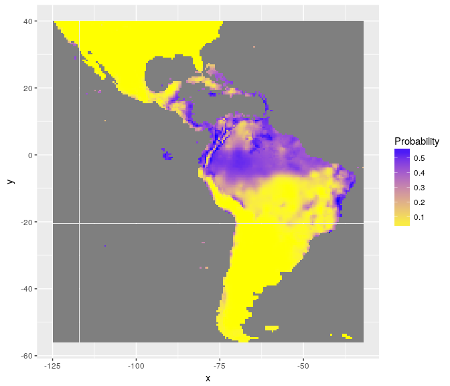
The R package SDMSelect is for species distribution modeling (SDM). It uses a forward model selection with cross-validation to select covariates and model specifications that allow for best predictions. This selection is not specifically for SDM. Results can then be used for mapping predicted species distribution and uncertainty of prediction.
Please give credit where credit is due and cite R and R packages when
you use them for data analysis. To cite SDMSelect properly, call the R
built-in command citation("SDMSelect"):
Sébastien Rochette. (2019, June 25). statnmap/SDMSelect: SDMSelect: A R-package for cross-validation model selection and species distribution mapping (Version v0.1.5). Zenodo. http://doi.org/10.5281/zenodo.3256536
Before installing SDMSelect, be sure to have updated versions of
mgcv (>= 1.8-19), dplyr (>= 0.7) and corrplot (>= 0.82).
To download the development version of the SDMSelect package, type the
following at the R command line:
install.packages("devtools")
devtools::install_github("statnmap/SDMSelect")Build with vignettes:
devtools::install_github("statnmap/SDMSelect", build_opts = c("--no-resave-data", "--no-manual"))To be able to test the vignettes, you will previously need to install
knitr, rmarkdown, dismo and rasterVis. Vignette are available on
{pkgdown} website for covariates
selection
and for species distribution model
selection
Note that spatial libraries like rgdal and sp may require additional
softwares to be installed on your computer if you work with Mac or
Linux. Look how to install proj4, geos and gdal on your system.
namespace 'mgcv' 1.xx is being loaded, but >= 1.8.19 is required. This means that you need to updatemgcvlibrary:install.packages("mgcv", force = TRUE).- Error when building vignette:
win.asp is not graphical parameter. This means you did not install the last version ofcorrplot:install.packages("corrplot", force = TRUE) there is no package called 'dismo' ... cannot create a RasterLayer object from this file (file does not exist): install librarydismo:install.packages("dismo")
Vignettes have been created to show how to use the library for covariate
selection on simple cases (vignette(package = "SDMSelect")).
-
First case is covariate selection procedure for classical dataset (not geographical data, nor species distribution data):
vignette("Covar_Selection", package = "SDMSelect"). See the vignette in the {pkgdown} website: https://statnmap.github.io/SDMSelect/articles/Covar_Selection.html -
The second case is for spatial data of species occurence to produce predicted species distribution maps and maps of uncertainties:
vignette("SDM_Selection", package = "SDMSelect"). See the vignette in the {pkgdown} website: https://statnmap.github.io/SDMSelect/articles/SDM_Selection.html
Note that most figures of the vignette are saved in “inst” so that
model selection is not run during vignette building. However, code in
the vignettes can be run on your own computer and should return the same
outputs. You can also find the path of the complete vignettes to be run
on your computer with:
system.file("Covar_Selection", "Covar_Selection.Rmd", package = "SDMSelect")
and
system.file("SDM_Selection", "SDM_Selection.Rmd", package = "SDMSelect").
Open and click knit button if you are on Rstudio. (This may require
5-10 minutes depending on the number of cores of your system).
Main functions are listed in the library general help: ?SDMSelect.
I decided to transform into a R package some R-scripts I have been using
for years for my studies with species distribution modelling. I know it
could be improved in many ways, but it works as is.
This library has originally been created for covariates selection to
predict species distribution (Biomass, density or presence/absence). Its
final aim is thus to produce maps of predicted distributions (Look at
vignette SDM_Selection). However, the core of the library is a N-times
k-fold cross-validation selection procedure that can be applied to any
kind of data, provided that model parameters are well defined (Look at
vignette Covar_selection).
This works with dataframe, SpatialPointsDataFrame and tibbles.
This has been designed to run in parallel in multiple cores computer,
using library parallel. The only steps missing in this library are
data exploration and data cleansing. These are important parts of
modelling and should be realized prior to model selection procedure.
The model selection procedure will test different combinations of
covariates with LM, GLM, GLM natural splines and GAM models, with
different distributions (Gaussian, Gamma, Log-Normal, Tweedie; Binomial)
and with different maximum degrees of freedom for GLM with polynoms or
natural splines. The multiple k-fold cross-validation realised on the
same folds for each model/submodel type allows their comparison using
cross-validated RMSE or AUC. Covariates correlation can be tested prior
to the selection procedure to avoid fitting models with correlated
covariates. However, the cross-validation procedure is coupled with a
forward stepwise procedure. This means that covariates are added to best
models selected at the previous step. This obviously not choose a new
covariate that add no predictive power to a model, thus avoiding
selecting correlated covariates in the same model.
Models are ordered according to RMSE (or AUC for presence-absence). The
cross-validation produces a distribution of (N*k) RMSE values for each
model at each step. Models are ordered according to mean RMSE. RMSE
distributions of model 2 to n are then statistically compared to the
best model. Models not statistically worse than the best model are
retained for the next step. After the stepwise procedure, the best model
among all is selected with the same method. Models not statistically
worse are also retained.
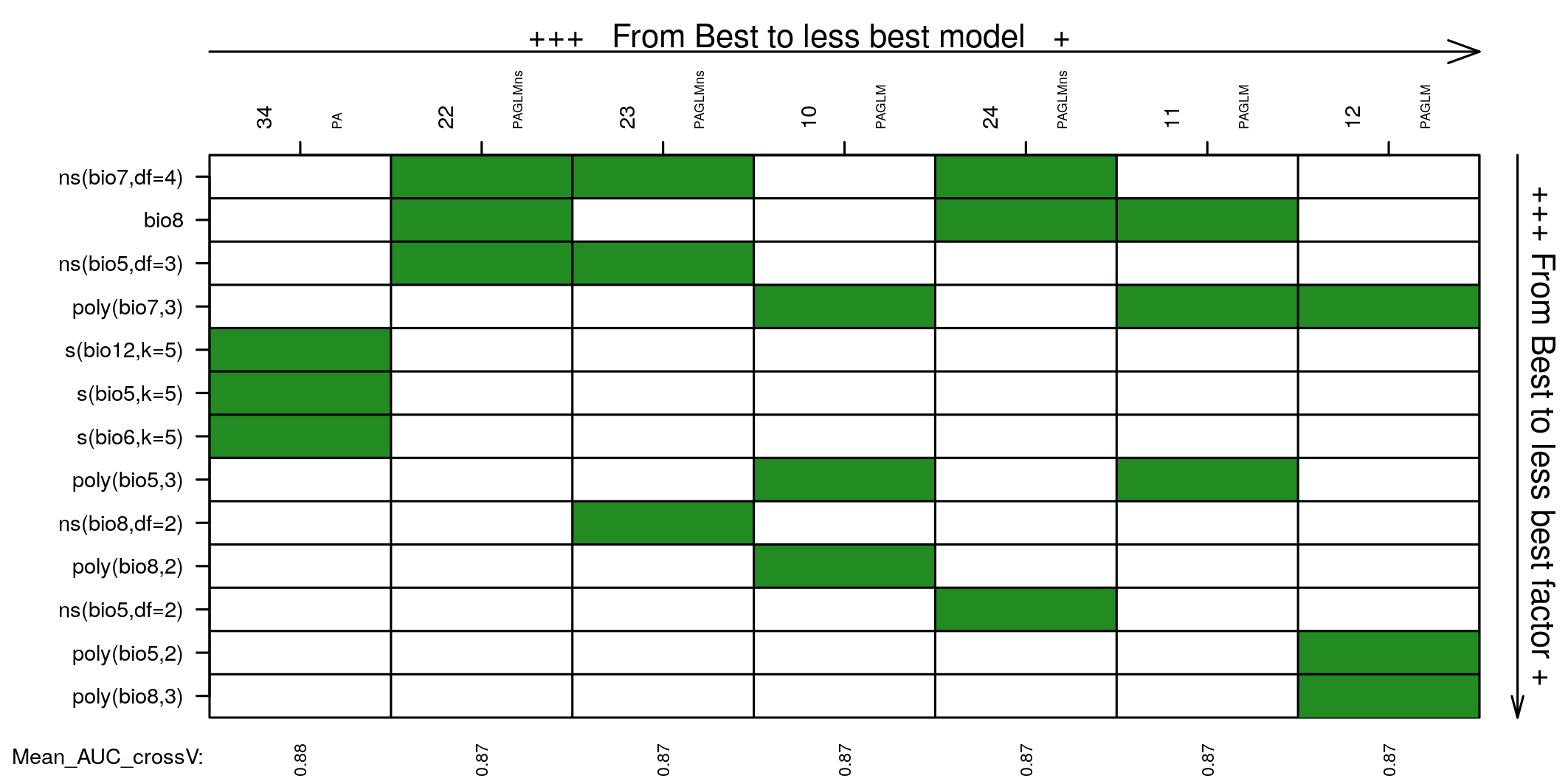
Models and covariates in models not statistically different from the best one. Covariates are ordered based on occurrence in the models selected.
Outputs of the selection procedure are numerous, allowing for summary of the model selection and the comparison of the different models alltogether (GAM, GLM; Gaussian, Gamma, Log-Normal, Tweedie; Binomial). All output files and figures are stored in a common folder and are not showing up in the R session directly. The final model retained by the user can then be analysed (residual analysis, variance analysis, effect of covariates). For spatial data, maps of prediction can be produced.
A particular attention has been given to assessment of uncertainty. Each prediction of a model is given with a standard error associated. This standard errors have been used to estimate possible minimum and maximum distributions of species (through estimations of quantiles).
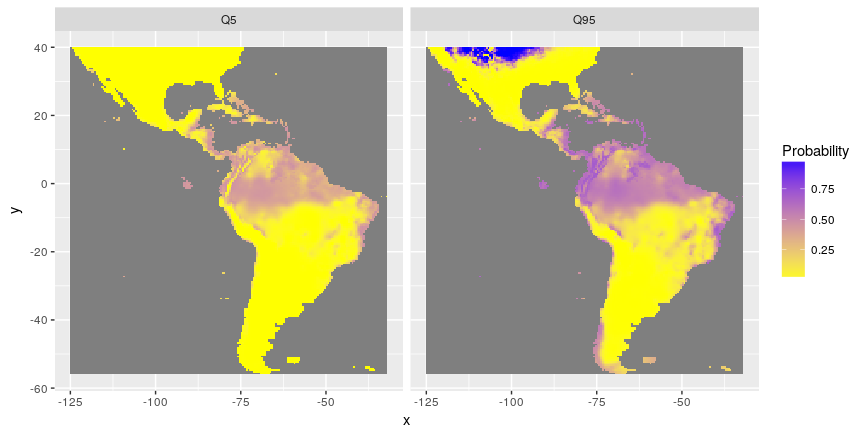
Distribution of minimum (quantile 5%) and maximum (quantile 95%) of probabilities of presence with SDMSelect library on Rstat
Concerning presence-absence data, the estimation of probability of presence is not enough. The balance between presences and absences in data may conduct to biased predictions. The best threshold value to classify a probability of presence in presence or absence is here explored in more details. Map of probability to be over the threshold value is calculated.
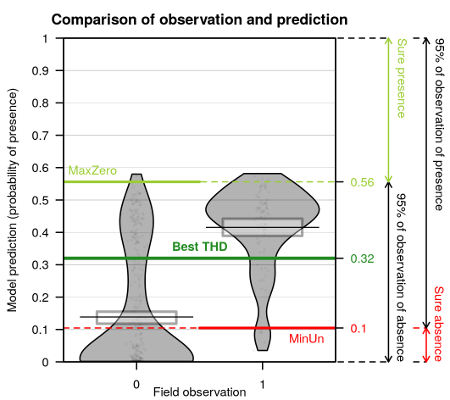
Comparison of predictions against observations in a presence-absence data model and thresholds values with SDMSelect library on Rstat
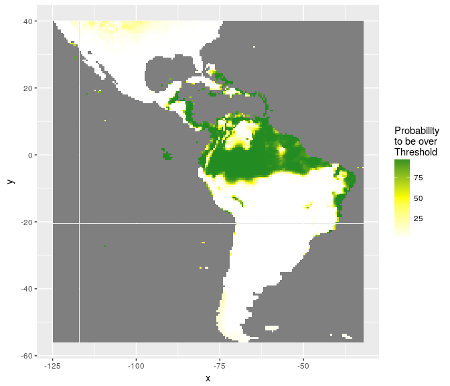
Distribution of probabilities to be over the best threshold value separating presences from absences with SDMSelect library on Rstat
This library relies on a lot of other R-packages, which means that any
modifications of those may prevent this library to work correctly. As I
continue to use it regularly, I may see if some updates broke some of my
functions and would try to fix them.
There are recent libraries like dplyr, tidyr, ggplot2 that did not
originally exist when I first created these R-scripts. I now try to
implement them from time to time.
This library is not as complete as can be library(caret) in terms of
model types available, however, as far as I know, caret model
selection procedure is designed for a unique combination of
model/distribution. Here, cross-validation outputs of each
model/distribution are kept, and can then be compared all together with
paired comparison. Not only the model with the best mean RMSE (or AUC)
is retained, but also all other models giving predictions statistically
as good as the best one. For biological purpose, knowing all models with
the same predictive power may modify outputs interpretation. Another
difference is that SDMSelect package is designed for predicting
species distribution models, which means that some outputs are maps of
species distributions as well as all maps of uncertainty that can be
deduced from model outputs.
For your work on covariates selection and species distribution modelling, you can contact me. Vignettes will give you a good starting point. I’ll be happy to participate to scientific collaborations based on this R-package. More information on my website https://statnmap.com.
If you want to participate in improving this library, please have a look
at my todo list (in Vignettes directory or in the pkgdown
site) and feel
free to clone, modify and provide pull requests. Please run the two Rmd
files in inst folder before submitting a pull request. These are the
complete versions of the two vignettes.
See full documentation realized using {pkgdown} at https://statnmap.github.io/SDMSelect/
This package is free and open source software, licensed under GPL.