-
Notifications
You must be signed in to change notification settings - Fork 61
Getting Started
The Ensembl REST API provides language agnostic programmatic access to data on the Ensembl database.
This API lets you:
-
Access Ensembl's gene, variation, comparative genomics and regulation data using a programming language of your choice.
-
Analyse variation data through the Linkage Disequilibrium (LD), Transcript Haplotypes and Variant Effect Predictor (VEP) endpoints.
-
Convert co-ordinates from one assembly to another.
The Ensembl REST API offers a stable service that is versioned with archives. It is read only, limited by network latency and does not cover our complete database. (For additional API access to the Ensembl database, you can use the Ensembl Perl API.)
About REST APIs. REST APIs use the HTTP protocol to perform request-and-response interactions between clients and servers (for example, your computer requests a resource and an API server responds to the request). The client making the request for the resource and the API server providing the response can use any programming language or platform — it doesn’t matter because the message request and response are made through a common HTTP web protocol.
REST APIs focus on resources (that is, things, rather than actions) and ways to access the resources. Resources are typically different types of information. You access the resources through URLs (Uniform Resource Locators), just like going to a URL in your browser retrieves an information resource. The URLs are accompanied by a method that specifies how you want to interact with the resource.
- A GET method retrieves a resource.
- A POST method posts information, such as a list of IDs, to the server. It allows you to run a query with multiple inputs at once.
- A PUT method updates an existing resource.
- A DELETE method removes a resource.
The Ensembl REST API uses only the GET and POST methods. It uses the GET method to obtain information from the Ensembl database and the POST method to write to the database.
The Ensembl REST API does not require authentication.
First, test your connection:
https://rest.ensembl.org/info/ping?content-type=application/json
If you get something that looks like this:
"ping":1
That means that everything is working properly.
Do two simple lookups:
Lookup 1:
First, find information for a symbol in a linked external database, using the syntax:
GET lookup/symbol/:species/:symbol
For the symbol and species parameters, use the gene symbol and species names provided by relevant external databases.
For example, using the species homo_sapiens and gene symbol BRCA2, you can use this cURL example:
curl 'https://rest.ensembl.org/lookup/symbol/homo_sapiens/BRCA2?expand=1' -H 'Content-type:application/json'
Lookup 2:
Now, dive a bit deeper into Ensembl resources by using the Ensembl stable ID to find the species and database for a single identifier, for example, a gene, transcript, or protein.
The syntax for this is:
GET lookup/id/:id
The one required id parameter is the Ensembl-generated stable ID.
Ensembl assigns stable IDs to features (such as genes, transcripts and proteins) to unambiguously identify these features in the Ensembl database. Although feature names can change, stable IDs continue to refer to the same genomic features.
If you don't know the stable ID of the feature you are interested in, you can use the search box on the main Ensembl website.
More details on getting the stable ID
For example, to get the Ensembl stable ID of the HGNC gene symbol ABCA1:
Point your browser to https://www.ensembl.org/index.html?redirect=no
In the search box, type in ABCA1.
You will get the following resulting page:
https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000165029;r=9:104781006-104928155
The page's heading lists the gene symbol ABCA1 followed by the Ensembl stable ID, which is
ENSG00000165029
Add in the stable ID to get the lookup results -- just type this into your browser:
https://rest.ensembl.org/lookup/id/ENSG00000165029?expand=1;content-type=application/json
Your results should look something like this:
{
"source": "ensembl_havana",
"display_name": "ABCA1",
"species": "homo_sapiens",
"object_type": "Gene",
"version": 16,
"description": "ATP binding cassette subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:29]",
"assembly_name": "GRCh38",
"start": 104781006,
"db_type": "core",
"Transcript": [
{
"version": 8,
"object_type": "Transcript",
"Translation": {
"id": "ENSP00000363868",
"end": 104903679,
"Parent": "ENST00000374736",
"object_type": "Translation",
"length": 2261,
"db_type": "core",
"start": 104784315,
"species": "homo_sapiens"
},
"display_name": "ABCA1-202",
"species": "homo_sapiens",
"Exon": [
{
"version": 2,
"end": 104928155,
"object_type": "Exon",
"strand": -1,
"seq_region_name": "9",
"id": "ENSE00001810407",
"db_type": "core",
"start": 104927935,
"species": "homo_sapiens",
"assembly_name": "GRCh38"
},
{
"end": 104903771,
"version": 1,
"object_type": "Exon",
"strand": -1,
"seq_region_name": "9",
"id": "ENSE00002201214",
"db_type": "core",
"species": "homo_sapiens",
"start": 104903614,
"assembly_name": "GRCh38"
},
...
],
"source": "havana",
"strand": -1,
"is_canonical": 0,
"seq_region_name": "9",
"end": 104928139,
"Parent": "ENSG00000165029",
"id": "ENST00000374733",
"biotype": "protein_coding",
"logic_name": "havana",
"start": 104861438,
"db_type": "core",
"assembly_name": "GRCh38"
}
],
"id": "ENSG00000165029",
"logic_name": "ensembl_havana_gene",
"biotype": "protein_coding",
"seq_region_name": "9",
"strand": -1,
"end": 104928155
}
Do this in cURL
$ curl 'https://rest.ensembl.org/lookup/id/ENSG00000165029?expand=1' -H 'Content-type:application/json' | json_pp
Do this in Python3
import requests, sys
server = "https://rest.ensembl.org"
ext = "/lookup/id/ENSG00000165029?"
r = requests.get(server+ext, headers={ "Content-Type" : "application/json"})
if not r.ok:
r.raise_for_status()
sys.exit()
decoded = r.json()
print(repr(decoded))
Additional endpoints. Here is the complete reference listing for all the Ensembl REST API endpoints.
An Ensembl REST URL has three main parts:
-
Base URL
-
Endpoint
-
Parameters
Base URL. The base URL is https:rest.ensembl.org.
Endpoint. An endpoint indicates which Ensembl resource you are interested in. Some examples:
-
The
/phenotype/accession/:species/:accessionendpoint indicates you are interested in phenotype annotations. -
The
/sequence/id/:idendpoint indicates you are interested in sequence information.
Parameters. Parameters specify details of how you want to interact with the resource. There are three main types of parameters:
-
Required
-
Optional
-
Message
Required parameters (also known as path parameters) are part of the endpoint itself. In the Ensembl REST documentation, path parameters are preceded by a colon. For example, the parameter for an Ensembl stable ID is :id.
Continuing with this example, the lookup/id/:id endpoint says that you want lookup information about the feature represented by a specific stable ID. In this case, you would replace :id with an actual stable ID like ENSG00000165029.
Optional parameters (also known as query and header parameters) are key-value pairs that are appended to the end of an endpoint using a question mark (?) to introduce the first parameter and a semi-colon (;) to introduce subsequent parameters.
Typically, you use these parameters to filter the information you want returned, as well as to specify the format.
For example, this endpoint uses the expand=1 query parameter to say that the response should include information not just about the gene, but also about its transcripts, translations and exons.
$ curl 'https://rest.ensembl.org/lookup/id/ENSG00000165029?expand=1'
Message parameters (also known as request body parameters) are typically used in POST operations. Depending on the language, they are sometimes preceded by the -d argument. In the Ensembl REST API, they often include an array of values.
Continuing with the simple GET lookup/id/:id example, first take a look at the API reference documentation for this operation:
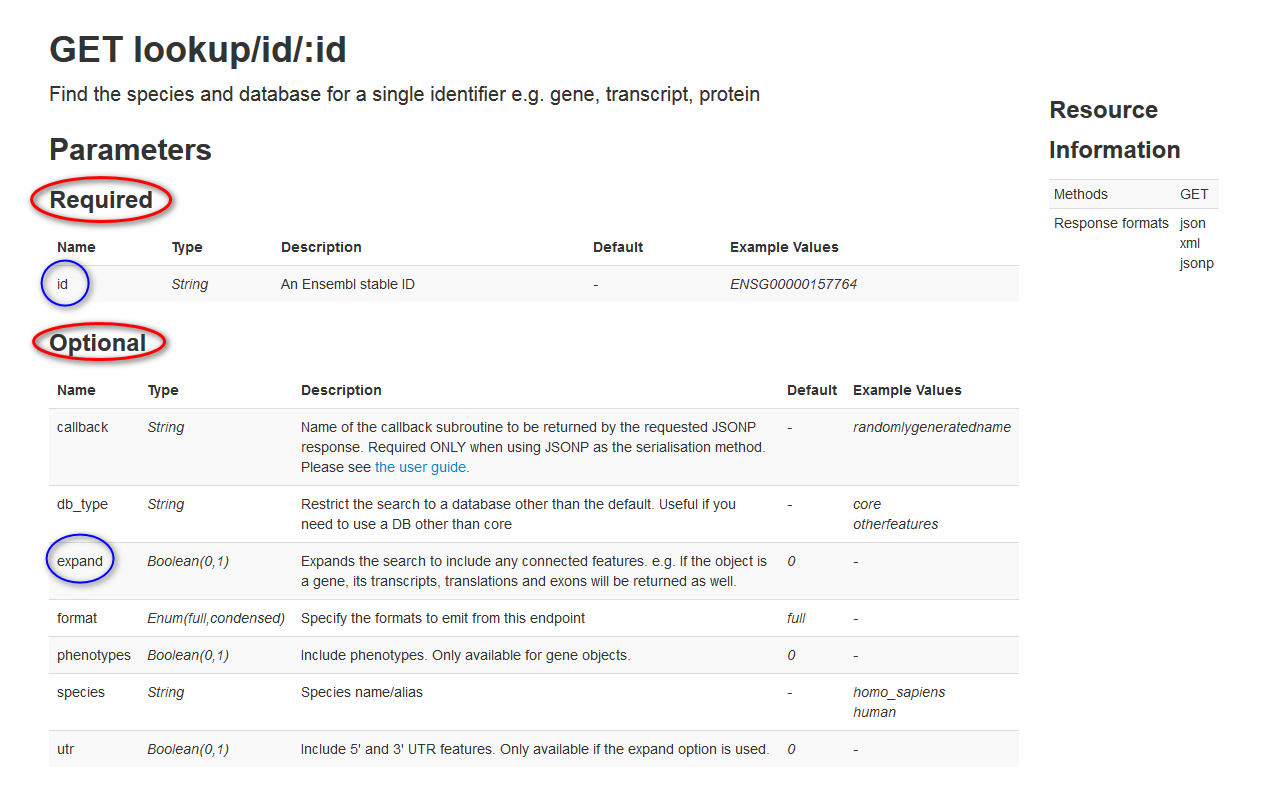
In this example, you need to supply the following parameters:
-
Required parameter: Replace the required
:idparameter with an Ensembl stable ID. In this example, use the stable ID ofENSG00000165029. -
Optional parameter: Set the optional
expandparameter to1. Settingexpandto1gives you information not just about the gene, but also about its transcripts, translations and exons. The syntax for this isexpand=1. -
Note on one other "generic" optional parameter. The
content-type=application/jsonparameter is used in many Ensembl endpoints to say that the response should be formatted in JSON. Because this parameter can be used in all the Ensembl endpoints, it is not explicitly called out in the documentation for each individual endpoint, but you will often see it in the sample requests, as shown below:
Sample cURL request
Here is the cURL request as you would type it in -- all on one line:
$ curl 'https://rest.ensembl.org/lookup/id/ENSG00000165029?expand=1' -H 'Content-type:application/json'

Now consider the POST vep/:species/id endpoint. This operation fetches variant consequences for multiple IDs. You provide these IDs in MESSAGE parameters.
Take a look at the reference API doc for the POST vep/:species/id endpoint:
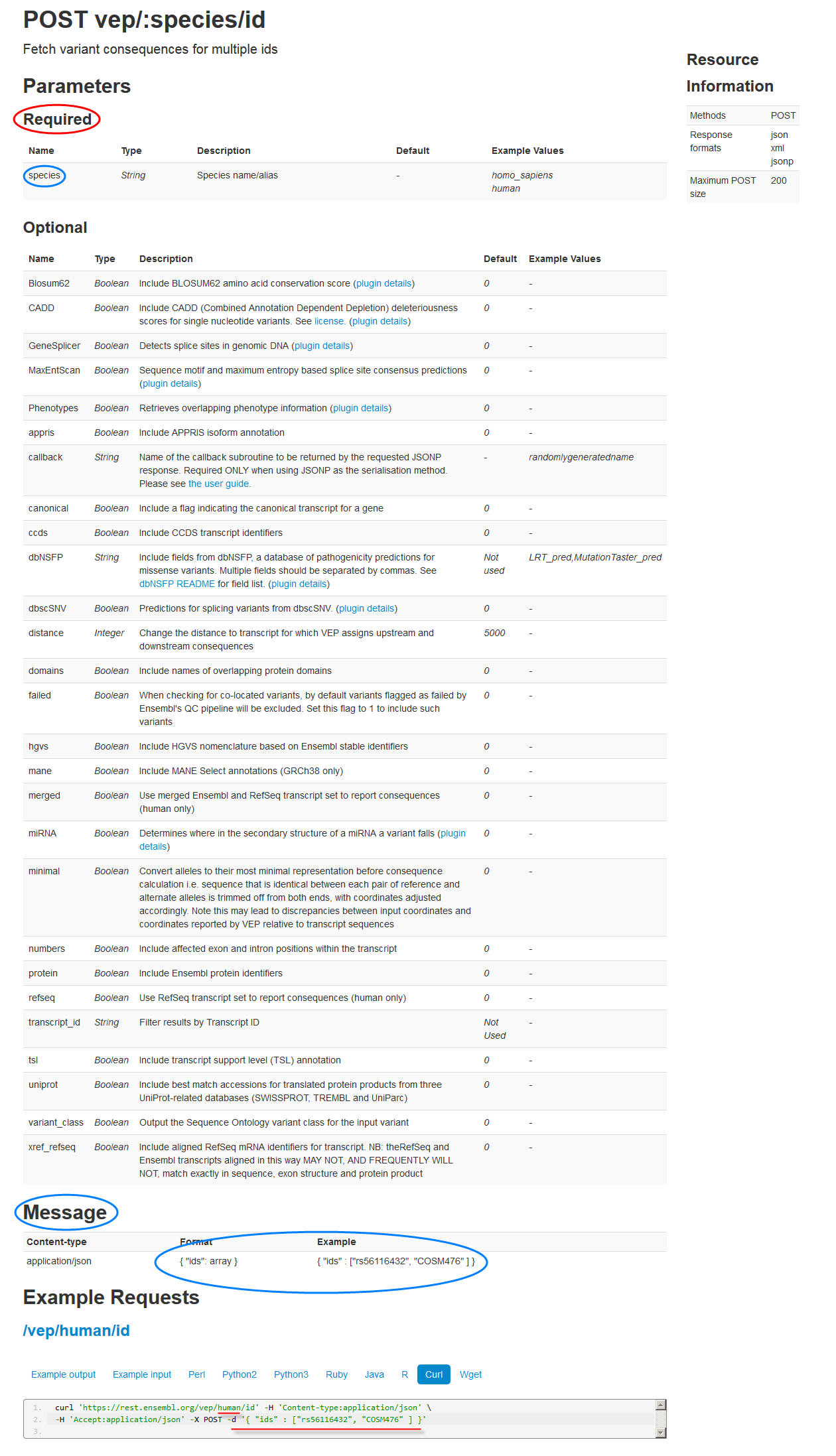
-
REQUIRED parameter: As you can see, the cURL example shows that you need to pass in the REQUIRED
:speciesparameter ofhuman. -
MESSAGE parameters: In addition, you need to use the cURL
-d '{ "ids" : ["rs56116432", "COSM476" ] }directive to pass in an array of the MESSAGE parameter IDs you want information for.In this case, the IDs are
rs56116432andCOSM476.
This section provides background information and examples on how to create useful Python, Perl and R scripts to access the Ensembl database.
At a high level, you:
-
Set variables to make requests
-
Handle errors
-
Decode responses
First step: Make sure you understand how to meet language dependencies, set request variables, handle errors and decode responses in either Python, Perl, or R.
Moving on: Assuming that you understand the basics of dependencies, request variables, error handling and response decoding, you can use the following "helper functions" at the start of each script to make things more efficient:
GET helper function
def fetch_endpoint(server, request, content_type):
"""
Fetch an endpoint from the server, allow overriding of default content-type
"""
r = requests.get(server+request, headers={ "Accept" : content_type})
if not r.ok:
r.raise_for_status()
sys.exit()
if content_type == 'application/json':
return r.json()
else:
return r.textPOST helper function
def fetch_endpoint_POST(server, request, data, content_type='application/json'):
r = requests.post(server+request,
headers={ "Content-Type" : content_type},
data=data )
if not r.ok:
r.raise_for_status()
sys.exit()
if content_type == 'application/json':
return r.json()
else:
return r.textGET helper function
# Fetch an endpoint from the server, allow overriding of the default content type
sub fetch_endpoint {
my $http = HTTP::Tiny->new();
my ($server, $extension, $content_type) = @_;
$content_type ||= 'application/json';
my $response = $http->get($server.$extension, { headers => { 'Accept' => $content_type } });
die "Error: ", $response->{status}, "\n" unless $response->{success};
if($content_type eq 'application/json') {
return decode_json($response->{content});
} else {
return $response->{content};
}
}
POST helper function
# Fetch an endpoint from the server, allow overriding of the default content type
sub fetch_endpoint_POST {
my $http = HTTP::Tiny->new();
my ($server, $extension, $data, $content_type) = @_;
$content_type ||= 'application/json';
my $response = $http->request( "POST", $server.$extension, { headers => { 'Accept' => $content_type }, content => $data });
die "Error: ", $response->{status}, "\n" unless $response->{success};
if($content_type eq 'application/json') {
return decode_json($response->{content});
} else {
return $response->{content};
}
}
GET helper function
Fetch_endpoint <- function(server, request, content_type){
"""
Fetch an endpoint from the server, allow overriding of default content-type
"""
r <- GET(paste(server, request, sep = ""), accept(content_type))
stop_for_status(r)
if (content_type == 'application/json'){
return (fromJSON(content(r, "text")))
} else {
return (content(r, "text"))
}
}
POST helper function
fetch_endpoint_POST <- function(server, request, content_type){
"""
Fetch an endpoint from the server, allow overriding of default content-type
"""
r <- POST(paste(server, request, sep = ""), content_type(content_type), accept(content_type), body = data)
stop_for_status(r)
if (content_type == 'application/json'){
return (fromJSON(content(r, "text")))
} else {
return (content(r, "text"))
}
}Problem: I get a ‘200’ HTTP status code, but no data
Possible Reason: A mis-spelt parameter, e.g.
https://rest.ensembl.org/info/analysis/homo_sapien?content-type=application/json
How to fix it: Correct the spelling, e.g.
https://rest.ensembl.org/info/analysis/homo_sapiens?content-type=application/json
Problem: I get an error: 'ERROR 404: Not Found' with wget or '{"error":"page not found. Please check your uri and refer to our documentation https://rest.ensembl.org/"}' in the browser
Possible Reason: A mis-spelt URL, e.g.
https://rest.ensembl.org/inf/analysis/homo_sapiens?content-type=application/json
How to fix it: Correct the spelling, e.g.
https://rest.ensembl.org/info/analysis/homo_sapiens?content-type=application/json
Problem: I get an error: '400 Bad Request' with wget or '{"error":"ID 'BRAF' not found"}' in the browser
Possible Reason: Gene symbol used for an endpoint that needs an Ensembl stable ID, e.g.
https://rest.ensembl.org/lookup/id/BRAF?content-type=application/json
How to fix it: Use the Ensembl stable ID, e.g.
https://rest.ensembl.org/lookup/id/ENSG00000157764?content-type=application/json
Problem: I get an error like '400 Bad Request' or an error like '{"error":"Variation?include_pubmed_id=1 is not a valid object type, valid types are: Gene, QTL, RegulatoryFeature, StructuralVariation, SupportingStructuralVariation, Variation"}'
Possible Reason: Incorrect use of ';' and '?', e.g.
https://rest.ensembl.org/phenotype/region/homo_sapiens/9:22125500-22136000?feature_type=Variation?include_pubmed_id=1;content-type=application/json
How to fix it: Separate optional parameters by ';', e.g.
https://rest.ensembl.org/phenotype/region/homo_sapiens/9:22125500-22136000?feature_type=Variation;include_pubmed_id=1;content-type=application/json
Write to the Ensembl helpdesk or join the developer (dev) mailing list:
http://www.ensembl.org/info/about/contact/index.html
Ensembl provides a training course that uses Jupyter Notebooks hosted by Microsoft Azure to walk you through the APIs and practise writing scripts to access Ensembl data.