iCellR is an interactive R package designed to facilitate the analysis and visualization of high-throughput single-cell sequencing data. It supports a variety of single-cell technologies, including scRNA-Seq, scVDJ-Seq, scATAC-Seq, CITE-Seq, and Spatial Transcriptomics (ST).
Maintainer: Alireza Khodadadi-Jamayran
Use the latest version of iCellR (v1.6.4) for scATAC-seq and Spatial Transcriptomics (ST) analyses. Leverage the i.score function for scoring cells based on gene signatures using methods such as Tirosh, Mean, Sum, GSVA, ssgsea, Zscore, and Plage.
Explore iCellR version 1.5.5, now featuring tools for cell cycle analysis (phases G0, G1S, G2M, M, G1M, and S). See example phase, New Pseudotime Abstract KNetL (PAK map) functionality added – visualize pseudotime progression (PAK map). Perform gene-gene correlation analysis using updated visualization tools. correlations.
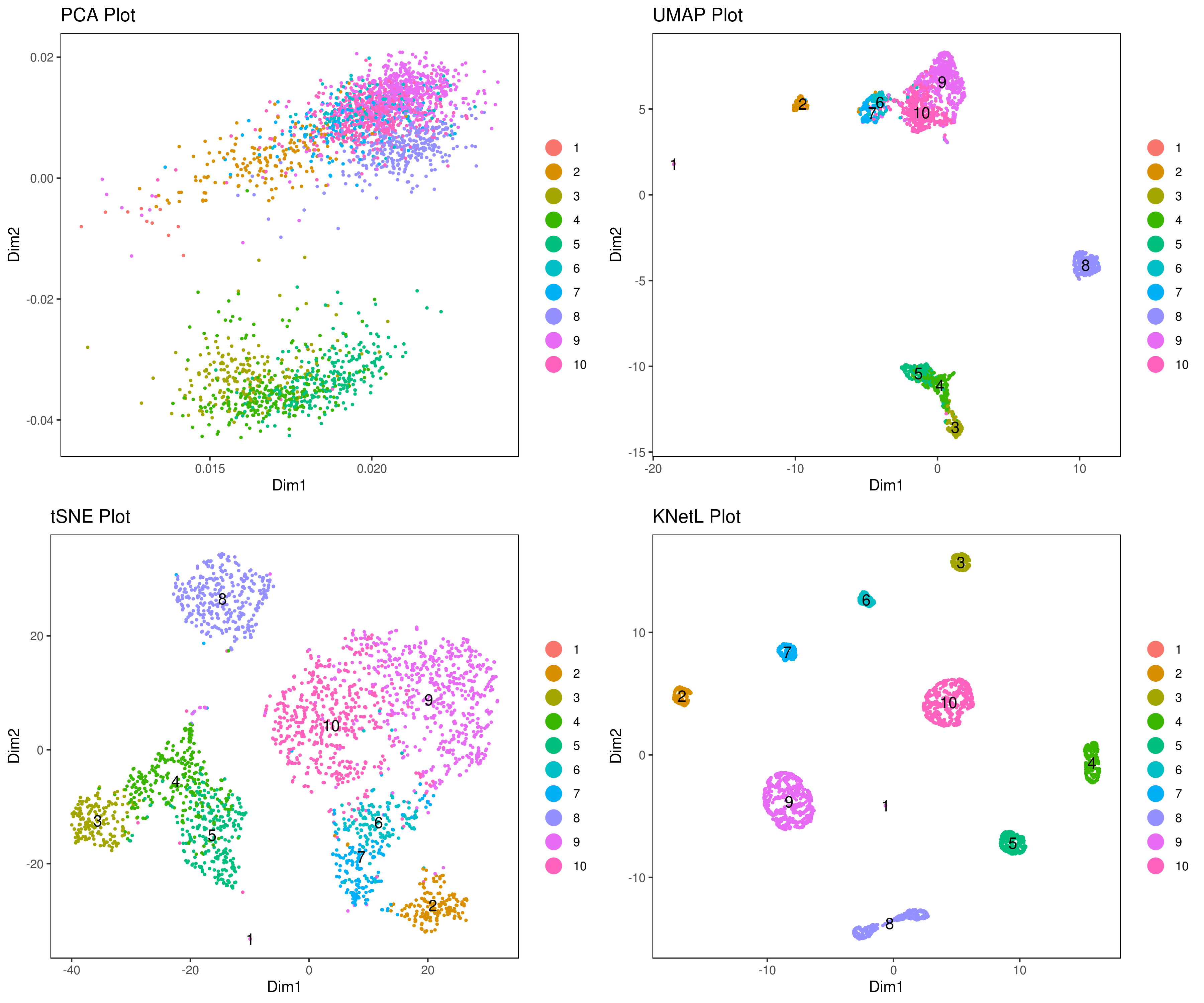
Explore the KNetL map, an advanced adjustable and dynamic dimensionality reduction method KNetL map ![]() KNetL (pronounced “nettle”) offers enhanced zooming capabilities KNetL to show significantly more detail compared to tSNE and UMAP.
KNetL (pronounced “nettle”) offers enhanced zooming capabilities KNetL to show significantly more detail compared to tSNE and UMAP.
Introducing imputation and coverage correction (CC) methods for improved gene-gene correlation analysis. (CC). Perform batch alignment using iCellR's CPCA and CCCA tools (CCCA and CPCA) methods. Expanded databases for cell type prediction now include ImmGen and MCA.
scSeqR has been renamed to iCellR, and scSeqR has been discontinued. Please use iCellR moving forward, as scSeqR is no longer supported. UMAP is added to iCellR. Interactive cell gating has been added, allowing users to select cells directly within HTML plots using Plotly.
- Link to
manualManual and Comprehensive R Archive Network (CRAN). - Gor
getting startedandtutorialgo to our Wiki page. - Link to a video tutorial for CITE-Seq and scRNA-Seq analysis: Video
- All you need to know about KNetL map: Video
- If you are using
FlowJoorSeqGeq, they offer plugins for iCellR and other single-cell analysis tools. You can find the list of all plugins here: https://www.flowjo.com/exchange/#/ . Specifically, the iCellR plugin can be found here: https://www.flowjo.com/exchange/#/plugin/profile?id=34. Additionally, a SeqGeq Differential Expression (DE) tutorial is available to guide you through the process: SeqGeq DE tutorial
For citing iCellR use this PMID: 34353854
iCellR publications: PMID: 35660135 (scRNA-seq/KNetL) PMID: 35180378 (CITE-seq/KNetL), PMID: 34911733 (i.score and cell ranking), PMID: 34963055 (scRNA-seq), PMID 31744829 (scRNA-seq), PMID: 31934613 (bulk RNA-seq from TCGA), PMID: 32550269 (scVDJ-seq), PMID: 34135081, PMID: 33593073, PMID: 34634466, PMID: 35302059, PMID: 34353854
Single (i) Cell R package (iCellR)
# Install the iCellR package
# Option 1: Install from CRAN (recommended for stable releases)
install.packages("iCellR")
# Option 2: Install the latest development version from GitHub
# Uncomment the lines below to use these steps:
# library(devtools)
# install_github("rezakj/iCellR")
# Option 3: Alternatively, clone the repository and install manually:
# Run this command in your terminal to clone the GitHub repository:
# git clone https://github.com/rezakj/iCellR.git
# Then, install the package manually from the cloned directory:
# install.packages('iCellR/', repos = NULL, type = "source")- Download and unzip a publicly available sample PBMC scRNA-Seq data.
# Set your working directory to the location where the file will be downloaded
setwd("/your/download/directory")
# Save the URL of the PBMC 3k dataset as an object
sample.file.url = "https://cf.10xgenomics.com/samples/cell/pbmc3k/pbmc3k_filtered_gene_bc_matrices.tar.gz"
# Download the file from the URL and save it in the working directory
download.file(url = sample.file.url,
destfile = "pbmc3k_filtered_gene_bc_matrices.tar.gz",
method = "auto")
# Unzip the downloaded tar.gz file
untar("pbmc3k_filtered_gene_bc_matrices.tar.gz")
# Check the contents of the unzipped folder to ensure successful extraction
list.files()# Step 1: Load the required library
library("iCellR")
# Step 2: Load the PBMC dataset from 10x Genomics processed files
# Specify the directory containing 10x Genomics files (barcodes.tsv, genes.tsv/features.tsv, and matrix.mtx)
my.data <- load10x(data.dir = "filtered_gene_bc_matrices/hg19/")
# Notes:
# - The directory ("filtered_gene_bc_matrices/hg19/") should include the following:
# - `barcodes.tsv` (cell barcodes)
# - `genes.tsv` or `features.tsv` (gene names or features)
# - `matrix.mtx` (sparse expression matrix)
# - The data can be zipped or unzipped; iCellR handles both.- If your data is in .csv or .tsv format
# Read the dataset directly from a .tsv.gz file
my.data <- read.delim("my_sample_RNA.tsv.gz", header = TRUE)
# For uncompressed .csv or .tsv files:
# my.data <- read.csv("my_sample_RNA.csv", header = TRUE)- If your data is in .h5 format:
# Load the hdf5r library to work with .h5 files
library(hdf5r)
# Load the dataset from an h5 file
my.data <- load.h5(file = "filtered_feature_bc_matrix.h5")-
If your data is in S3 or S4 object format types like (Seurat or iCellR objects, etc.)
Here we use a Seurat object as an exacple:
# my.Seurat.object is your Seurat 5 object name
# get the raw data from your Seurat object slots
my.data <- as.data.frame(as.matrix(my.Seurat.object@assays$RNA@layers$counts))
rownames(my.data) <- rownames(my.Seurat.object@assays$RNA@features@.Data)
colnames(my.data) <- rownames(my.Seurat.object@assays$RNA@cells@.Data)If you want to see the help page for any function in R, simply use a question mark (?) followed by the function name. Here's an example:
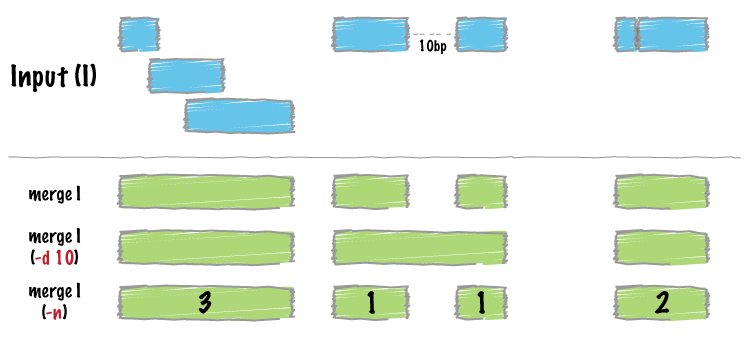
?load10xConditions in iCellR are defined or displayed in the column names of the data and are separated by an underscore (_) sign. If you want to merge multiple datasets (data frames/matrices) into one file and run iCellR in aggregate mode (combining all samples together), you can accomplish this using the data.aggregation function.
Example: Suppose you have divided your sample into four datasets and need to aggregate them into a single matrix. Let's say the samples are WT, KO, Ctrl, and KD. After aggregating these datasets into one matrix, iCellR will recognize the presence of four distinct samples for further analyses, such as batch alignment, plotting, differential expression (DE), and more. Here, I have divided this sample into four datasets for a test run.
# Check the dimensions of the dataset
dim(my.data)
# Output: [1] 32738 2700
# Divide your dataset into four separate samples for this example
sample1 <- my.data[1:900]
sample2 <- my.data[901:1800]
sample3 <- my.data[1801:2300]
sample4 <- my.data[2301:2700]
# Merge all samples into a single aggregated file
my.data <- data.aggregation(samples = c("sample1", "sample2", "sample3", "sample4"),
condition.names = c("WT", "KO", "Ctrl", "KD"))-
To check the head (the first few rows) of your file or dataset in R, use the head() function. Here's how you can do it.
This snippet shows how to inspect the header (column names) and the data for the first two cells in an aggregated data file:
# Display the head of your aggregated data and extract the first 2 columns (cells)
head(my.data)[, 1:2]
# Example Output:
# WT_AAACATACAACCAC-1 WT_AAACATTGAGCTAC-1
# A1BG 0 0
# A1BG.AS1 0 0
# A1CF 0 0
# A2M 0 0
# A2M.AS1 0 0
# as you see the header (column names) have the condition names added to the UMIs- Make an object of class iCellR.
my.obj <- make.obj(my.data)
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.6.0
Raw/original data dimentions (rows,columns): 32738,2700
Data conditions in raw data: Ctrl,KD,KO,WT (500,400,900,900)
Row names: A1BG,A1BG.AS1,A1CF ...
Columns names: WT_AAACATACAACCAC.1,WT_AAACATTGAGCTAC.1,WT_AAACATTGATCAGC.1 ...
###################################
QC stats performed:FALSE, PCA performed:FALSE
Clustering performed:FALSE, Number of clusters:0
tSNE performed:FALSE, UMAP performed:FALSE, DiffMap performed:FALSE
Main data dimensions (rows,columns): 0,0
Normalization factors:,...
Imputed data dimensions (rows,columns):0,0
############## scVDJ-seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-seq ############
ADT raw data dimensions (rows,columns):0,0
ADT main data dimensions (rows,columns):0,0
ADT columns names:...
ADT row names:...
############## scATAC-seq ############
ATAC raw data dimensions (rows,columns):0,0
ATAC main data dimensions (rows,columns):0,0
ATAC columns names:...
ATAC row names:...
############## Spatial ###########
Spatial data dimentions (rows,columns):0,0
########### iCellR object ##########my.obj <- qc.stats(my.obj)In iCellR, all plotting functions generate interactive HTML files by default.
These interactive plots are useful for exploring data visually in web browsers.
If you prefer static plots (e.g., for quick visualization or embedding in reports), you can disable interactivity by setting the parameter interactive = FALSE.
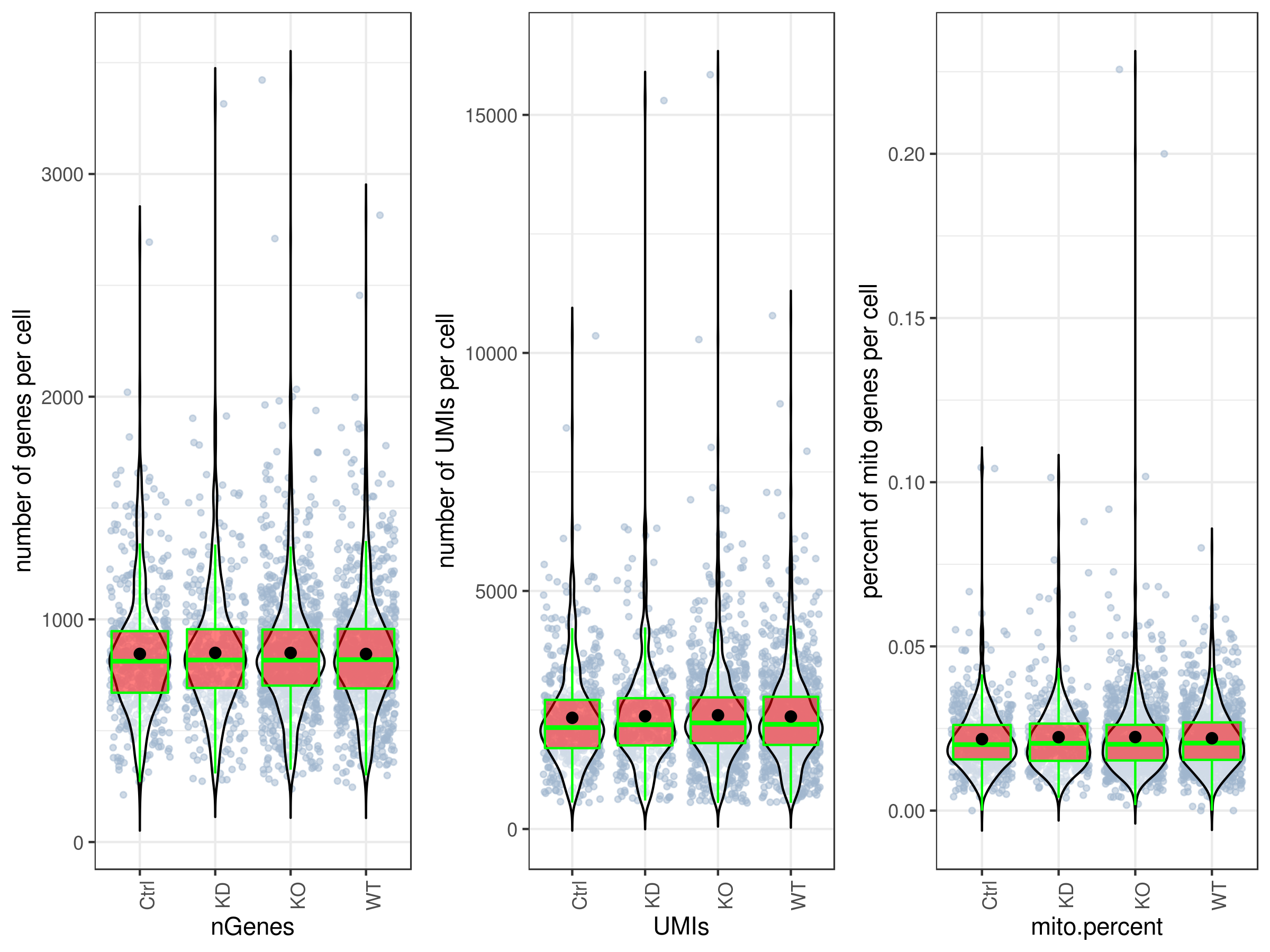
# plot UMIs, genes and percent mito all at once and in one plot.
# you can make them individually as well, see the arguments ?stats.plot.
stats.plot(my.obj,
plot.type = "three.in.one",
out.name = "UMI-plot",
interactive = FALSE,
cell.color = "slategray3",
cell.size = 1,
cell.transparency = 0.5,
box.color = "red",
box.line.col = "green")# Scatter plots
stats.plot(my.obj, plot.type = "point.mito.umi", out.name = "mito-umi-plot")
stats.plot(my.obj, plot.type = "point.gene.umi", out.name = "gene-umi-plot")The iCellR package provides flexibility to filter single-cell RNA-seq datasets based on various metrics, helping improve data quality and remove unwanted cells or genes from the analysis. You can filter your data using the following criteria:
Library Sizes (UMIs):
Filter cells based on the total library size (number of UMIs per cell). This can help exclude cells with very low UMI counts, which might indicate doublets or empty droplets.
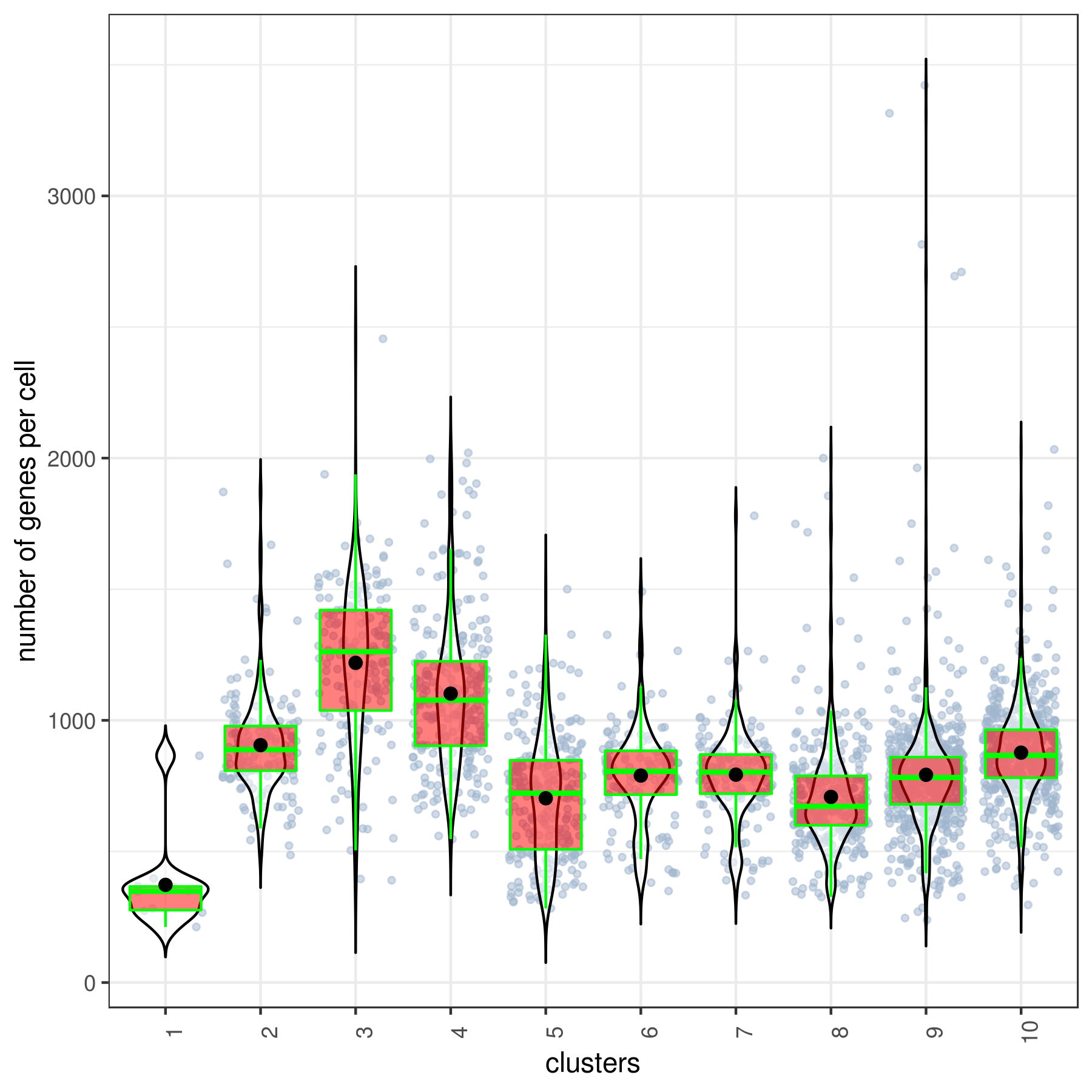
Number of Genes per Cell:
Filter cells by the number of detected genes. For example, you can remove cells with fewer than a certain threshold of expressed genes to exclude low-quality cells.
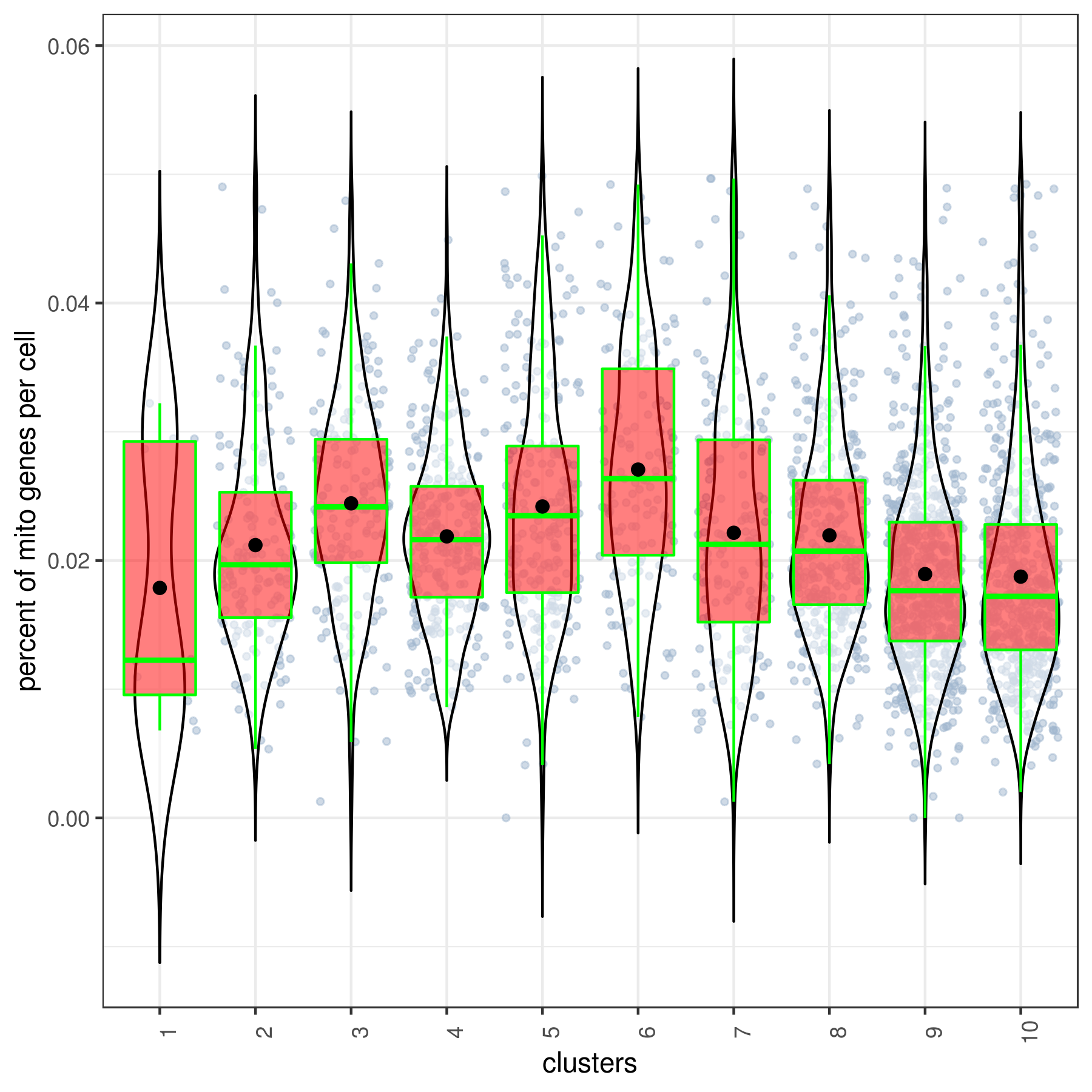
Percent Mitochondrial Content:
Filter cells by mitochondrial content. Typically, cells with excessively high mitochondrial expression (e.g., >10% of total expression) may indicate stressed or dying cells.
Based on One or More Genes:
Select cells whose expression levels meet criteria for one or more specific genes (e.g., filter based on marker gene expression).
Cell IDs:
Filter cells by specific cell IDs. This allows for targeted removal or selection of cells identified in previous analyses or metadata.
# Apply multiple filters to the iCellR object
my.obj <- cell.filter(
my.obj,
min.mito = 0, # Minimum fraction of mitochondrial content allowed
max.mito = 0.05, # Maximum fraction of mitochondrial content allowed
min.genes = 200, # Minimum number of detected genes per cell
max.genes = 2400, # Maximum number of detected genes per cell
min.umis = 0, # Minimum UMI count allowed
max.umis = Inf # Maximum UMI count (set to infinite to not limit)
)
# Example Output:
#[1] "cells with min mito ratio of 0 and max mito ratio of 0.05 were filtered."
#[1] "cells with min genes of 200 and max genes of 2400 were filtered."
#[1] "No UMI number filter"
#[1] "No cell filter by provided gene/genes"
#[1] "No cell id filter"
#[1] "filters_set.txt file has beed generated and includes the filters set for this experiment." You can add filters for specific genes. For example, the following line filters out cells that do not have counts for the genes "RPL13" or "RPL10":
# my.obj <- cell.filter(my.obj, filter.by.gene = c("RPL13","RPL10")) # filter our cell having no counts for these genesThis removes the cell WT_AAACATACAACCAC.1 from your dataset.
# my.obj <- cell.filter(my.obj, filter.by.cell.id = c("WT_AAACATACAACCAC.1")) # filter our cell cell by their cell ids.Check the dimensions after filtering
dim(my.obj@main.data)
# Output: [1] 32738 2637What is Down-Sampling?
Purpose: Down-sampling ensures that each condition (e.g., treatment groups like WT, KO, Ctrl, etc.) has the same number of cells for balanced comparisons.
Why?:
Prevent bias in downstream analyses caused by unequal cell counts across conditions.
This step is optional and should be used with caution, as it is generally not recommended. Down-sampling may result in the loss of important or rare cell populations, which can impact the accuracy of your analysis, especially for heterogeneous datasets with diverse cell types.
However, there are cases where down-sampling can be useful, such as:
When working with datasets containing an extremely large number of cells.
To speed up computational analysis for complex workflows or resource-limited environments.
For testing purposes or when uniform cell counts are needed across conditions.
Ultimately, the decision to down-sample should be made based on your specific experimental goals and the nature of your data. This option is available if necessary, but its use should be carefully weighed against the potential impact on downstream analyses.
# optional
# Perform down-sampling to equalize cells across conditions (optional)
# my.obj <- down.sample(my.obj)
#Before Down-Sampling:
#The dataset initially contains cells as follows:
#Ctrl: 877 cells
#KO: 877 cells
#WT: 883 cells
#After Down-Sampling:
#Down-sampling has equalized the number of cells across all conditions at 877 cells each.Normalization is an essential step in single-cell RNA sequencing analysis. iCellR provides several options for normalization, and you can choose the best approach depending on your study objectives and dataset characteristics.
Options for Normalization:
Use iCellR’s Built-in Normalization Methods:
iCellR offers multiple normalization techniques tailored to single-cell experiments. One highly recommended method is ranked.glsf.
External Tools for Normalization:
You can normalize your data using external tools, like:
DESeq2 (Geometric Normalization): Popular for bulk RNA-seq but adaptable for single-cell studies.
Scran: Computes size factors using clustering-based normalization for single-cell datasets.
After normalization, you can import the externally normalized data into iCellR for further analysis.
What is it?
Ranked Geometric Library Size Factor (ranked.glsf) is inspired by DESeq2's Geometric Mean Size Factor normalization, but adapted for single-cell challenges. The ranked component makes it better suited for sparse datasets by focusing on highly expressed genes.
- Designed to handle single-cell datasets with lots of zeros (dropouts) in the matrix.
- Because it uses a geometric approach, this normalization reduces batch-wise differences caused by variable library sizes.
my.obj <- norm.data(my.obj,
norm.method = "ranked.glsf",
top.rank = 500) # best for scRNA-Seq
# This focuses on the top 500 most highly expressed genes for calculating library size normalization.
# more examples
#my.obj <- norm.data(my.obj, norm.method = "ranked.deseq", top.rank = 500)
#my.obj <- norm.data(my.obj, norm.method = "deseq") # best for bulk RNA-Seq
#my.obj <- norm.data(my.obj, norm.method = "global.glsf") # best for bulk RNA-Seq
#my.obj <- norm.data(my.obj, norm.method = "rpm", rpm.factor = 100000) # best for bulk RNA-Seq
#my.obj <- norm.data(my.obj, norm.method = "spike.in", spike.in.factors = NULL)
#my.obj <- norm.data(my.obj, norm.method = "no.norm") # if the data is already normalized- Perform second QC (optioal) After initial filtering and normalization, a second QC step can be performed to further refine the dataset.
#my.obj <- qc.stats(my.obj,which.data = "main.data")
#stats.plot(my.obj,
# plot.type = "all.in.one",
# out.name = "UMI-plot",
# interactive = F,
# cell.color = "slategray3",
# cell.size = 1,
# cell.transparency = 0.5,
# box.color = "red",
# box.line.col = "green",
# back.col = "white")Why Scaling is Not Required in iCellR
In iCellR, scaling the data is handled dynamically or "on the fly" during tasks such as plotting or running dimensionality reduction methods like PCA.
Here's why this design is beneficial:
-
To
save storage: this eliminates the need for permanently scaling your main dataset beforehand. If you do choose to scale your data manually, scaling does not overwrite the main dataset. Instead, scaled data is saved into a separate slot in your iCellR object. iCellR automatically scales the data as needed during specific functions like plot.tsne() or run.pca(). -
Untransformed Data for Differential Expression Analysis: Untransformed data is used for generating accurate fold-change values during Differential Expression (DE) Analysis.
# my.obj <- data.scale(my.obj)Gene statistics typically involve summarizing the behavior or characteristics of genes across cells in your scRNA-seq dataset. iCellR provides tools to calculate and explore gene-level info, such as:
Gene Expression Levels:
Aggregate counts or normalized values for specific genes across all cells or conditions.
Gene Detection Frequency:
Proportion of cells in which each gene is expressed (non-zero counts).
Gene Variance:
Variability in expression levels across all cells or conditions to identify highly variable genes.
Top-Expressed Genes:
Identify genes that are most highly expressed across the dataset or within a condition.
my.obj <- gene.stats(my.obj, which.data = "main.data")
head(my.obj@gene.data[order(my.obj@gene.data$numberOfCells, decreasing = T),])
# genes numberOfCells totalNumberOfCells percentOfCells meanExp
#30303 TMSB4X 2637 2637 100.00000 38.55948
#3633 B2M 2636 2637 99.96208 45.07327
#14403 MALAT1 2636 2637 99.96208 70.95452
#27191 RPL13A 2635 2637 99.92416 32.29009
#27185 RPL10 2632 2637 99.81039 35.43002
#27190 RPL13 2630 2637 99.73455 32.32106
# SDs condition
#30303 7.545968e-15 all
#3633 2.893940e+01 all
#14403 7.996407e+01 all
#27191 2.783799e+01 all
#27185 2.599067e+01 all
#27190 2.661361e+01 allCreating a gene model for clustering in single-cell RNA-seq analysis involves selecting a subset of genes (e.g., highly variable genes, marker genes, or genes of interest) that are most informative for identifying cell clusters. This process helps reduce noise and focus on biologically relevant features for unsupervised clustering.
This function will help you find a good number of genes to use for running PCA.
# See model plot
make.gene.model(my.obj, my.out.put = "plot",
dispersion.limit = 1.5,
base.mean.rank = 1500,
no.mito.model = T,
mark.mito = T,
interactive = F,
out.name = "gene.model")
# Write the gene model data into the object
my.obj <- make.gene.model(my.obj, my.out.put = "data",
dispersion.limit = 1.5,
base.mean.rank = 1500,
no.mito.model = T,
mark.mito = T,
interactive = F,
out.name = "gene.model")
head(my.obj@gene.model)
# "ACTB" "ACTG1" "ACTR3" "AES" "AIF1" "ALDOA"
# get html plot (optional)
#make.gene.model(my.obj, my.out.put = "plot",
# dispersion.limit = 1.5,
# base.mean.rank = 1500,
# no.mito.model = T,
# mark.mito = T,
# interactive = T,
# out.name = "plot4_gene.model")To view an the html interactive plot click on this links: Dispersion plot
Principal Component Analysis (PCA) is a fundamental dimensionality reduction technique often used in single-cell RNA-seq analysis to represent high-dimensional gene expression data in a lower-dimensional space. However, PCA does not harmonize or integrate or batch align the data.
Skip the PCA step if you plan to perform batch correction, which typically realigns data across batches and conditions. For batch correction (sample alignment/harmonization/integration) see the sections; CPCA, CCCA, MNN or anchor alignment.
# When you run run.pca, iCellR uses raw or normalized data directly without correcting for batch-related artifacts. This results in principal components that may reflect technical variations (batch effects) rather than true biological signals.
# run PCA
my.obj <- run.pca(my.obj, method = "gene.model", gene.list = my.obj@gene.model,data.type = "main")
opt.pcs.plot(my.obj)2 round PCA (optional)
For finding top genes in the top principal components (PCs) and re-running PCA to achieve better segregation of cell populations. This is optional and not recommended except in certain cases.
#my.obj <- find.dim.genes(my.obj, dims = 1:10, top.pos = 20, top.neg = 20) # (optional)
#length([email protected])
# 211
# second round PC
#my.obj <- run.pca(my.obj, method = "gene.model", gene.list = [email protected],data.type = "main")# tSNE
my.obj <- run.pc.tsne(my.obj, dims = 1:10)# UMAP
my.obj <- run.umap(my.obj, dims = 1:10)Don't forget to set the zoom in the right range.
my.obj <- run.knetl(my.obj, dims = 1:20, zoom = 110)
KNetL works with a higher resolution; therfore using dims = 20 (2 times the number of PCs used for UMAP) usually produces the best results for most datasets.
- Recommended
ZoomSettings:
< 1,000 cells: Zoom range 5–50
1,000–5,000 cells: Zoom range 50–200
5,000–10,000 cells: Zoom range 100–300
10,000–30,000 cells: Zoom range 200–500
> 30,000 cells: Zoom range 400–600
Additional Notes: A zoom of 400 generally works well for large datasets, but adjustments might be needed for your desired resolution.
Remember:
Lower zoom numbers = zoom in.
Higher zoom numbers = zoom out (reverse logic).
# diffusion map
# this requires python packge phate or bioconductor R package destiny
# How to install destiny
# if (!requireNamespace("BiocManager", quietly = TRUE))
# install.packages("BiocManager")
# BiocManager::install("destiny")
# How to install phate
# pip install --user phate
# Install phateR version 2.9
# wget https://cran.r-project.org/src/contrib/Archive/phateR/phateR_0.2.9.tar.gz
# install.packages('phateR/', repos = NULL, type="source")
# or
# library(devtools)
# install_version("phateR", version = "0.2.9", repos = "http://cran.us.r-project.org")
# optional
# library(destiny)
# my.obj <- run.diffusion.map(my.obj, dims = 1:10)
# or
# library(phateR)
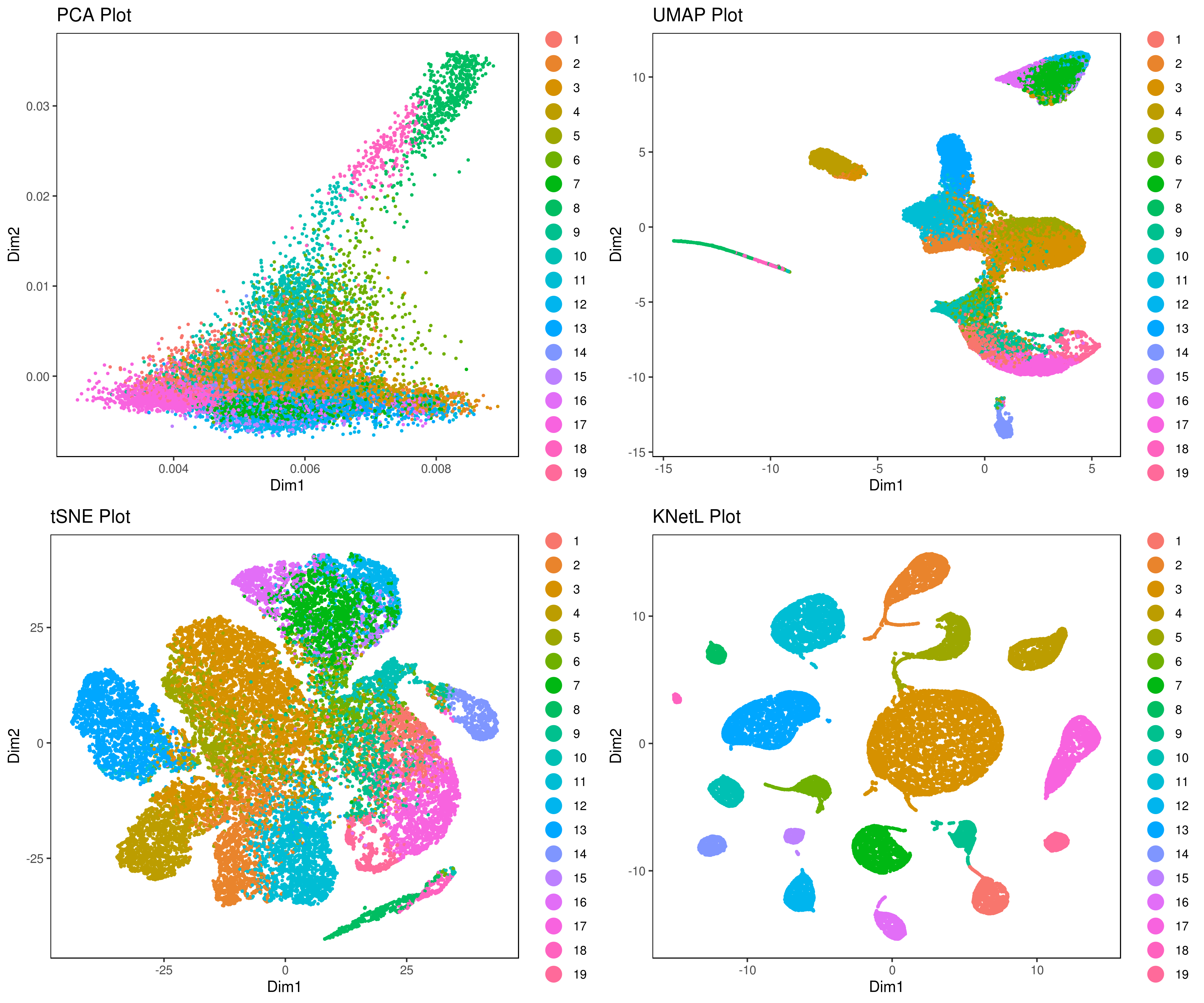
# my.obj <- run.diffusion.map(my.obj, dims = 1:10, method = "phate")# Generate cluster plots with different methods
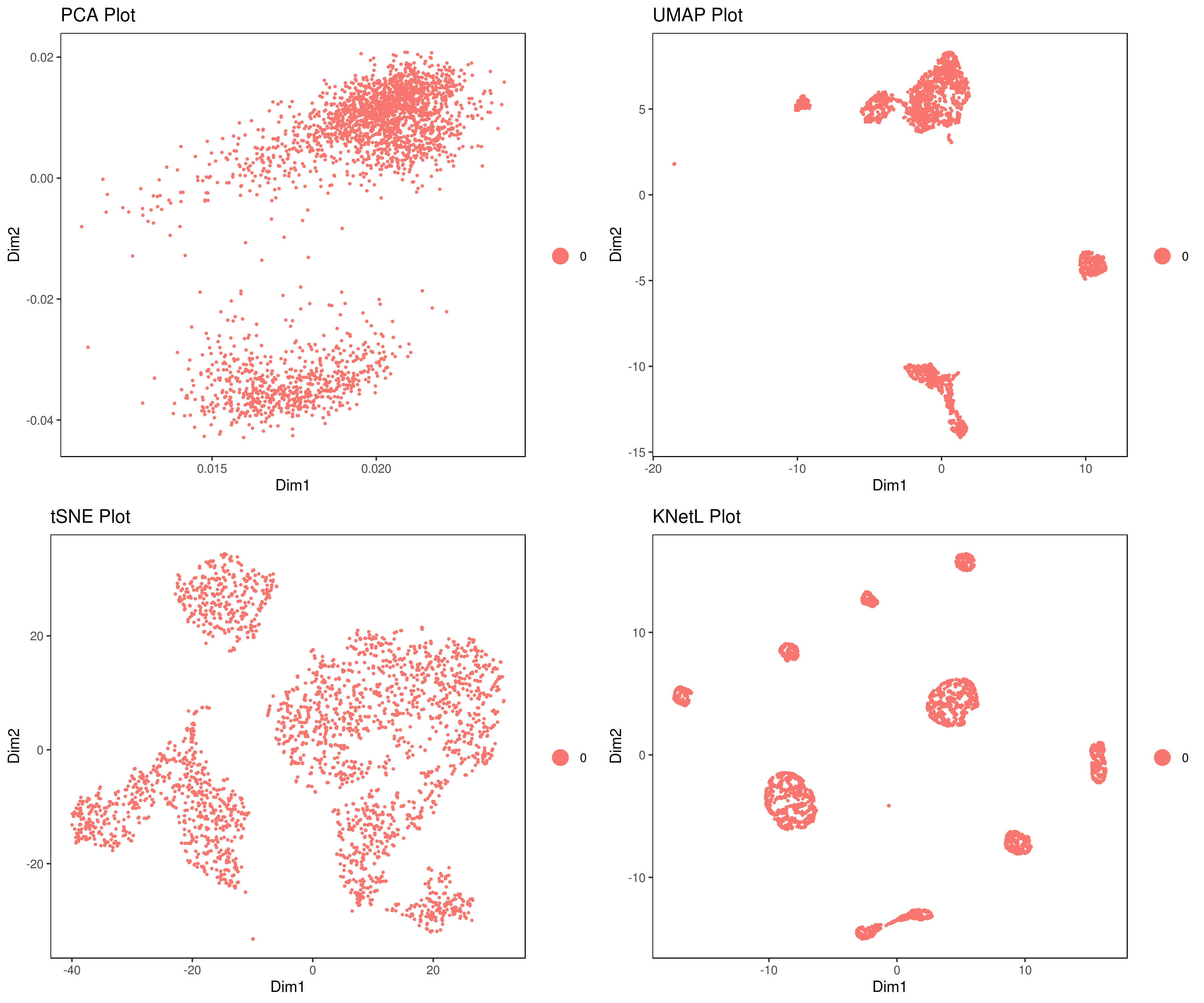
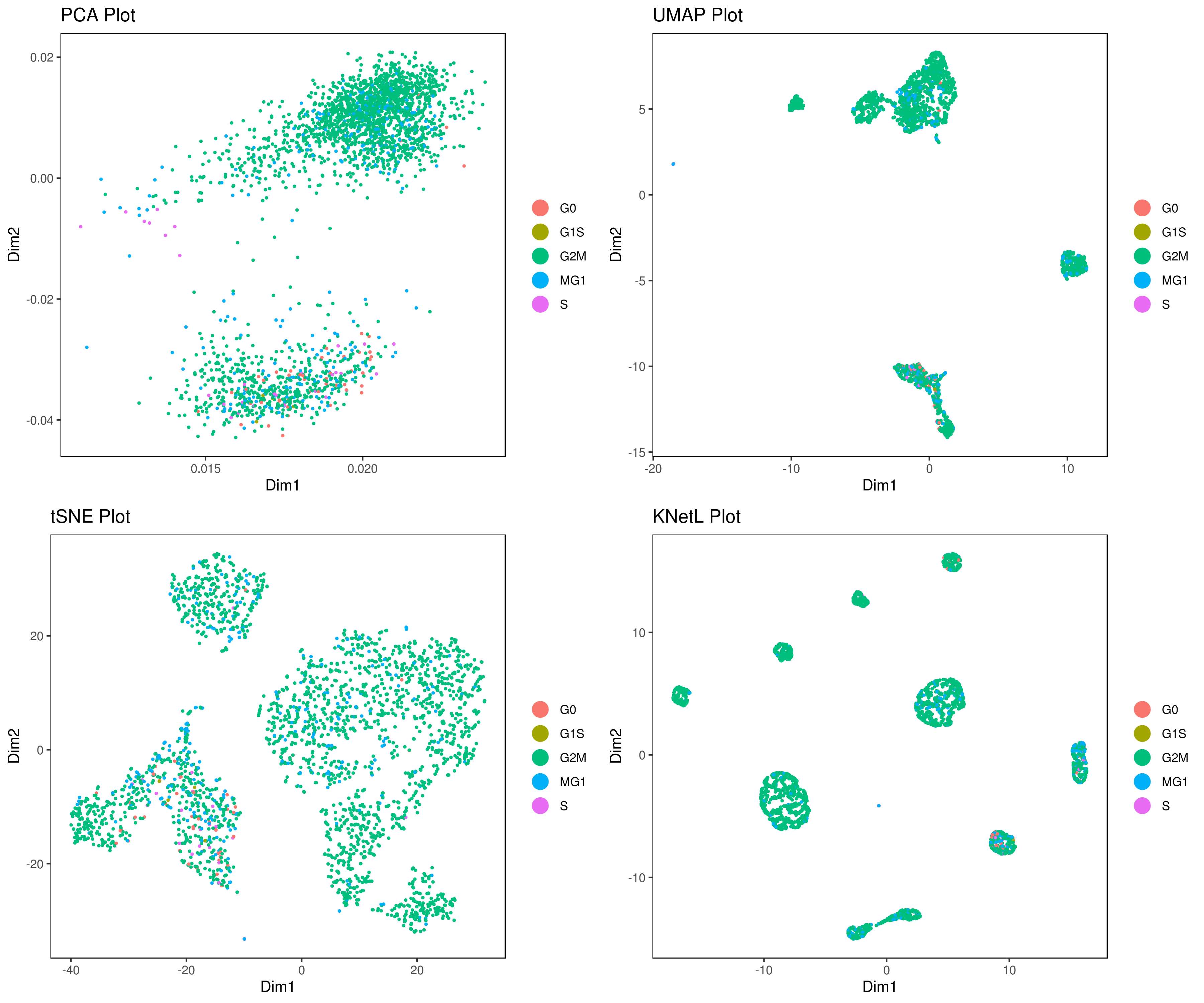
A <- cluster.plot(my.obj, plot.type = "pca", interactive = FALSE)
B <- cluster.plot(my.obj, plot.type = "umap", interactive = FALSE)
C <- cluster.plot(my.obj, plot.type = "tsne", interactive = FALSE)
D <- cluster.plot(my.obj, plot.type = "knetl", interactive = FALSE)
# Load the gridExtra library for arranging multiple plots
library(gridExtra)
# Combine and arrange the plots (PCA, UMAP, t-SNE, and KNetL) in a grid layout
grid.arrange(A, B, C, D)We provide three functions to run the clustering method of your choice:
This function is optimized for iCellR and supports PCA, UMAP, t-SNE, Destiny (diffusion map), PHATE, or KNetL maps as input. It utilizes the Louvain algorithm for clustering a graph constructed using k-Nearest Neighbor (KNN), similar to PhenoGraph (Levine et al., Cell, 2015). However, it employs distance values (Euclidean by default) as weights, instead of Jaccard similarity values.
R implementation of the PhenoGraph algorithm. Rphenograph wrapper (Levine et al., Cell, 2015).
This function offers a wide range of options to explore your data using various clustering and indexing methods. You can select any combination from the table below to experiment with different approaches and "flavors" of analysis.
| clustering methods | distance methods | indexing methods |
|---|---|---|
| ward.D, ward.D2, single, complete, average, mcquitty, median, centroid, kmeans | euclidean, maximum, manhattan, canberra, binary, minkowski or NULL | kl, ch, hartigan, ccc, scott, marriot, trcovw, tracew, friedman, rubin, cindex, db, silhouette, duda, pseudot2, beale, ratkowsky, ball, ptbiserial, gap, frey, mcclain, gamma, gplus, tau, dunn, hubert, sdindex, dindex, sdbw |
Conventionally, clustering is performed using PCA data (usually the first 10 dimensions). However, this function allows you to choose t-SNE, UMAP, or KNetL map dimensions as alternatives. If you have fine-tuned your KNetL map and are confident in its results, we recommend clustering based on the KNetL map.
Clustering can be one of the more challenging aspects of data analysis, and adjustments may be necessary based on marker genes. This might involve merging certain clusters, using gating tools (refer to our cell gating tools), or experimenting with different sensitivity values to identify a greater or smaller number of communities.
Notes:
- Adjust sensitivity for more or less clusters.
- Lower sensitivity numbers = more clusters.
- Higher sensitivity numbers = less clusters (reverse logic).
- 100-150 generally works best for most data.
my.obj <- iclust(my.obj, sensitivity = 150, data.type = "knetl")
# data.type could be umap or tsne, etc. Adjust sensitivity for more or less clusters.
Top 10 PCs generally works best for most data.
my.obj <- iclust(my.obj, sensitivity = 150, data.type = "pca", dims=1:10)
Other examples:
# my.obj <- iclust(my.obj,
# dist.method = "euclidean",
# sensitivity = 100,
# dims = 1:10,
# data.type = "pca")
# or
# run.phenograph
# my.obj <- run.phenograph(my.obj,k = 100,dims = 1:10)
# or
# run.clustering
# my.obj <- run.clustering(my.obj,
# clust.method = "kmeans",
# dist.method = "euclidean",
# index.method = "silhouette",
# max.clust = 25,
# min.clust = 2,
# dims = 1:10)
# If you want to manually set the number of clusters, and not used the predicted optimal number, set the minimum and maximum to the number you want:
#my.obj <- run.clustering(my.obj,
# clust.method = "ward.D",
# dist.method = "euclidean",
# index.method = "ccc",
# max.clust = 8,
# min.clust = 8,
# dims = 1:10)
# more examples
#my.obj <- run.clustering(my.obj,
# clust.method = "ward.D",
# dist.method = "euclidean",
# index.method = "kl",
# max.clust = 25,
# min.clust = 2,
# dims = 1:10)# plot clusters (in the figures below clustering is done based on KNetL)
# example: # my.obj <- iclust(my.obj, k = 150, data.type = "knetl")
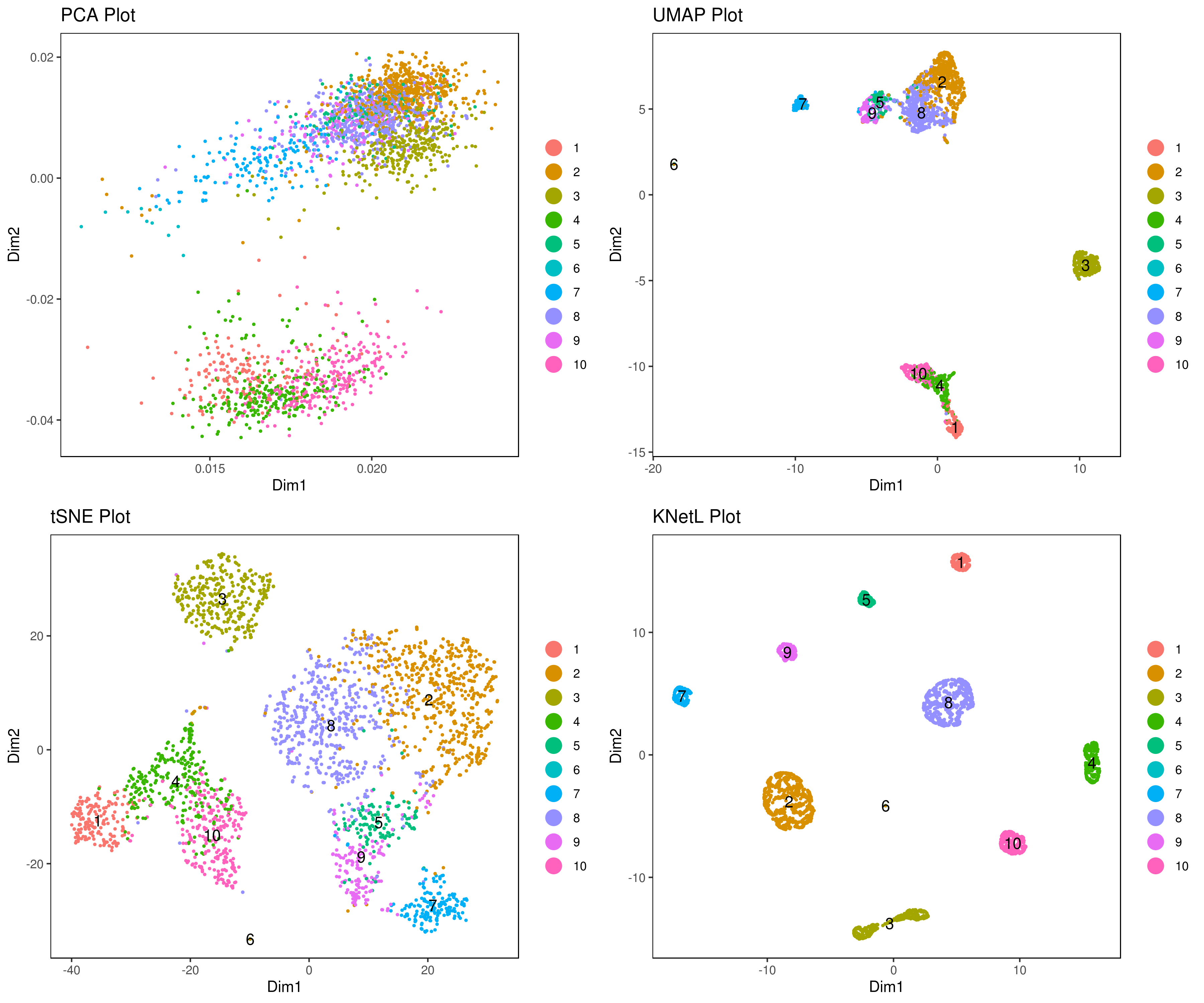
A <- cluster.plot(my.obj,plot.type = "pca",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
B <- cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
C <- cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
D <- cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
library(gridExtra)
grid.arrange(A,B,C,D)This step rearranges clusters so that they appear in a more consecutive order based on gene expression similarities.
This re-ordering can be visually beneficial when analyzing your heatmap after identifying marker genes. Similar cell communities will appear next to each other, making it easier to visually examine and compare them. Additionally, it can help in deciding which clusters may need merging or adjustment.
my.obj <- clust.ord(my.obj,top.rank = 500, how.to.order = "distance")
#my.obj <- clust.ord(my.obj,top.rank = 500, how.to.order = "random")Re-plot
A= cluster.plot(my.obj,plot.type = "pca",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
B= cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
C= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
D= cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
library(gridExtra)
grid.arrange(A,B,C,D)Example 1:
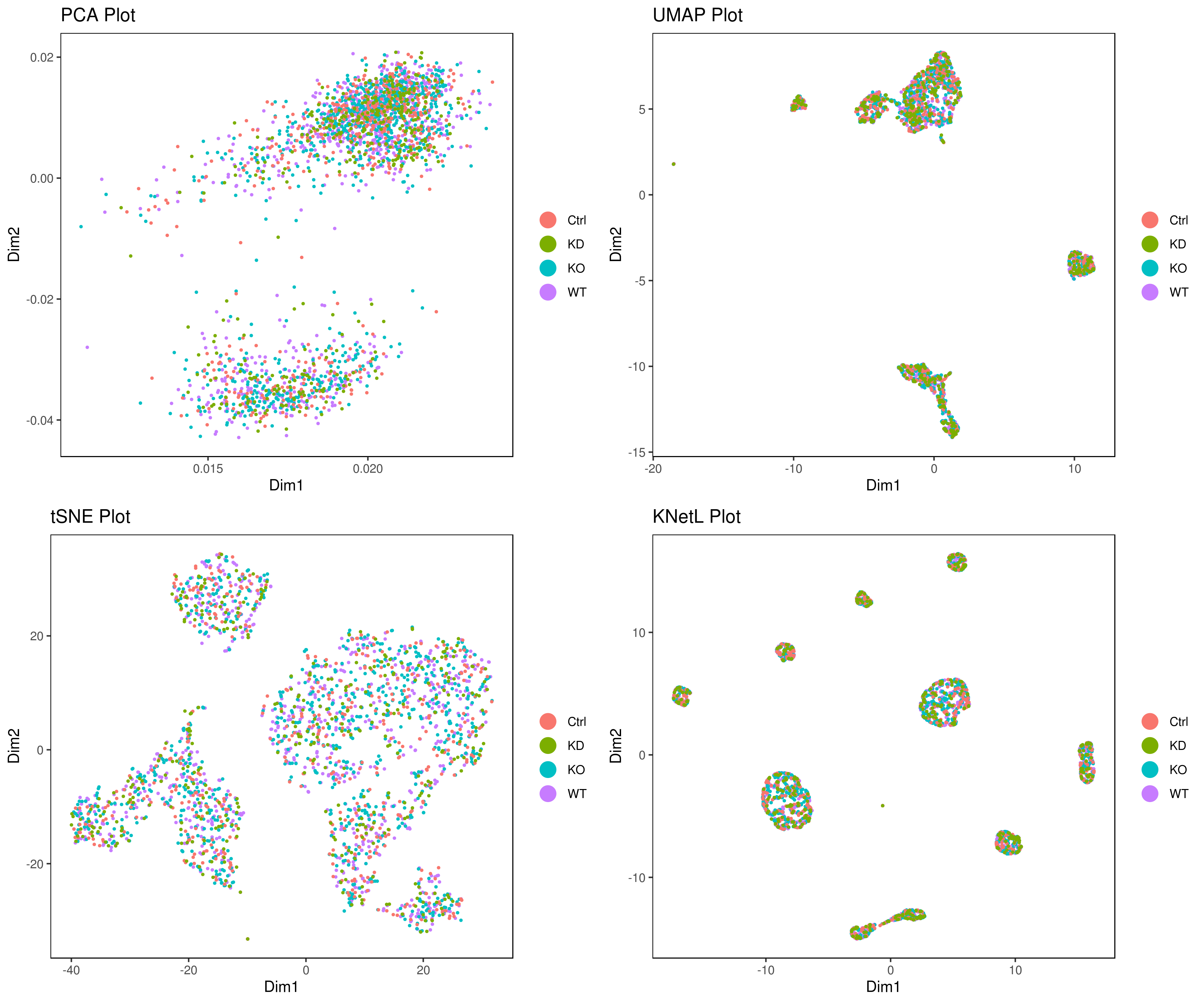
# conditions
A <- cluster.plot(my.obj,plot.type = "pca",col.by = "conditions",interactive = F,cell.size = 0.5)
B <- cluster.plot(my.obj,plot.type = "umap",col.by = "conditions",interactive = F,cell.size = 0.5)
C <- cluster.plot(my.obj,plot.type = "tsne",col.by = "conditions",interactive = F,cell.size = 0.5)
D <- cluster.plot(my.obj,plot.type = "knetl",col.by = "conditions",interactive = F,cell.size = 0.5)
library(gridExtra)
grid.arrange(A,B,C,D)Example 2:
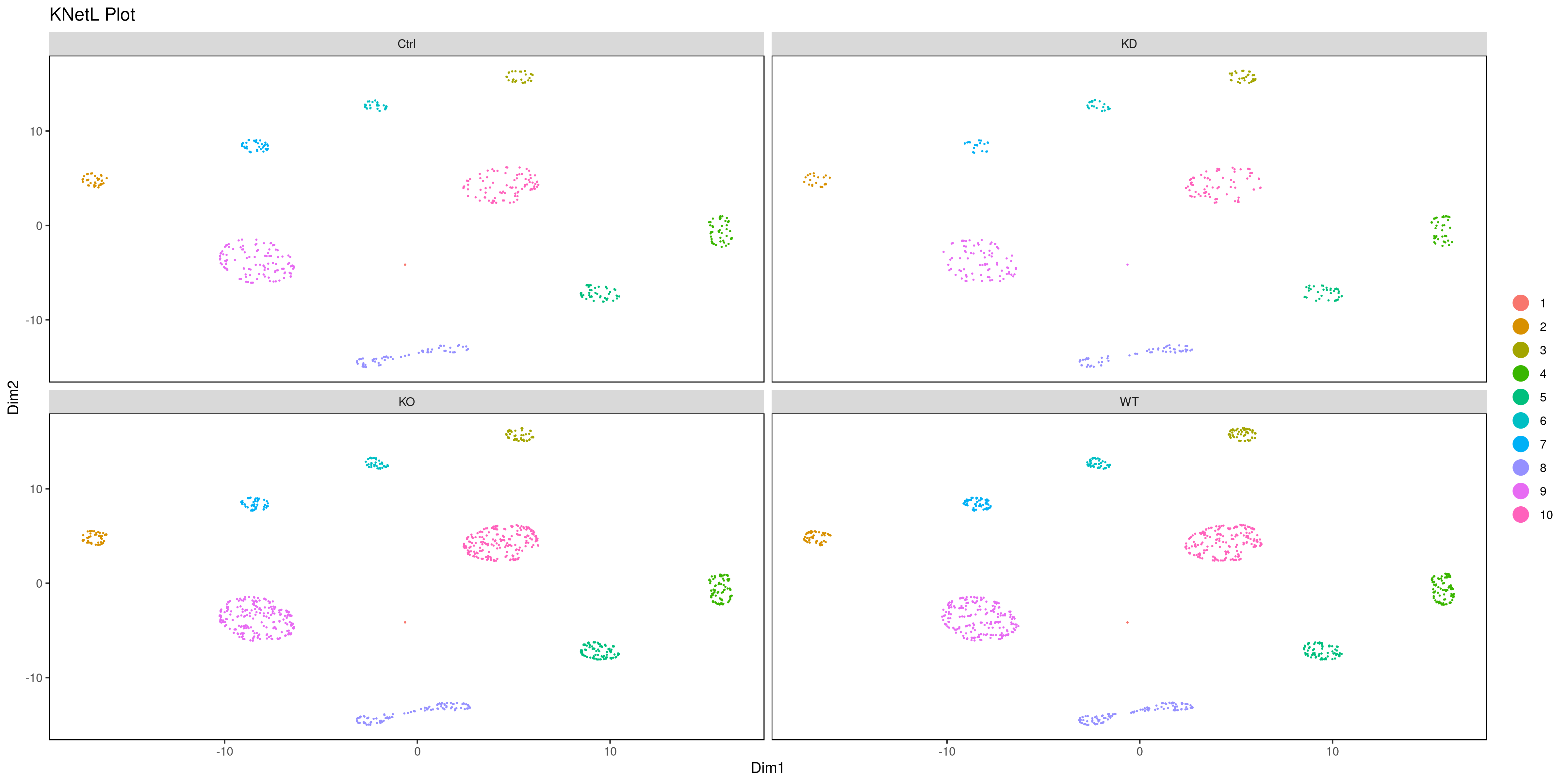
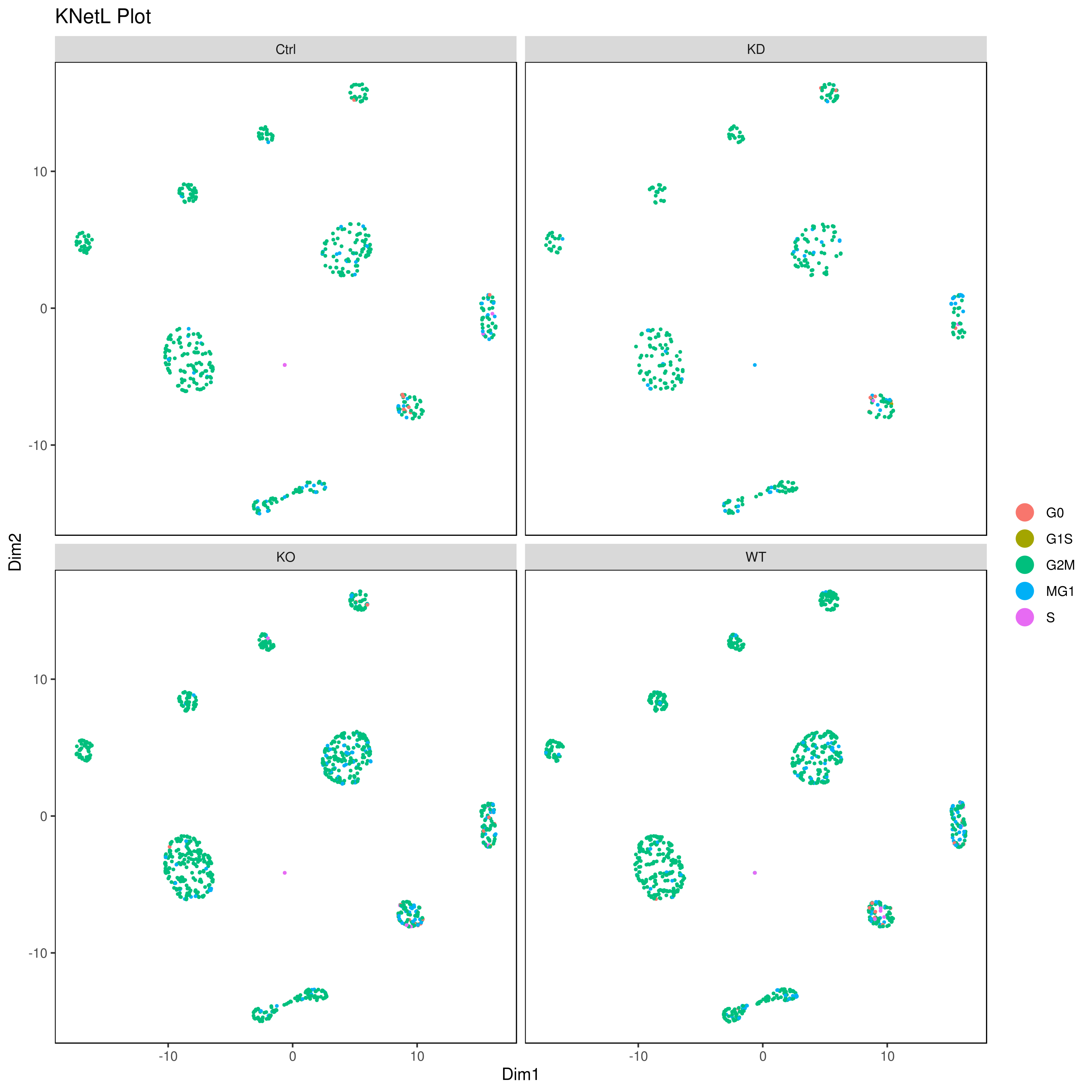
png('AllConds_clusts_knetl.png', width = 16, height = 8, units = 'in', res = 300)
cluster.plot(my.obj,
cell.size = 0.1,
plot.type = "knetl",
cell.color = "black",
back.col = "white",
cell.transparency = 1,
clust.dim = 2,
interactive = F,cond.facet = T)
dev.off()
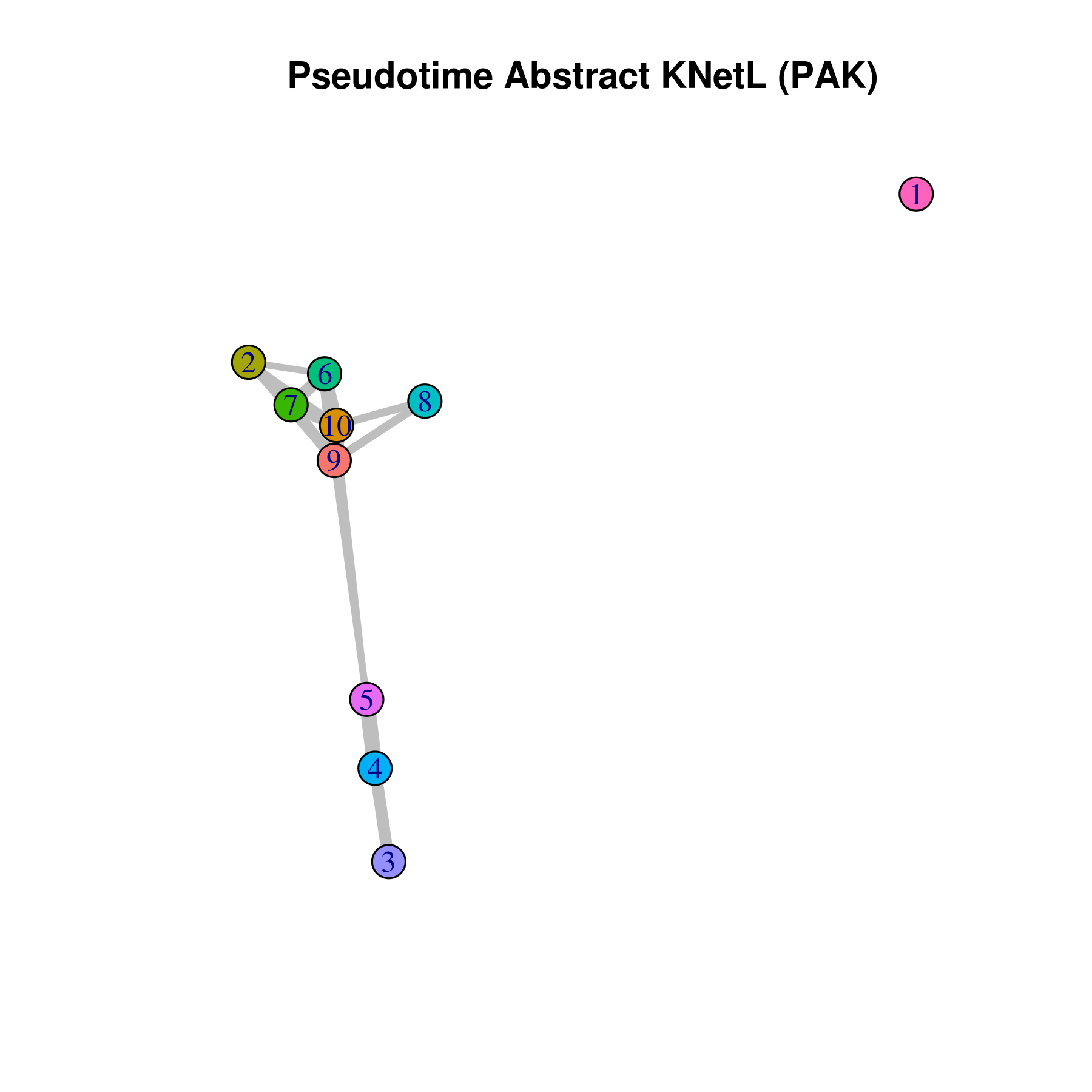
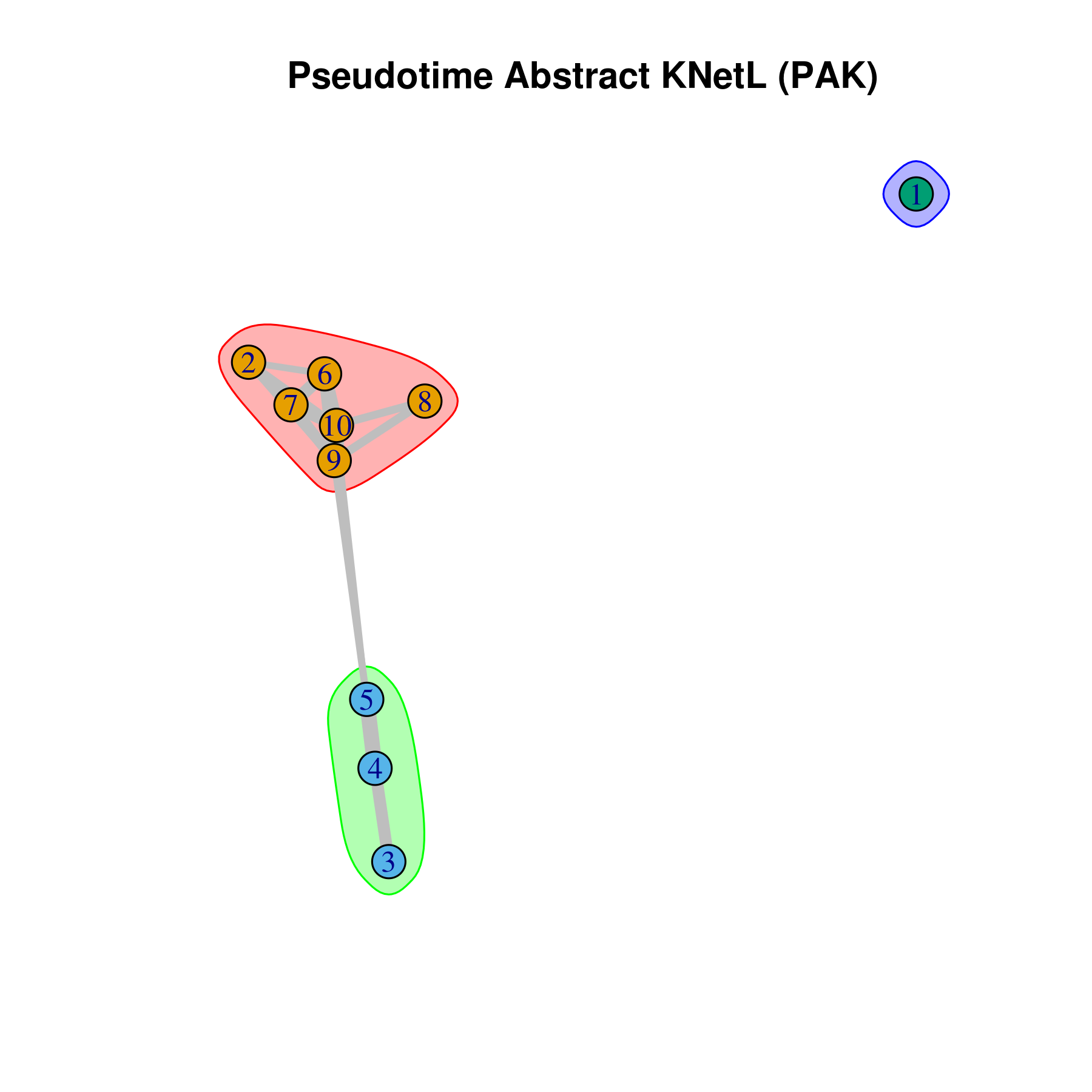
This approach is very useful for visualizing the distances or similarities between different communities. The shorter and thicker the lines or links (rubber bands) are, the more similar the communities. In this visualization:
- Nodes represent clusters, and
- Edges or links represent the distances between clusters.
pseudotime.knetl(my.obj,interactive = F,cluster.membership = F,conds.to.plot = NULL)
## with memberships
pseudotime.knetl(my.obj,interactive = F,cluster.membership = T,conds.to.plot = NULL)
### intractive plot
pseudotime.knetl(my.obj,interactive = T)This refers to the calculation of the mean gene expression values for each cluster. By averaging the expression of genes within a cluster, you can summarize the overall expression profile of cell populations, making it easier to compare clusters and identify distinctive marker genes or biological patterns.
- Option 1: all the cells in all the conditions/samples
# for all cunditions
my.obj <- clust.avg.exp(my.obj, conds.to.avg = NULL)- Option 2: choose condition/condition
s
# for one cundition
#my.obj <- clust.avg.exp(my.obj, conds.to.avg = "WT")
# for two or more cunditions
#my.obj <- clust.avg.exp(my.obj, conds.to.avg = c("WT","KO"))To examine the first few rows of the average expression data across clusters use the head() function.
head(my.obj@clust.avg)
# gene cluster_1 cluster_2 cluster_3 cluster_4 cluster_5
#1 A1BG 0 0.034248447 0.029590643 0.076486590 0.090270833
#2 A1BG.AS1 0 0.000000000 0.006274854 0.019724138 0.004700000
#3 A1CF 0 0.000000000 0.000000000 0.000000000 0.000000000
#4 A2M 0 0.006925466 0.003614035 0.000000000 0.000000000
#5 A2M.AS1 0 0.056155280 0.000000000 0.005344828 0.006795833
#6 A2ML1 0 0.000000000 0.000000000 0.000000000 0.000000000
# cluster_6 cluster_7 cluster_8 cluster_9 cluster_10
#1 0.074360294 0.07623494 0.04522321 0.088735057 0.065292818
#2 0.000000000 0.00000000 0.01553869 0.013072698 0.013550645
#3 0.000000000 0.00000000 0.00000000 0.000000000 0.000000000
#4 0.000000000 0.00000000 0.00000000 0.001810985 0.003200737
#5 0.008191176 0.06227108 0.00000000 0.011621971 0.012837937
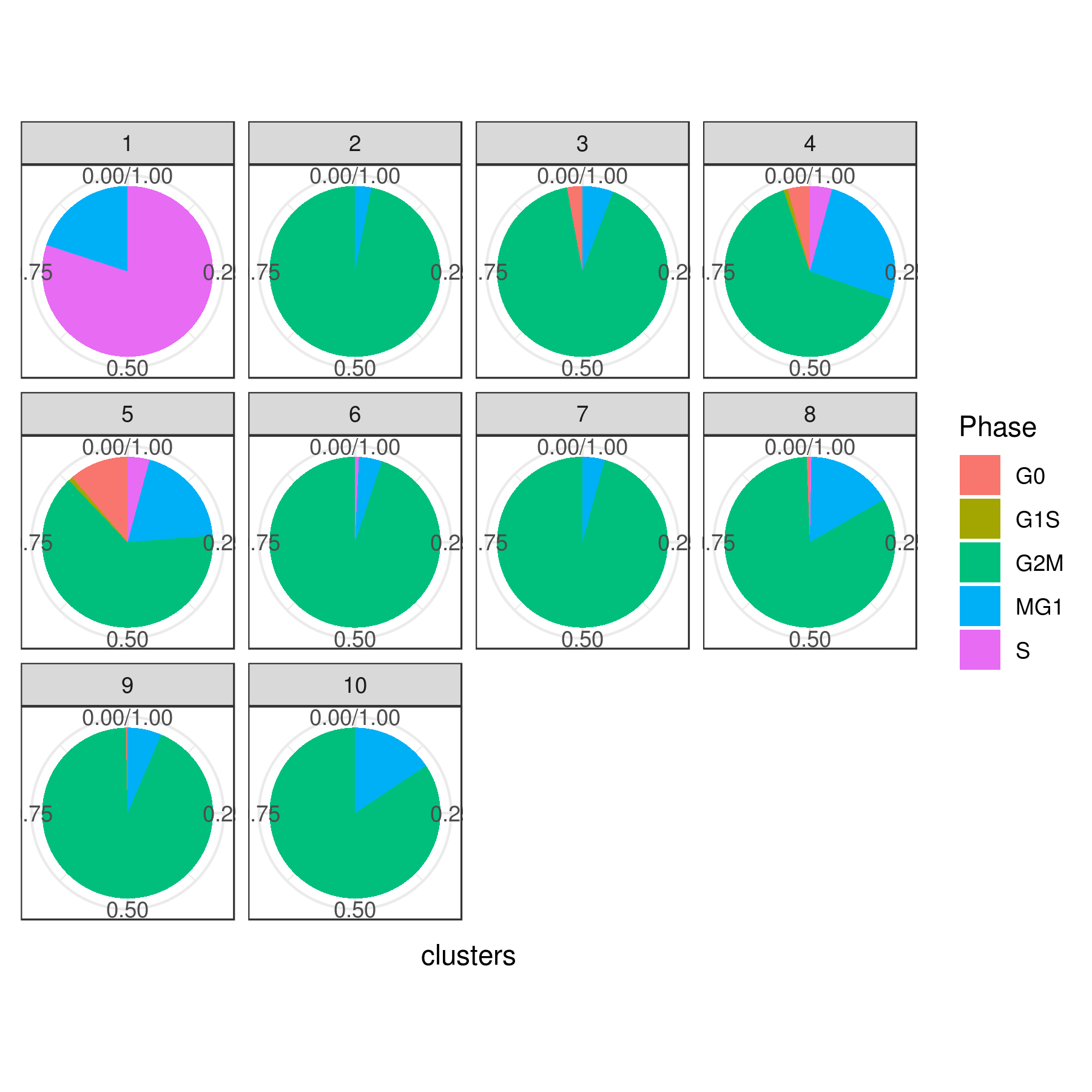
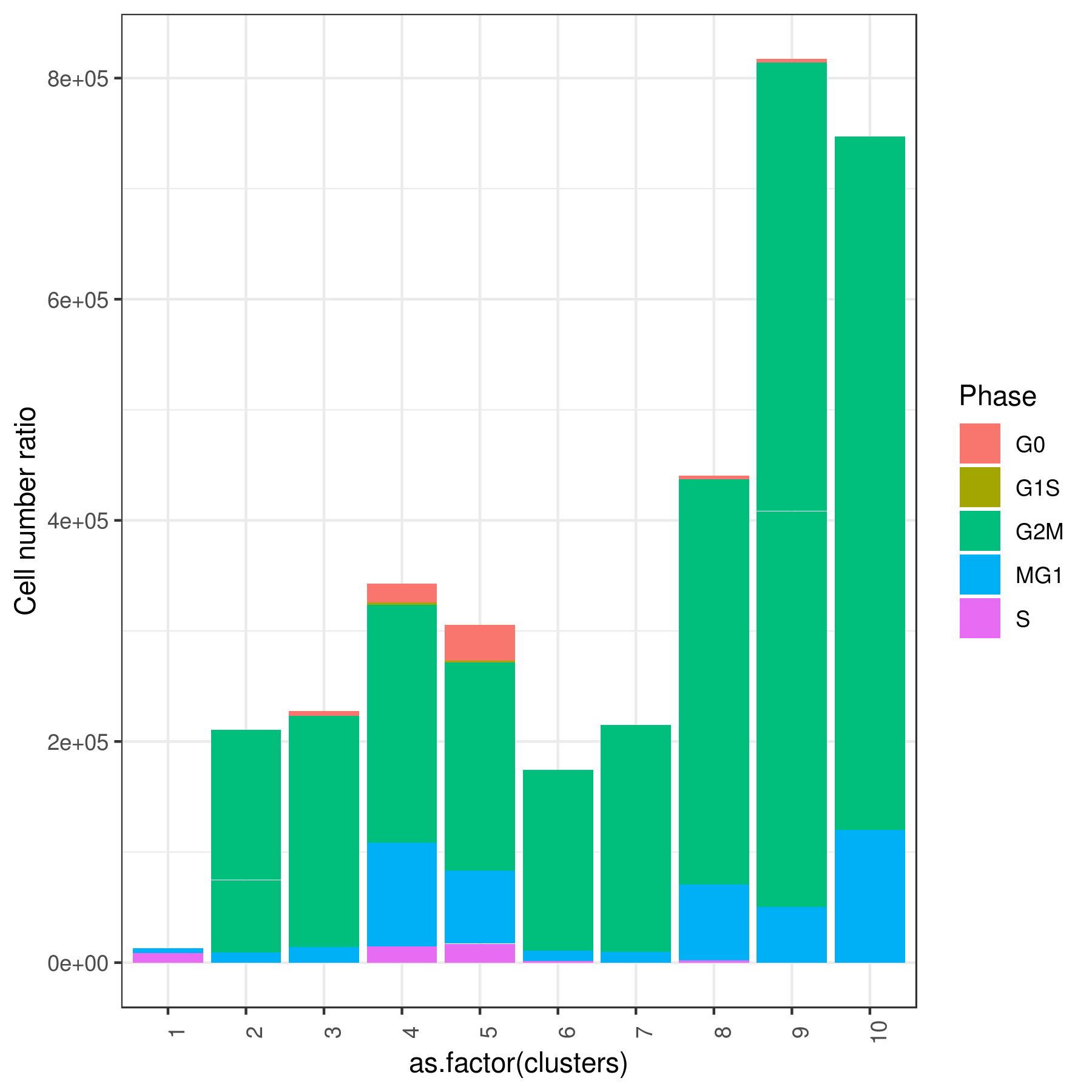
#6 0.000000000 0.00000000 0.00000000 0.000000000 0.000000000Tirosh scoring method Tirosh, et. al. 2016 (default) or coverage is used to calculate G0, G1S, G2M, M, G1M and S phase score. The gene lists for G0, G1S, G2M, M, G1M and S phase are chosen from previously published article Xue, et.al 2020
NOTE: These genes work best for cancer cells. You can use a different gene set for each category (G0, G1S, G2M, M, G1M and S).
# old method
# my.obj <- cc(my.obj, s.genes = s.phase, g2m.genes = g2m.phase)
# new method
G0 <- readLines(system.file('extdata', 'G0.txt', package = 'iCellR'))
G1S <- readLines(system.file('extdata', 'G1S.txt', package = 'iCellR'))
G2M <- readLines(system.file('extdata', 'G2M.txt', package = 'iCellR'))
M <- readLines(system.file('extdata', 'M.txt', package = 'iCellR'))
MG1 <- readLines(system.file('extdata', 'MG1.txt', package = 'iCellR'))
S <- readLines(system.file('extdata', 'S.txt', package = 'iCellR'))
# Tirosh scoring method (recomanded)
my.obj <- cell.cycle(my.obj, scoring.List = c("G0","G1S","G2M","M","MG1","S"), scoring.method = "tirosh")
# Coverage scoring method (recomanded)
# my.obj <- cell.cycle(my.obj, scoring.List = c("G0","G1S","G2M","M","MG1","S"), scoring.method = "coverage")
# plot cell cycle
A= cluster.plot(my.obj,plot.type = "pca",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T,col.by = "cc")
B= cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T, col.by = "cc")
C= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T, col.by = "cc")
D= cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T, col.by = "cc")
library(gridExtra)
grid.arrange(A,B,C,D)
## or
cluster.plot(my.obj,
cell.size = 0.5,
plot.type = "knetl",
col.by = "cc",
cell.color = "black",
back.col = "white",
cell.transparency = 1,
clust.dim = 2,
interactive = F,cond.facet = T)
# Pie
clust.stats.plot(my.obj, plot.type = "pie.cc", interactive = F, conds.to.plot = NULL)
dev.off()
# bar
clust.stats.plot(my.obj, plot.type = "bar.cc", interactive = F, conds.to.plot = NULL)
dev.off()
# or per condition
# clust.stats.plot(my.obj, plot.type = "pie.cc", interactive = F, conds.to.plot = "WT")-
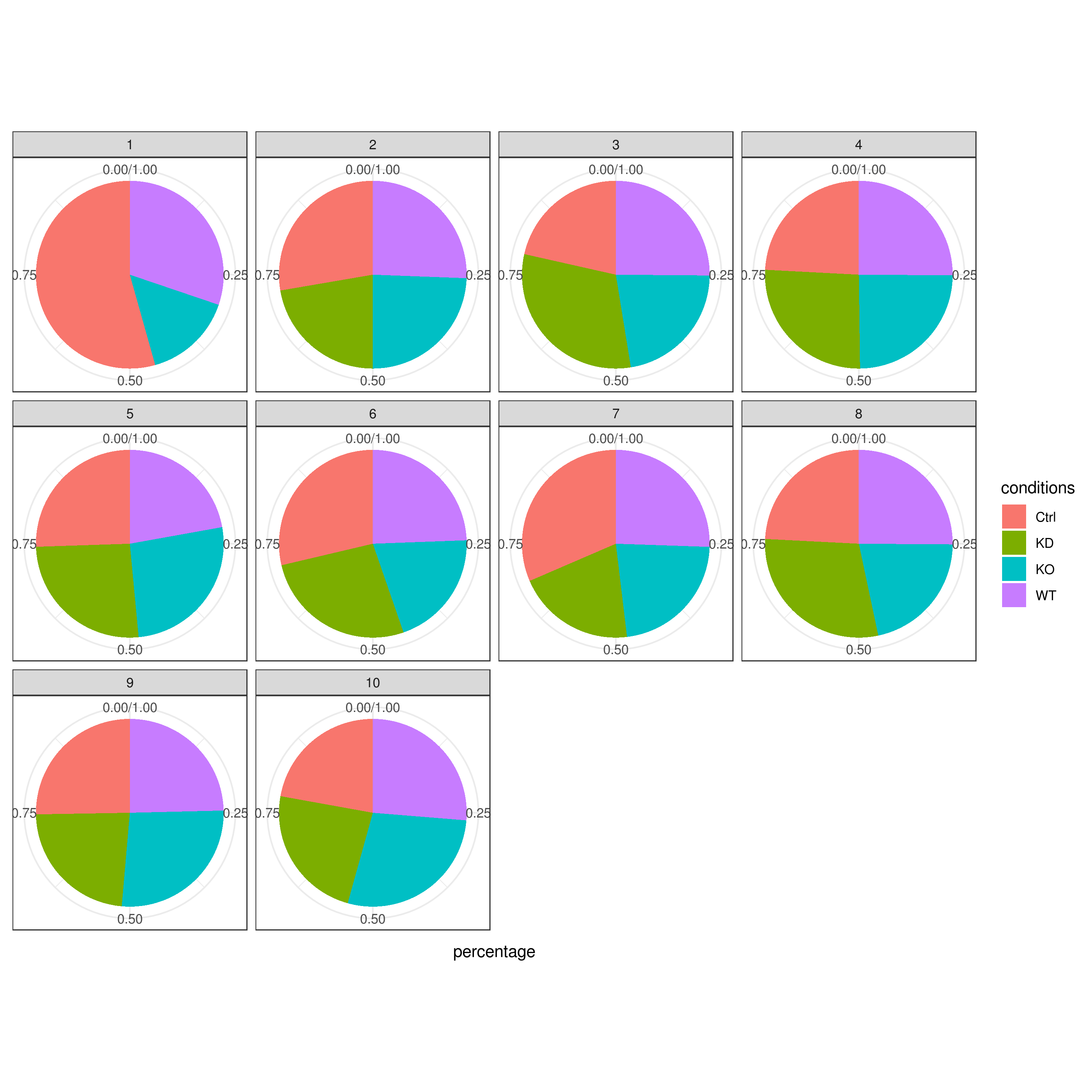
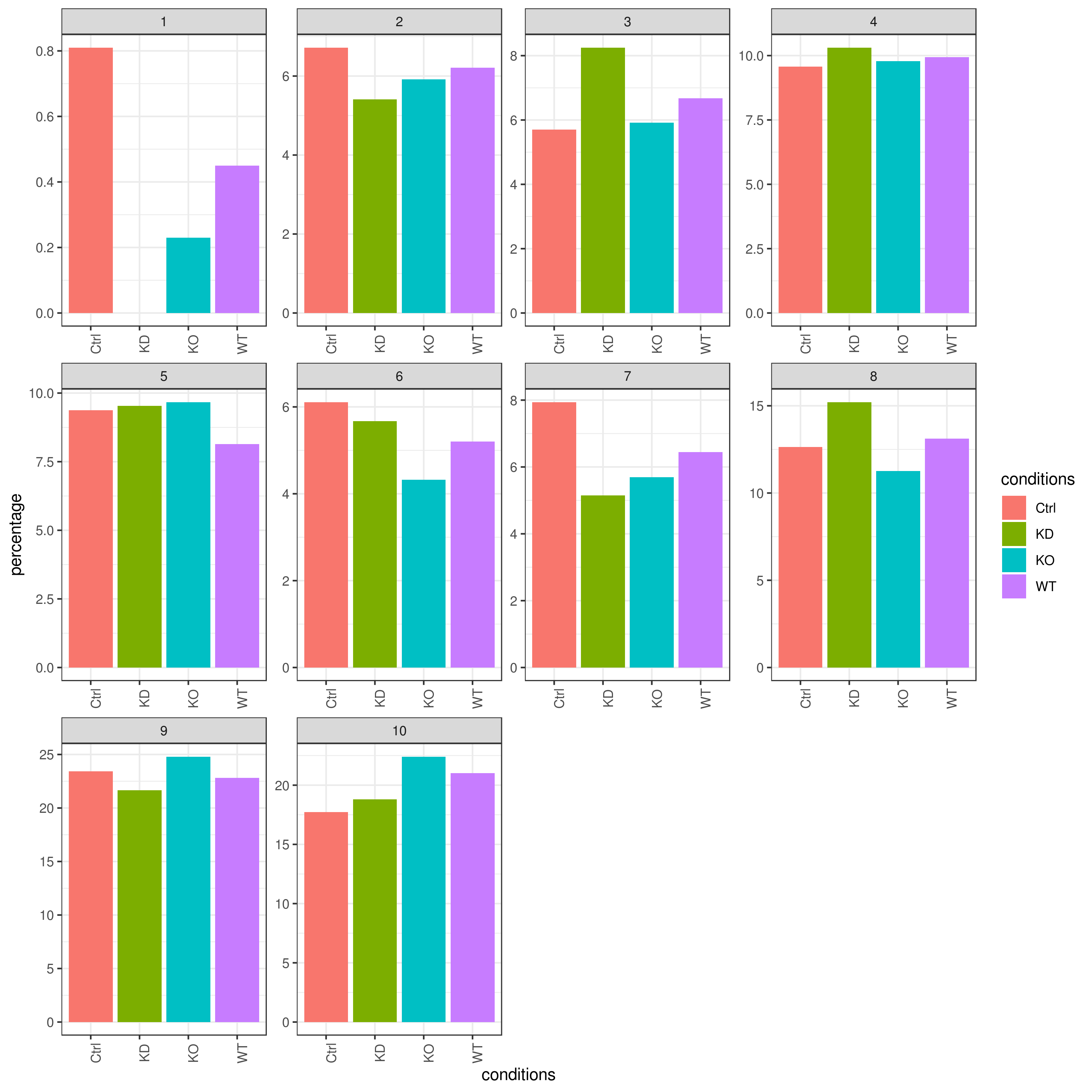
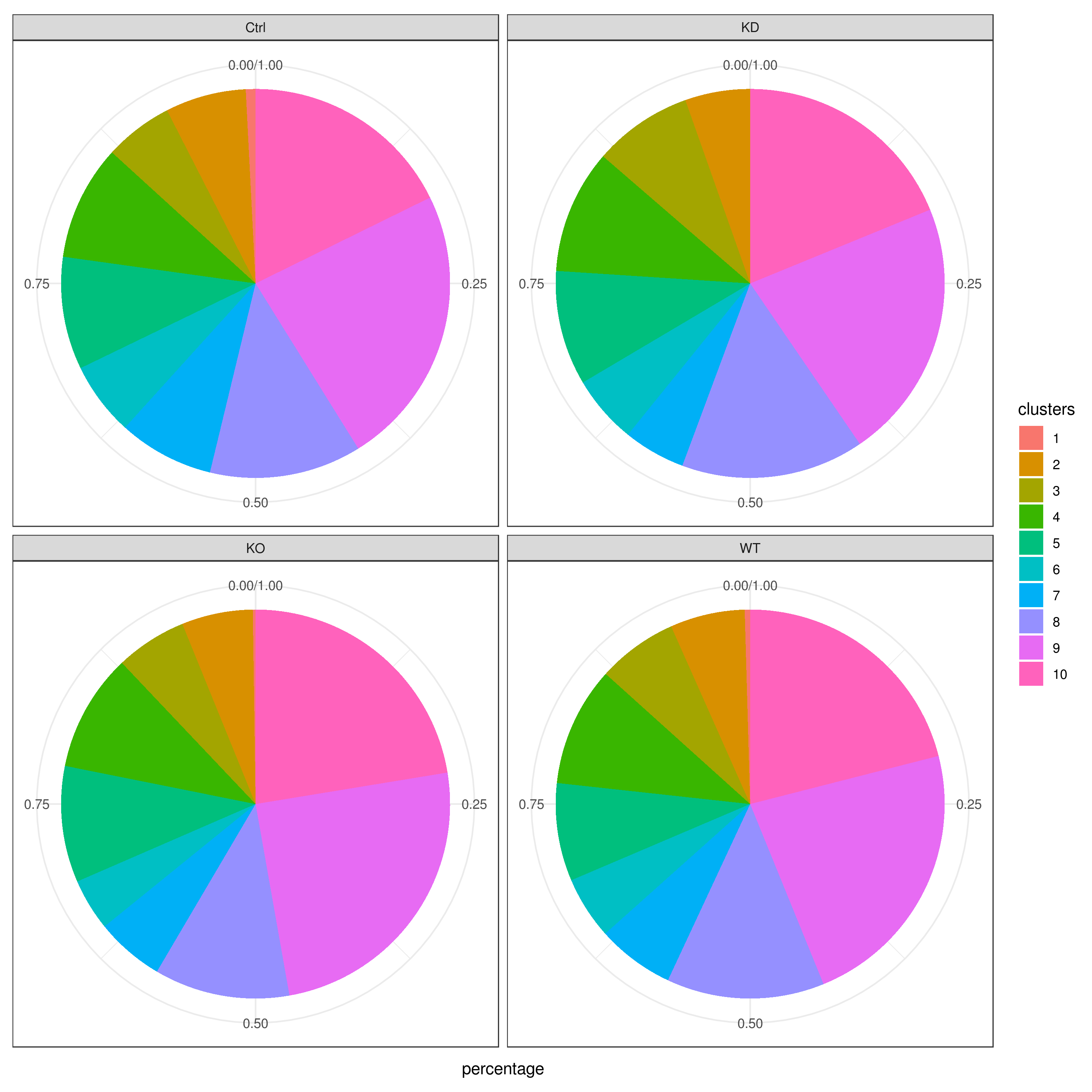
Cell frequencies refer to the number of cells present in each cluster or category, providing insights into how cells are distributed across the dataset.
-
Proportions, on the other hand, represent the relative size of each cluster or category compared to the total number of cells, expressed as a fraction or percentage.
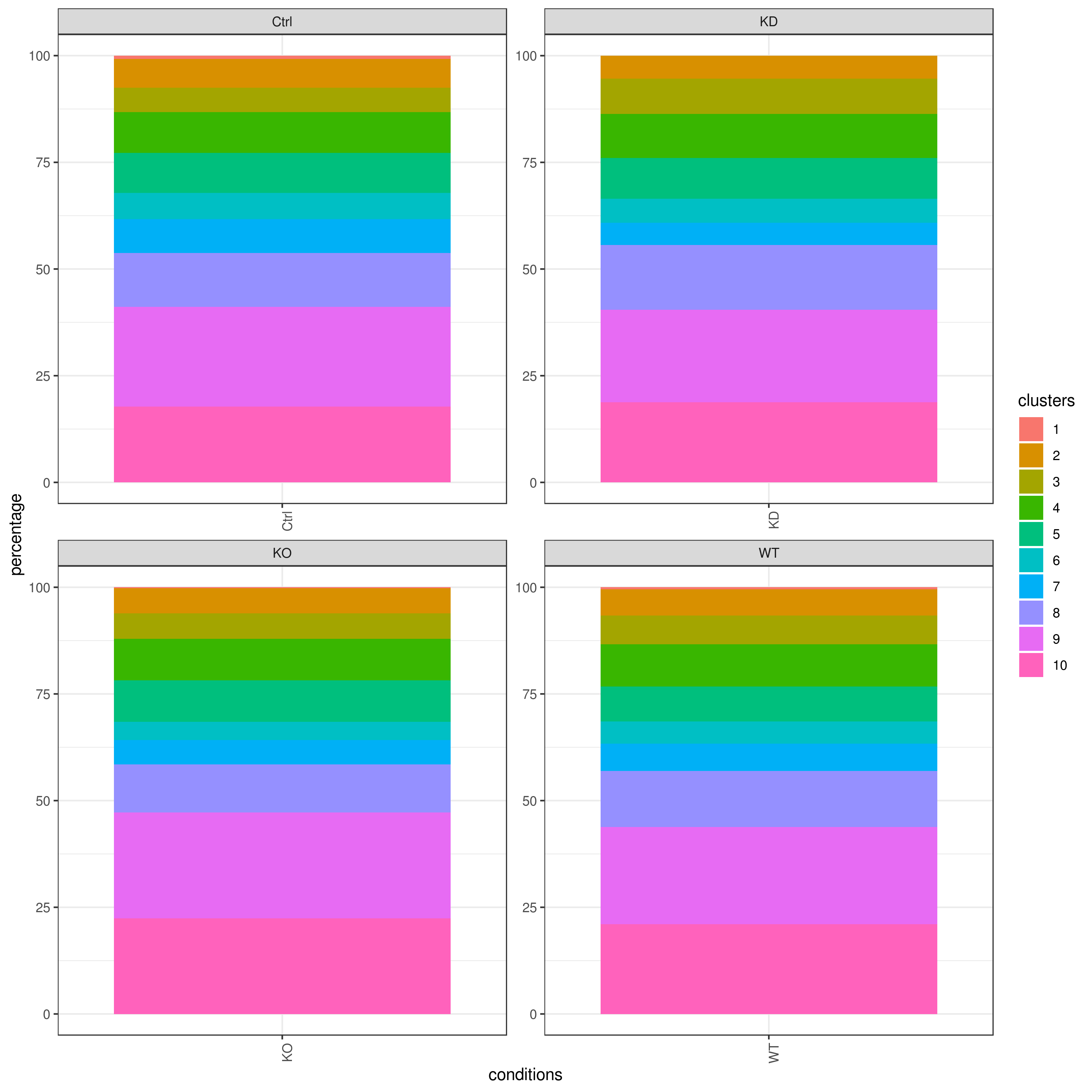
clust.cond.info(my.obj, plot.type = "pie", normalize.ncell = TRUE, my.out.put = "plot", normalize.by = "percentage")
clust.cond.info(my.obj, plot.type = "bar", normalize.ncell = TRUE,my.out.put = "plot", normalize.by = "percentage")
clust.cond.info(my.obj, plot.type = "pie.cond", normalize.ncell = T, my.out.put = "plot", normalize.by = "percentage")
clust.cond.info(my.obj, plot.type = "bar.cond", normalize.ncell = T,my.out.put = "plot", normalize.by = "percentage")
my.obj <- clust.cond.info(my.obj)
head(my.obj@my.freq)
# conditions TC SF clusters Freq Norm.Freq percentage
#1 Ctrl 491 1.265 1 4 3.162 0.81
#2 Ctrl 491 1.265 11 32 25.296 6.52
#3 Ctrl 491 1.265 8 114 90.119 23.22
#4 Ctrl 491 1.265 5 43 33.992 8.76
#5 Ctrl 491 1.265 2 33 26.087 6.72
#6 Ctrl 491 1.265 9 86 67.984 17.52clust.stats.plot(my.obj, plot.type = "box.mito", interactive = F)
clust.stats.plot(my.obj, plot.type = "box.gene", interactive = F)Data imputation is the process of inferring and filling in missing values in your dataset. This process is often used when dealing with single-cell RNA-seq data, where dropout events (zero or missing expression values) are common. Data imputation is generally not recommended to avoid introducing noise into your analysis. However, when applied correctly, proper data imputation can enhance downstream analyses such as clustering, visualization, and differential gene expression by generating a more complete and coherent dataset.
my.obj <- run.impute(my.obj, dims = 1:10, nn = 10, data.type = "pca")Saving your iCellR object allows you to preserve your analysis results and reload them later without having to rerun the pipeline.
- Option 1:
# Save the iCellR object to a file
save(my.obj, file = "my.obj.Robj")
# To load the object later
load("my.obj.Robj")
- Option 2:
# Save the iCellR object to a file
saveRDS(my.obj, file = "my_iCellR_object.rds")
# To load the object later
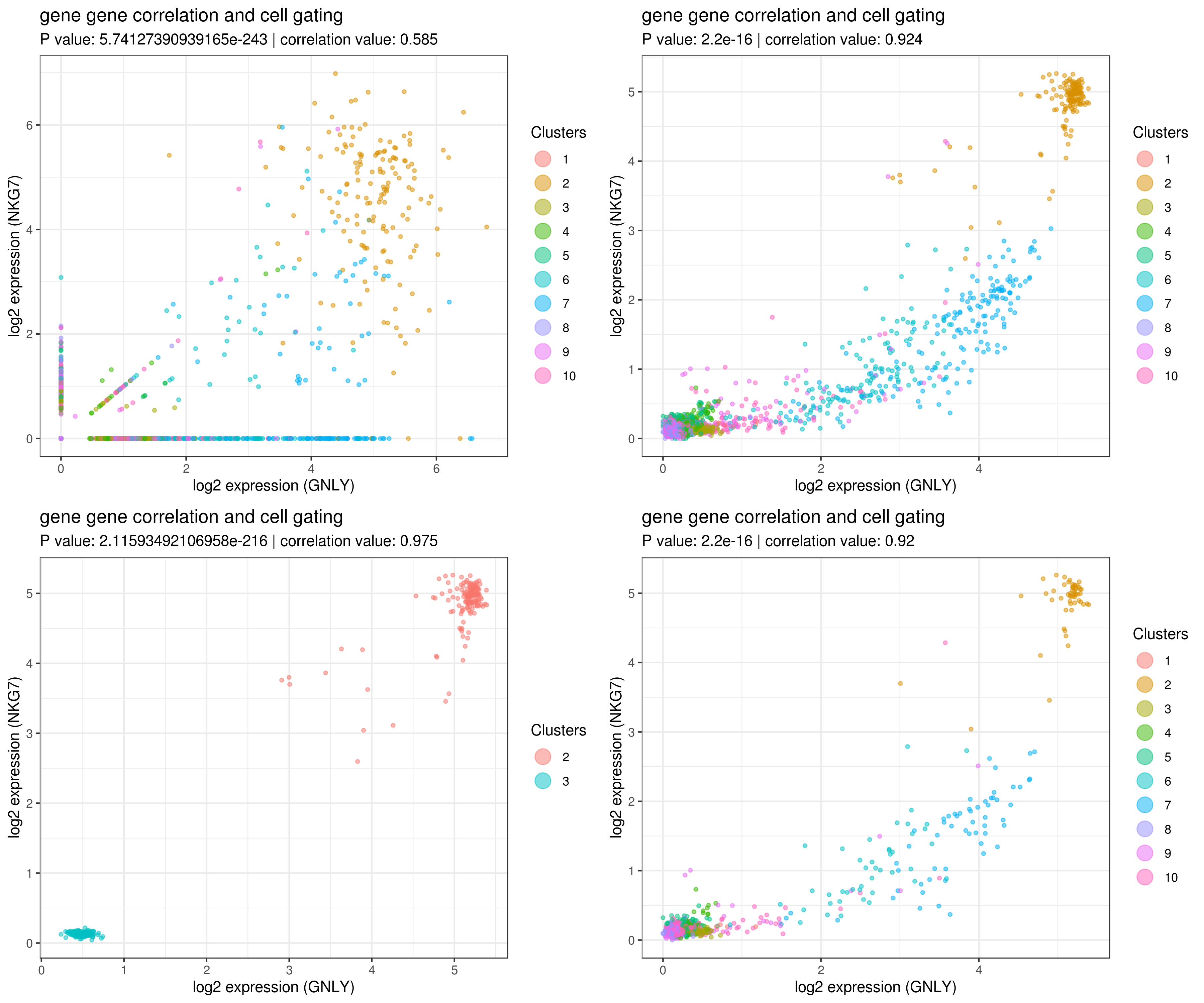
my.obj <- readRDS("my_iCellR_object.rds")Gene-gene correlation refers to the relationship or association between the expression levels of two genes across cells or samples. It helps identify patterns of co-expression, which can provide insights into cell type identification, biological pathways, regulatory networks, or functional relationships.
# impute more cells by increasing nn for better resulst.
my.obj <- run.impute(my.obj,dims = 1:10,data.type = "pca", nn = 50)
# main data
A <- gg.cor(my.obj,
interactive = F,
gene1 = "GNLY",
gene2 = "NKG7",
conds = NULL,
clusts = NULL,
data.type = "main")
# imputed data
B <- gg.cor(my.obj,
interactive = F,
gene1 = "GNLY",
gene2 = "NKG7",
conds = NULL,
clusts = NULL,
data.type = "imputed")
C <- gg.cor(my.obj,
interactive = F,
gene1 = "GNLY",
gene2 = "NKG7",
conds = NULL,
clusts = c(3,2),
data.type = "imputed")
# imputed data
D <- gg.cor(my.obj,
interactive = F,
gene1 = "GNLY",
gene2 = "NKG7",
conds = c("WT"),
clusts = NULL,
data.type = "imputed")
grid.arrange(A,B,C,D)Identifying marker genes for clusters helps define the unique biological characteristics of each cluster. Marker genes are genes whose expression is significantly enriched in a specific cluster compared to others, often showcasing distinct cell populations and functions.
marker.genes <- findMarkers(my.obj,
fold.change = 2,
padjval = 0.1)
Examin the marker genes:
dim(marker.genes)
# [1] 1070 17
head(marker.genes)
# baseMean baseSD AvExpInCluster AvExpInOtherClusters foldChange
#PPBP 0.8257760 12.144694 181.3945 0.1399852 1295.8120969
#GPX1 1.3989591 4.344717 57.4034 1.1862571 48.3903523
#CALM3 0.5469743 1.230942 10.7848 0.5080915 21.2260968
#OAZ1 4.9077851 5.979586 46.7867 4.7487311 9.8524635
#MYL6 3.0806167 3.562124 21.3690 3.0111584 7.0966045
#CD74 8.5523704 13.359205 2.6120 8.5749316 0.3046088
# log2FoldChange pval padj clusters gene cluster_1
#PPBP 10.339641 1.586683e-06 0.014786300 1 PPBP 181.3945
#GPX1 5.596648 1.107541e-07 0.001103775 1 GPX1 57.4034
#CALM3 4.407767 2.098341e-06 0.019415953 1 CALM3 10.7848
#OAZ1 3.300485 7.857814e-07 0.007464137 1 OAZ1 46.7867
#MYL6 2.827129 1.296112e-06 0.012156230 1 MYL6 21.3690
#CD74 -1.714970 9.505749e-06 0.083983296 1 CD74 2.6120
# cluster_2 cluster_3 cluster_4 cluster_5 cluster_6 cluster_7
#PPBP 0.0000000 0.1444327 0.2282912 0.0640625 0.01739706 0.1541084
#GPX1 0.2424969 1.2218772 3.9292720 4.4329583 0.25663235 0.2712831
#CALM3 0.6537205 0.8149415 0.6071034 0.5245625 0.44687500 0.5081867
#OAZ1 3.2077826 12.2072339 8.6080077 10.8738208 2.71288971 3.6402289
#MYL6 4.9660870 5.7945673 4.2813218 4.3046458 2.42854412 3.9030542
#CD74 2.9385839 8.9848538 15.7646245 5.9454250 2.19555882 3.8323072
# cluster_8 cluster_9 cluster_10
#PPBP 0.02478274 0.3668433 0.01026335
#GPX1 0.61210714 0.4635153 0.39311786
#CALM3 0.22591369 0.5210339 0.48856538
#OAZ1 3.67225595 2.3590420 2.53362063
#MYL6 1.72344048 1.6460420 2.59901289
#CD74 36.10877976 1.5638853 1.82587477
# baseMean: average expression in all the cells
# baseSD: Standard Deviation
# AvExpInCluster: average expression in cluster number (see clusters)
# AvExpInOtherClusters: average expression in all the other clusters
# foldChange: AvExpInCluster/AvExpInOtherClusters
# log2FoldChange: log2(AvExpInCluster/AvExpInOtherClusters)
# pval: P value
# padj: Adjusted P value
# clusters: marker for cluster number
# gene: marker gene for the cluster
# the rest are the average expression for each cluster# find top genes
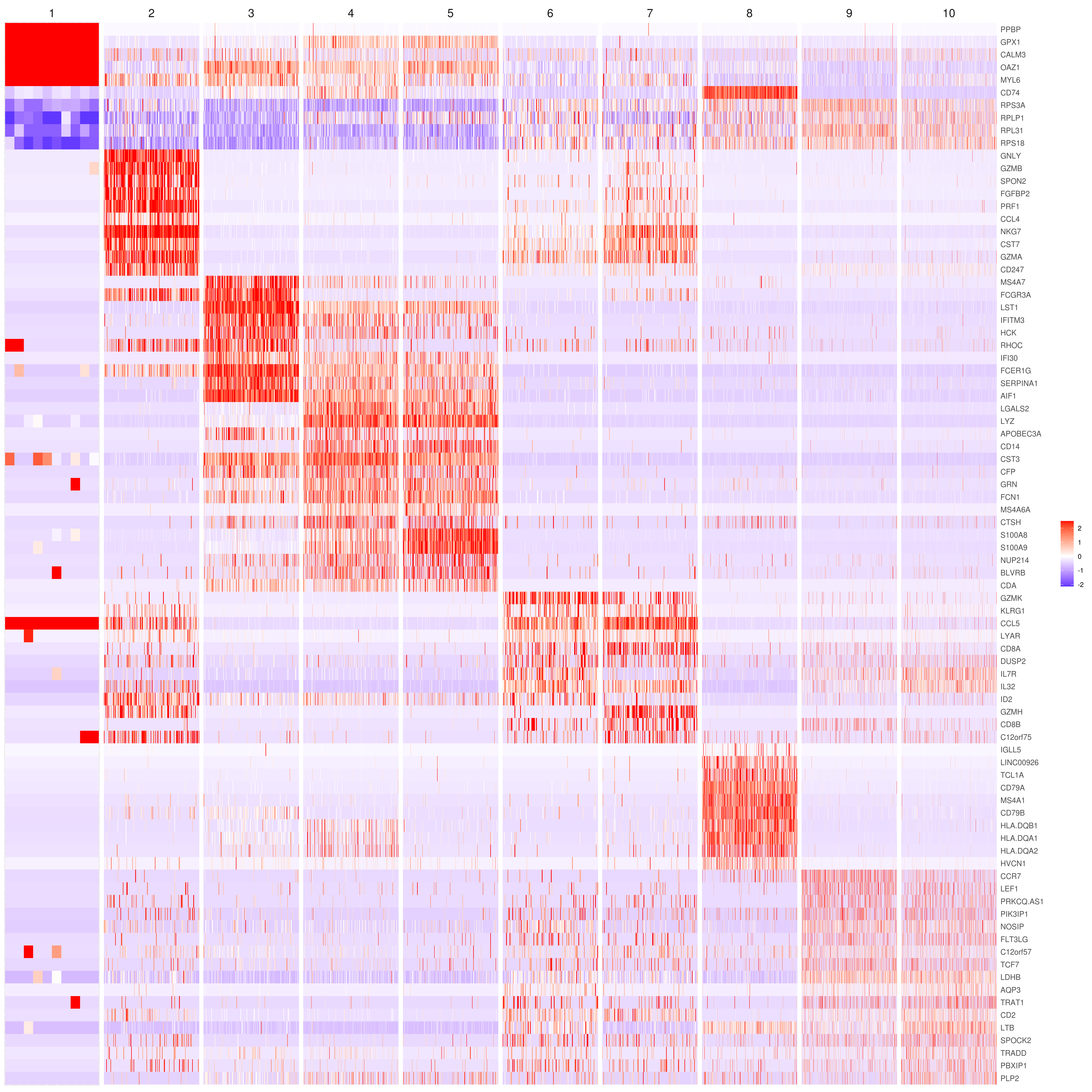
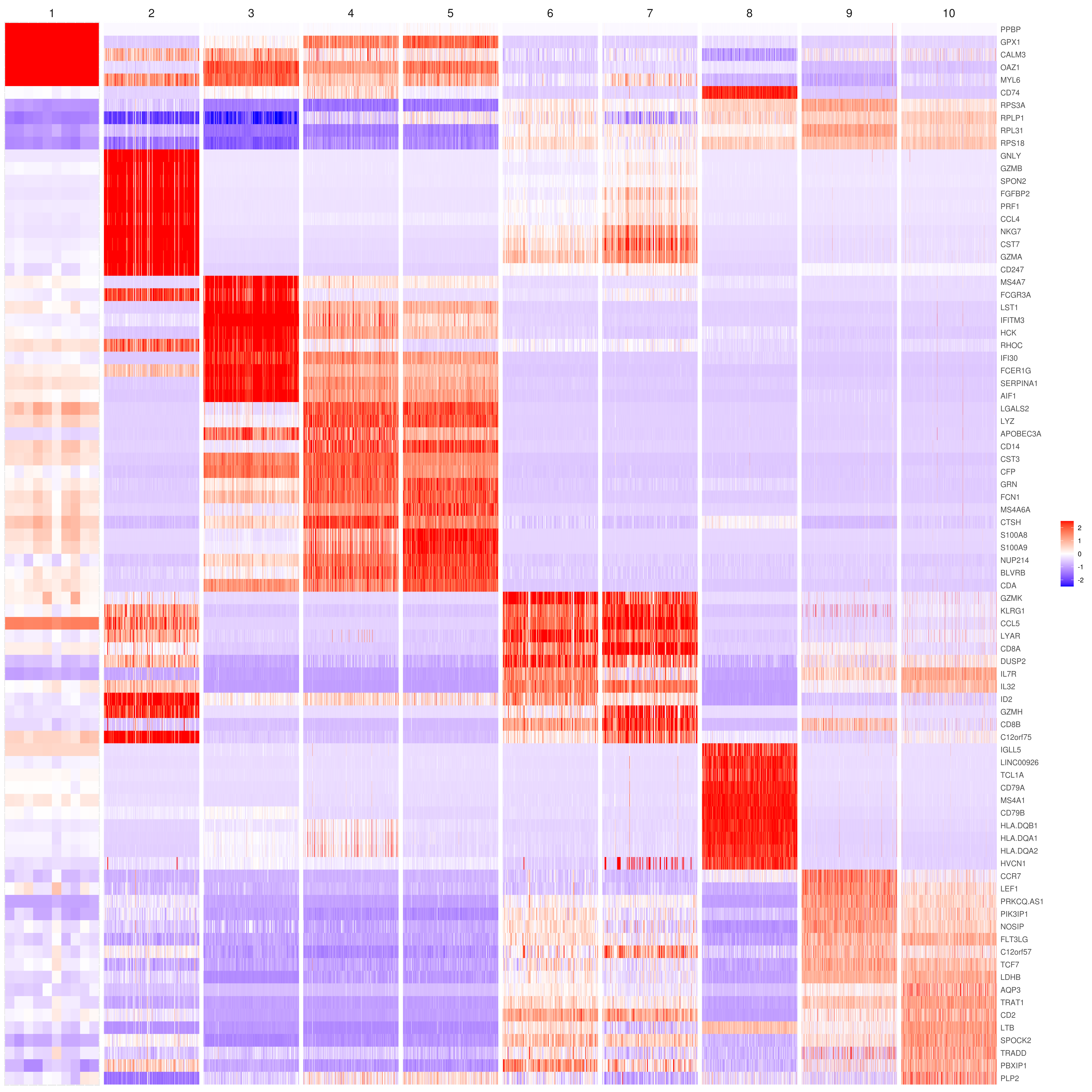
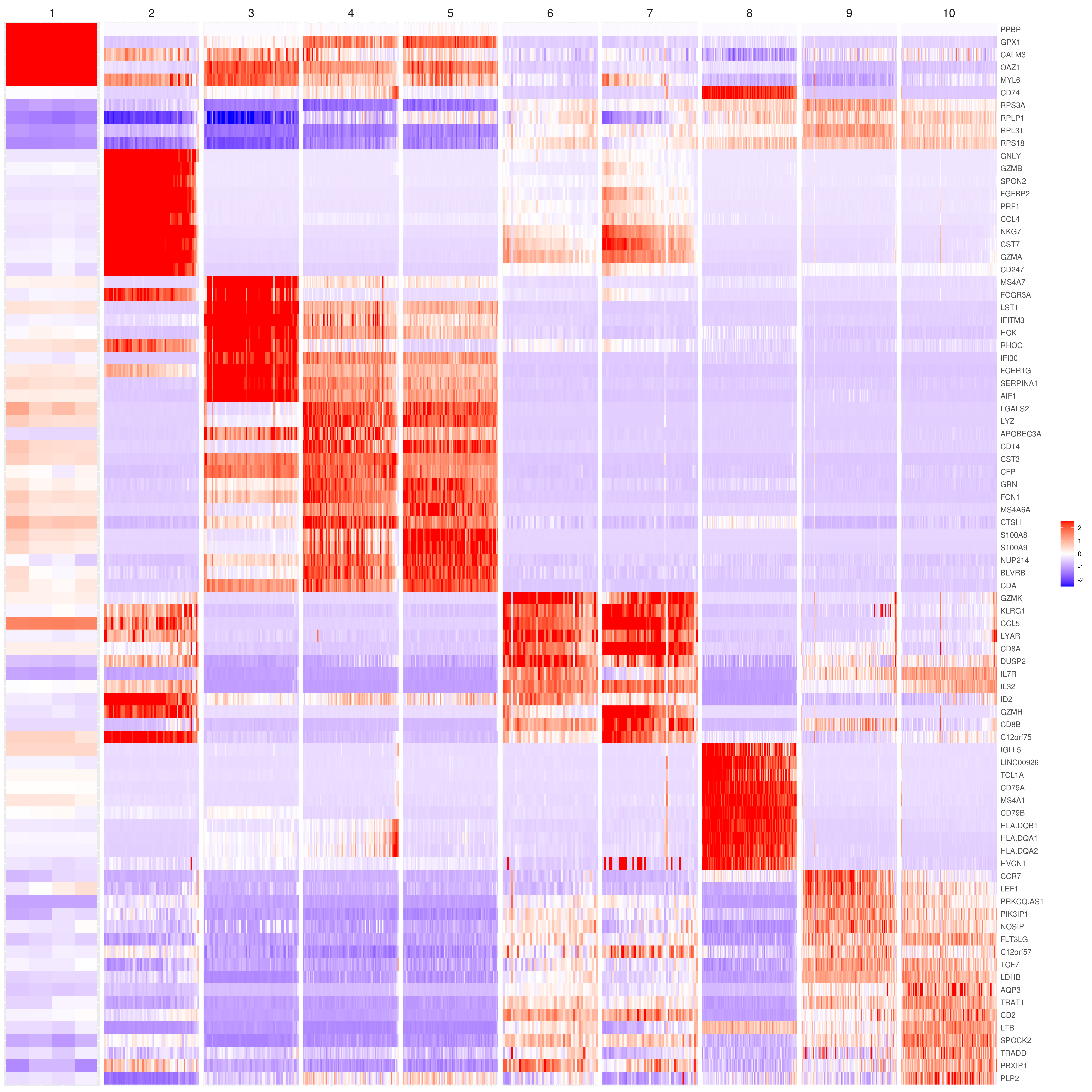
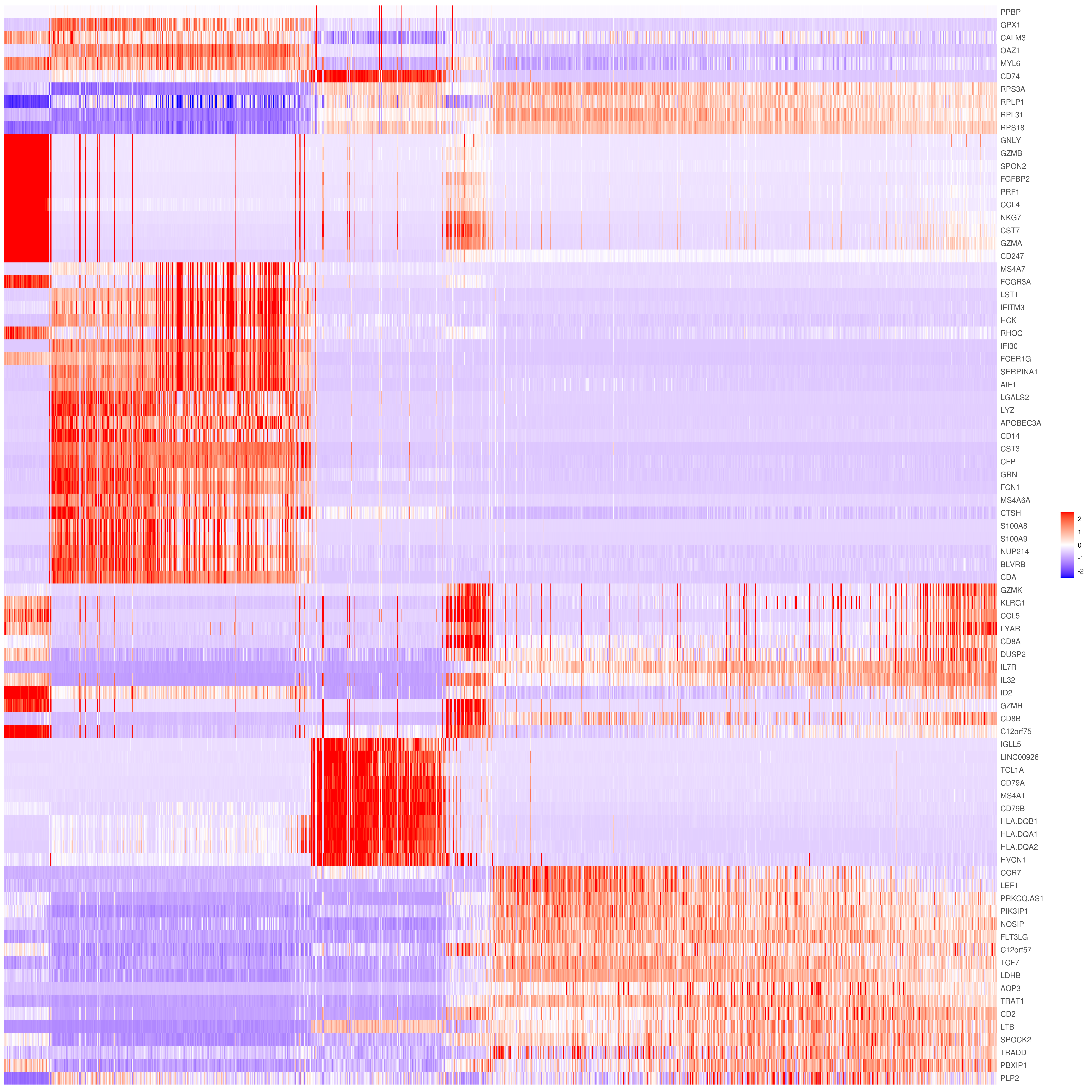
MyGenes <- top.markers(marker.genes, topde = 10, min.base.mean = 0.2,filt.ambig = F)
MyGenes <- unique(MyGenes)
# main data
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters", conds.to.plot = NULL)
# imputed data
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters", data.type = "imputed", conds.to.plot = NULL)
# sort cells and plot only one condition
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters", data.type = "imputed", cell.sort = TRUE, conds.to.plot = c("WT"))
# Pseudotime stile
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "none", data.type = "imputed", cell.sort = TRUE)
# intractive
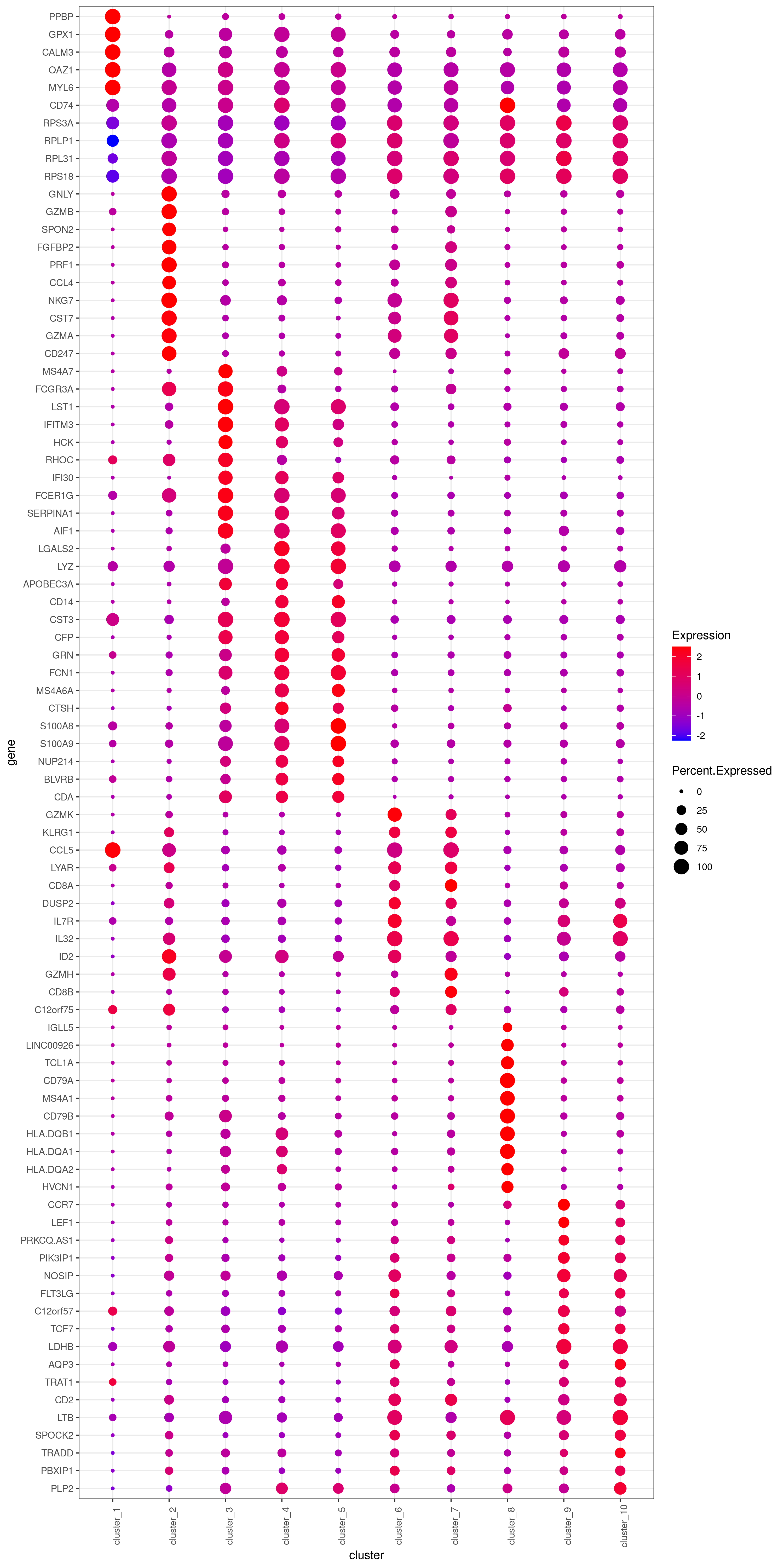
# heatmap.gg.plot(my.obj, gene = MyGenes, interactive = T, out.name = "heatmap_gg", cluster.by = "clusters")png('heatmap_bubble_gg_genes.png', width = 10, height = 20, units = 'in', res = 300)
bubble.gg.plot(my.obj, gene = MyGenes, interactive = F, conds.to.plot = NULL, size = "Percent.Expressed",colour = "Expression")
dev.off()- Plot genes
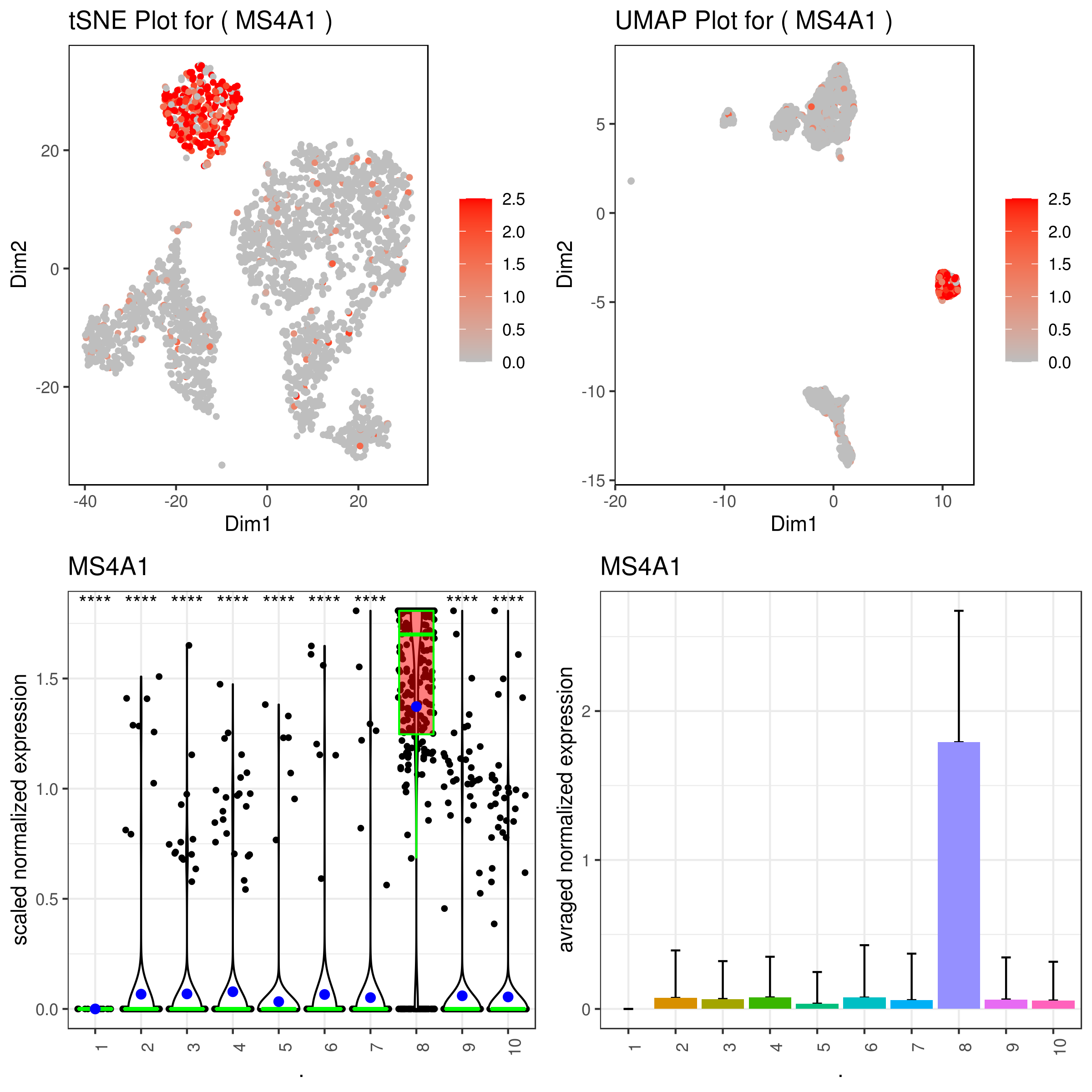
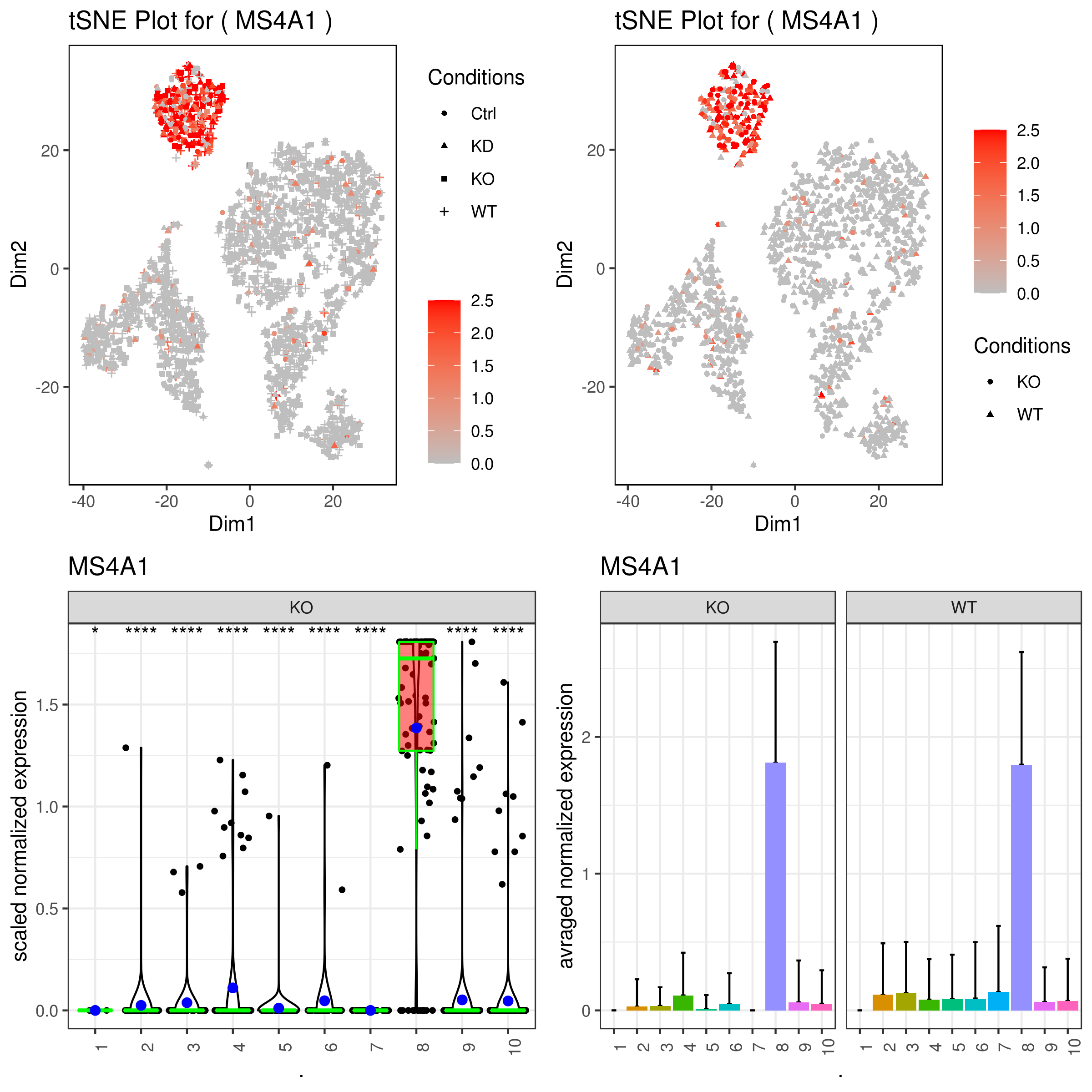
A <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "scatterplot",
interactive = F,
out.name = "scatter_plot")
# PCA 2D
B <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "scatterplot",
interactive = F,
out.name = "scatter_plot",
plot.data.type = "umap")
# Box Plot
C <- gene.plot(my.obj, gene = "MS4A1",
box.to.test = 0,
box.pval = "sig.signs",
col.by = "clusters",
plot.type = "boxplot",
interactive = F,
out.name = "box_plot")
# Bar plot (to visualize fold changes)
D <- gene.plot(my.obj, gene = "MS4A1",
col.by = "clusters",
plot.type = "barplot",
interactive = F,
out.name = "bar_plot")
library(gridExtra)
png('gene.plots.png', width = 8, height = 8, units = 'in', res = 300)
grid.arrange(A,B,C,D)
dev.off()
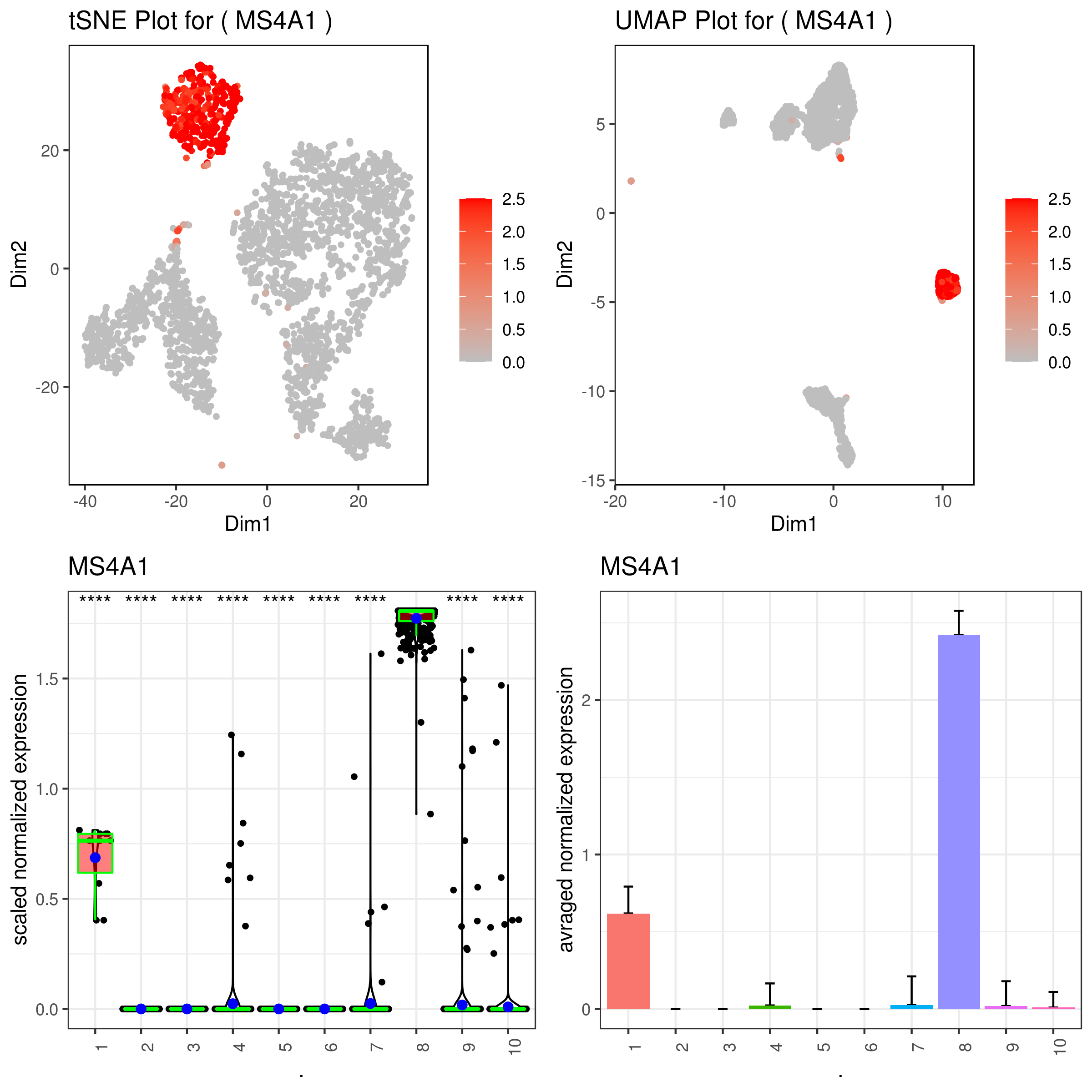
### same on imputed data
A <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "scatterplot",
interactive = F,
data.type = "imputed",
out.name = "scatter_plot")
# PCA 2D
B <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "scatterplot",
interactive = F,
out.name = "scatter_plot",
data.type = "imputed",
plot.data.type = "umap")
# Box Plot
C <- gene.plot(my.obj, gene = "MS4A1",
box.to.test = 0,
box.pval = "sig.signs",
col.by = "clusters",
plot.type = "boxplot",
interactive = F,
data.type = "imputed",
out.name = "box_plot")
# Bar plot (to visualize fold changes)
D <- gene.plot(my.obj, gene = "MS4A1",
col.by = "clusters",
plot.type = "barplot",
interactive = F,
data.type = "imputed",
out.name = "bar_plot")
library(gridExtra)
png('gene.plots_imputed.png', width = 8, height = 8, units = 'in', res = 300)
grid.arrange(A,B,C,D)
dev.off()Change the section in between #### signs for different plots (e.g. boxplot, bar, ...).
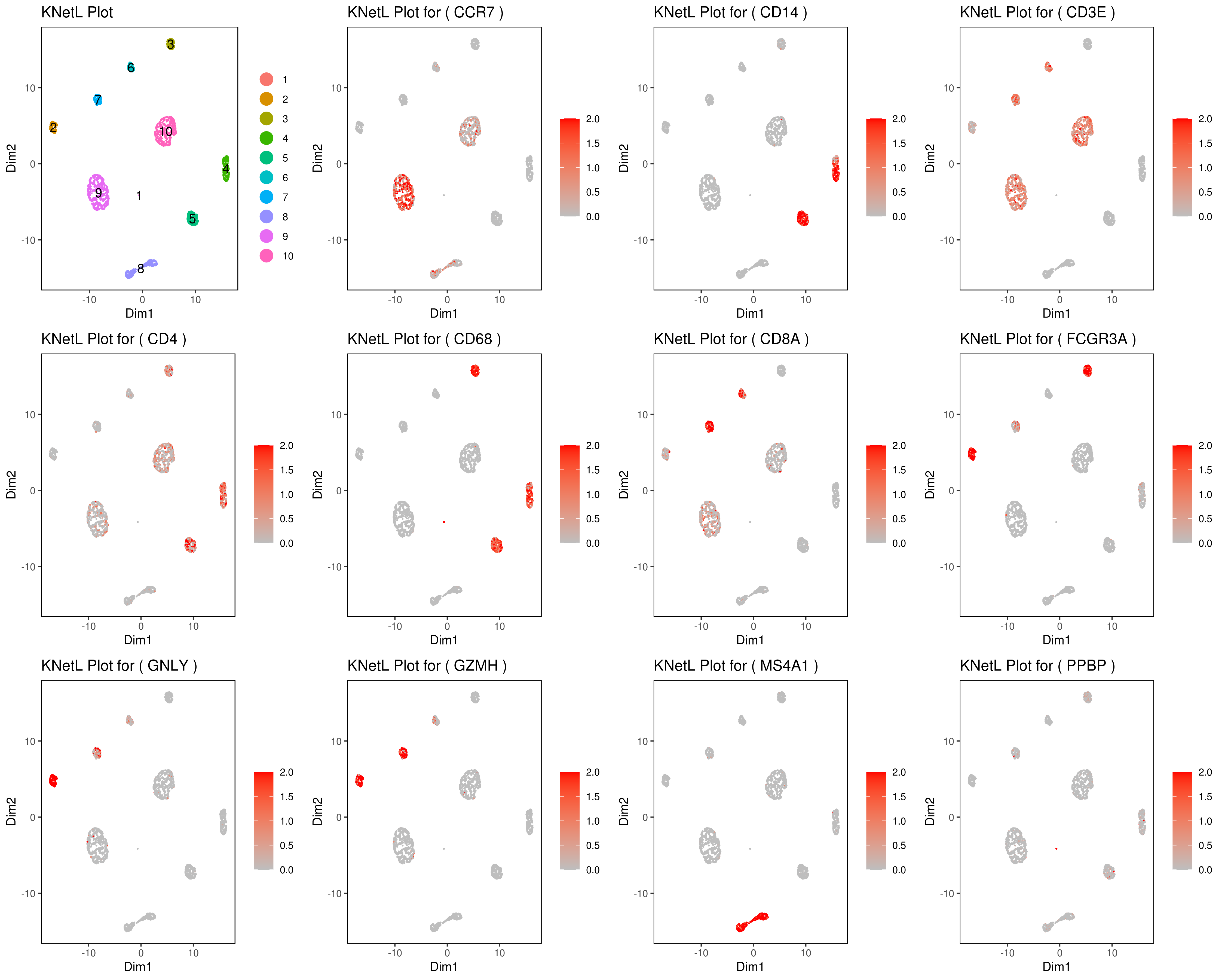
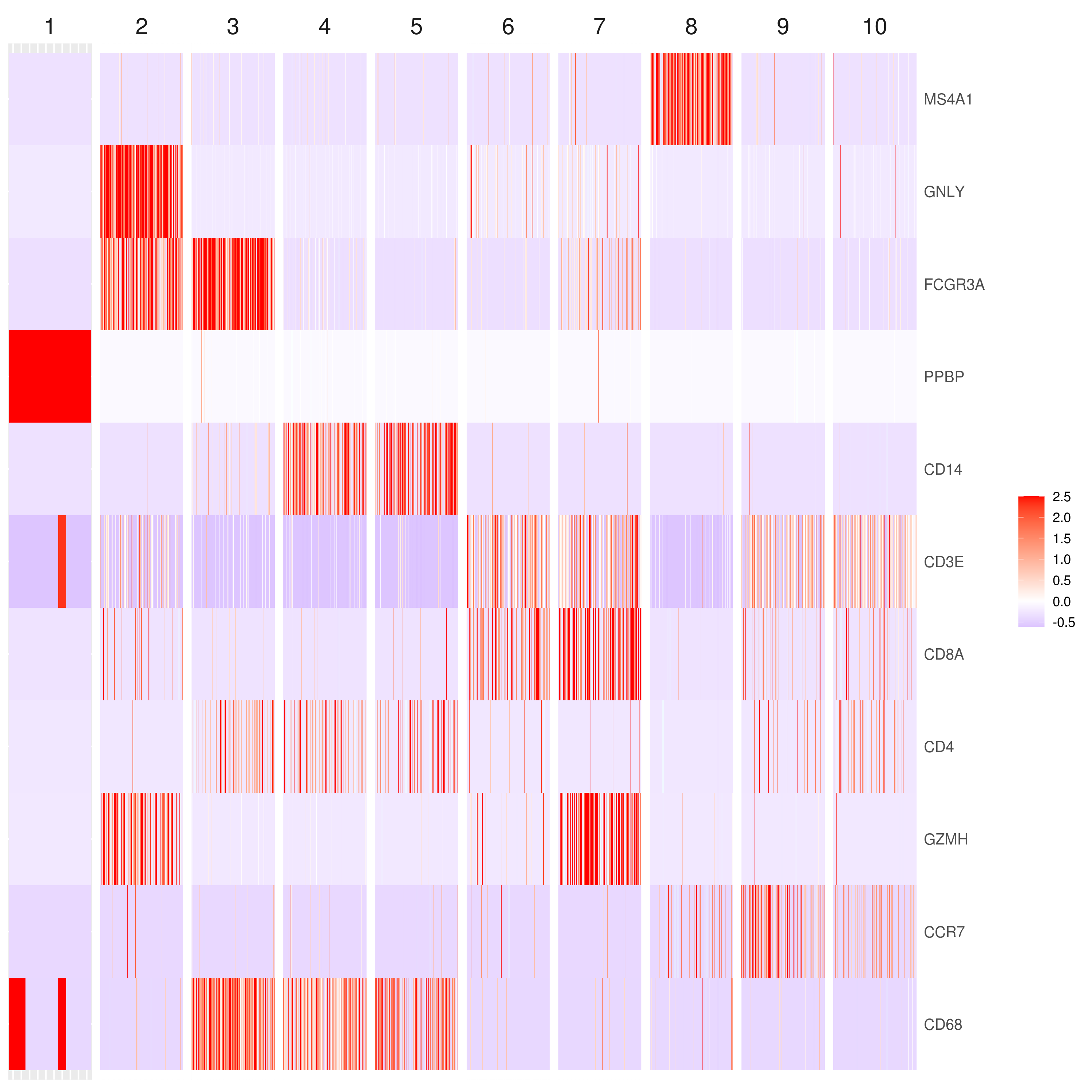
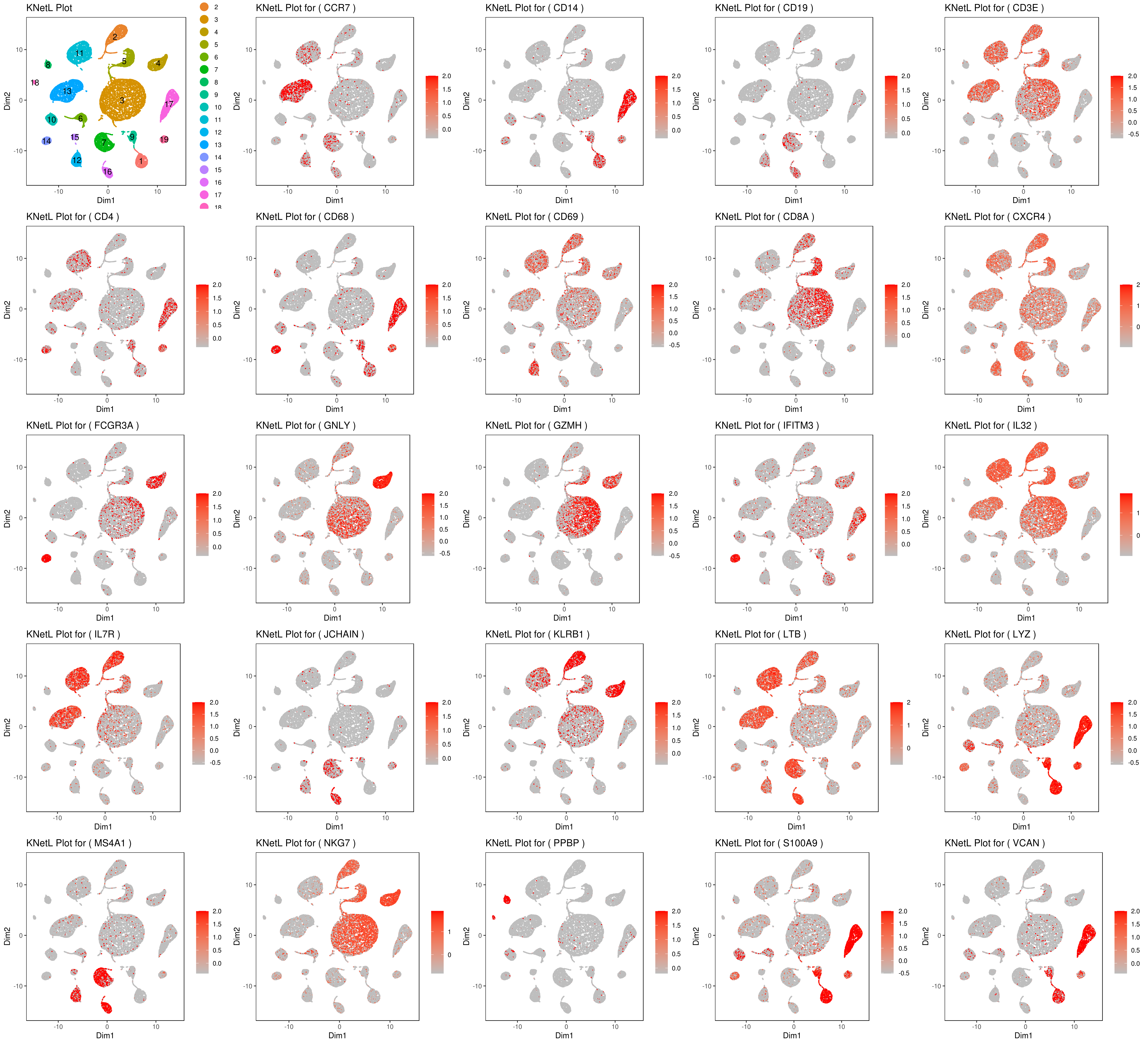
genelist = c("MS4A1","GNLY","FCGR3A","NKG7","CD14","CD3E","CD8A","CD4","GZMH","CCR7","CD68")
rm(list = ls(pattern="PL_"))
for(i in genelist){
####
MyPlot <- gene.plot(my.obj, gene = i,
interactive = F,
cell.size = 0.1,
plot.data.type = "knetl",
data.type = "main",
scaleValue = T,
min.scale = 0,max.scale = 2.0,
cell.transparency = 1)
####
NameCol=paste("PL",i,sep="_")
eval(call("<-", as.name(NameCol), MyPlot))
}
library(cowplot)
filenames <- ls(pattern="PL_")
B <- cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.1,cell.transparency = 1,anno.clust=T)
filenames <- c("B",filenames)
png('genes_KNetL.png',width = 15, height = 12, units = 'in', res = 300)
plot_grid(plotlist=mget(filenames))
dev.off()
# or heatmap
# heatmap.gg.plot(my.obj, gene = genelist, interactive = F, cluster.by = "clusters")# You can export the data using this command (one or multiple genes):
gene.plot(my.obj, gene = "MS4A1", write.data = T, scaleValue = F, data.type = "main")
# This would create a text file called "MS4A1.tsv".
head(read.table("MS4A1.tsv"))
# V1 V2 Expression Clusters Conditions
#WT_AAACATACAACCAC.1 12.499481 -11.436633 0.000000 9 WT
#WT_AAACATTGAGCTAC.1 -8.783793 24.417999 1.942233 8 WT
#WT_AAACATTGATCAGC.1 -2.650761 10.932273 0.000000 10 WT
#WT_AAACCGTGCTTCCG.1 -28.916702 -5.542731 0.000000 4 WT
#WT_AAACCGTGTATGCG.1 21.211557 -31.626822 0.000000 2 WT
#WT_AAACGCACTGGTAC.1 5.225419 -5.141192 0.000000 10 WT
# you use this to make your own plots in ggplot2 or other visualization packages. ###### Labeling the clusters
#CD3E: only in T Cells
#FCGR3A (CD16): in CD16+ monocytes and some expression NK cells
#GNLY: NK cells
#MS4A1: B cells
#GZMH: in GZMH+ T8 cells and some expression NK cells
#CD8A: in T8 cells
#CD4: in T4 and some myeloid cells
#CCR7: expressed more in memory cells
#CD14: in CD14+ monocytes
#CD68: in monocytes/MF
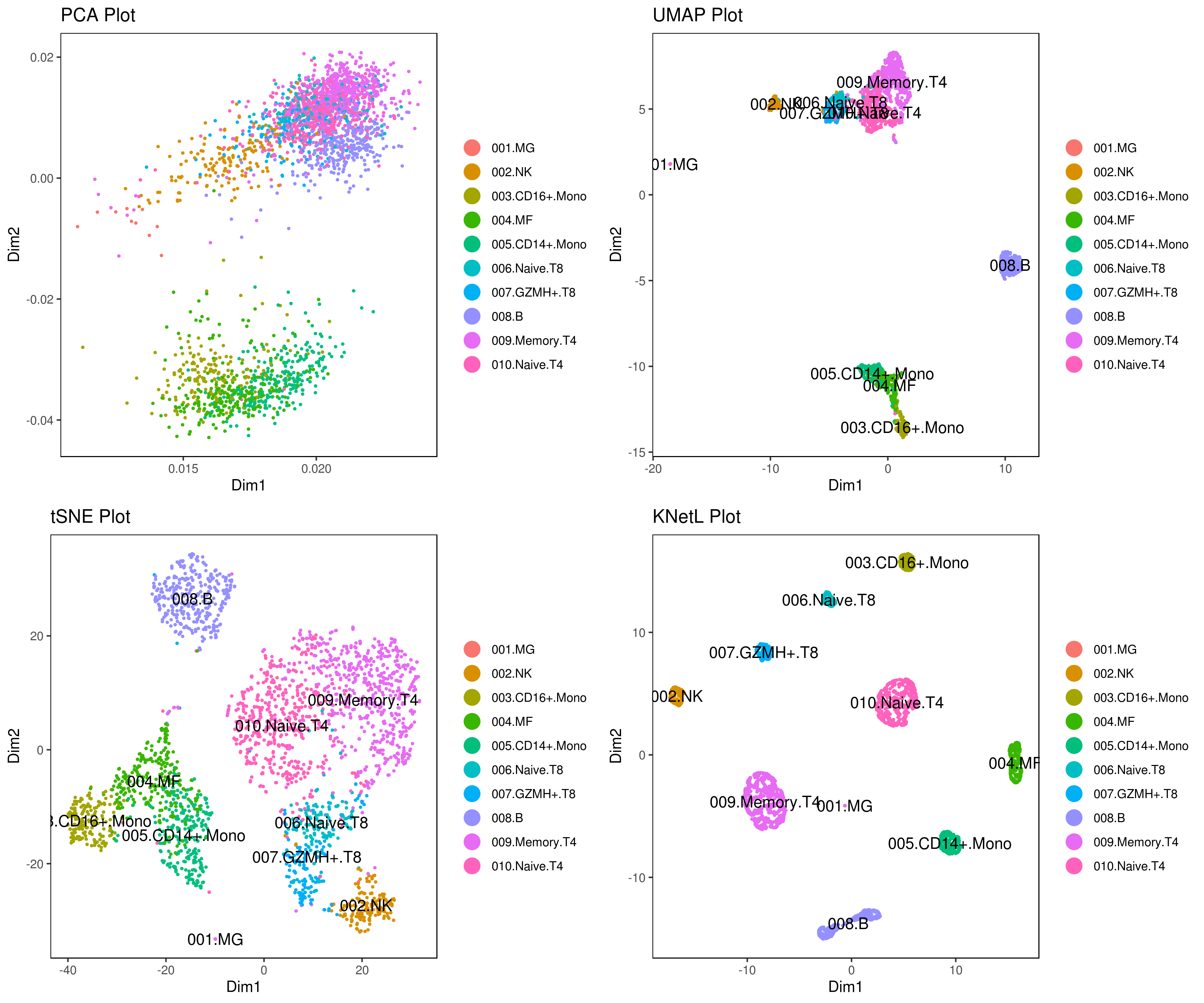
my.obj <- change.clust(my.obj, change.clust = 1, to.clust = "001.MG")
my.obj <- change.clust(my.obj, change.clust = 2, to.clust = "002.NK")
my.obj <- change.clust(my.obj, change.clust = 3, to.clust = "003.CD16+.Mono")
my.obj <- change.clust(my.obj, change.clust = 4, to.clust = "004.MF")
my.obj <- change.clust(my.obj, change.clust = 5, to.clust = "005.CD14+.Mono")
my.obj <- change.clust(my.obj, change.clust = 6, to.clust = "006.Naive.T8")
my.obj <- change.clust(my.obj, change.clust = 7, to.clust = "007.GZMH+.T8")
my.obj <- change.clust(my.obj, change.clust = 8, to.clust = "008.B")
my.obj <- change.clust(my.obj, change.clust = 9, to.clust = "009.Memory.T4")
my.obj <- change.clust(my.obj, change.clust = 10, to.clust = "010.Naive.T4")
A= cluster.plot(my.obj,plot.type = "pca",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
B= cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
C= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
D= cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=T)
grid.arrange(A,B,C,D)A <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "scatterplot",
interactive = F,
cell.transparency = 1,
scaleValue = TRUE,
min.scale = 0,
max.scale = 2.5,
back.col = "white",
cond.shape = TRUE)
B <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "scatterplot",
interactive = F,
cell.transparency = 1,
scaleValue = TRUE,
min.scale = 0,
max.scale = 2.5,
back.col = "white",
cond.shape = TRUE,
conds.to.plot = c("KO","WT"))
C <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "boxplot",
interactive = F,
back.col = "white",
cond.shape = TRUE,
conds.to.plot = c("KO"))
D <- gene.plot(my.obj, gene = "MS4A1",
plot.type = "barplot",
interactive = F,
cell.transparency = 1,
back.col = "white",
cond.shape = TRUE,
conds.to.plot = c("KO","WT"))
library(gridExtra)
grid.arrange(A,B,C,D)- Some example 2D and 3D plots and plotting clusters and conditions at the same time
# example
cluster.plot(my.obj,
cell.size = 1,
plot.type = "umap",
cell.color = "black",
back.col = "white",
col.by = "clusters",
cell.transparency = 0.5,
clust.dim = 2,
cond.shape = T,
interactive = T,
out.name = "2d_UMAP_clusters_conds")
# 2D
cluster.plot(my.obj,
cell.size = 1,
plot.type = "tsne",
cell.color = "black",
back.col = "white",
col.by = "clusters",
cell.transparency = 0.5,
clust.dim = 2,
interactive = F)
# interactive 2D
cluster.plot(my.obj,
plot.type = "tsne",
col.by = "clusters",
clust.dim = 2,
interactive = T,
out.name = "tSNE_2D_clusters")
# interactive 3D
cluster.plot(my.obj,
plot.type = "tsne",
col.by = "clusters",
clust.dim = 3,
interactive = T,
out.name = "tSNE_3D_clusters")
# Density plot for clusters
cluster.plot(my.obj,
plot.type = "pca",
col.by = "clusters",
interactive = F,
density=T)
# Density plot for conditions
cluster.plot(my.obj,
plot.type = "pca",
col.by = "conditions",
interactive = F,
density=T)
cluster.plot(my.obj,
cell.size = 1,
plot.type = "diffusion",
cell.color = "black",
back.col = "white",
col.by = "clusters",
cell.transparency = 0.5,
clust.dim = 2,
interactive = F)
cluster.plot(my.obj,
cell.size = 1,
plot.type = "diffusion",
cell.color = "black",
back.col = "white",
col.by = "clusters",
cell.transparency = 0.5,
clust.dim = 3,
interactive = F) The differential expression (DE) analysis function in iCellR provides users with flexibility to choose between various combinations of clusters and experimental conditions. This enables advanced comparisons and detailed insights into gene expression patterns across diverse biological contexts.
- Cluster vs. Cluster Comparison: Compare the gene expression profile of one cluster/clusters against another cluster/clusters.
Example: Comparing cluster 1 and 2 vs. cluster 4.
- Cluster Comparisons Within Specific Conditions: Compare clusters in one or more specific condition(s).
Example: Cluster 1 vs. Cluster 2 only in the "WT" (wild type) sample.
- Condition vs. Condition Comparison: Perform differential expression analysis between experimental conditions regardless of cluster assignment.
Example: Comparing samples labeled "WT" vs. "KO" (knockout).
- Condition Comparison Within Specific Clusters: Compare experimental conditions within specific cluster(s).
Example: Cluster 1 "WT" vs. Cluster 1 "KO".
diff.res <- run.diff.exp(my.obj, de.by = "clusters", cond.1 = c(1,4), cond.2 = c(2))
diff.res1 <- as.data.frame(diff.res)
diff.res1 <- subset(diff.res1, padj < 0.05)
head(diff.res1)
# baseMean 1_4 2 foldChange log2FoldChange pval
#AAK1 0.19554589 0.26338228 0.041792762 0.15867719 -2.655833 8.497012e-33
#ABHD14A 0.09645732 0.12708519 0.027038379 0.21275791 -2.232715 1.151865e-11
#ABHD14B 0.19132829 0.23177944 0.099644572 0.42991118 -1.217889 3.163623e-09
#ABLIM1 0.06901900 0.08749258 0.027148089 0.31029018 -1.688310 1.076382e-06
#AC013264.2 0.07383608 0.10584821 0.001279649 0.01208947 -6.370105 1.291674e-19
#AC092580.4 0.03730859 0.05112053 0.006003441 0.11743700 -3.090041 5.048838e-07
padj
#AAK1 1.294690e-28
#ABHD14A 1.708446e-07
#ABHD14B 4.636290e-05
#ABLIM1 1.540087e-02
#AC013264.2 1.950557e-15
#AC092580.4 7.254675e-03
# more examples
# Comparing a condition/conditions with different condition/conditions (e.g. WT vs KO)
diff.res <- run.diff.exp(my.obj, de.by = "conditions", cond.1 = c("WT"), cond.2 = c("KO"))
# Comparing a cluster/clusters with different cluster/clusters (e.g. cluster 1 and 2 vs. 4)
diff.res <- run.diff.exp(my.obj, de.by = "clusters", cond.1 = c(1,4), cond.2 = c(2))
# Comparing a condition/conditions with different condition/conditions only in one/more cluster/clusters (e.g. cluster 1 WT vs cluster 1 KO)
diff.res <- run.diff.exp(my.obj, de.by = "clustBase.condComp", cond.1 = c("WT"), cond.2 = c("KO"), base.cond = 1)
# Comparing a cluster/clusters with different cluster/clusters only in one/more condition/conditions (e.g. cluster 1 vs cluster 2 but only the WT sample)
diff.res <- run.diff.exp(my.obj, de.by = "condBase.clustComp", cond.1 = c(1), cond.2 = c(2), base.cond = "WT")# Volcano Plot
volcano.ma.plot(diff.res,
sig.value = "pval",
sig.line = 0.05,
plot.type = "volcano",
interactive = F)
# MA Plot
volcano.ma.plot(diff.res,
sig.value = "pval",
sig.line = 0.05,
plot.type = "ma",
interactive = F)# let's say you want to merge cluster 3 and 2.
my.obj <- change.clust(my.obj, change.clust = 3, to.clust = 2)
# to reset to the original clusters run this.
my.obj <- change.clust(my.obj, clust.reset = T)
# you can also re-name the cluster numbers to cell types. Remember to reset after this so you can ran other analysis.
my.obj <- change.clust(my.obj, change.clust = 7, to.clust = "B Cell")
# Let's say for what ever reason you want to remove acluster, to do so run this.
my.obj <- clust.rm(my.obj, clust.to.rm = 1)
# Remember that this would perminantly remove the data from all the slots in the object except frrom raw.data slot in the object. If you want to reset you need to start from the filtering cells step in the biginging of the analysis (using cell.filter function).
# To re-position the cells run tSNE again
my.obj <- run.tsne(my.obj, clust.method = "gene.model", gene.list = "my_model_genes.txt")
# Use this for plotting as you make the changes
cluster.plot(my.obj,
cell.size = 1,
plot.type = "tsne",
cell.color = "black",
back.col = "white",
col.by = "clusters",
cell.transparency = 0.5,
clust.dim = 2,
interactive = F)my.plot <- gene.plot(my.obj, gene = "GNLY",
plot.type = "scatterplot",
clust.dim = 2,
interactive = F)
cell.gating(my.obj, my.plot = my.plot, plot.type = "tsne")
# or
#my.plot <- cluster.plot(my.obj,
# cell.size = 1,
# cell.transparency = 0.5,
# clust.dim = 2,
# interactive = F)After downloading the cell ids, use the following command to rename their cluster.
my.obj <- gate.to.clust(my.obj, my.gate = "cellGating.txt", to.clust = 10)1- CPCA (iCellR)** recommended (faster than CCCA)
2- CCCA (iCellR)* recommended
3- MNN (scran wraper) optional
4- MultiCCA (Seurat wraper) optional
5- CPCA + ![]() KNetL based clustering (iCellR)*** recommended for best results!
KNetL based clustering (iCellR)*** recommended for best results!
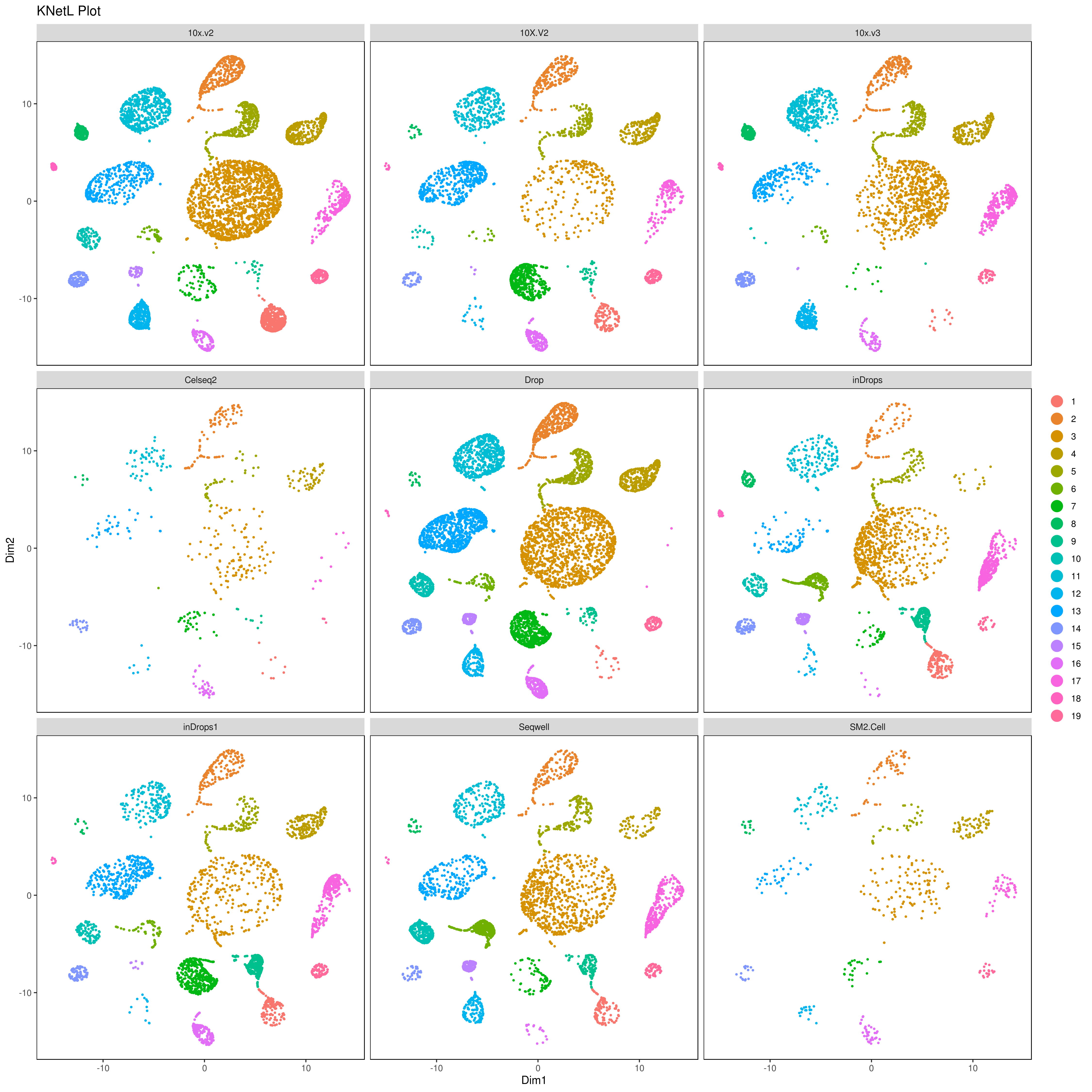
We analyzed nine PBMC sample datasets provided by the Broad Institute to detect batch differences. These datasets were generated using varying technologies, including 10x Chromium v2 (3 samples), 10x Chromium v3, CEL-Seq2, Drop-seq, inDrop, Seq-Well and SMART-Seq. For more info read: https://www.biorxiv.org/content/10.1101/2020.03.31.019109v1.full
## download an object of 9 PBMC samples
sample.file.url = "https://genome.med.nyu.edu/results/external/iCellR/data/pbmc_data/my.obj.Robj"
# download the file
download.file(url = sample.file.url,
destfile = "my.obj.Robj",
method = "auto")
### load iCellR and the object
library(iCellR)
load("my.obj.Robj")
### run PCA on top 2000 genes
my.obj <- run.pca(my.obj, top.rank = 2000)
### find best genes for second round PCA or batch alignment
my.obj <- find.dim.genes(my.obj, dims = 1:30,top.pos = 20, top.neg = 20)
length(my.obj@gene.model)
########### Batch alignment (CPCA method)
my.obj <- iba(my.obj,dims = 1:30, k = 10,ba.method = "CPCA", method = "gene.model", gene.list = my.obj@gene.model)
### impute data
my.obj <- run.impute(my.obj,dims = 1:10,data.type = "pca", nn = 10)
### tSNE and UMAP
my.obj <- run.pc.tsne(my.obj, dims = 1:10)
my.obj <- run.umap(my.obj, dims = 1:10)
### save object
save(my.obj, file = "my.obj.Robj")
### plot
library(gridExtra)
A= cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.1)
B= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.1)
C= cluster.plot(my.obj,plot.type = "umap",col.by = "conditions",interactive = F,cell.size = 0.1)
D=cluster.plot(my.obj,plot.type = "tsne",col.by = "conditions",interactive = F,cell.size = 0.1)
png('AllClusts.png', width = 12, height = 12, units = 'in', res = 300)
grid.arrange(A,B,C,D)
dev.off()
png('AllConds_clusts.png', width = 15, height = 15, units = 'in', res = 300)
cluster.plot(my.obj,
cell.size = 0.5,
plot.type = "umap",
cell.color = "black",
back.col = "white",
cell.transparency = 1,
clust.dim = 2,
interactive = F,cond.facet = T)
dev.off()
genelist = c("PPBP","LYZ","MS4A1","GNLY","FCGR3A","NKG7","CD14","S100A9","CD3E","CD8A","CD4","CD19","IL7R","FOXP3","EPCAM")
for(i in genelist){
MyPlot <- gene.plot(my.obj, gene = i,
interactive = F,
conds.to.plot = NULL,
cell.size = 0.1,
data.type = "main",
plot.data.type = "umap",
scaleValue = T,
min.scale = -2.5,max.scale = 2.0,
cell.transparency = 1)
NameCol=paste("PL",i,sep="_")
eval(call("<-", as.name(NameCol), MyPlot))
}
UMAP = cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.1, anno.size=5)
library(cowplot)
filenames <- ls(pattern="PL_")
filenames <- c("UMAP", filenames)
png('genes.png',width = 18, height = 15, units = 'in', res = 300)
plot_grid(plotlist=mget(filenames))
dev.off()
# same as above only change the option to CCCA
my.obj <- iba(my.obj,dims = 1:30, k = 10,ba.method = "CCCA", method = "gene.model", gene.list = my.obj@gene.model)# same as above only use run.mnn function instead of iba.
###### Run MNN
# This would automatically run all the samples in your experiment
library(scran)
my.obj <- run.mnn(my.obj, k=20, d=50, method = "gene.model", gene.list = my.obj@gene.model)
# detach the scran pacakge after MNN as it masks some of the functions
detach("package:scran", unload=TRUE)# same as above only use run.anchor function instead of iba.
###### Run Anchor
# This would automatically run all the samples in your experiment
library(Seurat)
my.obj <- run.anchor(my.obj,
normalization.method = "SCT",
scale.factor = 10000,
selection.method = "vst",
nfeatures = 2000,
dims = 1:20)## download an object of 9 PBMC samples
sample.file.url = "https://genome.med.nyu.edu/results/external/iCellR/example2/my.obj.Robj"
# download the file
download.file(url = sample.file.url,
destfile = "my.obj.Robj",
method = "auto")
### load iCellR and the object
library(iCellR)
load("my.obj.Robj")
### run PCA on top 2000 genes
my.obj <- run.pca(my.obj, top.rank = 2000)
### find best genes for second round PCA or batch alignment
my.obj <- find.dim.genes(my.obj, dims = 1:30,top.pos = 20, top.neg = 20)
length(my.obj@gene.model)
########### Batch alignment (CPCA method)
my.obj <- iba(my.obj,dims = 1:30, k = 10,ba.method = "CPCA", method = "gene.model", gene.list = my.obj@gene.model)
### impute data
my.obj <- run.impute(my.obj,dims = 1:10,data.type = "pca", nn = 10)
### tSNE and UMAP
my.obj <- run.pc.tsne(my.obj, dims = 1:10)
my.obj <- run.umap(my.obj, dims = 1:10)
### run KNetL
my.obj <- run.knetl(my.obj, dims = 1:20, k = 400)
### cluster based on KNetL coordinates
# The object is already clustered but here is an example:
# my.obj <- iclust(my.obj, k = 300, data.type = "knetl")
### save object
save(my.obj, file = "my.obj.Robj")
### plot 1
A= cluster.plot(my.obj,plot.type = "pca",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=F)
B= cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=F)
C= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=F)
D= cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=F)
library(gridExtra)
grid.arrange(A,B,C,D)
### plot 2
cluster.plot(my.obj,
cell.size = 0.5,
plot.type = "knetl",
cell.color = "black",
back.col = "white",
cell.transparency = 1,
clust.dim = 2,
interactive = F,cond.facet = T)
### plot 3
genelist = c("LYZ","MS4A1","GNLY","FCGR3A","NKG7","CD14","S100A9","CD3E","CD8A","CD4","CD19","KLRB1","LTB","IL7R","GZMH","CD68","CCR7","CD68","CD69","CXCR4","IFITM3","IL32","JCHAIN","VCAN","PPBP")
rm(list = ls(pattern="PL_"))
for(i in genelist){
MyPlot <- gene.plot(my.obj, gene = i,
interactive = F,
cell.size = 0.1,
plot.data.type = "knetl",
data.type = "main",
scaleValue = T,
min.scale = -2.5,max.scale = 2.0,
cell.transparency = 1)
NameCol=paste("PL",i,sep="_")
eval(call("<-", as.name(NameCol), MyPlot))
}
library(cowplot)
filenames <- ls(pattern="PL_")
B <- cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.1,cell.transparency = 1,anno.clust=T)
filenames <- c("B",filenames)
plot_grid(plotlist=mget(filenames)) MyGenes <- top.markers(marker.genes, topde = 50, min.base.mean = 0.2)
MyGenes <- unique(MyGenes)
pseudotime.tree(my.obj,
marker.genes = MyGenes,
type = "unrooted",
clust.method = "complete")
# or
pseudotime.tree(my.obj,
marker.genes = MyGenes,
type = "classic",
clust.method = "complete")
pseudotime.tree(my.obj,
marker.genes = MyGenes,
type = "jitter",
clust.method = "complete")
library(monocle)
MyMTX <- my.obj@main.data
GeneAnno <- as.data.frame(row.names(MyMTX))
colnames(GeneAnno) <- "gene_short_name"
row.names(GeneAnno) <- GeneAnno$gene_short_name
cell.cluster <- (my.obj@best.clust)
Ha <- data.frame(do.call('rbind', strsplit(as.character(row.names(cell.cluster)),'_',fixed=TRUE)))[1]
clusts <- paste("cl.",as.character(cell.cluster$clusters),sep="")
cell.cluster <- cbind(cell.cluster,Ha,clusts)
colnames(cell.cluster) <- c("Clusts","iCellR.Conds","iCellR.Clusts")
Samp <- new("AnnotatedDataFrame", data = cell.cluster)
Anno <- new("AnnotatedDataFrame", data = GeneAnno)
my.monoc.obj <- newCellDataSet(as.matrix(MyMTX),phenoData = Samp, featureData = Anno)
## find disperesedgenes
my.monoc.obj <- estimateSizeFactors(my.monoc.obj)
my.monoc.obj <- estimateDispersions(my.monoc.obj)
disp_table <- dispersionTable(my.monoc.obj)
unsup_clustering_genes <- subset(disp_table, mean_expression >= 0.1)
my.monoc.obj <- setOrderingFilter(my.monoc.obj, unsup_clustering_genes$gene_id)
# tSNE
my.monoc.obj <- reduceDimension(my.monoc.obj, max_components = 2, num_dim = 10,reduction_method = 'tSNE', verbose = T)
# cluster
my.monoc.obj <- clusterCells(my.monoc.obj, num_clusters = 10)
## plot conditions and clusters based on iCellR analysis
A <- plot_cell_clusters(my.monoc.obj, 1, 2, color = "iCellR.Conds")
B <- plot_cell_clusters(my.monoc.obj, 1, 2, color = "iCellR.Clusts")
## plot clusters based monocle analysis
C <- plot_cell_clusters(my.monoc.obj, 1, 2, color = "Cluster")
# get marker genes from iCellR analysis
MyGenes <- top.markers(marker.genes, topde = 30, min.base.mean = 0.2)
my.monoc.obj <- setOrderingFilter(my.monoc.obj, MyGenes)
my.monoc.obj <- reduceDimension(my.monoc.obj, max_components = 2,method = 'DDRTree')
# order cells
my.monoc.obj <- orderCells(my.monoc.obj)
# plot based on iCellR analysis and marker genes from iCellR
D <- plot_cell_trajectory(my.monoc.obj, color_by = "iCellR.Clusts")
## heatmap genes from iCellR
plot_pseudotime_heatmap(my.monoc.obj[MyGenes,],
cores = 1,
cluster_rows = F,
use_gene_short_name = T,
show_rownames = T)# Read an example file
# my.hto <- read.table(file = system.file('extdata', 'dense_umis.tsv', package = 'iCellR'), as.is = TRUE)
# or
my.data <- load10x("filtered_feature_bc_matrix",gene.name = 2)
# Your HTOs are usually in the end of all the gene names
# tail(row.names(my.data),5)
# [1] "TotalSeq.C0254_anti.human_Hashtag_4_Antibody"
# [2] "TotalSeq.C0255_anti.human_Hashtag_5_Antibody"
# [3] "TotalSeq.C0256_anti.human_Hashtag_6_Antibody"
# [4] "TotalSeq.C0257_anti.human_Hashtag_7_Antibody"
# [5] "TotalSeq.C0258_anti.human_Hashtag_8_Antibody"
# your HTOs are usually in the matrix and have names that are different than gene names
# Your HTO names
HTOs <- grep("^TotalSeq",row.names(my.data),value=T)
# your gene names
RNAs <- subset(row.names(my.data), !(row.names(my.data) %in% HTOs))
MyHTOs <- subset(my.data, row.names(my.data) %in% HTOs)
MyRNAs <- subset(my.data, row.names(my.data) %in% RNAs)
dim(MyHTOs)
dim(MyRNAs)
# run annotation
data <- hto.anno(hto.data = MyHTOs, cov.thr = 3, assignment.thr = 80)
data <- (cbind(ID = rownames(data),data))
write.table((data),"HTOs_annotated_HSThigh.tsv",sep="\t", row.names =F)
head(data)
# Hashtag1-GTCAACTCTTTAGCG Hashtag2-TGATGGCCTATTGGG
#TGACAACAGGGCTCTC 3 18
#AAGGAGCGTCATTAGC 7 24
#AGTGAGGAGACTGTAA 7 1761
#ATCCACCCATGTTCCC 753 20
#AAACGGGCAGGACCCT 728 24
#ATGTGTGAGTCTTGCA 4 25
# Hashtag3-TTCCGCCTCTCTTTG Hashtag4-AGTAAGTTCAGCGTA
#TGACAACAGGGCTCTC 7 0
#AAGGAGCGTCATTAGC 8 0
#AGTGAGGAGACTGTAA 5 0
#ATCCACCCATGTTCCC 3 0
#AAACGGGCAGGACCCT 3 0
#ATGTGTGAGTCTTGCA 370 0
# Hashtag5-AAGTATCGTTTCGCA Hashtag7-TGTCTTTCCTGCCAG unmapped
#TGACAACAGGGCTCTC 890 5 17
#AAGGAGCGTCATTAGC 2 3 3
#AGTGAGGAGACTGTAA 11 3 87
#ATCCACCCATGTTCCC 5 6 18
#AAACGGGCAGGACCCT 9 3 16
#ATGTGTGAGTCTTGCA 9 1011 25
# assignment.annotation percent.match coverage low.cov
#TGACAACAGGGCTCTC Hashtag5-AAGTATCGTTTCGCA 94.68085 940 FALSE
#AAGGAGCGTCATTAGC Hashtag2-TGATGGCCTATTGGG 51.06383 47 TRUE
#AGTGAGGAGACTGTAA Hashtag2-TGATGGCCTATTGGG 93.97012 1874 FALSE
#ATCCACCCATGTTCCC Hashtag1-GTCAACTCTTTAGCG 93.54037 805 FALSE
#AAACGGGCAGGACCCT Hashtag1-GTCAACTCTTTAGCG 92.97573 783 FALSE
#ATGTGTGAGTCTTGCA Hashtag7-TGTCTTTCCTGCCAG 70.01385 1444 FALSE
# assignment.threshold
#TGACAACAGGGCTCTC good.assignment
#AAGGAGCGTCATTAGC unsure
#AGTGAGGAGACTGTAA good.assignment
#ATCCACCCATGTTCCC good.assignment
#AAACGGGCAGGACCCT good.assignment
#ATGTGTGAGTCTTGCA unsure
# plot
A = ggplot(data, aes(assignment.annotation,percent.match)) +
geom_jitter(alpha = 0.25, color = "blue") +
geom_boxplot(alpha = 0.5) +
theme_bw() +
theme(axis.text.x=element_text(angle=90))
B = ggplot(data, aes(low.cov,percent.match)) +
geom_jitter(alpha = 0.25, color = "blue") +
geom_boxplot(alpha = 0.5) +
theme_bw() +
theme(axis.text.x=element_text(angle=90))
library(gridExtra)
Name="HTO_stats.png"
png(Name, width = 8, height = 8, units = 'in', res = 300)
grid.arrange(A,B,ncol=2)
dev.off()- Filtering HTOs and merging the samples
# let's see how many cells are there
dim(data)
# let's say you want to have the cells that are above 80 % likelihood of belonging to an HTO
data <- subset(data, percent.match > 80)
# let's see how many cells are left
dim(data)
# Take the HTO IDs that passed filtering
bestHTOs <- as.character(unique(data$assignment.annotation))
####################
# create new files (matrices) for each HTO (with number of cells added to the folder names)
####################
library(Matrix)
for(i in bestHTOs){
sample <- row.names(subset(data,data$assignment.annotation == i))
message(paste(" getting sample",i,"..."))
sample <- MyRNAs[ , which(names(MyRNAs) %in% sample)]
message(paste(" number of cells",dim(sample)[2]))
Name=paste("RNAs",i,dim(sample)[2],sep="_")
message(paste(" writing sample",i,"..."))
dir.create(Name)
COLs <- colnames(sample)
ROWs <- row.names(sample)
colnames(sample) <- NULL
row.names(sample) <- NULL
sparse.gbm <- Matrix(as.matrix.data.frame(sample), sparse = T )
Name1=paste(Name,"matrix.mtx",sep="/")
writeMM(obj = sparse.gbm, file=Name1)
Name1=paste(Name,"barcodes.tsv.gz",sep="/")
write.table((COLs),gzfile(Name1), row.names =FALSE, quote = FALSE, col.names = FALSE)
MY.ROWs <- cbind(ROWs,ROWs)
Name1=paste(Name,"genes.tsv.gz",sep="/")
write.table((MY.ROWs),gzfile(Name1),sep="\t", row.names =F, quote = FALSE, col.names = FALSE)
}
####################
####################
# example data aggregation for 2 samples/HTOs
my.data <- data.aggregation(samples = c("HTO1","HTO2"),
condition.names = c("HTO1","HTO2"))
# make iCellR object
my.obj <- make.obj(my.data)
# The rest is as above :)This data is from this publication (GEO number: GSE156246 and PMID: 34911733)
This is a how to guide to run i.score function in iCellR and to reproduce the above published data for G0 and non G0 cells.
Download the sample iCellR objects (used in the publication) from here: https://genome.med.nyu.edu/results/external/iCellR/i.score/ ([email protected] in these objects are log normalized)
Download sample gene signatures from here: https://genome.med.nyu.edu/results/external/iCellR/i.score/gene_signatures.tar.gz (gene signatures used in the publication are in the supplementary data of the paper)
# load sample gene signature that are in iCellR
# (these cell cycle signatures are from here: https://www.nature.com/articles/s41586-019-1884-x)
library(iCellR)
G0 <- readLines(system.file('extdata', 'G0.txt', package = 'iCellR'))
G1S <- readLines(system.file('extdata', 'G1S.txt', package = 'iCellR'))
G2M <- readLines(system.file('extdata', 'G2M.txt', package = 'iCellR'))
M <- readLines(system.file('extdata', 'M.txt', package = 'iCellR'))
MG1 <- readLines(system.file('extdata', 'MG1.txt', package = 'iCellR'))
S <- readLines(system.file('extdata', 'S.txt', package = 'iCellR'))
# load all the gene signatures
Melnick_10_GILMORE_CORE_NFKB_PATHWAY.txt <- readLines("10_GILMORE_CORE_NFKB_PATHWAY.txt")
Melnick_11_HALLMARK_MYC_TARGETS_V1.txt <- readLines("11_HALLMARK_MYC_TARGETS_V1.txt")
Melnick_12_GO_BETA_CATENIN_BINDING.txt <- readLines("12_GO_BETA_CATENIN_BINDING.txt")
Melnick_13_PID_BETA_CATENIN_NUC_PATHWAY.txt <- readLines("13_PID_BETA_CATENIN_NUC_PATHWAY.txt")
Melnick_14_PID_WNT_SIGNALING_PATHWAY.txt <- readLines("14_PID_WNT_SIGNALING_PATHWAY.txt")
Melnick_15_PID_WNT_CANONICAL_PATHWAY.txt <- readLines("15_PID_WNT_CANONICAL_PATHWAY.txt")
Melnick_16_Pribluda_SENESCENCE_INFLAMMATORY_GENES.txt <- readLines("16_Pribluda_SENESCENCE_INFLAMMATORY_GENES.txt")
Melnick_17_FRIDMAN_SENESCENCE_DN.txt <- readLines("17_FRIDMAN_SENESCENCE_DN.txt")
Melnick_18_FRIDMAN_SENESCENCE_UP.txt <- readLines("18_FRIDMAN_SENESCENCE_UP.txt")
Melnick_19_DeJONGE_LSC_TOP50_genes.txt <- readLines("19_DeJONGE_LSC_TOP50_genes.txt")
Melnick_1_AML1566_AraC_UP.txt <- readLines("1_AML1566_AraC_UP.txt")
Melnick_20_GAL_LEUKEMIC_STEM_CELL_UP.txt <- readLines("20_GAL_LEUKEMIC_STEM_CELL_UP.txt")
Melnick_21_GAL_LEUKEMIC_STEM_CELL_DN.txt <- readLines("21_GAL_LEUKEMIC_STEM_CELL_DN.txt")
Melnick_22_EPPERT_CE_HSC_LSC.txt <- readLines("22_EPPERT_CE_HSC_LSC.txt")
Melnick_23_JAATINEN_HEMATOPOIETIC_STEM_CELL_UP.txt <- readLines("23_JAATINEN_HEMATOPOIETIC_STEM_CELL_UP.txt")
Melnick_24_JAATINEN_HEMATOPOIETIC_STEM_CELL_DN.txt <- readLines("24_JAATINEN_HEMATOPOIETIC_STEM_CELL_DN.txt")
Melnick_25_INFLAMMATORY_RESPONSE.txt <- readLines("25_INFLAMMATORY_RESPONSE.txt")
Melnick_26_RAMALHO_STEMNESS_DN.txt <- readLines("26_RAMALHO_STEMNESS_DN.txt")
Melnick_27_RAMALHO_STEMNESS_UP.txt <- readLines("27_RAMALHO_STEMNESS_UP.txt")
Melnick_28_REACTOME_REGULATION_OF_MITOTIC_CELL_CYCLE.txt <- readLines("28_REACTOME_REGULATION_OF_MITOTIC_CELL_CYCLE.txt")
Melnick_2_AML1566_AraC_DN.txt <- readLines("2_AML1566_AraC_DN.txt")
Melnick_3_DUY_CISG_UP.txt <- readLines("3_DUY_CISG_UP.txt")
Melnick_4_DUY_CISG_DN.txt <- readLines("4_DUY_CISG_DN.txt")
Melnick_5_DIAPAUSE_UP_BOROVIAK.txt <- readLines("5_DIAPAUSE_UP_BOROVIAK.txt")
Melnick_6_BOROVIAK_DIAPAUSE_DN.txt <- readLines("6_BOROVIAK_DIAPAUSE_DN.txt")
Melnick_7_SASP_COPPE.txt <- readLines("7_SASP_COPPE.txt")
Melnick_8_SALDIVAR_ATR_SUPPRESSED_TARGETS.txt <- readLines("8_SALDIVAR_ATR_SUPPRESSED_TARGETS.txt")
Melnick_9_BIOCARTA_NFKB_PATHWAY.txt <- readLines("9_BIOCARTA_NFKB_PATHWAY.txt")
diapause_neg.txt <- readLines("diapause_neg.txt")
diapause_pos_and_neg.txt <- readLines("diapause_pos_and_neg.txt")
diapause_pos.txt <- readLines("diapause_pos.txt")
DTP_sig_150_Down.txt <- readLines("DTP_sig_150_Down.txt")
DTP_sig_150_up.txt <- readLines("DTP_sig_150_up.txt")
Lum_uniq_down.txt <- readLines("Lum_uniq_down.txt")
Lum_uniq_up.txt <- readLines("Lum_uniq_up.txt")
Mes_uniq_down.txt <- readLines("Mes_uniq_down.txt")
Mes_uniq_up.txt <- readLines("Mes_uniq_up.txt")
panDTP_DN.txt <- readLines("new_panDTP_DN.txt")
panDTP_up.txt <- readLines("new_panDTP_up.txt")
mes_DTP_included_DEG_DN.txt <- readLines("new_mes_DTP_included_DEG_DN.txt")
mes_DTP_included_DEG_UP.txt <- readLines("new_mes_DTP_included_DEG_UP.txt")
lum_DTP_included_DEG_DN.txt <- readLines("new_lum_DTP_included_DEG_DN.txt")
lum_DTP_included_DEG_UP.txt <- readLines("new_lum_DTP_included_DEG_UP.txt")
lum_DTP_specific_UP_noCC.txt <- readLines("new_lum_DTP_specific_UP_noCC_.txt")
mes_DTP_specific_UP_noCC.txt <- readLines("new_mes_DTP_specific_UP_noCC_.txt")Group all the signatures in one character object:
All <- c("Melnick_10_GILMORE_CORE_NFKB_PATHWAY.txt","Melnick_11_HALLMARK_MYC_TARGETS_V1.txt","Melnick_12_GO_BETA_CATENIN_BINDING.txt","Melnick_13_PID_BETA_CATENIN_NUC_PATHWAY.txt","Melnick_14_PID_WNT_SIGNALING_PATHWAY.txt","Melnick_15_PID_WNT_CANONICAL_PATHWAY.txt","Melnick_16_Pribluda_SENESCENCE_INFLAMMATORY_GENES.txt","Melnick_17_FRIDMAN_SENESCENCE_DN.txt","Melnick_18_FRIDMAN_SENESCENCE_UP.txt","Melnick_19_DeJONGE_LSC_TOP50_genes.txt","Melnick_1_AML1566_AraC_UP.txt","Melnick_20_GAL_LEUKEMIC_STEM_CELL_UP.txt","Melnick_21_GAL_LEUKEMIC_STEM_CELL_DN.txt","Melnick_22_EPPERT_CE_HSC_LSC.txt","Melnick_23_JAATINEN_HEMATOPOIETIC_STEM_CELL_UP.txt","Melnick_24_JAATINEN_HEMATOPOIETIC_STEM_CELL_DN.txt","Melnick_25_INFLAMMATORY_RESPONSE.txt","Melnick_26_RAMALHO_STEMNESS_DN.txt","Melnick_27_RAMALHO_STEMNESS_UP.txt","Melnick_28_REACTOME_REGULATION_OF_MITOTIC_CELL_CYCLE.txt","Melnick_2_AML1566_AraC_DN.txt","Melnick_3_DUY_CISG_UP.txt","Melnick_4_DUY_CISG_DN.txt","Melnick_5_DIAPAUSE_UP_BOROVIAK.txt","Melnick_6_BOROVIAK_DIAPAUSE_DN.txt","Melnick_7_SASP_COPPE.txt","Melnick_8_SALDIVAR_ATR_SUPPRESSED_TARGETS.txt","Melnick_9_BIOCARTA_NFKB_PATHWAY.txt","diapause_neg.txt","diapause_pos_and_neg.txt","diapause_pos.txt","DTP_sig_150_Down.txt","DTP_sig_150_up.txt","Lum_uniq_down.txt","Lum_uniq_up.txt","Mes_uniq_down.txt","Mes_uniq_up.txt","G0","G1S","G2M","M","MG1","S","panDTP_DN.txt","panDTP_up.txt","mes_DTP_included_DEG_DN.txt","mes_DTP_included_DEG_UP.txt","lum_DTP_included_DEG_DN.txt","lum_DTP_included_DEG_UP.txt","lum_DTP_specific_UP_noCC.txt","mes_DTP_specific_UP_noCC.txt")Load your sample iCellR object
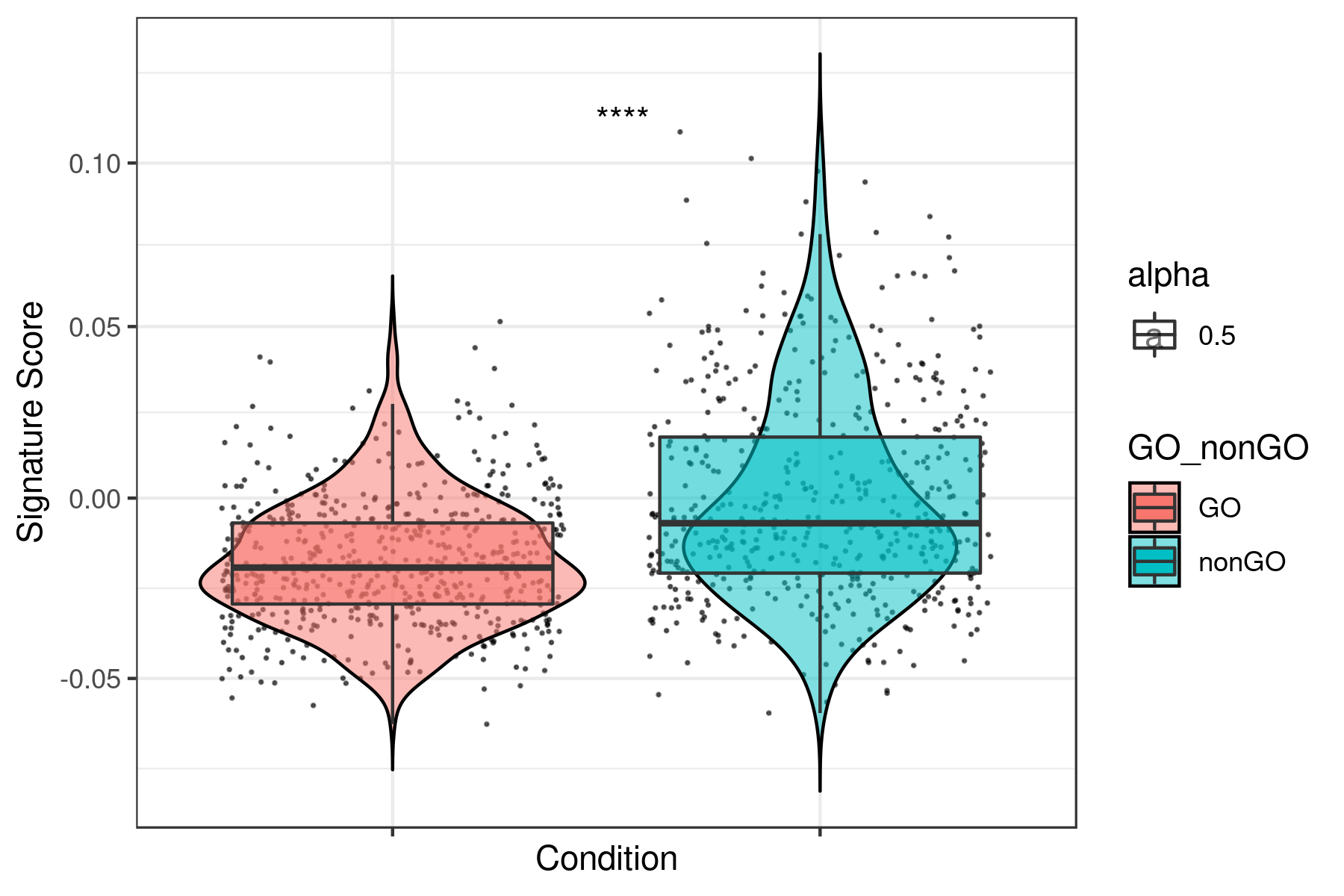
load("BT474_DTP.Robj")Score for cell cycle gene signatures with any of the following scoring methods: tirosh, mean, sum, gsva, ssgsea, zscore and plage. (tirosh and zscore methods are recommended to perform best)
dat1 <- i.score(my.obj, scoring.List = c("G0","G1S","G2M","M","MG1","S") ,scoring.method = "tirosh",return.stats = TRUE, data.type = "raw.data")
write.table(dat1,"tirosh_G0.tsv",sep="\t")Score for all the other signatures (tirosh, mean, sum, gsva, ssgsea, zscore and plage)
dat2 <- i.score(my.obj, scoring.List = All ,scoring.method = "tirosh",return.stats = TRUE, data.type = "raw.data")
write.table(dat2,"tirosh_all.tsv",sep="\t")Prepare data to plot (marge dat1 and dat2)
dir.create("boxplots_tirosh")
setwd("boxplots_tirosh")
data <- read.table("../tirosh_all.tsv",sep="\t",header=T)
dataCC <- read.table("../tirosh_G0.tsv",sep="\t",header=T)
df = as.character(dataCC$assignment.annotation) == "G0"
df[ df == "TRUE" ] <- "GO"
df[ df == "FALSE" ] <- "nonGO"
data <- cbind(cond = rep("sample",length(df)),
ID = rownames(data),
assignment.annotation = dataCC$assignment.annotation,
GO_nonGO = df,
data)
write.table((data),file="data.xls",sep="\t", row.names =F)Plot all the signatures individually:
data <- read.table("data.xls",sep="\t",header=T)
g <- head(data)[5:55]
g <- colnames(g)
library(ggpubr)
for(i in g){
name <- paste("boxplot_",i,".png",sep="")
png(name,width = 6, height = 4, units = 'in', res = 300)
print(ggplot(data, aes(x= GO_nonGO,y=data[, i],fill = GO_nonGO, alpha = 0.5)) +
geom_jitter(size = 0.2, color="black") +
geom_violin(trim=FALSE, col = "black", alpha = 0.5) +
geom_boxplot(outlier.color = NA) +
theme_bw() +
xlab("Condition") +
ylab("Signature Score") +
scale_y_continuous(trans = "log1p") +
stat_compare_means(aes(group = GO_nonGO), label = "p.signif", label.x = 1.5) +
theme(axis.text.x = element_blank()))
dev.off()
}Example for "lum_DTP_included_DEG_DN.txt"
To see all the plots made as above go to this link: https://genome.med.nyu.edu/results/external/iCellR/i.score/test/boxplots_tirosh/
- Download test samples
sample.file.url = "https://genome.med.nyu.edu/results/external/iCellR/data/CITE-Seq_sample_RNA.tsv.gz"
# download RNA file
download.file(url = sample.file.url,
destfile = "CITE-Seq_sample_RNA.tsv.gz",
method = "auto")
sample.file.url = "https://genome.med.nyu.edu/results/external/iCellR/data/CITE-Seq_sample_ADT.tsv.gz"
# download ADT file
download.file(url = sample.file.url,
destfile = "CITE-Seq_sample_ADT.tsv.gz",
method = "auto") - Read the files and make your object
# Read RNA file
rna.data <- read.delim("CITE-Seq_sample_RNA.tsv.gz",header=TRUE)
# see the head
head(rna.data)[1:3]
# CTGTTTACACCGCTAG CTCTACGGTGTGGCTC AGCAGCCAGGCTCATT
#A1BG 0 0 0
#A1BG-AS1 0 0 0
#A1CF 0 0 0
#A2M 0 0 0
#A2M-AS1 0 0 0
#A2ML1 0 0 0
# Read ADT file
adt.data <- read.delim("CITE-Seq_sample_ADT.tsv.gz",header=TRUE)
# see the head
head(adt.data)[1:3]
# CTGTTTACACCGCTAG CTCTACGGTGTGGCTC AGCAGCCAGGCTCATT
#CD3 60 52 89
#CD4 72 49 112
#CD8 76 59 61
#CD45RA 575 3943 682
#CD56 64 68 87
#CD16 161 107 117
# if you had multiple sample use the data.aggregation function for both RNA and ADT data.
# make iCellR object
my.obj <- make.obj(rna.data)
# check object
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.1.4
Raw/original data dimentions (rows,columns): 20501,8617
Data conditions: no conditions/single sample
Row names: A1BG,A1BG-AS1,A1CF ...
Columns names: CTGTTTACACCGCTAG,CTCTACGGTGTGGCTC,AGCAGCCAGGCTCATT ...
###################################
QC stats performed:FALSE, PCA performed:FALSE, CCA performed:FALSE
Clustering performed:FALSE, Number of clusters:0
tSNE performed:FALSE, UMAP performed:FALSE, DiffMap performed:FALSE
Main data dimentions (rows,columns):0,0
Normalization factors:,...
Imputed data dimentions (rows,columns):0,0
############## scVDJ-Seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-Seq ############
ADT raw data dimentions (rows,columns):0,0
ADT main data dimentions (rows,columns):0,0
ADT columns names:...
ADT row names:...
########### iCellR object ##########- add ADT data
my.obj <- add.adt(my.obj, adt.data = adt.data)
# check too see
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.1.4
Raw/original data dimentions (rows,columns): 20501,8617
Data conditions: no conditions/single sample
Row names: A1BG,A1BG-AS1,A1CF ...
Columns names: CTGTTTACACCGCTAG,CTCTACGGTGTGGCTC,AGCAGCCAGGCTCATT ...
###################################
QC stats performed:FALSE, PCA performed:FALSE, CCA performed:FALSE
Clustering performed:FALSE, Number of clusters:0
tSNE performed:FALSE, UMAP performed:FALSE, DiffMap performed:FALSE
Main data dimentions (rows,columns):0,0
Normalization factors:,...
Imputed data dimentions (rows,columns):0,0
############## scVDJ-Seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-Seq ############
- ADT raw data dimentions (rows,columns):10,8617
ADT main data dimentions (rows,columns):0,0
ADT columns names:...
ADT row names:...
########### iCellR object ##########- QC, filter, normalize, merge ADT and RNA data, run PCA and UMAP
# QC
my.obj <- qc.stats(my.obj,
s.phase.genes = s.phase,
g2m.phase.genes = g2m.phase)
# plot as mentioned above
# filter
my.obj <- cell.filter(my.obj,
min.mito = 0,
max.mito = 0.07 ,
min.genes = 500,
max.genes = 4000,
min.umis = 0,
max.umis = Inf)
# normalize RNA
my.obj <- norm.data(my.obj, norm.method = "ranked.glsf", top.rank = 500)
# normalize ADT
my.obj <- norm.adt(my.obj)
# gene stats
my.obj <- gene.stats(my.obj, which.data = "main.data")
# find genes for PCA
my.obj <- make.gene.model(my.obj, my.out.put = "data",
dispersion.limit = 1.5,
base.mean.rank = 500,
no.mito.model = T,
mark.mito = T,
interactive = F,
no.cell.cycle = T,
out.name = "gene.model")
# merge RNA and ADT data
my.obj <- adt.rna.merge(my.obj, adt.data = "main")
# run PCA and the rest is as above
my.obj <- run.pca(my.obj, method = "gene.model", gene.list = my.obj@gene.model,data.type = "main")
# 2 pass PCA
my.obj <- find.dim.genes(my.obj, dims = 1:20,top.pos = 20, top.neg = 20)
# second round PC
my.obj <- run.pca(my.obj, method = "gene.model", gene.list = my.obj@gene.model,data.type = "main")
my.obj <- run.umap(my.obj, dims = 1:10)
# check your object
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.1.4
Raw/original data dimentions (rows,columns): 20501,8617
Data conditions: no conditions/single sample
Row names: A1BG,A1BG-AS1,A1CF ...
Columns names: CTGTTTACACCGCTAG,CTCTACGGTGTGGCTC,AGCAGCCAGGCTCATT ...
###################################
QC stats performed:TRUE, PCA performed:TRUE, CCA performed:FALSE
Clustering performed:TRUE, Number of clusters:14
tSNE performed:FALSE, UMAP performed:TRUE, DiffMap performed:FALSE
Main data dimentions (rows,columns):20511,8305
Normalization factors:8.448547776071,...
Imputed data dimentions (rows,columns):0,0
############## scVDJ-Seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-Seq ############
ADT raw data dimentions (rows,columns):10,8617
ADT main data dimentions (rows,columns):10,8617
ADT columns names:CTGTTTACACCGCTAG...
ADT row names:ADT_CD3...
########### iCellR object ##########- plot
# find ADT gene names
grep("^ADT_", rownames(my.obj@main.data),value=T)
# [1] "ADT_CD3" "ADT_CD4" "ADT_CD8" "ADT_CD45RA" "ADT_CD56"
# [6] "ADT_CD16" "ADT_CD11c" "ADT_CD14" "ADT_CD19" "ADT_CD34"
A = gene.plot(my.obj,
gene = "ADT_CD3",
plot.data.type = "umap",
interactive = F,
cell.transparency = 0.5)
B = gene.plot(my.obj,
gene = "CD3E",
plot.data.type = "umap",
interactive = F,
cell.transparency = 0.5)
C = gene.plot(my.obj,
gene = "ADT_CD16",
plot.data.type = "umap",
interactive = F,
cell.transparency = 0.5)
D = gene.plot(my.obj,
gene = "FCGR3A",
plot.data.type = "umap",
interactive = F,
cell.transparency = 0.5)
library(gridExtra)
grid.arrange(A,B,C,D)Here is an example of how to add VDJ data.
###### an example file
my.vdj <- read.csv(file = system.file('extdata', 'all_contig_annotations.csv',
package = 'iCellR'),
as.is = TRUE)
###
head(my.vdj)
# barcode is_cell contig_id high_confidence length
#1 AAACCTGTCCGAACGC-1 True AAACCTGTCCGAACGC-1_contig_1 True 654
#2 AAACCTGTCCGAACGC-1 True AAACCTGTCCGAACGC-1_contig_2 True 697
#3 AAACCTGTCCGAACGC-1 True AAACCTGTCCGAACGC-1_contig_3 False 496
#4 AAACCTGTCCGAACGC-1 True AAACCTGTCCGAACGC-1_contig_4 True 539
#5 AAACCTGTCGATGAGG-1 True AAACCTGTCGATGAGG-1_contig_1 True 705
#6 AAACCTGTCGATGAGG-1 True AAACCTGTCGATGAGG-1_contig_2 True 491
# chain v_gene d_gene j_gene c_gene full_length productive cdr3
#1 TRB TRBV4-1 None TRBJ2-7 TRBC2 True True CASSQGVEQYF
#2 TRA TRAV8-1 None TRAJ42 TRAC True True CAVKGGSQGNLIF
#3 TRB None None TRBJ1-4 TRBC1 False None None
#4 Multi None None TRAJ10 TRBC1 False None None
#5 TRB TRBV5-5 TRBD1 TRBJ2-7 TRBC1 True True CASSLVSGGNEQYF
#6 TRB None None TRBJ1-2 TRBC1 False None None
# cdr3_nt reads umis raw_clonotype_id
#1 TGCGCCAGCAGCCAAGGGGTCGAGCAGTACTTC 42610 19 clonotype150
#2 TGTGCCGTGAAGGGAGGAAGCCAAGGAAATCTCATCTTT 12297 4 clonotype150
#3 None 4314 1 clonotype150
#4 None 2212 1 clonotype150
#5 TGTGCCAGCAGCTTGGTCTCAGGGGGAAACGAGCAGTACTTC 21148 8 clonotype2
#6 None 17717 16 clonotype2
# raw_consensus_id
#1 clonotype150_consensus_1
#2 clonotype150_consensus_2
#3 None
#4 None
#5 clonotype2_consensus_1
#6 None
#### Prepare the vdj file
My.VDJ <- prep.vdj(vdj.data = my.vdj, cond.name = "NULL")
###
head(My.VDJ)
# raw_clonotype_id barcode is_cell contig_id
#1 clonotype1 ACGCCAGCAAGCGCTC.1 True ACGCCAGCAAGCGCTC-1_contig_2
#2 clonotype1 AACGTTGAGTACGATA.1 True AACGTTGAGTACGATA-1_contig_2
#3 clonotype1 AACTCTTGTCAAAGCG.1 True AACTCTTGTCAAAGCG-1_contig_1
#4 clonotype1 AACGTTGAGTACGATA.1 True AACGTTGAGTACGATA-1_contig_1
#5 clonotype1 ACGCCAGCAAGCGCTC.1 True ACGCCAGCAAGCGCTC-1_contig_1
#6 clonotype1 ACGATGTTCTGGTATG.1 True ACGATGTTCTGGTATG-1_contig_2
# high_confidence length chain v_gene d_gene j_gene c_gene full_length
#1 True 571 TRA TRAV27 None TRAJ37 TRAC True
#2 True 730 TRA TRAV27 None TRAJ37 TRAC True
#3 True 722 TRB TRBV6-3 TRBD2 TRBJ1-1 TRBC1 True
#4 True 723 TRB TRBV6-3 TRBD2 TRBJ1-1 TRBC1 True
#5 True 722 TRB TRBV6-3 TRBD2 TRBJ1-1 TRBC1 True
#6 True 726 TRA TRAV27 None TRAJ37 TRAC True
# productive cdr3 cdr3_nt reads
#1 True CAGGRSSNTGKLIF TGTGCAGGAGGACGCTCTAGCAACACAGGCAAACTAATCTTT 14241
#2 True CAGGRSSNTGKLIF TGTGCAGGAGGACGCTCTAGCAACACAGGCAAACTAATCTTT 27679
#3 True CASRTGAGATEAFF TGTGCCAGCAGGACCGGGGCGGGAGCCACTGAAGCTTTCTTT 51844
#4 True CASRTGAGATEAFF TGTGCCAGCAGGACCGGGGCGGGAGCCACTGAAGCTTTCTTT 38120
#5 True CASRTGAGATEAFF TGTGCCAGCAGGACCGGGGCGGGAGCCACTGAAGCTTTCTTT 24635
#6 True CAGGRSSNTGKLIF TGTGCAGGAGGACGCTCTAGCAACACAGGCAAACTAATCTTT 13720
# umis raw_consensus_id my.raw_clonotype_id clonotype.Freq proportion
#1 8 clonotype1_consensus_2 clonotype1 43 0.1572212
#2 10 clonotype1_consensus_2 clonotype1 43 0.1572212
#3 24 clonotype1_consensus_1 clonotype1 43 0.1572212
#4 23 clonotype1_consensus_1 clonotype1 43 0.1572212
#5 11 clonotype1_consensus_1 clonotype1 43 0.1572212
#6 7 clonotype1_consensus_2 clonotype1 43 0.1572212
# total.colonotype
#1 109
#2 109
#3 109
#4 109
#5 109
#6 109
####
png('vdj.stats.png',width = 16, height = 8, units = 'in', res = 300)
vdj.stats(My.VDJ)
dev.off()
### add vdj data to you object
my.obj <- add.vdj(demo.obj, vdj.data = My.VDJ)Another example with multiple files
# First read the vdj data
File="all_contig_annotations.csv"
my.vdj.data <- read.csv(File)
# then see the conditions
my.obj
# For each condition (WT,KO, ...) subset from the VDJ data
Get="WT"
#######
dat <- colnames(my.obj@main.data)
name <- paste(Get,".tsv",sep="")
do <- grep(Get,dat, value=T)
do <- as.character(as.matrix(data.frame(do.call('rbind', strsplit(as.character(do),'_',fixed=TRUE)))[2]))
do <- gsub("\\.","-",do)
do <- subset(my.vdj.data, my.vdj.data$barcode %in% do)
write.table((do),file=name,sep="\t", row.names =F)
#######
Get="KO"
#######
dat <- colnames(my.obj@main.data)
name <- paste(Get,".tsv",sep="")
do <- grep(Get,dat, value=T)
do <- as.character(as.matrix(data.frame(do.call('rbind', strsplit(as.character(do),'_',fixed=TRUE)))[2]))
do <- gsub("\\.","-",do)
do <- subset(my.vdj.data, my.vdj.data$barcode %in% do)
write.table((do),file=name,sep="\t", row.names =F)
#######
#### read and prep all conditions
Get="WT"
name <- paste(Get,".tsv",sep="")
do <- read.table(name, header=T)
WT <- prep.vdj(vdj.data = do, cond.name = Get)
Get="KO"
name <- paste(Get,".tsv",sep="")
do <- read.table(name, header=T)
KO <- prep.vdj(vdj.data = do, cond.name = Get)
# concatenate all the conditions
my.vdj.data <- rbind(WT, KO)
# see head of the file
head(my.vdj.data)
# raw_clonotype_id barcode is_cell contig_id
#1 clonotype1 WT_AAACCTGAGCTAACTC-1 True AAACCTGAGCTAACTC-1_contig_1
#2 clonotype1 WT_AAACCTGAGCTAACTC-1 True AAACCTGAGCTAACTC-1_contig_2
#3 clonotype1 WT_AGTTGGTTCTCGCATC-1 True AGTTGGTTCTCGCATC-1_contig_3
#4 clonotype1 WT_TGACAACCAACTGCTA-1 True TGACAACCAACTGCTA-1_contig_1
#5 clonotype1 WT_TGTCCCAGTCAAACTC-1 True TGTCCCAGTCAAACTC-1_contig_1
#6 clonotype1 WT_TGTCCCAGTCAAACTC-1 True TGTCCCAGTCAAACTC-1_contig_2
# high_confidence length chain v_gene d_gene j_gene c_gene full_length
#1 True 693 TRA TRAV8-1 None TRAJ21 TRAC True
#2 True 744 TRB TRBV28 TRBD1 TRBJ2-1 TRBC2 True
#3 True 647 TRA TRAV8-1 None TRAJ21 TRAC True
#4 True 508 TRB TRBV28 TRBD1 TRBJ2-1 TRBC2 True
#5 True 660 TRA TRAV8-1 None TRAJ21 TRAC True
#6 True 770 TRB TRBV28 TRBD1 TRBJ2-1 TRBC2 True
# productive cdr3 cdr3_nt
#1 True CAVKDFNKFYF TGTGCCGTGAAAGACTTCAACAAATTTTACTTT
#2 True CASSLFSGTGTNEQFF TGTGCCAGCAGTTTATTTTCCGGGACAGGGACGAATGAGCAGTTCTTC
#3 True CAVKDFNKFYF TGTGCCGTGAAAGACTTCAACAAATTTTACTTT
#4 True CASSLFSGTGTNEQFF TGTGCCAGCAGTTTATTTTCCGGGACAGGGACGAATGAGCAGTTCTTC
#5 True CAVKDFNKFYF TGTGCCGTGAAAGACTTCAACAAATTTTACTTT
#6 True CASSLFSGTGTNEQFF TGTGCCAGCAGTTTATTTTCCGGGACAGGGACGAATGAGCAGTTCTTC
# reads umis raw_consensus_id my.raw_clonotype_id clonotype.Freq
#1 1241 2 clonotype1_consensus_1 clonotype1 120
#2 2400 4 clonotype1_consensus_2 clonotype1 120
#3 1090 2 clonotype1_consensus_1 clonotype1 120
#4 2455 4 clonotype1_consensus_2 clonotype1 120
#5 1346 2 clonotype1_consensus_1 clonotype1 120
#6 3073 8 clonotype1_consensus_2 clonotype1 120
# proportion total.colonotype
#1 0.04098361 1292
#2 0.04098361 1292
#3 0.04098361 1292
#4 0.04098361 1292
#5 0.04098361 1292
#6 0.04098361 1292
# add it to iCellR object
my.obj <- add.vdj(my.obj, vdj.data = my.vdj.data)How to plot clonotypes
# once you have imported your clonotype data to your iCellR object, in order to plot them you need to have the following parapmeters:
# -1 clonotype name (e.g. clono = "clonotype1")
# -2 which column number has the clonotype names (e.g. clonotype.column = 2)
# -3 which column number has the cell barcode names (e.g. barcode.column = 1)
# In order to plot you need 2 things a- cell barcodes that match the barcodes in UMAP,PCA,tSNE or KNetL data and b- clonotype names.
# to check your clonotype data do this (example):
head(my.obj@vdj.data)
# raw_clonotype_id_SampleID MyBarcodes V1
#1 S5_clonotype98 Nor2.A_AAACCTGAGACAGACC.1 AAACCTGAGACAGACC.1
#2 S5_clonotype98 Nor2.A_AAACCTGAGACAGACC.1 AAACCTGAGACAGACC.1
#3 S4_clonotype100 Nor2.B_AAACCTGAGAGACTAT.1 AAACCTGAGAGACTAT.1
#4 S4_clonotype100 Nor2.B_AAACCTGAGAGACTAT.1 AAACCTGAGAGACTAT.1
#5 S3_clonotype3 Nor1.B_AAACCTGAGAGTCGGT.1 AAACCTGAGAGTCGGT.1
#6 S5_clonotype99 Nor2.A_AAACCTGAGATATGGT.1 AAACCTGAGATATGGT.1
# barcode SampleID raw_clonotype_id is_cell
#1 S5_AAACCTGAGACAGACC.1 5 clonotype98 True
#2 S5_AAACCTGAGACAGACC.1 5 clonotype98 True
#3 S4_AAACCTGAGAGACTAT.1 4 clonotype100 True
#4 S4_AAACCTGAGAGACTAT.1 4 clonotype100 True
#5 S3_AAACCTGAGAGTCGGT.1 3 clonotype3 True
#6 S5_AAACCTGAGATATGGT.1 5 clonotype99 True
# contig_id high_confidence length chain v_gene d_gene
#1 AAACCTGAGACAGACC-1_contig_2 True 514 TRB TRBV14 None
#2 AAACCTGAGACAGACC-1_contig_1 True 495 TRB TRBV20-1 None
#3 AAACCTGAGAGACTAT-1_contig_2 True 496 TRB TRBV9 None
#4 AAACCTGAGAGACTAT-1_contig_1 True 529 TRA TRAV26-1 None
#5 AAACCTGAGAGTCGGT-1_contig_1 True 512 TRB TRBV6-5 None
#6 AAACCTGAGATATGGT-1_contig_2 True 544 TRA TRAV12-2 None
# j_gene c_gene full_length productive cdr3
#1 TRBJ1-5 TRBC1 True True CASSFEGGSTQPQHF
#2 TRBJ2-7 TRBC2 True True CSARVRGRSSYEQYF
#3 TRBJ2-2 TRBC2 True True CASSVGVNTGELFF
#4 TRAJ52 TRAC True True CIVRGAGGTSYGKLTF
#5 TRBJ1-1 TRBC1 True True CASSYRPNTEAFF
#6 TRAJ33 TRAC True True CAVKRDSNYQLIW
# cdr3_nt reads umis
#1 TGTGCCAGCAGTTTTGAGGGGGGATCGACTCAGCCCCAGCATTTT 886 1
#2 TGCAGTGCTAGAGTAAGGGGACGGAGCTCCTACGAGCAGTACTTC 1912 3
#3 TGTGCCAGCAGCGTGGGCGTAAACACCGGGGAGCTGTTTTTT 10804 12
#4 TGCATCGTCAGGGGGGCTGGTGGTACTAGCTATGGAAAGCTGACATTT 960 4
#5 TGTGCCAGCAGTTACCGCCCGAACACTGAAGCTTTCTTT 4286 6
#6 TGTGCCGTGAAAAGGGATAGCAACTATCAGTTAATCTGG 1244 2
# raw_consensus_id my.raw_clonotype_id clonotype.Freq proportion
#1 clonotype98_consensus_1 S5_clonotype98 1 0.0001983930
#2 clonotype98_consensus_2 S5_clonotype98 1 0.0001983930
#3 clonotype100_consensus_2 S4_clonotype100 1 0.0001923817
#4 clonotype100_consensus_1 S4_clonotype100 1 0.0001923817
#5 clonotype3_consensus_1 S3_clonotype3 49 0.0070635721
#6 clonotype99_consensus_1 S5_clonotype99 1 0.0001983930
# total.colonotype
#1 5096
#2 5096
#3 5280
#4 5280
#5 5943
#6 5096
# In this example column number 1 and 2 have the clonotype and barcode info needed to plot.
# Sort clonotype names with highset frequency:
clonotype.frequency <- as.data.frame(sort(table(as.character(as.matrix((my.obj@vdj.data)[1]))),decreasing = TRUE))
head(clonotype.frequency)
# Var1 Freq
#1 S2_clonotype1 306
#2 S1_clonotype1 242
#3 S3_clonotype1 232
#4 S4_clonotype1 216
#5 S5_clonotype1 210
#6 S2_clonotype2 113
# let's plot S1_clonotype1 which is seen in 242 cells in all the conditions.
# if you want to plot only in one condtion or few conditions use this option "conds.to.plot" (e.g. conds.to.plot = c("WT","KO"))
# If conds.to.plot = NULL it would plot all of them (all 242 cells).
# Plot colonotype 1
clono.plot(my.obj, plot.data.type = "knetl",
clonotype.column = 1,
barcode.column = 2,
clono = "S1_clonotype1",
conds.to.plot = NULL,
cell.transparency = 1,
clust.dim = 2,
interactive = F)
# plot multiple clonotypes
ordered.clonotypes <- as.character(as.matrix((clonotype.frequency)[1]))
# let's plot top 19 clonotypes with highest frequency:
clonolist <- (ordered.clonotypes)[1:19]
clonolist
# [1] "S2_clonotype1" "S1_clonotype1" "S3_clonotype1" "S4_clonotype1"
# [5] "S5_clonotype1" "S2_clonotype2" "S3_clonotype2" "S1_clonotype2"
# [9] "S2_clonotype4" "S1_clonotype4" "S3_clonotype4" "S2_clonotype3"
#[13] "S4_clonotype2" "S1_clonotype3" "S4_clonotype3" "S5_clonotype2"
#[17] "S3_clonotype3" "S2_clonotype9" "S3_clonotype6"
rm(list = ls(pattern="PL_"))
for(i in clonolist){
MyPlot <- clono.plot(my.obj, plot.data.type = "knetl",
clonotype.column = 1,
barcode.column = 2,
clono = i,
conds.to.plot = NULL,
cell.transparency = 1,
clust.dim = 2,
interactive = F)
NameCol=paste("PL",i,sep="_")
eval(call("<-", as.name(NameCol), MyPlot))
}
library(cowplot)
filenames <- ls(pattern="PL_")
B= cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1,anno.clust=TRUE)
filenames <- c("B",filenames)
png("19_clonotypes.png",width = 20, height = 20, units = 'in', res = 300)
plot_grid(plotlist=mget(filenames))
dev.off()In this example the samples are normalized using DESeq2 so no normalization is needed.
sample.file.url = "https://genome.med.nyu.edu/results/external/iCellR/data/TCGA_sample_Normalized_data.tsv.gz"
download.file(url = sample.file.url,
destfile = "TCGA_sample_Normalized_data.tsv.gz",
method = "auto")
TCGA.data <- read.table("TCGA_sample_Normalized_data.tsv.gz")
head(TCGA.data)[1:3]
# Basal_TCGA.A1.A0SK.txt Basal_TCGA.A1.A0SP.txt Basal_TCGA.A2.A04P.txt
#TSPAN6 5823.4300 4318.034382 5265.733258
#TNMD 0.0000 6.049079 6.763079
#DPM1 3248.1536 2528.515113 1183.538813
#SCYL3 1059.7135 965.836315 1109.144945
#C1orf112 1251.3155 1070.687022 485.589067
#FGR 106.2438 933.574559 512.641383
library(iCellR)
my.obj <- make.obj(TCGA.data)
my.obj@main.data <- my.obj@raw.data
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.2.4
Raw/original data dimentions (rows,columns): 69797,882
Data conditions in raw data: Basal,Her2,LumA,LumB,Normal (131,64,404,170,113)
Row names: TSPAN6,TNMD,DPM1 ...
Columns names: Basal_TCGA.A1.A0SK.txt,Basal_TCGA.A1.A0SP.txt,Basal_TCGA.A2.A04P.txt ...
###################################
QC stats performed:FALSE, PCA performed:FALSE, CCA performed:FALSE
Clustering performed:FALSE, Number of clusters:0
tSNE performed:FALSE, UMAP performed:FALSE, DiffMap performed:FALSE
Main data dimentions (rows,columns):69797,882
Normalization factors:,...
Imputed data dimentions (rows,columns):0,0
############## scVDJ-Seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-Seq ############
ADT raw data dimentions (rows,columns):0,0
ADT main data dimentions (rows,columns):0,0
ADT columns names:...
ADT row names:...
########### iCellR object ##########
my.obj <- run.pca(my.obj)
my.obj <- run.clustering(my.obj,
clust.method = "kmeans",
dist.method = "euclidean",
index.method = "silhouette",
max.clust =25,
min.clust = 2,
dims = 1:10)
my.obj <- run.pc.tsne(my.obj, dims = 1:10)
my.obj <- run.umap(my.obj, dims = 1:10, method = "umap-learn")
cluster.plot(my.obj,plot.type = "pca",cell.color = "black",col.by = "conditions",cell.transparency = 0.5,interactive = F)
cluster.plot(my.obj,plot.type = "umap",cell.color = "black",col.by = "conditions",cell.transparency = 0.5,interactive = F)
cluster.plot(my.obj,plot.type = "tsne",cell.color = "black",col.by = "conditions",cell.transparency = 0.5,interactive = F)
cluster.plot(my.obj,plot.type = "umap",cell.color = "black",cell.transparency = 1,interactive = F)To do this you need to download the following databse files from our iCellR data link (more data to come soon).
# download the .rda files from here: https://genome.med.nyu.edu/results/external/iCellR/data/
# Load the .rda files as below
load("Immgen.GSE109125.205.rda")
load("Immgen.GSE122108.412.rda")
load("Immgen.GSE122597.83.rda")
load("Immgen.GSE124829.190.rda")
load("Immgen.microarray.GSE15907.653.rda")
load("Immgen.microarray.GSE37448.189.rda")
load("immgen.rna.rda")
load("immgen.uli.rna.rda")
load("mouse.cell.atlas.rda") | Key | Source | Samples | Description | Cell Types |
|---|---|---|---|---|
| GSE109125 | ImmGen | 205 | 83 populations representing all lineages and several differentiation cascades prepared from unchallenged mice and after LPS, anti-CD3, viral infection cell activation. | B Cells, Stromal Cells, Dendritic Cells, Granulocytes, Innate Lymphocytes, Stem Cells, Macrophages, ab T Cells, gd T Cells |
| GSE122108 | ImmGen | 412 | 130 populations comprising progenitors, residents, and stimulated (C.alb, LPS, injury, APAP+ starved overnight and pIC) mononuclear phagocytes for OpenSource MNP Project. | Macrophages, Kupffer Cell/Macrophages, Dendritic Cells, Microglia, Monocytes. |
| GSE122597 | ImmGen | 83 | Five highly purified immunocyte populations profiled to unusual depth as multiple replicates (8 to 16). Suitable for exploration of genes expressed at very low levels. | NK Cells, Follicular B, Naive CD4+ abT, gdT cells and peritoneal macrophages. |
| GSE124829 | ImmGen | 190 | 11 diverse immunocyte populations from male and female mice of varying ages stimulated with different dose of IFN to understand the immune system's sexual differences. | B Cells, Dendritic Cells, Neutrophils, Macrophages, Natural Killer T Cells, ab T Cells, gd T Cells, Microglia, Regulatory T Cells. |
| GSE15907 | ImmGen | 653 | 178 populations compromiing of gene-expression microarray datasets ("version1" labeling) from primary cells from multiple immune lineages are isolated ex-vivo, primarily from 6weeks B6 male mice. | gd T Cells, ab T Cells, Dendritic Cells, Macrophages, Stem Cells, B Cells, Stromal Cells, Neutrophils, Fibroblast, NK Cells, NK T Cells, Monocytes, CD4 Naive T Cell. |
| GSE37448 | ImmGen | 189 | 80 populations compromising of gene-expression microarray datasets ("version2" labeling) from primary cells from multiple immune lineages are isolated ex-vivo, primarily from 6weeks B6 male mice. Complements the V1 compendium with additional cells. Unfortunately, the version change in the labeling process, while more efficient, introduced some biases such that the two sections of the data can be compared grossly, but not at fine resolution (we tried...). | gd T Cells, ab T Cells, Dendritic Cells, Macrophages, Stem Cells, B Cells, Stromal Cells, Neutrophils, Fibroblast, NK Cells, NK T Cells, Monocytes, CD4 Naive T Cell. |
| rna | ImmGen | 23 | Full depth directional RNA sequencing was performed on the core ImmGen populations to generate reference datasets for the tissues from 5 week-old C57BL/6J (Jackson Laboratory) males and females, double-sorted by flow cytometry, per ImmGen cell preparation SOP. | B, CD4T, CD8T, DC, MQ,NK, NKT, Treg |
| uli.rna | ImmGen | 157 | ||
| mca | Mouse Cell Atlas | 43 tissues | Constructed as a basic scheme for the Mouse Cell Atlas using Microwell-seq. | Uterus, TrophoblastStemCells, Thymus, Testis, Stomach, Spleen, SmallIntestine, Prostate, Placenta, PeripheralBlood, Pancreas, Ovary, NeontalBrain, NeonatalSkin, NeonatalRib, NeonatalMuscle, NeonatalHeart, NeonatalCalvaria, Muscle, Mouse3T3, MesenchymalStemCellsPrimary, MesenchymalStemCells, MammaryGland.Virgin, MammaryGland.Pregnancy, MammaryGland.Lactation, MammaryGland.Involution, Male.fetal.Gonad, Lung, Liver, Kidney, FetalStomach, FetalLung, FetalLiver, FetalKidney, FetalIntestine, FetalBrain, Female.fetal.Gonad, EmbryonicStemCells, EmbryonicMesenchyme, Brain, BoneMarrowcKit, BoneMarrow, Bladder |
Choose a cluster and take for example top 10 genes for that cluster and then choose one of the databases that is best for you from the above list and predict your cell type. Note that if you have B cells for example and the database of your choice dose not have B cells, it would predict the closest looking cells to B cells. So it's important to use the right database for the right type of data.
# Choose top 40 genes for cluster 8 for example
MyGenes <- top.markers(marker.genes, topde = 40, min.base.mean = 0.2, cluster = 8)
####### predict
# plot
cell.type.pred(immgen.data = "rna", gene = MyGenes, plot.type = "point.plot")
cell.type.pred(immgen.data = "uli.rna", gene = MyGenes, plot.type = "point.plot", top.cell.types = 50)
cell.type.pred(immgen.data = "rna", gene = MyGenes, plot.type = "heatmap")
cell.type.pred(immgen.data = "uli.rna", gene = MyGenes, plot.type = "heatmap")
# As you can see cluster 8 is most likely to be B-cells.
# more examples
cell.type.pred(immgen.data = "GSE109125", gene = MyGenes, plot.type = "point.plot", top.cell.types = 50)
cell.type.pred(immgen.data = "GSE37448", gene = MyGenes, plot.type = "heatmap", top.cell.types = 50)
# for tissue type prediction use this:
cell.type.pred(immgen.data = "mca", gene = MyGenes, plot.type = "point.plot")
# And finally check the genes in the cells and find the common ones to predict
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters") You can automate this for all the clusters as below. Add as many plot as you wish.
Clusters = sort(unique(my.obj@best.clust$clusters))
for(i in Clusters){
Cluster = i
MyGenes <- top.markers(marker.genes, topde = 10, min.base.mean = 0.2, cluster = Cluster)
# first plot
Name <- paste("ImmGen_Cluster_",Cluster,"_pointPlot_RNA.pdf",sep="")
pdf(Name, width = 10, height = 10)
print(cell.type.pred(immgen.data = "rna", gene = MyGenes, plot.type = "point.plot"))
dev.off()
# second plot
Name <- paste("ImmGen_Cluster_",Cluster,"_check.pdf",sep="")
pdf(Name, width = 10, height = 10)
print(heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters"))
dev.off()
}- Pathway analysis
# Pathway
# pathways.kegg(my.obj, clust.num = 7)
# this function is being improved and soon will be availableIn this example, we have downloaded 2 samples from 10X genomics website. You can get the data from these links: Anterior and Posterior. To make it easier you can also use the commands below to download from our server.
# download sample data
url = "https://genome.med.nyu.edu/results/external/iCellR/example7_Spatial_Transcriptomic/V1_Mouse_Brain_Sagittal_Anterior_Section_2_filtered_feature_bc_matrix.tar.gz"
# download the file
download.file(url = url,
destfile = "V1_Mouse_Brain_Sagittal_Anterior_Section_2_filtered_feature_bc_matrix.tar.gz",
method = "auto")
url ="https://genome.med.nyu.edu/results/external/iCellR/example7_Spatial_Transcriptomic/V1_Mouse_Brain_Sagittal_Anterior_Section_2_spatial.tar.gz"
# download the file
download.file(url = url,
destfile = "V1_Mouse_Brain_Sagittal_Anterior_Section_2_spatial.tar.gz",
method = "auto")
url ="https://genome.med.nyu.edu/results/external/iCellR/example7_Spatial_Transcriptomic/V1_Mouse_Brain_Sagittal_Posterior_Section_2_filtered_feature_bc_matrix.tar.gz"
# download the file
download.file(url = url,
destfile = "V1_Mouse_Brain_Sagittal_Posterior_Section_2_filtered_feature_bc_matrix.tar.gz",
method = "auto")
url ="https://genome.med.nyu.edu/results/external/iCellR/example7_Spatial_Transcriptomic/V1_Mouse_Brain_Sagittal_Posterior_Section_2_spatial.tar.gz"
# download the file
download.file(url = url,
destfile = "V1_Mouse_Brain_Sagittal_Posterior_Section_2_spatial.tar.gz",
method = "auto")
#########################
##### untar
untar("V1_Mouse_Brain_Sagittal_Anterior_Section_2_filtered_feature_bc_matrix.tar.gz")
untar("V1_Mouse_Brain_Sagittal_Anterior_Section_2_spatial.tar.gz")
file.rename("spatial","spatial_Anterior2")
file.rename("filtered_feature_bc_matrix","filtered_feature_bc_matrix_Anterior2")
untar("V1_Mouse_Brain_Sagittal_Posterior_Section_2_filtered_feature_bc_matrix.tar.gz")
untar("V1_Mouse_Brain_Sagittal_Posterior_Section_2_spatial.tar.gz")
file.rename("spatial","spatial_Posterior2")
file.rename("filtered_feature_bc_matrix","filtered_feature_bc_matrix_Posterior2")- Load the data
library(iCellR)
Anterior2 <- load10x("filtered_feature_bc_matrix_Anterior2",gene.name = 2)
Posterior2 <- load10x("filtered_feature_bc_matrix_Posterior2",gene.name = 2)
# if you want to analyze both samples
Samples <- c("Anterior2","Posterior2")
my.data <- data.aggregation(samples = Samples, condition.names = Samples)
# if you want to analyze 1 sample
# my.data <- load10x("filtered_feature_bc_matrix_Posterior2",gene.name = 2)
my.obj <- make.obj(my.data)
Anterior2 <- capture.image.10x("spatial_Anterior2")
Posterior2 <- capture.image.10x("spatial_Posterior2")
# if you want to analyze both samples
Samples <- c("Anterior2","Posterior2")
my.obj <- add.10x.image(my.obj,
image.data.list = Samples, condition.names = Samples)
# if one sample
# My.image <- image.capture.10x("Post2_spatial")
# my.obj <- add.10x.image(my.obj, image.data.list = "My.image")
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.6.0
Raw/original data dimentions (rows,columns): 31053,6118
Data conditions in raw data: Anterior2,Posterior2 (2825,3293)
Row names: A030001D20Rik,A030003K21Rik,A030005K14Rik ...
Columns names: Anterior2_AAACAAGTATCTCCCA.1,Anterior2_AAACACCAATAACTGC.1,Anterior2_AAACAGAGCGACTCCT.1 ...
###################################
QC stats performed:FALSE, PCA performed:FALSE
Clustering performed:FALSE, Number of clusters:0
tSNE performed:FALSE, UMAP performed:FALSE, DiffMap performed:FALSE
Main data dimensions (rows,columns): 0,0
Normalization factors:,...
Imputed data dimensions (rows,columns):0,0
############## scVDJ-seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-seq ############
ADT raw data dimensions (rows,columns):0,0
ADT main data dimensions (rows,columns):0,0
ADT columns names:...
ADT row names:...
############## scATAC-seq ############
ATAC raw data dimensions (rows,columns):0,0
ATAC main data dimensions (rows,columns):0,0
ATAC columns names:...
ATAC row names:...
############## Spatial ###########
Spatial data dimentions (rows,columns):9984,5
########### iCellR object ##########The rest of the analysis is just like regular scRNA-Seq. Filter, normalize, run PCA, tSNE, UMAP, KNetL map and cluster. Then you can start ploting as below:
A=spatial.plot(my.obj,col.by = "clusters",conds.to.plot = "Anterior2",interactive= F)
B=spatial.plot(my.obj,col.by = "clusters",conds.to.plot = "Posterior2",interactive= F)
C= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
D= cluster.plot(my.obj,plot.type = "tsne",col.by = "conditions",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
E=spatial.plot(my.obj,col.by = "gene", gene = c("Cd4"), conds.to.plot = "Anterior2",interactive= F, scaleValue = TRUE)
F=spatial.plot(my.obj,col.by = "gene", gene = c("Cd4"), conds.to.plot = "Posterior2",interactive= F, scaleValue = TRUE)
library(gridExtra)
png('AllClusts.png', width = 8, height = 8, units = 'in', res = 300)
grid.arrange(A,B,C,D,E,F)
dev.off()library("iCellR")
my.data <- load10x("filtered_gene_bc_matrices/")
# see the row names
row.names(my.data)
# get peak names
ATAC <- grep("^chr",row.names(my.data),value=T)
# get scATAC data
MyATAC <- subset(my.data, row.names(my.data) %in% ATAC)
head(MyATAC)[1:3]
# AAACAGCCAAGTGAAC.1 AAACAGCCACTGACCG.1 AAACAGCCATGATTGT.1
#chr1.181218.181695 0 0 1
#chr1.191296.191699 0 0 0
#chr1.629770.630129 0 0 0
#chr1.633806.634251 0 0 0
#chr1.778422.779040 0 0 0
#chr1.827306.827702 0 0 0
dim(MyATAC)
# [1] 21923 6326
# get RNA data
MyRNAs <- subset(my.data, !row.names(my.data) %in% ATAC)
head(MyRNAs)[1:3]
# AAACAGCCAAGTGAAC.1 AAACAGCCACTGACCG.1 AAACAGCCATGATTGT.1
#MIR1302.2HG 0 0 0
#FAM138A 0 0 0
#OR4F5 0 0 0
#AL627309.1 0 0 0
#AL627309.3 0 0 0
#AL627309.2 0 0 0
dim(MyRNAs)
#[1] 36633 6326
# make iCellR object
my.obj <- make.obj(MyRNAs)
# add ATAC-Seq data
my.obj@atac.raw <- MyATAC
my.obj@atac.main <- MyATAC
# check your object
my.obj
###################################
,--. ,-----. ,--.,--.,------.
`--'' .--./ ,---. | || || .--. '
,--.| | | .-. :| || || '--'.'
| |' '--'\ --. | || || |
`--' `-----' `----'`--'`--'`--' '--'
###################################
An object of class iCellR version: 1.6.2
Raw/original data dimentions (rows,columns): 24127,6326
Data conditions: no conditions/single sample
Row names: MIR1302.2HG,TTLL10.AS1,MRPL20.AS1 ...
Columns names: AAACAGCCAAGTGAAC.1,AAACAGCCACTGACCG.1,AAACAGCCATGATTGT.1 ...
###################################
QC stats performed:FALSE, PCA performed:FALSE
Clustering performed:FALSE, Number of clusters:0
tSNE performed:FALSE, UMAP performed:FALSE, DiffMap performed:FALSE
Main data dimensions (rows,columns): 0,0
Normalization factors:,...
Imputed data dimensions (rows,columns):0,0
############## scVDJ-seq ###########
VDJ data dimentions (rows,columns):0,0
############## CITE-seq ############
ADT raw data dimensions (rows,columns):0,0
ADT main data dimensions (rows,columns):0,0
ADT columns names:...
ADT row names:...
############## scATAC-seq ############
ATAC raw data dimensions (rows,columns):21923,6326
ATAC main data dimensions (rows,columns):21923,6326
ATAC columns names:AAACAGCCAAGTGAAC.1...
ATAC row names:chr1.181218.181695...
############## Spatial ###########
Spatial data dimentions (rows,columns):0,0
########### iCellR object ##########
From here do the regular scRNA-seq as expleind above. See example below
# QC
my.obj <- qc.stats(my.obj,
s.phase.genes = s.phase,
g2m.phase.genes = g2m.phase)
# plot as mentioned above
# filter
my.obj <- cell.filter(my.obj,
min.mito = 0,
max.mito = 0.07 ,
min.genes = 500,
max.genes = 4000,
min.umis = 0,
max.umis = Inf)
# normalize RNA
my.obj <- norm.data(my.obj, norm.method = "ranked.glsf", top.rank = 500)
# normalize ADT
my.obj <- norm.adt(my.obj)
# gene stats
my.obj <- gene.stats(my.obj, which.data = "main.data")
# find genes for PCA
my.obj <- make.gene.model(my.obj, my.out.put = "data",
dispersion.limit = 1.5,
base.mean.rank = 500,
no.mito.model = T,
mark.mito = T,
interactive = F,
no.cell.cycle = T,
out.name = "gene.model")
# run PCA and the rest is as above
my.obj <- run.pca(my.obj, method = "gene.model", gene.list = my.obj@gene.model,data.type = "main")
# tSNE
my.obj <- run.pc.tsne(my.obj, dims = 1:10)
# UMAP
my.obj <- run.umap(my.obj, dims = 1:10)
# KNetL
my.obj <- run.knetl(my.obj, dims = 1:20, zoom = 200, dim.redux = "umap")
# clustering based on KNetL
my.obj <- iclust(my.obj, sensitivity = 200, data.type = "knetl")
# clustering based on PCA
# my.obj <- iclust(my.obj, sensitivity = 100, data.type = "pca", dims=1:10)
# check clusters and adjust if needed (optinal)
# cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
# my.obj <- change.clust(my.obj, change.clust = 3, to.clust = 4)
# my.obj <- change.clust(my.obj, change.clust = 3, to.clust = 10)
# order clusters
my.obj <- clust.ord(my.obj,top.rank = 500, how.to.order = "distance")
# plot
A= cluster.plot(my.obj,plot.type = "pca",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
B= cluster.plot(my.obj,plot.type = "umap",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
C= cluster.plot(my.obj,plot.type = "tsne",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
D= cluster.plot(my.obj,plot.type = "knetl",interactive = F,cell.size = 0.5,cell.transparency = 1, anno.clust=T)
library(gridExtra)
png('AllClusts.png', width = 12, height = 10, units = 'in', res = 300)
grid.arrange(A,B,C,D)
dev.off()
# save object
save(my.obj, file = "my.obj.Robj")
# find markers
marker.genes <- findMarkers(my.obj,
data.type = "main",
fold.change = 2,
padjval = 0.1,
uniq = F,
positive = T)
marker.genes1 <- cbind(row = rownames(marker.genes), marker.genes)
write.table((marker.genes1),file="marker.genes.tsv", sep="\t", row.names =F)
MyGenes <- top.markers(marker.genes, topde = 10, min.base.mean = 0.2, filt.ambig = F)
MyGenes <- unique(MyGenes)
png('heatmap_gg_genes.png', width = 10, height = 10, units = 'in', res = 300)
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters",cell.sort = F, conds.to.plot = NULL)
dev.off()Work on scATAC data (normalize and find marker peaks for each cluster)
# normalize ACAT
my.obj <- norm.data(my.obj, norm.method = "ranked.glsf", top.rank = 500, ATAC.data = TRUE, ATAC.filter = TRUE)
marker.peaks <- findMarkers(my.obj,
data.type = "atac",
fold.change = 2,
padjval = 0.1,
uniq = F,
positive = T)
marker.peaks1 <- cbind(row = rownames(marker.peaks), marker.peaks)
write.table((marker.peaks1),file="marker.peaks.tsv", sep="\t", row.names =F)
head(marker.peaks1)
# row baseMean baseSD
#chr17.64986035.64986113 chr17.64986035.64986113 0.01217359 0.18257818
#chr1.26542287.26542678 chr1.26542287.26542678 0.05828764 0.80077656
#chr4.8199063.8199275 chr4.8199063.8199275 0.04280424 0.56205649
#chr20.50274929.50275237 chr20.50274929.50275237 0.04684509 0.63361490
#chr2.218382038.218382236 chr2.218382038.218382236 0.03122394 0.31153105
#chr11.1760469.1760814 chr11.1760469.1760814 0.07050175 0.63322284
# AvExpInCluster AvExpInOtherClusters foldChange
#chr17.64986035.64986113 0.07868394 0.002346603 33.530999
#chr1.26542287.26542678 0.33849093 0.016887273 20.044144
#chr4.8199063.8199275 0.24803497 0.012481148 19.872769
#chr20.50274929.50275237 0.27007513 0.013862584 19.482308
#chr2.218382038.218382236 0.17736269 0.009631770 18.414340
#chr11.1760469.1760814 0.39043782 0.023230813 16.806894
# log2FoldChange pval padj clusters
#chr17.64986035.64986113 5.067424 1.187697e-05 2.875415e-02 1
#chr1.26542287.26542678 4.325109 3.653916e-05 8.539202e-02 1
#chr4.8199063.8199275 4.312721 6.059691e-06 1.489472e-02 1
#chr20.50274929.50275237 4.284093 2.871301e-05 6.779143e-02 1
#chr2.218382038.218382236 4.202758 6.359572e-10 1.677019e-06 1
#chr11.1760469.1760814 4.070981 6.813447e-11 1.800794e-07 1
# gene cluster_1 cluster_2
#chr17.64986035.64986113 chr17.64986035.64986113 0.0786839378 0.000000000
#chr1.26542287.26542678 chr1.26542287.26542678 0.3384909326 0.008895062
#chr4.8199063.8199275 chr4.8199063.8199275 0.2480349741 0.038672840
#chr20.50274929.50275237 chr20.50274929.50275237 0.2700751295 0.028703704
#chr2.218382038.218382236 chr2.218382038.218382236 0.1773626943 0.000000000
#chr11.1760469.1760814 chr11.1760469.1760814 0.3904378238 0.004537037
# cluster_3 cluster_4 cluster_5 cluster_6
#chr17.64986035.64986113 0.000000000 0.0000000000 0.0006934750 0.0010038760
#chr1.26542287.26542678 0.031485714 0.0029244992 0.0052261002 0.0041264535
#chr4.8199063.8199275 0.000000000 0.0007226502 0.0027450683 0.0113212209
#chr20.50274929.50275237 0.027092857 0.0121741140 0.0041820941 0.0051516473
#chr2.218382038.218382236 0.004292857 0.0042095532 0.0006722307 0.0038561047
#chr11.1760469.1760814 0.071678571 0.0177288136 0.0141820941 0.0061099806
# cluster_7 cluster_8
#chr17.64986035.64986113 0.003099029 0.009080357
#chr1.26542287.26542678 0.047110680 0.044484375
#chr4.8199063.8199275 0.014819417 0.028651786
#chr20.50274929.50275237 0.025499029 0.028765625
#chr2.218382038.218382236 0.011988350 0.035508929
#chr11.1760469.1760814 0.042093204 0.050708705
MyGenes <- top.markers(marker.peaks, topde = 10, min.base.mean = 0.2, filt.ambig = F)
MyGenes <- unique(MyGenes)
png('heatmap_gg_peaks.png', width = 10, height = 10, units = 'in', res = 300)
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters",cell.sort = F, conds.to.plot = NULL, data.type = "atac")
dev.off()
my.obj <- run.impute(my.obj,data.type = "knetl", nn = 10, ATAC.data = FALSE)
png('heatmap_gg_peaks.png', width = 10, height = 10, units = 'in', res = 300)
heatmap.gg.plot(my.obj, gene = MyGenes, interactive = F, cluster.by = "clusters",cell.sort = F, conds.to.plot = NULL, data.type = "atac.imputed")
dev.off()
## you can also find avarage peak intensity per cluster
my.obj <- clust.avg.exp(my.obj, data.type = "atac")
head(my.obj@clust.avg)
#gene cluster_1 cluster_2 ...
#chr1.100037799.100038931 0.38238731 0.36750000 ...
#chr1.100132733.100133298 0.11195725 1.13593827 ...
#chr1.100249637.100250160 0.09851425 0.09511728 ...
#chr1.100265992.100266479 0.06768394 0.17707407 ...
#chr1.10032488.10033387 0.35273705 0.14885802 ...
#chr1.100352150.100352921 0.12006088 0.00000000 ...
# find out which cluster has the highest number
dat <- as.data.frame(t((my.obj@clust.avg)[,-1]))
dat <- hto.anno(hto.data = dat)
head(dat$assignment.annotatio)
#[1] cluster_1 cluster_2 cluster_4 cluster_2 cluster_3 cluster_4
#8 Levels: cluster_1 cluster_2 cluster_3 cluster_4 cluster_5 ... cluster_8Peak analysis
# make a bed file per cluster from the marker.peaks file you made up here
make.bed(marker.peaks)
# load packages
library(ChIPseeker)
library(clusterProfiler)
# load genome
require(TxDb.Hsapiens.UCSC.hg38.knownGene)
txdb <- TxDb.Hsapiens.UCSC.hg38.knownGene
Anno="org.Hs.eg.db"
# load bed files
Mylist1 = list.files(pattern=".bed")
Mylist1
Mylist <- as.list(Mylist1)
NAMES <- gsub('_peaks.bed','',Mylist1)
names(Mylist) <- NAMES
files <- Mylist
files
# perform analysis (example)
promoter <- getPromoters(TxDb=txdb, upstream=3000, downstream=3000)
tagMatrixList <- lapply(files, getTagMatrix, windows=promoter)
pdf("Plot_ProfileLineAll.pdf")
plotAvgProf(tagMatrixList, xlim=c(-3000, 3000))
dev.off()
pdf('Plot_ProfileLine.pdf', width = 8, height = 10)
plotAvgProf(tagMatrixList, xlim=c(-3000, 3000), facet="row")
dev.off()
pdf("Plot_heatmaps.pdf", width = 50, height = 6)
tagHeatmap(tagMatrixList, xlim=c(-3000, 3000), color=NULL)
dev.off()
# annotate
peakAnnoList <- lapply(files, annotatePeak, TxDb=txdb,
tssRegion=c(-3000, 3000), verbose=FALSE)
# plot annotatin
pdf("Plot_AnnoBar.pdf")
plotAnnoBar(peakAnnoList)
dev.off()
############### peak annotation
peakAnnoList <- lapply(files, annotatePeak, TxDb=txdb,
tssRegion=c(-3000, 3000), verbose=FALSE, annoDb=Anno)
capture.output(peakAnnoList, file = "peakAnnoList.txt")
genes = lapply(peakAnnoList, function(i) as.data.frame(i))
lapply(1:length(genes), function(i) write.table(genes[[i]],
file = paste0(names(genes[i]), ".xls"),
row.names = FALSE, sep="\t"))
Merging scATAC files with different intervals (as dipicted in bedtools website)
# Let's say you have 2 files that you need to merege
# example file
C <- load10x("count-JJsn_C_cDNA/",gene.name = 2)
M <- load10x("count-JJsn_M_cDNA/",gene.name = 2)
ATAC.C <- grep("^chr",row.names(C),value=T)
ATAC.M <- grep("^chr",row.names(M),value=T)
MyATAC.C <- subset(C, row.names(C) %in% ATAC.C)
MyATAC.M <- subset(M, row.names(M) %in% ATAC.M)
head(MyATAC.C)[1:3]
C <- MyATAC.C
M <- MyATAC.M
dim(C)
#[1] 58678 4211
dim(M)
#[1] 57776 4736
f1 <- row.names(C)
f2 <- row.names(M)
all.peaks <- c(f1,f2)
length(all.peaks)
#[1] 116454
# make a bed file
chr <- as.character(as.matrix(data.frame(do.call('rbind', strsplit(as.character(all.peaks),'.',fixed=TRUE)))[1]))
start <- data.frame(do.call('rbind', strsplit(as.character(all.peaks),'.',fixed=TRUE)))[2]
end <- data.frame(do.call('rbind', strsplit(as.character(all.peaks),'.',fixed=TRUE)))[3]
DAT <- as.data.frame(chr)
DAT$start <- as.numeric(as.matrix(start))
DAT$end <- as.numeric(as.matrix(end))
head(DAT)
# chr start end
#1 chr1 181218 181695
#2 chr1 191296 191699
#3 chr1 629770 630129
#4 chr1 633806 634251
#5 chr1 778422 779040
#6 chr1 827306 827702
# make Genomic Ranges
library("GenomicRanges")
all.gr <- GRanges(seqnames=DAT$chr,ranges=IRanges(start=DAT$start,end=DAT$end))
all.gr
#GRanges object with ?? ranges and 0 metadata columns:
# seqnames ranges strand
# <Rle> <IRanges> <Rle>
# [1] chr1 181218-181695 *
# [2] chr1 191296-191699 *
# [3] chr1 629770-630129 *
# [4] chr1 633806-634251 *
# [5] chr1 778422-779040 *
# ... ... ... ...
# [52] chr1 1303892-1306216 *
# [53] chr1 1307242-1309359 *
# [54] chr1 1324425-1325236 *
# [55] chr1 1348940-1349958 *
# [56] chr1 1372031-1372220 *
# -------
# seqinfo: 1 sequence from an unspecified genome; no seqlengths
################## sort and merge the peaks
mrg <- reduce(all.gr)
#Before merge
length(all.gr)
#[1] 116454
#after merge
length(mrg)
#[1] 71426
########################## choose file and give name
MyFile <- f1
name="f1_new.bed"
########################## copy paste the code here to make a new bed file
########################## the new bed has the old and new intervals (new intervals to be replaced with old)
chr <- as.character(as.matrix(data.frame(do.call('rbind', strsplit(as.character(MyFile),'.',fixed=TRUE)))[1]))
start <- data.frame(do.call('rbind', strsplit(as.character(MyFile),'.',fixed=TRUE)))[2]
end <- data.frame(do.call('rbind', strsplit(as.character(MyFile),'.',fixed=TRUE)))[3]
# make a bed file
DAT <- as.data.frame(chr)
DAT$start <- as.numeric(as.matrix(start))
DAT$end <- as.numeric(as.matrix(end))
MyFile <- DAT
# make intrval file to replace to new regions
MyFile.gr <- GRanges(seqnames=MyFile$chr,ranges=IRanges(start=MyFile$start,end=MyFile$end))
OverLap <- findOverlaps(MyFile.gr, mrg)
OLD1 <- (OverLap@from)
NEW1 <- (OverLap@to)
OLD = MyFile.gr[OLD1]
NEW = mrg[NEW1]
chr <- as.character(OLD@seqnames)
DAT <- as.data.frame(chr)
DAT$start <- OLD@ranges@start
DAT$end <- (OLD@ranges@start + OLD@ranges@width) - 1
DAT$new.chr<- as.character(NEW@seqnames)
DAT$new.start <- NEW@ranges@start
DAT$new.end <- (NEW@ranges@start + NEW@ranges@width) - 1
head(DAT)
# chr start end new.chr new.start new.end
#1 chr1 3247563 3248453 chr1 3247563 3248453
#2 chr1 3360706 3361554 chr1 3360706 3361554
#3 chr1 3552372 3553230 chr1 3552372 3553230
#4 chr1 3645171 3646034 chr1 3645093 3646034
#5 chr1 3670318 3671081 chr1 3670318 3671090
#6 chr1 3671326 3672230 chr1 3671314 3672230
dim(DAT)
#58678 6
length(MyFile.gr)
#58678
# diff
have = mrg[unique(NEW1)]
dontHave = mrg[-unique(NEW1)]
ADD <- dontHave
L <- length(as.character(ADD@seqnames))
chr <-rep("NA",L)
DAT1 <- as.data.frame(chr)
DAT1$start <- rep("NA",L)
DAT1$end <- rep("NA",L)
DAT1$new.chr<- as.character(ADD@seqnames)
DAT1$new.start <- ADD@ranges@start
DAT1$new.end <- (ADD@ranges@start + ADD@ranges@width) - 1
Final.DAT <- rbind(DAT,DAT1)
##### Write
write.table(Final.DAT,name,row.names=FALSE,sep="\t", quote = FALSE)
### reapeat this process for f2 (M) as well
# The first 3 columns are the original peaks and the last 3 are the ones that need to be replaced with original one. The NA peaks would also get the new peak ids but in the matrix the cells will have 0 for expressions. To do this use the iCellR function replace.peak.id.
MyATAC.C <- replace.peak.id(atac.data=MyATAC.C, bed.file = Final.DAT.C)
MyATAC.M <- replace.peak.id(atac.data=MyATAC.M, bed.file = Final.DAT.M)
# finally aggregate the samples and add to iCellR object
my.atac.data <- data.aggregation(samples = c("MyATAC1","MyATAC2","MyATAC3"),
condition.names = c("WT","KO","Ctrl"))
# add ATAC-Seq data
[email protected] <- my.atac.data
[email protected] <- my.atac.data