-
Notifications
You must be signed in to change notification settings - Fork 30
Evolutionary variability GCIs
Status: initial draft
Authors and contributors:
- Chris Mungall (author)
Date: 2012
Document Type: ontology_design_pattern
Multi-species ontologies may be axiomatically weaker than dedicated species ontologies, as all statements in an ontology must be universal. Here we describe how axioms can be scoped to a particular species or taxon using OWL general axioms
This is also described in the section 'Managing taxonomic variation' in the Uberon paper.
Frequently we are confronted with situations where we want to make a statement such as 'A REL some B', but this statement doesn't universally hold for all instances of A, as A encompasses structures present in a variety of taxa that vary in properties such as developmental lineage.
One possibility is to make subclasses of A and attach the axiom to
those subclasses. For example, we can make subclasses of a generic
eye, such as camera-type eye' or compound eye`, and attach
relationships to these. However, this approach can lead to ontology
inflation where the subclasses are trivially distinguished.
Our solution here is to use general class axioms (aka General Class Inclusion axioms, or GCIs). This is logically equivalent to creating the subclass, but we use an OWL class expression in place of a named class:
(A and part_of some TAXON-CLASS) SubClassOf REL some B
So if we want to make a statement that the adenohypophysis develops from Rathke's pouch in tetrapods, we would say:
(adenohypophysis and part_of some Tetrapoda) SubClassOf develops_from some 'Rathkes pouch'
Note that we do not need to taxonomically scope the target class.
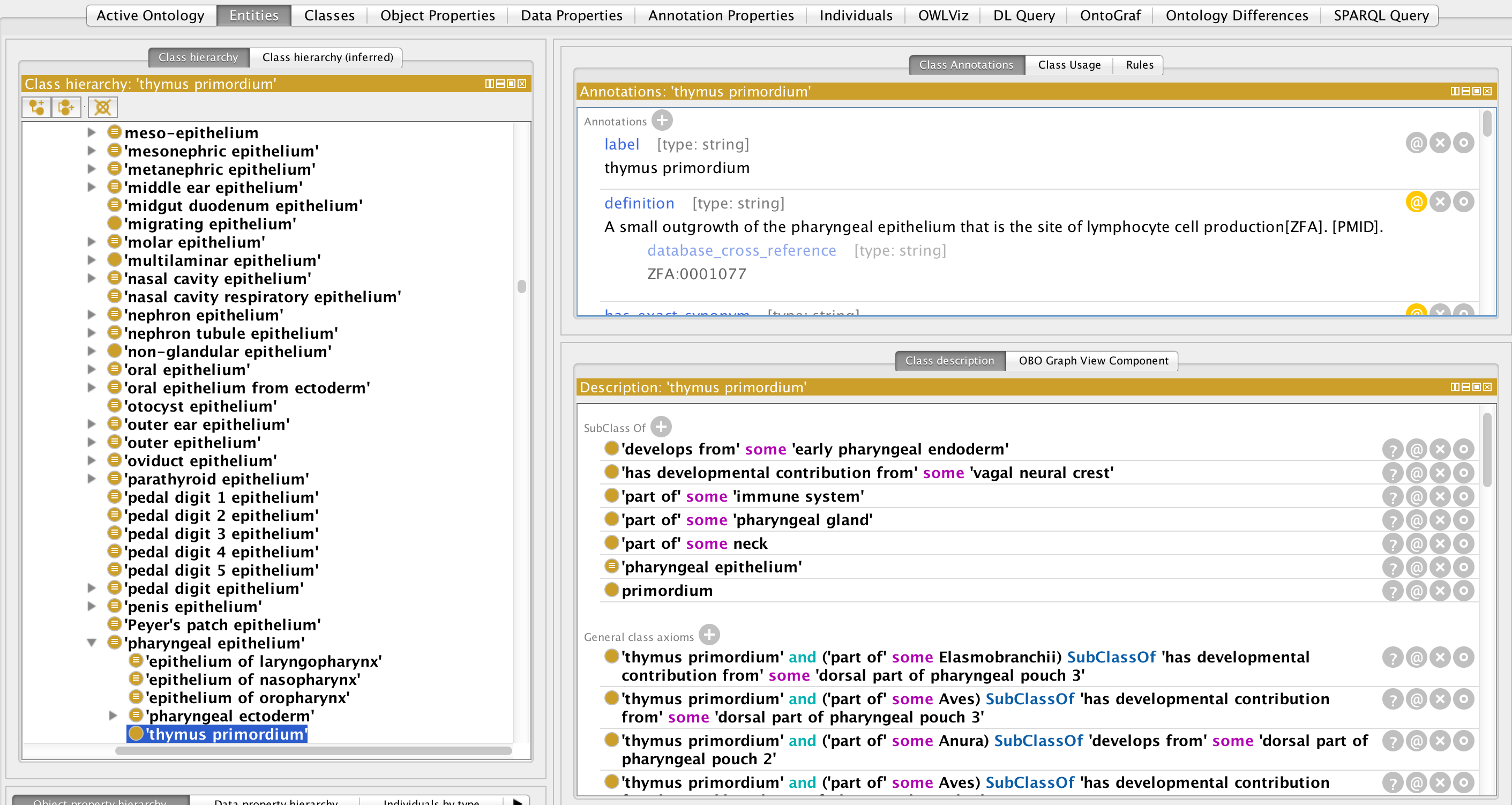
See thymus primordium as an example (note: GCIs may not be visible in OntoBee). For best results view in Protege version 5:
(the GCI viewer in Protege 4 is not great, you have to look at the 'about' tab)
This contains all the taxon-specific developmental relationships from table 13.1 of Kardong
OBO format has a very limited form of GCI support, but this is sufficient for taxon GCIs. In obo-format we would write:
id: A
relationship: REL B {gci_relation=part_of, gci_filler=NCBITaxon:nnnn}
This is provided as an authoring convenience. These relationships should be removed from obo-basic versions of the ontology, as they can possibly confuse some obo tools.
Currently these axioms of this form in Uberon come from two sources:
- Manually curated, often tagged with evidence from journal articles, textbooks and reviews
- Automatically generated, using axioms from species-specific ontologies
The former is available in standard editions of the ontology (with the exception of the 'basic' edition). The latter is only present in the 'composite' editions, such as 'composize-metazoan'. Here when we merge classes from a species-specific ontology (say ZFA, which is zebrafish specific) it isn't safe to merge in all of their relations, as they may not hold more generically. A classic case is the relationships to stages indicating when each structure arises (and possibly when it disappears) - these are highly specific. In these scenarios, safe taxon GCIs are added to the merge ontology.
For more details, see: Multi-species-composite-ontologies
Uberon is a multi-species anatomy ontology and knowledge base, find out more on the home page