-
Notifications
You must be signed in to change notification settings - Fork 21
MoBILAB
See Notes about current meetings and test data.
Mobilab is a multimodal data browser and motion capture data preprocessor for XDF format data.
A new paradigm for human brain imaging, mobile brain/body imaging (MoBI), involves synchronous collection of human brain activity (via electroencephalography, EEG), behavior (via body motion capture, eye tracking, etc.) and environmental events (via scene and experimental event recording) to study the joint brain / body dynamics supporting natural human cognition, i.e., motivated human actions and interactions in natural 3-D environments (Makeig et al., 2009). Processing complex, concurrent, multi-modal, and multi-rate data streams requires a signal processing environment quite different from environments designed to process single-modality time series data.
MoBILAB Toolbox for MATLAB was designed in 2011 by Alejandro Ojeda with Nima Bigdley Shamlo, and Christian Kothe in consultation with Arnaud Delorme and Scott Makeig, to support the Mobile Brain/Body imaging (MoBI) paradigm (Makeig et al., 2009). Since then, the software has evolved from a simple plug-in that bridged multi-modal data collection systems with EEGLAB software environment (Delorme & Makeig, 2004), to a standalone, open source, cross platform toolbox operating on MATLAB (The Mathworks, Inc.) that supports analysis and visualization of any combination of synchronously recorded brain, body, behavioral, and environmental data, designed primarily to enable joint analyses of EEG, body motion, and other behavioral data streams.
To make working with concurrent data streams of different modalities as simple as possible, MoBILAB exploits object-oriented programming (oop) to ease supporting new data types. In contrast to the common practice of representing data streams as in-memory variables in MATLAB workspace, MoBILAB uses memory-mapped data files to make it possible to process data streams of virtually any size without requiring large amounts of random access memory (RAM) and without compromising performance. Currently MoBILAB includes classes designed for holding, analyzing and visualizing EEG, motion capture, eye tracking, and force plate data as well as for recorded audio and video. These classes include methods (functions acting on class properties) for filters, time derivatives, wavelet time-frequency decomposition, independent component analysis (ICA), principal component analysis (PCA), and realistic electrical forward head and inverse EEG source modeling.
Makeig S, Gramann K, Jung T-P, Sejnowski TJ, Poizner (2009). Linking brain, mind and behavior: The promise of mobile brain/body imaging (MoBI). International Journal of Psychophysiology 73:985-100.
Delorme A, Makeig S, EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis (2004) Journal of Neuroscience Methods 134:9-21.
A Ojeda, N Bigdely-Shamlo, S Makeig. MoBILAB: An open source toolbox for analysis and visualization of mobile brain/body imaging data. Frontiers in Human Neuroscience, 2014.
See also:
- the EEGLAB Home pages,
- the LabStreamingLayer software distribution, and
- its mulitmodal, extensible XDF data format.
The following tutorials are available: Run MoBILAB from the MoBILAB GUI: MoBILAB operation Run MoBILAB from the Matlab command line: MoBILAB commands Head model module: https://github.com/aojeda/headModel
MoBILAB toolbox is hosted on GitHub: https://github.com/aojeda/mobilab
If you use this toolbox please cite our paper.
The MATLAB language enables the creation of programs using both procedural and object-oriented techniques.
Procedural programming typically represents data as individual variables or field of a structure and implements operations as functions that take the variables as arguments. Programs typically call a sequence of functions, each one of which is passed data and then returns modified data. As functions become too large and complex, a common approach is to break them into smaller functions. However, as the number of functions becomes large, designing and managing the data passed to functions becomes difficult and error prone.
Object oriented programming or OOP tries to overcome this issue identifying commonalities, differences, and interacting pieces of the software and encapsulates them in classes. As a real-life system can be decomposed into objects (example: car -> engine -> fuel system -> oxygen sensor), one piece of software can be decoupled into modules defined by natural logical boundaries. These modules (objects) provide interfaces by means of which they can interact with other modules (other objects or functions).
For more information about MATLAB OOP see MATLAB reference.
A class describes a set of objects with common characteristics. Objects are specific instances of a class as variable a may be from the built-in class single, double, or char (see example here). Objects from the same class may differ in the data contained in their properties but the functions defining how they operate on them and interact with external parameters may be common for all instances of a given class, these functions are called methods.
In MATLAB one might organize classes in hierarchies, this facilitates reusing the code and solutions to design problems that have been already solved. MoBILAB is been implemented using the following class hierarchies: 1) data sources: for importing data from different acquisition systems, 2) stream objects: for analysis of different types of MoBI data on continuous time, 3) epoch objects: for more traditional ERP analysis, and 4) stream browsers: for visualization and annotation of the different data-types.
Note for users not familiar with OOP: I heard somewhere a remark that I found very true -- paraphrased goes like this: It is OK if at first glance a new language looks alien to you -- every new language does until we get used to it (unless that language is Perl). Fortunately, MATLAB oop is much easier than Perl.
MoBILAB software is built from three independent functional modules: 1) data-specific objects that "know" what to do with their data set, 2) a datasource object that reads different data formats (locally or remotely) and serves the independent files to the data-specific objects, and 3) an object called "mobilab" that lives in MATLAB's workspace, this object is in charge of controlling the GUI and drawing data-dependent menus depending on the information provided by the datasource. The design concept is to decouple modules that are independent (such as the gui, file formats, and data processing) so each of them can be replaced, extended, or re-implemented without making dramatic changes to the whole system, e.g., preserving very large, still-stable parts of the code. The figure below shows a schematic of this architecture:

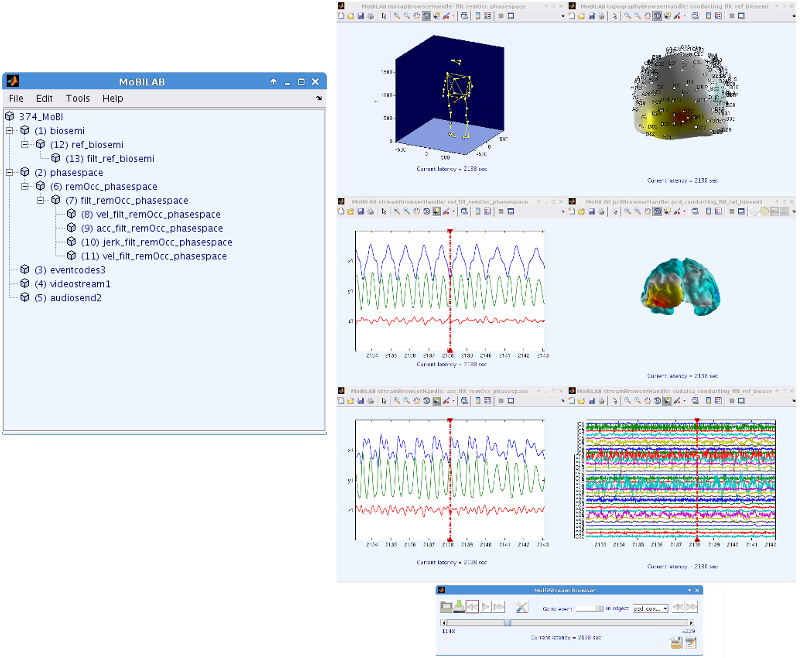
Figure 1: This figure shows the architecture of the MoBILAB toolbox. From left to right: Through the MoBILAB GUI new or existent datasets can be loaded, depending on their format (i.e., LSL, EEGLAB, MoBILAB, DataRiver, etc.) . A corresponding datasource is instantiated. This object "knows" how to read system-specific formats; it identifies the different streams of data and creates data-specific objects to handle them. Each data-specific object (or stream object) defines methods for processing and visualization of the data it handles. The arrows show that each module can communicate bi-directionally with any other it interfaces with. Each stream object handles two files, a header that provides the metadata that informs its properties and a binary file that is memory mapped, allowing its data (no matter how large) to be accessed through its data field.
This section presents a brief definition of each class used in MoBILAB plus an example showing how to use and extend them. These classes are listed in the table below; clicking on a class will jump to its documentation.
| Class name | Inherit from | Description |
|---|---|---|
| mobilabApplication | handle | The role of the class mobilabApplication is to control the graphic user interface (gui). The gui is created at time of execution by embedding a Java JTree component in the main window. This interactive tree is created to display parent-child relationships between the objects provided by the dataSource. Then a context menu is constructed for each object (element on the tree); this exposes different options for computation, annotation , and visualization. |
| dataSource | handle | This abstract class serves as a base for classes that implement interfaces between data acquisition systems and MoBILAB toolbox. |
| dataSourceXDF | dataSource | Imports into MoBILAB files recorded by the Lab Streaming Layer software framework (LSL) library and saved in xdf or xdfz format (XDF format) |
| dataSourceSET | dataSource | Imports EEGLAB set files into MoBILAB. If the set file contains an ICA decomposition it creates two objects, one for EEG and the other (its child) for the ICA data. |
| dataSourceMoBI | dataSource | Reads into MoBILAB the content of a folder containing MoBILAB files. |
| dataSourceDRF | dataSource | Imports into MoBILAB (earlier) files recorded by Datariver saved in drf format. |
| coreStreamObject | handle | Serves as a base to all objects that represent data in continuous time (i.e., not 'epoched' into a concatenated set of event-related data epochs). |
| dataStream | coreStreamObject | Serves as base class to all objects representing any modality of time series data. Defines methods for re-sampling, smoothing, filtering, plotting, fft and time frequency analysis. |
| eeg | dataStream, headModel | Defines analysis methods that operate exclusively on EEG data. |
| headModel | handle | Implements methods for solving EEG forward and inverse problems. |
| icaFields | handle | Defines the the ICA fields used in the EEGLAB EEG data structure. |
| icaEEG | eeg, icaFields | Serves as a placeholder for ICA analysis of EEG data. Derived from the eeg class incorporating its ICA fields. |
The role of the class mobilabApplication is to control the graphical user interface (gui). The gui is created on execution time by embedding a Java JTree component in the main window. This interactive tree is created to display parent-child relationships between the objects provided by the dataSource. Then, a context menu is constructed per object (element on the tree) exposing different options for computation, annotation, and visualization. MoBILAB toolbox can be launched running the command runmobilab or if it is working as a plugin for EEGLAB, clicking on the menu File - Import data -> Into MoBILAB, which also runs underneath the command runmobilab. This command creates an instance of the class on the workspace called mobilab. Although when an object, as a variable, could have any name, we will stick to the convention of calling 'mobilab' to objects from this class.
| Property name | Type | Description |
|---|---|---|
| allStreams | dataSource | Data source object containing a list of stream objects. |
| doc | string | URL of the documentation online. |
| path | string | Path to the directory where MoBILAB toolbox is being installed. |
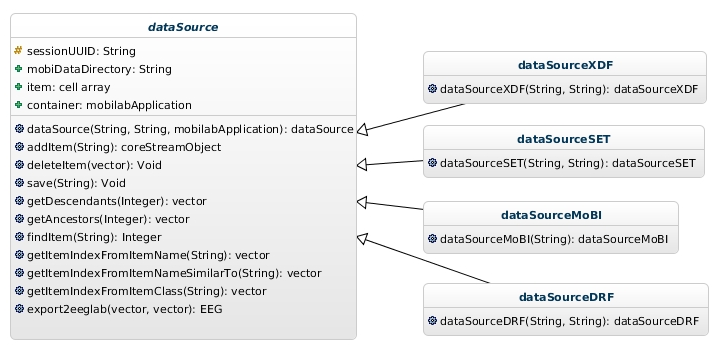 Figure 2: Data source hierarchy of classes.
Figure 2: Data source hierarchy of classes.
This is an abstract class that serves as a base for classes that implement interfaces between data acquisition systems or intermediate data formats and MoBILAB toolbox.
Property name |
Type |
Description |
|---|---|---|
sessionUUID |
string |
Universal unique identifier that links all the data sets belonging to the same session. A session is defined as a raw file or collection of files recorded the same day and its derived processed files. To guarantee that files within the same session are traceable, the sessionUUID code is attached at the end of each file name.
|
mobiDataDirectory |
string |
Path to the directory where the collection of files of one session are stored. |
item |
cell array |
Cell array (list) of objects (handles), where each object represents one single data set. The objects connect to binary and header files in the mobiDataDirectory and come up to live as functional entities that "know" what to do with the data sets they handle. |
container |
mobilabApplication |
The term container is used to refer to an object that can perform certain operations on the contained object but not the other way around. In this case, for a dataSource object the container is an object from the class mobilabApplication. The role of mobilabApplication is controlling the GUI, but because GUI needs change rapidly, a different class can be implemented to satisfy the new requirements in the front-end and still serve as container to the dataSource. This is a way of separating GUI programming from the rest of the application that may be more stable. |
Method name |
Description |
Input arguments |
Output arguments |
Usage |
|---|---|---|---|---|
dataSource |
Creates a dataSource object. As this is an abstract class it cannot be instantiated on its own. Therefore, this constructor must be call only within a child class constructor. |
|
dataSource (handle) |
|
export2eeglab |
Export an EEG structure combining multiple streams and event markers. This method takes care of aligning and re-sampling objects with different sampling rates. The first argument is a vector with the indices of the objects whose data will be concatenated (by the channel dimension) one after the other to make EEG.data. The second argument is a vector with the indices of the objects whose event markers will be used to populate EEG.event. |
|
EEG (EEGLAB structure) |
|
This class imports into MoBILAB files recorder by the Lab Streaming Layer (LSL) library saved in xdf or xdfz format.
Method name |
Description |
Input arguments |
Output arguments |
Usage |
|---|---|---|---|---|
dataSourceXDF |
Creates a dataSourceXDF object. |
|
|
|
This class imports EEGLAB .set files into MoBILAB. If the .set file contains an ICA decomposition it creates two objects, one for EEG and the other one (a child of the former) for the ICA data.
Method name |
Description |
Input arguments |
Output arguments |
Usage |
|---|---|---|---|---|
dataSourceSET |
Creates a dataSourceSET object. |
|
|
|
This class reads into MoBILAB the content of a folder containing MoBILAB files.
Method name |
Description |
Input arguments |
Output arguments |
Usage |
|---|---|---|---|---|
dataSourceMoBI |
Creates a dataSourceMoBI object. |
|
|
|
This class imports into MoBILAB files recorder by Datariver saved in drf format.
Method name |
Description |
Input arguments |
Output arguments |
Usage |
|---|---|---|---|---|
dataSourceDRF |
Creates a dataSourceDRF object. |
|
|
|
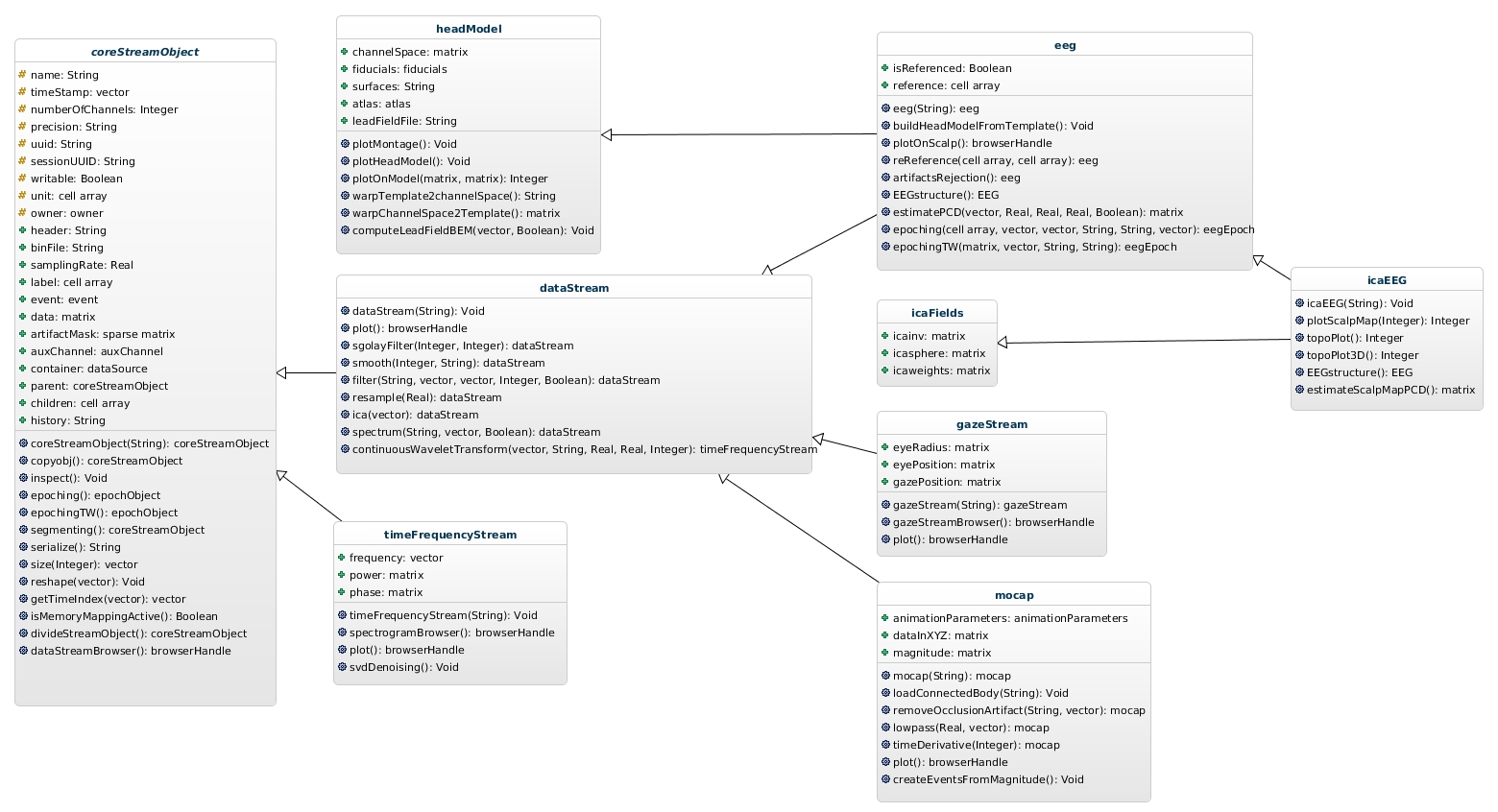 Figure 3: Data stream hierarchy of classes.
Figure 3: Data stream hierarchy of classes.
The class coreStreamObject serves as base to all stream objects.
| "Property name | Type | Description |
|---|---|---|
| name | string | Name of the object. It usually reflects the operation that has created the object. |
| timeStamp | vector of doubles | Array of numbers containing the time stamps of each sample in seconds. |
| numberOfChannels | integer | Number of channels. |
| precision | string | Data precision. |
| uuid | string | Universal unique identifier. Each object has its own uuid. |
| sessionUUID | string | UUID common to all the objects belonging to the same session. A session is defined as a raw file or collection of files recorded the same day and its derived processed objects. |
| writable | logical | If true the object can be modified and even deleted, otherwise is considered raw data and it cannot be modified. |
| unit | cell array | Cell array of strings specifying the unit of each channel. |
| owner | struct | Contact information of the person who has generated/recorded the data set. It has the fields username, email, and organization. Even when supplying this information is optional it is highly recommended to do so. This info will keep other users from modifying a data set created by somebody else, for instance objects can be deleted only by its owner. |
| header | string | Pointer to the file containing the metadata, it has the following structure:path + object name + _ uuid _ sessionUUID .hdr |
| binFile | string | Pointer to the file containing the data that is going to be memory mapped. It has the following structure:path + object name + _ uuid _ sessionUUID .bin |
| samplingRate | double | Sampling rate in hertz. |
| label | cell array | Cell array of strings specifying the label of each channel. |
| event | event | Event object that contain event markers relative to the time base of the object that serves as container. |
| data | matrix | Matrix size number of samples (same as time stamps) by numberOfChannels. Data is a dependent property that access the time series stored in a binary file through a memory mapping file object. This makes possible working with data sets of any size, even if they don't fit in memory. |
| artifactMask | sparse matrix | Sparse matrix with the same dimensions of "data". Non-zero entries contain a number between 0 and 1 with the probability of the correspondent sample to be an artifact. |
| auxChannel | structure | Auxiliary channels whose type may be not the same as the data contained in obj.data. For instance a Trigger channel will go here. auxChannel.data contains the data and auxChannel.label the channel labels. |
| container | dataSource | Pointer to the dataSource object. |
| pointer | coreStreamObject | Pointer to the parent object. Object containing raw data have no parent, in this case it returns empty. |
| children | cell array | Cell array containing immediate descendant objects. |
| history | string | Command who has created the object. Calling this command will produce exactly the same data set. This field is very useful for the creation of scripts. |
This class serves as base class to all objects that representing some modality of time series data. It defines methods for re-sampling, smoothing, filtering, plotting, fft, and time frequency analysis.
The class eeg defines analysis methods exclusively for EEG data. It inherits signal processing methods from dataStream and incorporates routines for custom artifact rejection, filtering, and source localization. It also inherits from headModel fields and methods for co-registration of head shape and sensor coordinates, anatomic labeling of the cortical surface, and fast computation of the lead field matrix of realistic geometries by means of OpenMEEG.
Property name |
Type |
Description |
|---|---|---|
channelSpace |
number of sensors by 3 matrix |
xyz coordinates of the sensors. Inherited from headModel. |
fiducials |
structure |
xyz of the fiducial landmarks: nassion, lpa, rpa, vertex, and inion. Inherited from headModel. |
surfaces |
string |
Pointer to the file where the surfaces representing different layers of tissue are stored. The surfaces |
atlas |
structure |
Atlas correspondent to the most internal of the surfaces (gray matter). Inherited from headModel. |
leadFieldFile |
string |
Pointer to the file where the lead field matrix was stored. Inherited from headModel. |
isReferenced |
logical |
It reflects whether an EEG data set has been re-referenced of not. |
reference |
cell array |
Labels of the channels used to compute the reference. |
style="width: 15%; | Method name |
Description |
Input arguments |
Output arguments |
Usage |
|---|---|---|---|---|
readMontage |
Reads a file containing the sensor positions and its labels. If recorder, it also extracts fiducial landmarks. If the number of channels in the file are less than the number of channels in the object, a new object is created with the common set. If more than one file is supplied the program creates separate objects containing the different set of channels selected by each file. |
|
None |
|
buildHeadModelFromTemplate |
Builds an individualized head model that matches the channel space (sensor positions) of the subject. This methodology represents an alternative to cases where the MRI of the subject is not available. If no arguments are passed the program uses a default head model specified in mobilab.preferences.eeg.headModel |
|
None |
For plotting the resulting head model it uses the method plotHeadModel, explained below in this table. |
plotMontage |
Plots a figure with the xyz distribution of sensors, fiducial landmarks, and coordinate axes. |
None |
|
|
plotHeadModel |
Plots the different layers of tissue, the sensor positions, and their labels. It colors different regions of the cortical surface according to a defined anatomical atlas. Several interactive options for customizing the figure are available. |
None |
|
|
artifactsRejection |
Automatic artifact rejection routines developed by Christian Kothe. It is highly recommended to run this pipeline before running ICA. No segments will be removed from the resulting stream object, although hopeless segments will be marked as artifacts in the field artifactMask. The pipeline implements different cleaning strategies in the following order: |
|
|
|
plotOnScalp |
Plot EEG data on the surface of the scalp. |
None |
None |
|
computeLeadFieldBEM |
Solves the forward problem of the EEG using Boundary Element Method on a realistic geometry. It uses OpenMEEG toolbox [1]. The computed lead field matrix is stored as a file in the field leadFieldFile. |
|
None |
|
estimatePCD |
Computes the posterior distribution of the Primary Current Density (PCD) given a topographic voltage. Being J a vector of PCD at each cortical dipole (vertices in the cortical surface) and V the EEG signal in channel space (measured on the scalp), the program computes the posterior distribution of the parameters J given the data V solving two levels of inference: 1) optimization of parameters J, and 2) optimization of hyper-parameters α and β. Level of inference 1 is addressed maximizing the posterior probability of the parameters J given the data V and hyper-parameters α and β: |
|
|
|
plotOnModel |
Plots cortical/topographical maps onto the cortical/scalp surface. |
|
|
|
The headModel class encapsulates common routines used for solving the forward and inverse problem of the EEG and it is now hosted in a separate repository.
https://github.com/sccn/mobilab/wiki/files/MotionTrackerCorrectionTool1.00.zip












