This repository provides a simplified particle physics analysis example for the REANA reusable research data analysis plaftorm. The example mimics a typical particle physics analysis where the signal and background data is processed and fitted against a model. The example will use the RooFit package of the ROOT framework.
Making a research data analysis reproducible means to provide "runnable recipes" addressing (1) where the input datasets are, (2) what software was used to analyse the data, (3) which computing environment was used to run the software, and (4) which workflow steps were taken to run the analysis.
In this example the signal and background data will be generated; see below. Therefore there is no explicit input file to be taken care of.
Our analysis will consist of two stages. In the first stage, signal and background are generated. In the second stage, a fit will be made for the signal and background.
For the first generation stage, gendata.C is a ROOT macro that generates signal and background data. The code was taken from the RooFit tutorial rf502_wspacewrite.C and it was slightly modified. One could run it locally for 20000 events as follows:
$ root -b -q 'gendata.C(20000,"data.root")'
Note that this generates a temporary data.root data file:
$ ls -l data.root -rw-r--r-- 1 root root 153295 Jun 1 17:01 data.root
For the second fitting stage, fitdata.C is a ROOT macro that makes a fit for the signal and the background data. The code was taken from the RooFit tutorial rf503_wspaceread.C and it was slightly modified. One could run it locally as follows:
$ root -b -q 'fitdata.C("data.root","plot.png")'
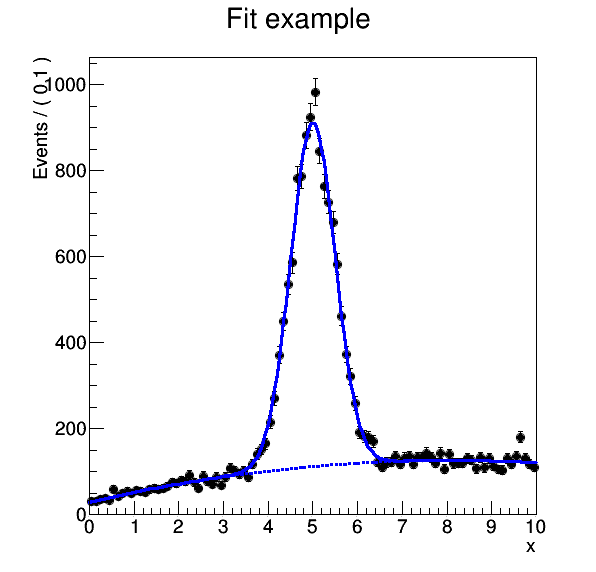
This generates a final plot representing the result of our analysis:
Let us now try to provide runnable recipes so that our analysis can be run in a reproducible manner on the REANA cloud.
First we need to take care of expressing our runtime environment in a reusable
manner. Our example analysis is completely done within the ROOT6 analysis framework. The computing environment can be
therefore easily encapsulated by using the upstream reana-env-root6 base image. (See there how it
was created.) We can actually use this base image "as is", because our two
macros gendata.C and fitdata.C can be mounted into the container via
code volume. We don't need to create any specially customised environment.
Secondly we need to capture the analysis workflow and the commands we have run to obtain the final plot.
As mentioned above, the analysis workflow had two stages, the generation stage and the fitting stage. We can represent these steps in a structured YAML manner using the Yadage workflow engine and the Common Workflow Language specification. The corresponding workflow descriptions can be found here:
Our example analysis is now fully described in the REANA-compatible reusable analysis manner and is prepared to be run on the REANA cloud.
Let us test whether everything works well locally in our containerised
environment. We shall use Docker locally. Note how we mount our local
directories inputs, code and outputs into the containerised
environment:
$ mkdir -p inputs
$ rm -rf outputs && mkdir outputs
$ docker run -i -t --rm \
-v `pwd`/code:/code \
-v `pwd`/inputs:/inputs \
-v `pwd`/outputs:/outputs \
reanahub/reana-env-root6 \
root -b -q '/code/gendata.C(20000,"/outputs/data.root")'
$ docker run -i -t --rm \
-v `pwd`/code:/code \
-v `pwd`/inputs:/inputs \
-v `pwd`/outputs:/outputs \
reanahub/reana-env-root6 \
root -b -q '/code/fitdata.C("/outputs/data.root","/outputs/plot.png")'Let us check whether the resulting plot is the same as the one showed in the documentation:
$ diff outputs/plot.png ./docs/plot.pngLet us test whether the Yadage workflow engine execution works locally.
Since Yadage only accepts one input directory as parameter, we are going to
create a wrapper directory which will contain links to inputs and code
directories:
$ mkdir -p yadage-local-run/yadage-inputs
$ cd yadage-local-run
$ cp -a ../code ../inputs yadage-inputsWe can now run Yadage locally as follows:
$ yadage-run . ../workflow/yadage/workflow.yaml \
-p events=20000 \
-p gendata=code/gendata.C \
-p fitdata=code/fitdata.C \
-d initdir=`pwd`/yadage-inputs
2018-02-19 16:01:34,297 - yadage.utils - INFO - setting up backend multiproc:auto with opts {}
2018-02-19 16:01:34,299 - packtivity.asyncbackends - INFO - configured pool size to 4
2018-02-19 16:01:34,311 - yadage.utils - INFO - local:. {u'initdir': '/home/simko/private/src/reana-demo-root6-roofit/yadage-local-run/yadage-inputs'}
2018-02-19 16:01:34,357 - yadage.steering_object - INFO - initializing workflow with {u'gendata': 'code/gendata.C', u'fitdata': 'code/fitdata.C', u'events': 20000}
2018-02-19 16:01:34,357 - adage.pollingexec - INFO - preparing adage coroutine.
2018-02-19 16:01:34,357 - adage - INFO - starting state loop.
2018-02-19 16:01:34,413 - yadage.handlers.scheduler_handlers - INFO - initializing scope from dependent tasks
2018-02-19 16:01:34,435 - yadage.wflowview - INFO - added node <YadageNode init DEFINED lifetime: 0:00:00.000253 runtime: None (id: 23855c9fe3d01cc568e891af020be486cb0eac17) has result: True>
2018-02-19 16:01:34,619 - yadage.wflowview - INFO - added node <YadageNode gendata DEFINED lifetime: 0:00:00.000127 runtime: None (id: 3075a77f855645a5556f5355ff66952a3c03b58f) has result: True>
2018-02-19 16:01:34,780 - yadage.wflowview - INFO - added node <YadageNode fitdata DEFINED lifetime: 0:00:00.000128 runtime: None (id: 6908bd540badcabce2d97fa095a7772a5d577210) has result: True>
2018-02-19 16:01:34,865 - packtivity_logger_init.step - INFO - publishing data: <TypedLeafs: {u'gendata': u'/home/simko/private/src/reana-demo-root6-roofit/yadage-local-run/yadage-inputs/code/gendata.C', u'fitdata': u'/home/simko/private/src/reana-demo-root6-roofit/yadage-local-run/yadage-inputs/code/fitdata.C', u'events': 20000}>
2018-02-19 16:01:34,897 - adage.node - INFO - node ready <YadageNode init SUCCESS lifetime: 0:00:00.462261 runtime: 0:00:00.031310 (id: 23855c9fe3d01cc568e891af020be486cb0eac17) has result: True>
2018-02-19 16:01:34,922 - packtivity_logger_gendata.step - INFO - starting file loging for topic: step
2018-02-19 16:01:34,981 - packtivity_logger_gendata.step - INFO - prepare pull
2018-02-19 16:01:39,672 - adage.node - INFO - node ready <YadageNode gendata SUCCESS lifetime: 0:00:05.053356 runtime: 0:00:04.751996 (id: 3075a77f855645a5556f5355ff66952a3c03b58f) has result: True>
2018-02-19 16:01:39,695 - packtivity_logger_fitdata.step - INFO - starting file loging for topic: step
2018-02-19 16:01:39,733 - packtivity_logger_fitdata.step - INFO - prepare pull
2018-02-19 16:01:45,540 - adage.node - INFO - node ready <YadageNode fitdata SUCCESS lifetime: 0:00:10.759921 runtime: 0:00:05.846398 (id: 6908bd540badcabce2d97fa095a7772a5d577210) has result: True>
2018-02-19 16:01:45,547 - adage.controllerutils - INFO - no nodes can be run anymore and no rules are applicable
2018-02-19 16:01:45,547 - adage.pollingexec - INFO - exiting main polling coroutine
2018-02-19 16:01:45,548 - adage - INFO - adage state loop done.
2018-02-19 16:01:45,548 - adage - INFO - execution valid. (in terms of execution order)
2018-02-19 16:01:45,555 - adage.controllerutils - INFO - no nodes can be run anymore and no rules are applicable
2018-02-19 16:01:45,555 - adage - INFO - workflow completed successfully.Let us check whether the resulting plot is the same as the one showed in the documentation:
$ diff outputs/plot.png ./docs/plot.pngLet us test whether the CWL workflow execution works locally as well.
To prepare the execution, we are creating a working directory called cwl-local-run which will contain both
inputs and code directory content. Also, we need to copy the workflow input file:
$ mkdir cwl-local-run
$ cd cwl-local-run
$ cp ../code/* ../workflow/cwl/input.yml .We can now run the corresponding commands locally as follows:
$ cwltool --quiet --outdir="../outputs" ../workflow/cwl/workflow.cwl input.yml
{
"plot": {
"checksum": "sha1$adc52c16836ac4cc385aab7aeddf492fe83c45e2",
"basename": "plot.png",
"location": "file:///path/to/reana-demo-root6-roofit/outputs/plot.png",
"path": "/path/to/reana-demo-root6-roofit/outputs/plot.png",
"class": "File",
"size": 16273
}
}Let us check whether the resulting plot is the same as the one showed in the documentation:
$ diff outputs/plot.png ./docs/plot.pngPutting all together, we can now describe our ROOT6 RooFit physics analysis example, its runtime environment, the inputs, the code, the workflow and its outputs by means of the following REANA specification file:
version: 0.2.0
metadata:
authors:
- Ana Trisovic <[email protected]>
- Lukas Heinrich <[email protected]>
- Tibor Simko <[email protected]>
title: ROOT6 and RooFit physics analysis example
date: 19 February 2018
repository: https://github.com/reanahub/reana-demo-root6-roofit/
code:
files:
- code/gendata.C
- code/fitdata.C
inputs:
parameters:
events: 20000
gendata: code/gendata.C
fitdata: code/fitdata.C
outputs:
files:
- outputs/plot.png
environments:
- type: docker
image: reanahub/reana-env-root6
workflow:
type: yadage
file: workflow/yadage/workflow.yamlWe can now install the REANA client and submit the ROOT6 RooFit analysis example to run on some particular REANA cloud instance. We start by installing the client:
$ mkvirtualenv reana-client -p /usr/bin/python2.7
$ pip install reana-clientand connect to the REANA cloud instance where we will run this example:
$ export REANA_SERVER_URL=http://192.168.99.100:32658If you run REANA cluster locally as well, then:
$ eval $(reana-cluster env)Let us check the connection:
$ reana-client ping
Server is running.We can now initialise workflow and upload our ROOT macros as input code:
$ reana-client workflow create
workflow.4
$ export REANA_WORKON=workflow.4
$ reana-client code upload ./code
/home/simko/private/project/reana/src/reana-demo-root6-roofit/code/gendata.C was uploaded successfully.
/home/simko/private/project/reana/src/reana-demo-root6-roofit/code/fitdata.C was uploaded successfully.
$ reana-client code list
NAME SIZE LAST-MODIFIED
fitdata.C 1648 2018-04-20 15:31:08.108119+00:00
gendata.C 1937 2018-04-20 15:31:08.095119+00:00Start workflow execution and enquire about its running status:
$ reana-client workflow start
workflow.4 has been started.
$ reana-client workflow status
NAME RUN_NUMBER ID USER ORGANIZATION STATUS
workflow 4 826da1cc-ea96-4eef-9bac-85f21c954293 00000000-0000-0000-0000-000000000000 default running
$ reana-client workflow status
NAME RUN_NUMBER ID USER ORGANIZATION STATUS
workflow 4 826da1cc-ea96-4eef-9bac-85f21c954293 00000000-0000-0000-0000-000000000000 default finishedAfter the workflow execution successfully finished, we can retrieve its output:
$ reana-client outputs list
NAME SIZE LAST-MODIFIED
gendata/data.root 153467 2018-04-20 15:33:02.601120+00:00
fitdata/plot.png 16273 2018-04-20 15:33:02.600120+00:00
_yadage/yadage_snapshot_backend.json 773 2018-04-20 15:33:02.600120+00:00
_yadage/yadage_snapshot_workflow.json 16135 2018-04-20 15:33:02.600120+00:00
_yadage/yadage_template.json 1843 2018-04-20 15:33:02.600120+00:00
$ reana-client outputs download fitdata/plot.png
File fitdata/plot.png downloaded to ./outputs/Let us check whether the resulting plot is the same as the one showed in the documentation:
$ ls -l outputs/fitdata/plot.png
-rw-r--r-- 1 simko simko 16273 Apr 20 17:33 outputs/fitdata/plot.png
$ diff outputs/fitdata/plot.png ./docs/plot.pngNote that this example demonstrated the use of the Yadage workflow engine. If
you would like to use the CWL workflow engine, please just use -f
reana-cwl.yaml option with the reana-client commands.
Thank you for using the REANA reusable analysis platform.