This repository is a collection of notebooks that analyzes thousand datasets on heart-disease in order to try and predict
if a user has heart disease. The dataset was downloaded from Kaggle.

What problem are we trying to solve?
This data set dates from 1988 and consists of four databases: Cleveland, Hungary, Switzerland, and Long Beach V. It contains 76 attributes, including the predicted attribute, but all published experiments refer to using a subset of 14 of them. The "target" field refers to the presence of heart disease in the patient. It is integer valued 0 = no disease and 1 = disease.
The objective of this model creation is to classify if our incoming patients have heart disease based on the same set of tests.
- Type of Machine Learning: Supervised Learning
- Type of Learning: Classification
Our main dataset for this project was downloaded from Kaggle. It is very important to have knowledge on what our data looks like and what it represents. Furthermore, it would also be important for our analysis that we get to categorize the different variables of our data so we can check the data accuracy. We will also be showing how to filter and sort our data in this section.
- __Type of Data __
- Based on Structure:
- Structured
- Unstructured
- Based on Frequency:
- Static
- Streaming
- Based on Structure:
What defines accuracy?
We calculate accuracy by dividing the number of correct predictions (the corresponding diagonal in the matrix) by the total number of samples.

from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
y_actual = [1,1,1,1,0,0,0,0,0,0]
y_predicted = [1,1,1,0,1,1,1,1,0,0]
cm = confusion_matrix(y_actual, y_predicted)
cmd = ConfusionMatrixDisplay(cm)
cmd.plot();What features does our data have and which ones to use? The features that we have are:
-
Binary
- sex (Male or Female)
- 0 = female
- 1 = male
- fbs (Fasting Blood Sugar > 120 mg/dl)
- 0 = no
- 1 = yes
- exang (Exercise Induced Angina)
- 0 = no
- 1 = yes
- target (Heart Disease / Target Field)
- 0 = disease
- 1 = no disease
- sex (Male or Female)
-
Categorical
- cp (Chest Pain Type)
- 0: asymptomatic
- 1: atypical angina
- 2: non-anginal pain
- 3: typical angina
- restecg (Resting ECG)
- 0: showing probable or definite left ventricular hypertrophy by Estes’ criteria
- 1: normal
- 2: having ST-T wave abnormality
- slope (the slope of the peak exercise ST segment)
- 0: downsloping
- 1: flat
- 2: upsloping
- ca (number of major vessels)
- (0–3)
- thal (Thalassemia)
- 1 = normal
- 2 = fixed defect
- 3 = reversible defect
- cp (Chest Pain Type)
-
Continuous
- age (Age of the individual)
- trestbps (Resting Blood Pressure in mm/hg)
- chol (Serum Cholesterol in mg/dl)
- thalac (Maximum heart rate achieved)
- oldpeak (ST depression induced by exercise relative to rest)
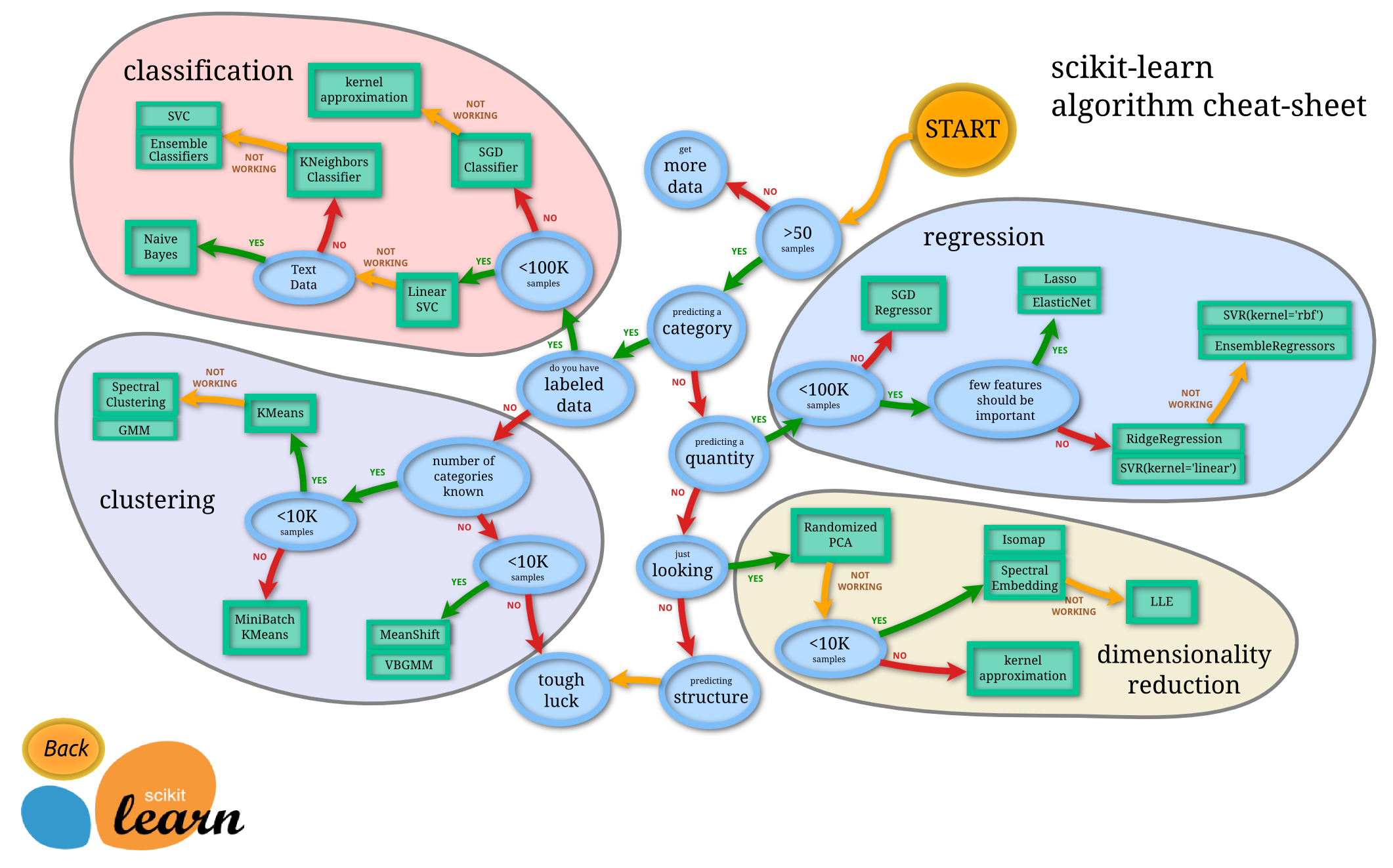
What kind of model should we use? How to use a model?
Finding the best estimator for the task might frequently be the most difficult step in tackling a machine learning challenge. For various data kinds and issues, other estimators are more appropriate.
The Linear SVM produced an accuracy of 79.51% and we can optimize our features or modify hyperparamters but let us try and look at the sci-kit learn estimator chart.

Our naive-bayes classification has produced a good accuracy of 80% already but having 20 incorrect heart disease prediction out of 100 patients seems a little bit high, let us try implementing another model.

Our random forest model, produced 92.98% accuracy. Here is the last tree from our random forest.

How can we share our model?
import pickle
pickle.dump(clf, open("heart_disease_random_forest_model.pkl", "wb"))We can export our models into a pickle file and from there, deploy it on the web. For deployment, you may access the heart-disease repository which was deployed to Heroku.