All materials associated with the chapter for Methods in Molecular Biology entitled,
"A practical guide to reproducible modeling for signaling networks" are contained in this
repository.
Explore the docs »
View Demo
·
Report Bug
·
Request Feature
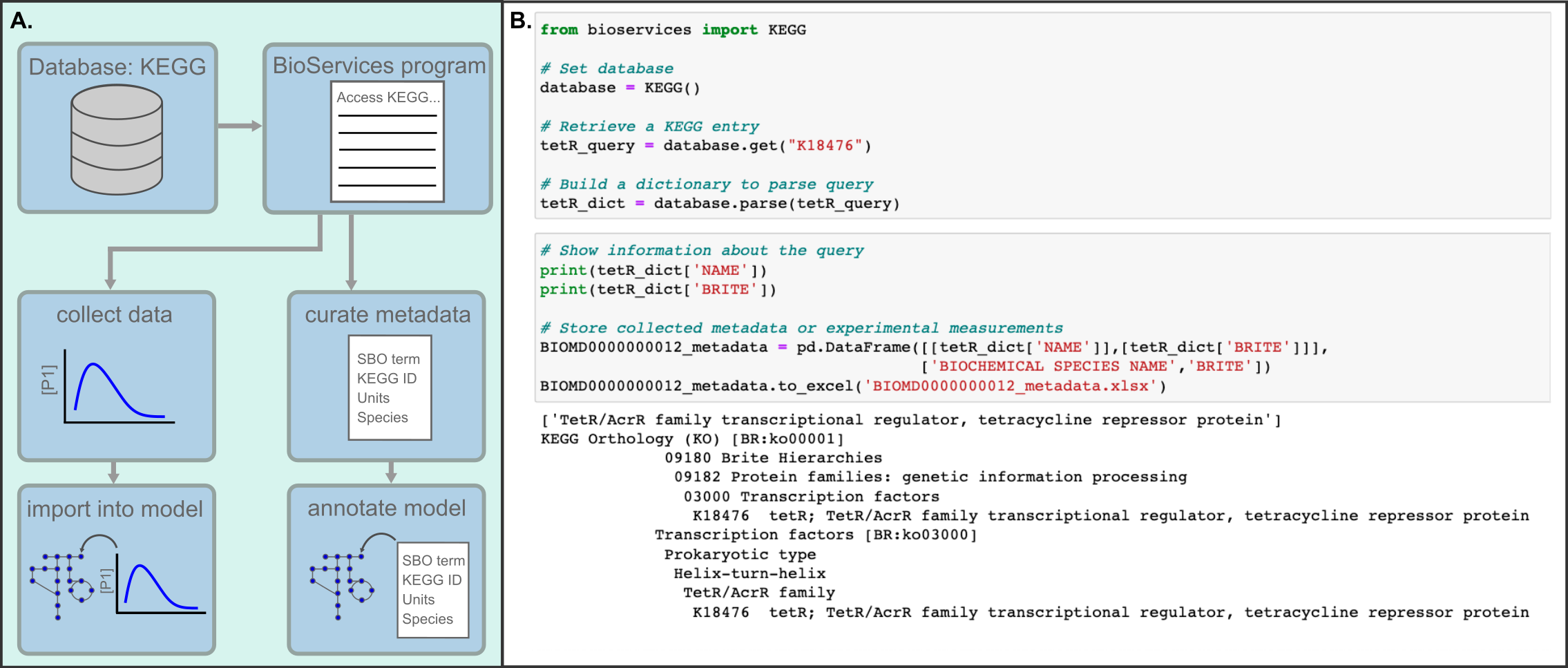
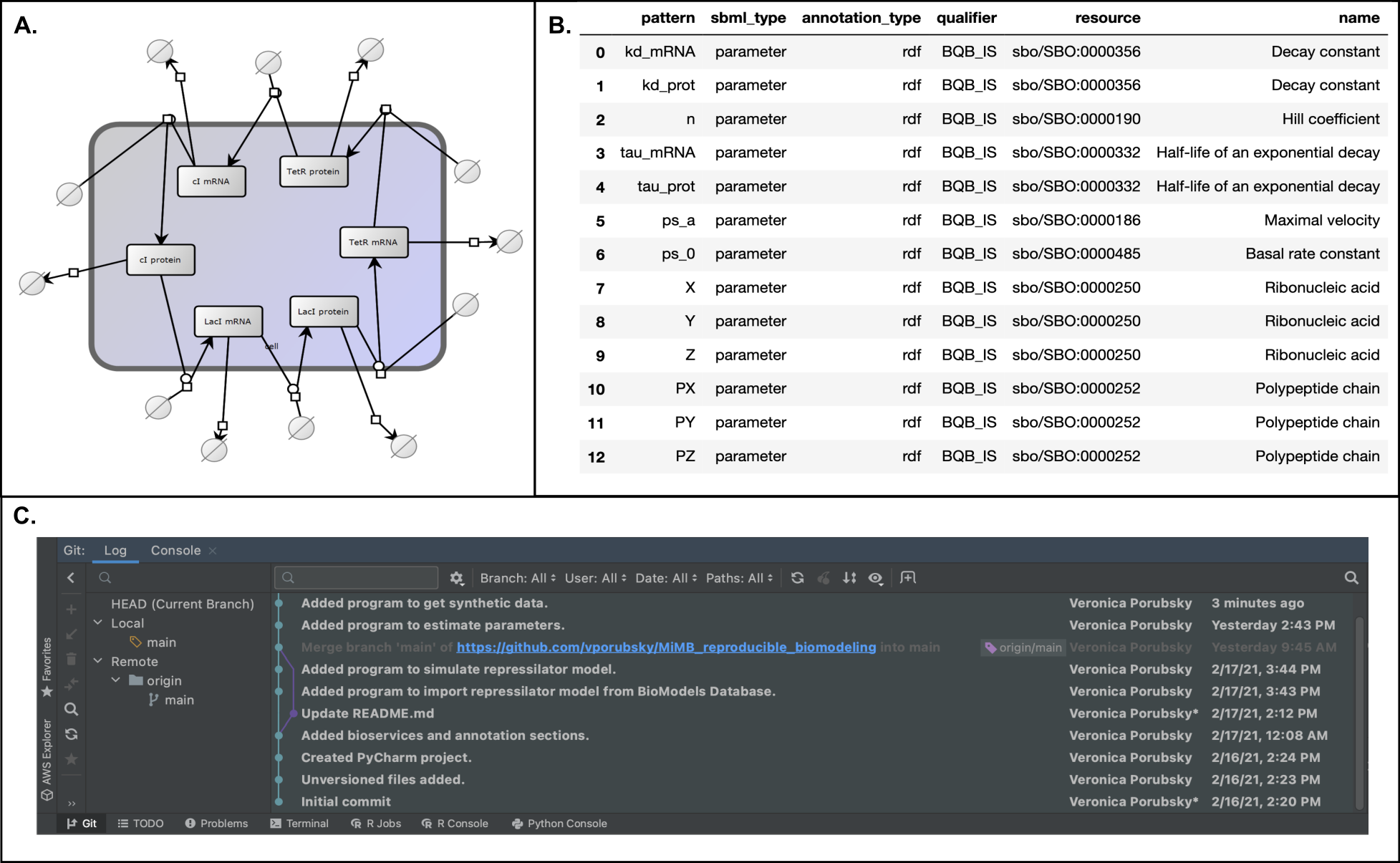
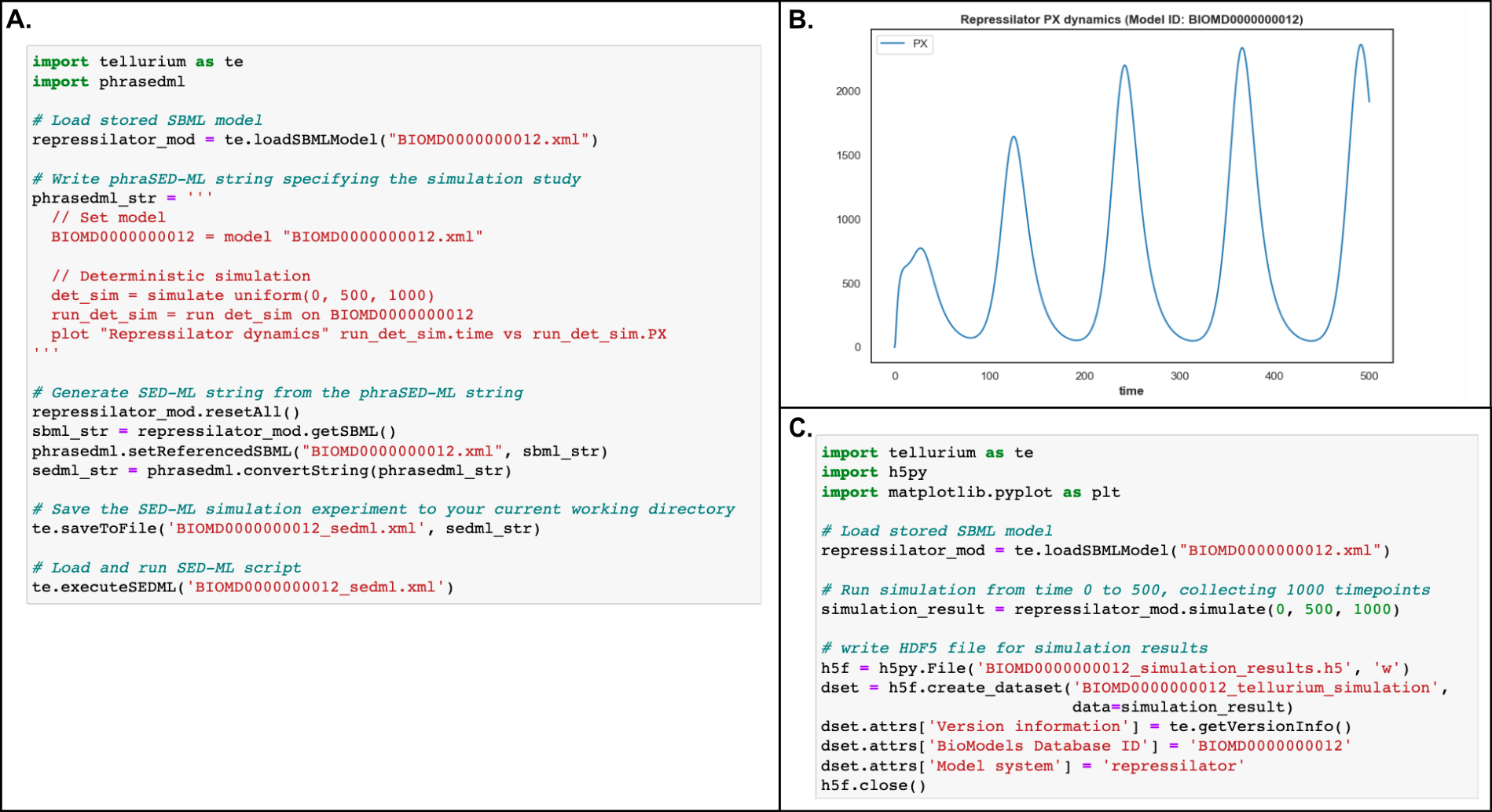
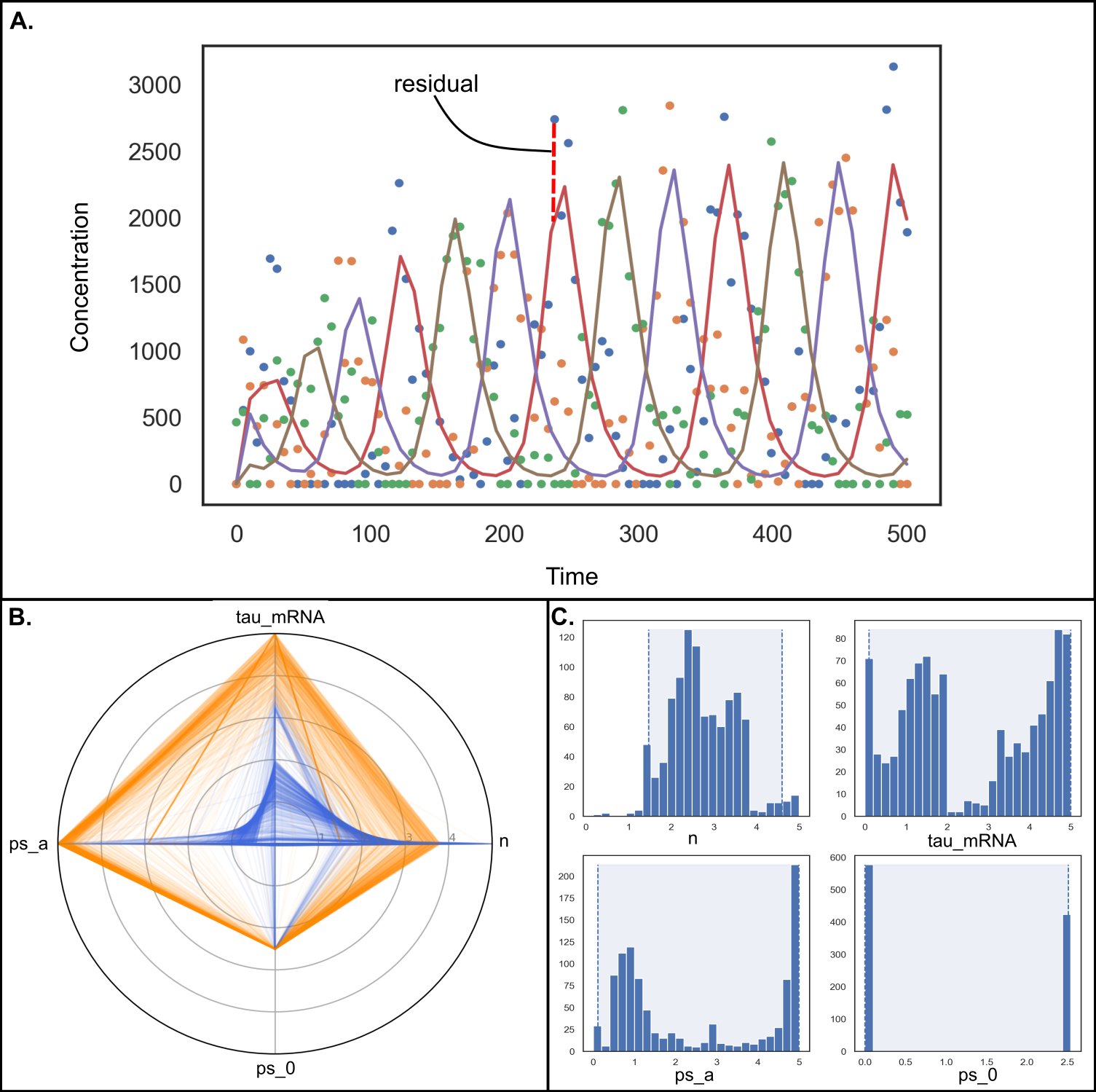
While scientific disciplines revere reproducibility, many studies - experimental and computational alike - fall short of this ideal and cannot be reproduced or even repeated when the model is shared. For computational modeling of biochemical networks, there is a dearth of formal training and resources available describing how to practically implement reproducible methods, despite a wealth of existing tools and formats which could be used to support reproducibility. This chapter points the reader to useful software tools and standardized formats that support reproducible modeling of biochemical networks and provides suggestions on how to implement reproducible methods in practice. Many of the suggestions encourage readers to use best practices from the software development community in order to automate, test, and version control their model components. A Jupyter Notebook demonstrating several of the key steps in building a reproducible biochemical network model is included to supplement the recommendations in the text.
You can also try out the study on Google Colab: https://colab.research.google.com/drive/1HgorZNOUyE_3wVNJdsR3ZQrG5qdk0tVi?usp=sharing
To get a local copy up and running follow these simple steps.
All software dependencies required to run the code contained in this repository using the same versions employed during development.
- Python 3.8.2
- pip packages
pip install bioservices==1.7.11
pip install tellurium==2.1.6
pip install matplotlib==3.3.4
pip install IPython==7.12.0
pip install libsbgnpy
pip install sbmllint
pip install sbmlutils~=0.4.9
pip install phrasedml~=1.1.1
pip install h5py~=3.1.0
pip install pandas~=1.2.2
pip install seaborn~=0.11.0
pip install scikit-learn~=0.24.1
pip install lmfit~=1.0.1
Clone the repo
git clone https://github.com/vporubsky/MiMB_reproducible_biomodeling.gitThis is intended as an introduction to reproducible biochemical modeling in Python.
Distributed under the MIT License. See LICENSE for more information.
Veronica Porubksy - [email protected]
Project Link: https://github.com/vporubsky/MiMB_reproducible_biomodeling