-
Notifications
You must be signed in to change notification settings - Fork 8
Continuous Data
Matt Wheeler Ph.D. 4/26/2023
This file shows ToxicR for dichotomous benchmark dose analyses using both single model and multiple model fits. The first thing we need to do is load the data. From an excel file this can be done using the following R commands:
cont_data <- matrix(0,nrow=5,ncol=4)
colnames(cont_data) <- c("Dose","Mean","N","SD")
cont_data[,1] <- c(0,50,100,200,400)
cont_data[,2] <- c(5.26,5.76,6.13,8.24,9.23)
cont_data[,3] <- c(20,20,20,20,20)
cont_data[,4]<- c(2.23,1.47,2.47,2.24,1.56)
Y <- cont_data[,2:4]In what follows, I describe fitting the models using summary statistics.
One can also fit these models using the original data with no change to
the function call except the Y is a
now one column vector, and the dose vector is also
and corresponds to each entry in the data vector.
When summary data are used, ToxicR expects
to be a
matrix. The first column of
is the the mean, the second colunn is the number of units on test, and
the third column is the observed standard deviation. In what follows,
lets look at the Hill model fit, where the Hill model is
library(ToxicR)
#>
#>
#> _______ _____
#> |__ __| 🤓 | __ \
#> | | _____ ___ ___| |__) |
#> | |/ _ \ \/ / |/ __| _ /
#> | | (_) > <| | (__| | \ \
#> |_|\___/_/\_\_|\___|_| \_\
#> 23.4.1.1.0
#> ___
#> | |
#> / \ ____()()
#> /☠☠☠\ / xx
#> (_____) `~~~~~\_;m__m.__>o
#>
#>
#> THE SOFTWARE IS PROVIDED AS IS, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED,
#> INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A
#> PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT
#> HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF
#> CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE
#> OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
library(ggplot2)
hill_fit <- single_continuous_fit(cont_data[,"Dose"],Y,
model_type="hill")The last line fits the Hill Bayesian Laplace model and puts all of the information one needs in the `hill_fit’ object variable. This variable has the same structure as its dichotomous counterpart. By default the procedure assumes normal variance proportional to the mean. To see what model you fit, simply type the following line:
hill_fit$full_model
#> [1] "Model: Hill Distribution: Normal-NCV"
hill_fit$prior
#> Hill Model [normal-ncv] Parameter Priors
#> ------------------------------------------------------------------------
#> Prior [a]:Normal(mu = 1.00, sd = 1.000) 1[-100.00,100.00]
#> Prior [b]:Normal(mu = 0.00, sd = 1.000) 1[-100.00,100.00]
#> Prior [c]:Log-Normal(log-mu = 0.00, log-sd = 2.000) 1[0.00,100.00]
#> Prior [d]:Log-Normal(log-mu = 0.47, log-sd = 0.421) 1[0.00,18.00]
#> Prior [rho]:Log-Normal(log-mu = 0.00, log-sd = 1.000) 1[0.00,100.00]
#> Prior [log(sigma^2)]:Normal(mu = 0.83, sd = 1.000) 1[-18.00,18.00]Here, we see the Hill model is fit using the Normal-NCV, or normal variance proportional to the mean. For the Hill model, you can also use ‘normal’ as a distribution option.
hill_fit <- single_continuous_fit(cont_data[,"Dose"],
cbind(cont_data[,"Mean"],cont_data[,"N"],cont_data[,"SD"]),
model_type="hill",distribution = "normal",
fit_type = "mcmc")
hill_fit$full_model
#> [1] "Model: Hill Distribution: Normal"
plot(hill_fit)
Notice how the `distribution’ option controls the distribution. For the “hill”, “power”, and “polynomial” DR models you can choose either “normal” or “normal-ncv.” or the “exp-3” and “exp-5” models you can also choose “lognormal.”
exp5_fit <- single_continuous_fit(cont_data[,"Dose"],Y,
model_type="exp-5",distribution = "lognormal",fit_type="laplace")
exp5_fit$full_model
#> [1] "Model: Exponential-5 Distribution: Log-Normal"
plot(exp5_fit)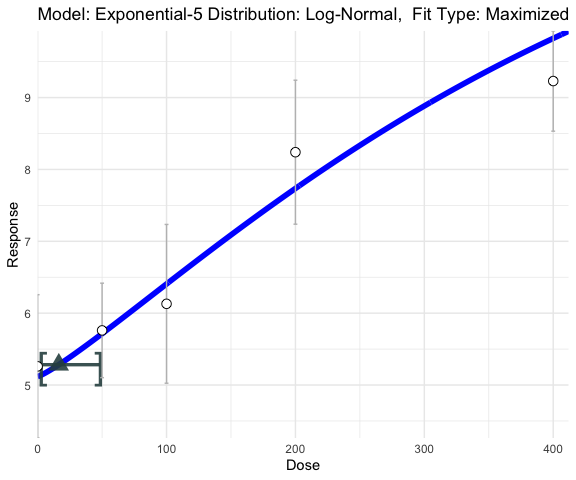
There are also other types of BMDs you can choose. Here one can specify
absolute deviation as ‘abs,’ which solves where
is a specific cutoff value. In the example below, the abosolute
difference of
hill_sd_fit <- single_continuous_fit(cont_data[,"Dose"],Y,
model_type="hill",distribution = "normal-ncv", fit_type="mcmc",
BMD_TYPE="abs",BMR = 2)
#> Warning in single_continuous_fit(cont_data[, "Dose"], Y, model_type = "hill", :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
hill_sd_fit$full_model
#> [1] "Model: Hill Distribution: Normal-NCV"
plot(hill_sd_fit)
The standard deviation approach is the default approach, and this is the
value that solves and in this definition the
is the number of standard deviations the mean changes from no-exposure.
In the example below, the
hill_sd_fit <- single_continuous_fit(cont_data[,"Dose"],
cbind(cont_data[,"Mean"],cont_data[,"N"],cont_data[,"SD"]),
model_type="hill",distribution = "normal-ncv",fit_type="laplace",
BMD_TYPE="sd",BMR = 1.5)
#> Warning in single_continuous_fit(cont_data[, "Dose"], cbind(cont_data[, :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
hill_sd_fit$full_model
#> [1] "Model: Hill Distribution: Normal-NCV"
plot(hill_sd_fit)
The hybrid approach is a probabilistic approach that mimics risk for
dichotomous data. Here, the BMD is the value that solves Here,
is the probability that an ‘adverse’ response is observed at background
dose, and
is the increase in probability of seeing an adverse response at dose
.
For this approach
must be specified using the “point_p” option. This option is only used
when “hybrid” is specified. The following shows a Hill fit, where the
BMD with only a
chance of being observed at no dose, but has a
probability of being observed at dose
Note:
For more information on this approach see Crump
[@crump1995calculation].
hill_hybrid_fit <- single_continuous_fit(cont_data[,"Dose"],
cbind(cont_data[,"Mean"],cont_data[,"N"],cont_data[,"SD"]),
model_type="hill",distribution = "normal-ncv",fit_type="mcmc",
BMD_TYPE="hybrid",point_p = 0.025,BMR = 0.1)
#> Warning in single_continuous_fit(cont_data[, "Dose"], cbind(cont_data[, :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
hill_hybrid_fit$full_model
#> [1] "Model: Hill Distribution: Normal-NCV"
plot(hill_hybrid_fit)
The relatived deviation approach [@slob2002dose] is the value that solves
where
is the value of increase/decrease from the background.
hill_rd_fit <- single_continuous_fit(cont_data[,"Dose"],
cbind(cont_data[,"Mean"],cont_data[,"N"],cont_data[,"SD"]),
model_type="hill",distribution = "normal-ncv", fit_type="mcmc",
BMD_TYPE="rel",BMR = 0.1,samples = 50000)
#> Warning in single_continuous_fit(cont_data[, "Dose"], cbind(cont_data[, :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
hill_rd_fit$full_model
#> [1] "Model: Hill Distribution: Normal-NCV"
plot(hill_rd_fit) Note:
The above definitions are for for increasing data, `ToxicR’ will
determine the direction of adversity and adjust computations
accordingly.
Note:
The above definitions are for for increasing data, `ToxicR’ will
determine the direction of adversity and adjust computations
accordingly.
Like the dichotomous case you can modify the priors, to see what prior you want to modify Again, this is easy we can type the following code to see what our current prior is:
hill_sd_fit$prior
#> Hill Model [normal-ncv] Parameter Priors
#> ------------------------------------------------------------------------
#> Prior [a]:Normal(mu = 1.00, sd = 1.000) 1[-100.00,100.00]
#> Prior [b]:Normal(mu = 0.00, sd = 1.000) 1[-100.00,100.00]
#> Prior [c]:Log-Normal(log-mu = 0.00, log-sd = 2.000) 1[0.00,100.00]
#> Prior [d]:Log-Normal(log-mu = 0.47, log-sd = 0.421) 1[0.00,18.00]
#> Prior [rho]:Log-Normal(log-mu = 0.00, log-sd = 1.000) 1[0.00,100.00]
#> Prior [log(sigma^2)]:Normal(mu = 0.83, sd = 1.000) 1[-18.00,18.00]
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(0,1, 0, 100),
lnormprior(log(1),0.4215,0,18),
lnormprior(0,1,0,100),
normprior(0, 10,-100,100));
p_hill_ncv = create_continuous_prior(prior,"hill","normal-ncv")
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(0,1, 0, 100),
lnormprior(log(1),0.4215,0,18),
normprior(0, 10,-100,100));
p_hill_norm = create_continuous_prior(prior,"hill","normal")When specifying a prior, it is slightly different than the dichotomous
case. Here, we need to specify what distribution we are using too. This
adds one or two parameters depending on the parameters. All models have
a parameter called
which is the variance term, and it is ALWAYS the last parameter in the
model, and the
prior is placed over the log of this value. For the ‘normal-NCV’ option,
the variance is
i.e., it has an additional parameter
The parameter
must have a prior, which is
in the above code.
hill_sd_a_fit <- single_continuous_fit(cont_data[,"Dose"],
cbind(cont_data[,"Mean"],cont_data[,"N"],cont_data[,"SD"]),
prior=p_hill_ncv,
fit_type="laplace",
BMD_TYPE="sd",BMR = 1.5)
#> Warning in single_continuous_fit(cont_data[, "Dose"], cbind(cont_data[, :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
hill_sd_b_fit <- single_continuous_fit(cont_data[,"Dose"],
cbind(cont_data[,"Mean"],cont_data[,"N"],cont_data[,"SD"]),
prior=p_hill_norm,
fit_type="laplace",
BMD_TYPE="sd",BMR = 1.5)
#> Warning in single_continuous_fit(cont_data[, "Dose"], cbind(cont_data[, :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
library(ggpubr)
figure <- ggarrange(plot(hill_sd_a_fit)+ggtitle(""),
plot(hill_sd_b_fit)+ggtitle(""),
labels = c("Prior NCV", "Prior Normal"),
ncol = 1, nrow = 2)
figure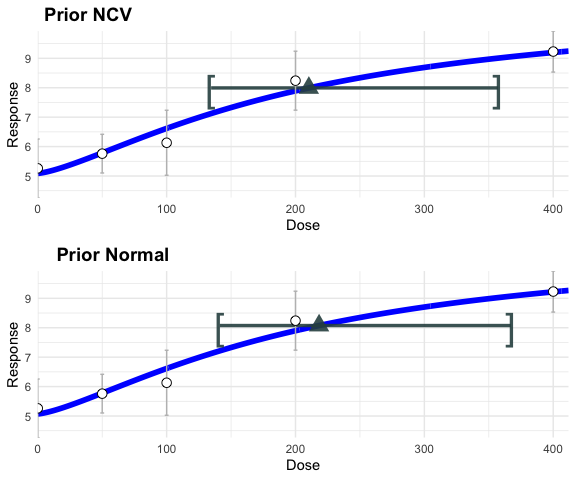
In the above code, we didn’t need to specify the ‘model_type’ or ‘distribution’ because this is done implicitly when you specify the prior. The above code works for the hill model, lets try it for a polynomial model.
poly_sd <- single_continuous_fit(cont_data[,"Dose"],Y,
distribution="normal",model_type="polynomial",
degree = 4,
fit_type="laplace",
BMD_TYPE="sd",BMR = 0.5)
#> WARNING: Polynomial models may provide unstable estimates because of possible non-monotone behavior.
#> Warning in single_continuous_fit(cont_data[, "Dose"], Y, distribution =
#> "normal", : BMD_TYPE is deprecated. Please use BMR_TYPE instead
plot(poly_sd)
Beyond single model fits, one can also use model averging for continuous models. As a default 10 models are fit. Here, the ‘hill’ and ‘power’ options are fit under the ‘normal’ and ‘normal-ncv’ distribution assumptions. Additionally to the ‘normal’ and ‘normal-ncv’ distributions the ‘exp-3’ and ‘exp-5’ also use the ‘lognormal’ distribution. Note that the polynomial model is not used because of constraint issues.
ma_sd_mcmc <- ma_continuous_fit(cont_data[,"Dose"],Y, fit_type="mcmc",
BMD_TYPE="sd",BMR = 0.5,samples = 50000)
#> Warning in ma_continuous_fit(cont_data[, "Dose"], Y, fit_type = "mcmc", :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
ma_sd_laplace <- ma_continuous_fit(cont_data[,"Dose"],Y, fit_type="laplace",
BMD_TYPE="sd",BMR = 0.5)
#> Warning in ma_continuous_fit(cont_data[, "Dose"], Y, fit_type = "laplace", :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
plot(ma_sd_mcmc)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
summary(ma_sd_laplace)
#> Summary of single MA BMD
#>
#> Individual Model BMDS
#> Model BMD (BMDL, BMDU) Pr(M|Data)
#> ___________________________________________________________________________________________
#> Logistic-Aerts Distribution: Normal 83.83 (45.11 ,135.72) 0.178
#> Probit-Aerts Distribution: Normal 86.91 (47.50 ,134.81) 0.144
#> LMS Distribution: Log-Normal 80.72 (42.66 ,172.08) 0.112
#> Logistic-Aerts Distribution: Log-Normal 70.06 (32.38 ,130.14) 0.101
#> Exponential-Aerts Distribution: Log-Normal 70.86 (32.02 ,136.19) 0.068
#> Probit-Aerts Distribution: Log-Normal 75.77 (36.90 ,125.07) 0.068
#> Gamma-EFSA Distribution: Log-Normal 55.65 (21.31 ,111.81) 0.052
#> Gamma-EFSA Distribution: Normal 67.07 (29.97 ,122.22) 0.046
#> Exponential-Aerts Distribution: Normal 82.46 (42.23 ,142.67) 0.044
#> Hill-Aerts Distribution: Log-Normal 70.01 (34.68 ,118.87) 0.040
#> Inverse Exponential-Aerts Distribution: Normal 106.96 (67.79 ,145.12) 0.032
#> Lognormal-Aerts Distribution: Normal 98.03 (56.37 ,152.66) 0.031
#> Lognormal-Aerts Distribution: Log-Normal 87.75 (46.03 ,147.16) 0.029
#> Inverse Exponential-Aerts Distribution: Log-Normal 106.12 (55.00 ,145.95) 0.027
#> Hill-Aerts Distribution: Normal 80.76 (43.98 ,127.61) 0.027
#> LMS Distribution: Normal 133.08 (124.95 ,142.68) 0.001
#> ___________________________________________________________________________________________
#> Model Average BMD: 80.46 (37.81, 143.23) 90.0% CI
cleveland_plot(ma_sd_laplace)
MAdensity_plot(ma_sd_mcmc)
#> Picking joint bandwidth of 2.94
Here, ‘cleveland_plot’ and ‘MAdensity_plot’ can be used in the same way as in dichotomous plot. Like the dichotomous case, we can create a prior list for our model average.
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(0,1, 0, 100),
lnormprior(log(1),0.4215,0,18),
lnormprior(0,1,0,100),
normprior(0, 10,-100,100));
p_hill_ncv = create_continuous_prior(prior,"hill","normal-ncv")
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(0,1, 0, 100),
lnormprior(log(1),0.4215,0,18),
normprior(0, 10,-100,100));
p_hill_norm = create_continuous_prior(prior,"hill","normal")
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(0,1, 0, 100),
lnormprior(log(1),0.4215,0,18),
normprior(0, 10,-100,100));
p_exp5_norm = create_continuous_prior(prior,"exp-5","normal")
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(log(1),0.4215,0,18),
normprior(0, 10,-100,100));
p_power_norm = create_continuous_prior(prior,"power","normal")
prior <- create_prior_list(normprior(0,1,-100,100),
normprior(0,1,-1e4,1e4),
lnormprior(log(1),0.4215,0,18),
normprior(0, 10,-100,100));
p_exp3_norm = create_continuous_prior(prior,"exp-3","normal")
#> NOTE: Parameter 'c' added to prior list. It is not used in the analysis.
prior_list = list(p_exp3_norm,p_hill_norm,p_exp5_norm,p_power_norm)With a list of priors one can then run a model average based upon a user specified model space. Here, all of the same benchmark dose options of ‘single_continuous_fit’ apply.
ma_sd_mcmc_2 <- ma_continuous_fit(cont_data[,"Dose"],Y, fit_type= "laplace",
BMD_TYPE="sd",BMR = 0.5,samples = 50000,model_list=prior_list)
#> Warning in ma_continuous_fit(cont_data[, "Dose"], Y, fit_type = "laplace", :
#> BMD_TYPE is deprecated. Please use BMR_TYPE instead
plot(ma_sd_mcmc_2)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
Wout Slob, Dose-Response Modeling of Continuous Endpoints, Toxicological Sciences, Volume 66, Issue 2, April 2002, Pages 298–312
Crump, Kenny S., Calculation of benchmark doses from continuous data, Risk Analysis, Volume 15, Issue 1, February 1995, Pages 79-89
