig1, subfig2) = fig.subfigures(1, 2, facecolor='black')
# On subfig1, add two axis (subplots)
(ax11, ax12) = subfig1.subplots(2, 1)
# Make the axis (subplots) black
ax11.set_facecolor('black')
ax12.set_facecolor('black')
# Generate line plots, iterating over number of runs
for i in range(len(os.listdir(v3_5_file_path))):
# We need something to push the lines up on the plot, otherwise each line will be plotted on top of one another and
# it will look messy, create this y-axis offset (this is just made up and will be individual to the graph, but works
# for this graph at least)
y_offset = i * max([max(j) for j in v3_5_perc_cases_lines]) / len(os.listdir(v3_5_file_path)) * 2
# Do same for deaths
y_deaths_offset = i * max([max(j) for j in v3_5_perc_deaths_lines]) / len(os.listdir(v3_5_file_path)) * 2
# Plot the cases on axis 11 in white
ax11.plot(np.arange(len(v3_5_perc_cases_lines[i])), np.add(y_offset, v3_5_perc_cases_lines[i]), color="w", lw=2)
# Plot the deaths on axis 12 in red
ax12.plot(np.arange(len(v3_5_perc_deaths_lines[i])),
np.add(y_deaths_offset, v3_5_perc_deaths_lines[i]), color="r", lw=2)
# Currently the axis for these plots are invisible, say we want to see the number of cases and deaths each day, then we
# have to alter parts of the axis so they show on a black background (their default colour is black, hence them not
# appearing)
# On the top subplot, we only don't want to see any information so turn the x and y ticks off
ax11.set_xticks([])
ax11.set_yticks([])
# On the bottom subplot, we only want to see the x axis, so specify the axis, and make the ticks black and labels red
ax12.tick_params(axis='x', color='black', labelcolor='r')
# Set x and y labels to say what we are plotting on each subplot
ax11.set_ylabel('Cases', color='w', fontweight='bold', fontsize=15)
ax12.set_ylabel('Deaths', color='r', fontweight='bold', fontsize=15)
# let people know the sample size for this plot by using the suptitle function
subfig1.suptitle('5%', fontweight='bold', color='w')
# Do the same for the 50 percent sample
# On subfig2, add two axis (subplots)
(ax21, ax22) = subfig2.subplots(2, 1)
# Make the axis (subplots) black
ax21.set_facecolor('black')
ax22.set_facecolor('black')
# Generate line plots, iterating over number of runs
for i in range(len(os.listdir(v3_50_file_path))):
# We need something to push the lines up on the plot, otherwise each line will be plotted on top of one another and
# it will look messy, create this y-axis offset (this is just made up and will be individual to the graph, but works
# for this graph at least)
y_offset = i * max([max(j) for j in v3_50_perc_cases_lines]) / len(os.listdir(v3_50_file_path)) * 2
# Do same for deaths
y_deaths_offset = i * max([max(j) for j in v3_50_perc_deaths_lines]) / len(os.listdir(v3_50_file_path)) * 2
# Plot the cases on axis 21 in white
ax21.plot(np.arange(len(v3_50_perc_cases_lines[i])), np.add(y_offset, v3_50_perc_cases_lines[i]), color="w", lw=2)
# Plot the deaths on axis 22 in red
ax22.plot(np.arange(len(v3_50_perc_deaths_lines[i])),
np.add(y_deaths_offset, v3_50_perc_deaths_lines[i]), color="r", lw=2)
# we don't want either to show on the top plot so turn off the axis
ax21.set_xticks([])
ax21.set_yticks([])
# On the bottom subplot, we only want to see only the x axis, so specify that axis, and make the ticks black and labels
# red
ax22.tick_params(axis='x', color='black', labelcolor='r')
# let people know the sample size for this plot by using the suptitle function
subfig2.suptitle('50%', fontweight='bold', color='w')
# Label the figure's x-axis
fig.supxlabel('Time', color='w', fontweight='bold', fontsize=15)
# Create a figure title
fig.suptitle('The shape of the epidemic curves for cases and deaths,\n for each model run', color='w',
fontweight='bold', fontsize=15)
plt.show()
Example of a Sankey diagram
from matplotlib.sankey import Sankey
from matplotlib import pyplot as plt
# first flow into the diagram, the first value is the total quantity introduced into the
# flow, the remaining, the subsequent terms remove a certain quantity from the first flow.
# In this example we will plot the percent health care budget consumed by various conditions, 50% is spent on HIV, 30%
# on road traffic injuries and 20% on epilepsy, we store this information in two arrays, flows1 which houses the
# data and labels1 which gives each percentage a label.
# I want the total budget to go in a straight line from left to right, the hiv budget to go up from the total budget,
# the road traffic budget to carry on straight and the epilepsy budget to go down, I will store these directions in an
# array orientations1, the first entry is 0 as we don't want to change the orientations from the default direction,
# the second entry is 1 as we want the HIV budget to go up, the third entry is 0 as we want the entry to go on straight,
# the fourth entry is -1 as we want the epilepsy budget to go down.
flow1 = [100, -30, -50, -20]
labels1 = ['Total expenditure on health', '% spent on HIV', "% spent on road "
"\n"
"traffic injuries",
'% spent on epilepsy']
orientations1 = [0, 1, 0, -1]
# The second flow breaks down the what the road traffic injuries budget was spent on, 10% of
# the total budget was spent on bandages, 15% on plaster of paris and 5% on surgery, we store data in flows 2 and the
# labelling info in labels2, leaving the first entry blank as this is where the '% spent on road traffic injuries'
# flow links to the breakdown flow.
# In the orientations, the first entry is zero as we want this flow to carry on in the same direction, the second entry
# for bandage expenditure is 1 as we want this to head up from the flow, the second entry is zero as we want the
# plaster of paris expenditure to go on straight and finally the fourth entry is -1 to make the surgery expenditure go
# down
flow2 = [50, -10, -15, -25]
labels2 = ['', 'bandages', 'plaster of paris', 'surgery']
orientations2 = [0, 1, 0, -1]
# Now we have created the flows and set the labels and directions the arrows go in, we can create the sankey diagram.
# The sankey object needs to be scaled (controls how much space the diagram takes up), but if it's not scaled properly
# it can look pretty terrible, I found that a fairly reasonable scale to use is to use 1/a where a is the first entry of
# the first flow (in this example 100).
# The offset zooms into and out from the diagram
fig = plt.figure(figsize=(20, 10)) # create figure
ax = fig.add_subplot(1, 1, 1, xticks=[], yticks=[])
ax.axis('off') # Turn off the box for the plot
plt.title('Budget example') # create title
sankey = Sankey(ax=ax,
scale=1 / flow1[0],
offset=0.2,
unit='') # create sankey object
sankey.add(flows=flow1, # add the first flow
labels=labels1,
orientations=orientations1,
pathlengths=[0.1, 0.1, 0.1, 0.1],
trunklength=0.605, # how long this flow is
edgecolor='#027368', # choose colour
facecolor='#027368')
sankey.add(flows=flow2,
labels=labels2,
trunklength=0.5,
pathlengths=[0.25, 0.25, 0.25, 0.25],
orientations=[0, 1, 0, -1],
prior=0, # which sankey are you connecting to (0-indexed)
connect=(2, 0), # flow number to connect: this is the index of road traffic injury portion of the budget in
# the first flow (third entry, python index 2) which connects to the first entry in the second flow (python
# index 0).
edgecolor='#58A4B0', # choose colour
facecolor='#58A4B0')
diagrams = sankey.finish()
plt.show()
plt.close()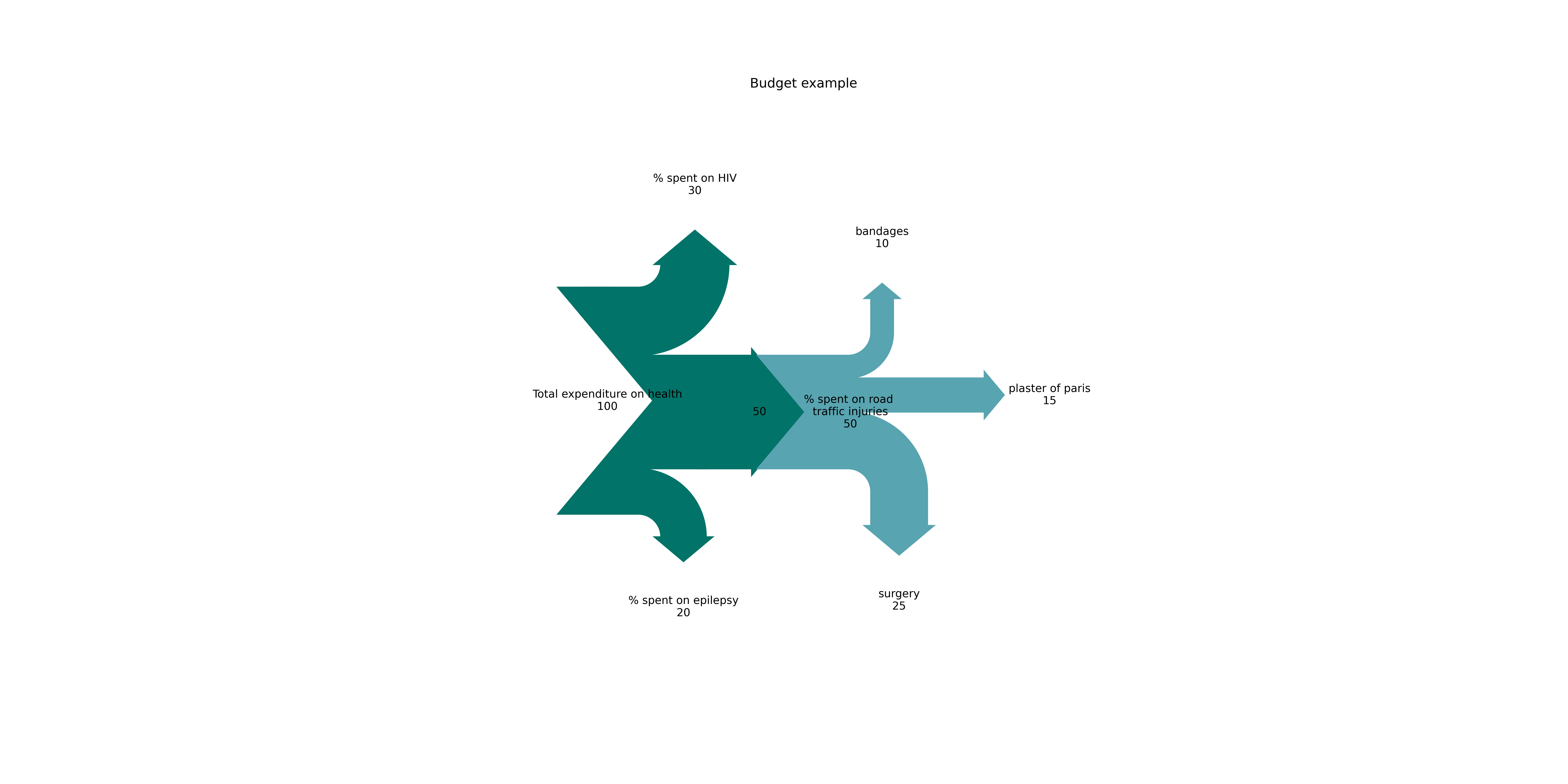
Gifs are really useful to convey information in a presentation, or just to help you understand model for yourself. Essentially they are a bunch of images played one after the other, so to create the gif, we first need to have to created a bunch of images. Fortunately I have a ton of pictures of my dog which we can use to create a gif.
The PIL library can be accessed by downloading the PILLOW package (use pip install PILLOW for example)
I've made a little function to create a gif, if you are creating a gif of something which needs to be in a particular order (e.g. moving forward in time) then make sure to label the date in the figure title, you can then set the 'files_numbered' parameter to be true and then you will be set!
import os
import re
from PIL import Image, ImageDraw
def pictures_to_gif(filepath, savepath, savename, gif_duration, files_numbered=False):
"""
:param filepath: A folder where you have the images you want to make a gif from
:param savepath: A filepath where you want to save the gif
:param savename: The filename of your gif
:param gif_duration: How long you want your gif to have between images (no idea what units this is in)
:param files_numbered: If the order in which the images is shown is important you need to set this to true and make
sure that your file names are saved in the order you want to make a gif with e.g. cases_day_1.png etc...
:return:
"""
# Find the files
files = os.listdir(filepath)
# If necessary order the files
if files_numbered:
files.sort(key=lambda f: int(re.sub('\D', '', f)))
# Create a list to store the images in
images = []
for file in files:
im = Image.open(filepath + file)
draw = ImageDraw.Draw(im)
images.append(im)
# Some kind of saving operation, I never dug in to the details to be honest, but it works
images[0].save(savepath + savename, save_all=True, append_images=images[1:], optimize=False,
duration=gif_duration, loop=0)
# get the pictures of the dog
picture_folder = "/Users/robbiework/PycharmProjects/spacialEpidemiologyAnalysis/Barney/"
# Choose where to save the gif
save_folder = "/Users/robbiework/PycharmProjects/spacialEpidemiologyAnalysis/crappyPlots/"
# give the gif a name
save_name = 'barney.gif'
# make sure that each image goes by slowly enough for everyone to enjoy
gif_duration = 400
# Use the function
pictures_to_gif(picture_folder, save_folder, save_name, gif_duration, files_numbered=False)
Colours in python can be specified in a number of ways, you can use strings to call certain colours as shown here:

You can specify RGB (red, green, blue) values to create colours. This is useful if you are plotting a bunch of lines/bars/whatevers and you need to create a number of distinct colours and you don't want to specify exactly the colours used for each line by name. Note that the RGB numbers need to be between 1 and 0. An example is shown in the bar plot below:
x = np.arange(1, 10)
y = np.multiply(x, 5)
reds = [[1 / i, 0, 0] for i in x]
greens = [[0, 1 / i, 0] for i in x]
blues = [[0, 0, 1 / i] for i in x]
plt.bar(x, y, color=reds)
plt.show()
plt.clf()
plt.bar(x, y, color=greens)
plt.show()
plt.clf()
plt.bar(x, y, color=blues)
plt.show()
plt.clf()


When choosing colour schemes for maps you need to specify a colour map. There are loads to choose from and examples can be found here https://matplotlib.org/stable/gallery/color/colormap_reference.html
If you need to distinguish between lines beyond just colour, you can do so with linestyle controls
import matplotlib.pyplot as plt
# Plot a horizontal line spanning the x axis, specify different linestyles
plt.axhline(y=1, linestyle='solid', color='r', label='solid')
plt.axhline(y=2, linestyle='dotted', color='r', label='dotted')
plt.axhline(y=3, linestyle='dashed', color='r', label='dashed')
plt.axhline(y=4, linestyle='dashdot', color='r', label='dashdot')
plt.legend()
plt.xticks([])
plt.yticks([])
plt.title('Different linestyles!')
plt.show()