Implementation of the Loop Extrusion Model using OpenMM - a Python library for molecular dynamics simulations.
The goal was to replicate simulations as seen in the video:
http://symposium.cshlp.org/content/82/45.full
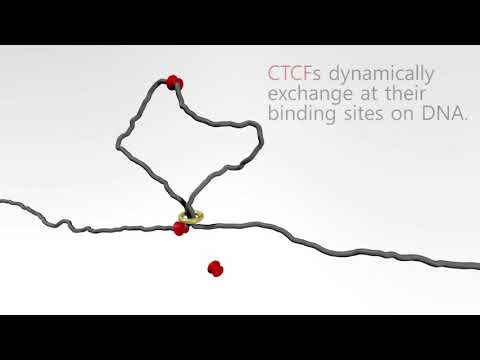
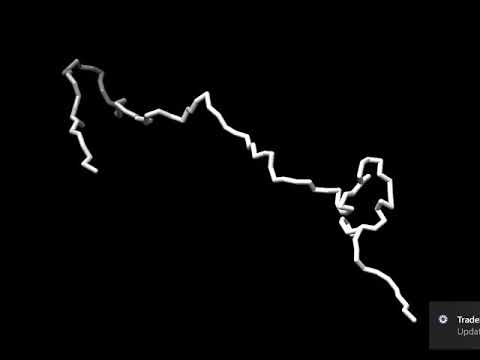
The human genome consists of DNA strands totaling 2 meters in length. These strands are packaged into chromatin within cell nuclei, with a diameter of approximately 6-12 micrometers. Despite its large size confined to a small volume, the DNA remains untangled. Furthermore, its structure is highly organized on multiple levels of organization. Chromatin can actively change its conformation during various biological processes such as gene expression and replication. One of the proposed models for chromatin folding is the Loop Extrusion Model (LEM), in which chromatin is extruded by a protein ring called cohesin, and this process halts upon encountering the CTCF protein attached to the strand. The goal of the student project is to replicate the results of previous work, specifically an animated trajectory from simulations visualizing the loop extrusion process. To accomplish this task, students will utilize the Python library OpenMM dedicated to molecular dynamics calculations, create their own force field representing coarse-grained polymers, and conduct necessary simulations and visualizations. A long-term objective that could potentially be pursued after completing the project is the utilization of developed methods for simulating entire cell nuclei.
[1] Fudenberg, Geoffrey, et al. "Formation of chromosomal domains by loop extrusion." Cell reports 15.9 (2016): 2038-2049.
[2] Fudenberg, Geoffrey, et al. "Emerging evidence of chromosome folding by loop extrusion." Cold Spring Harbor symposia on quantitative biology. Vol. 82. Cold Spring Harbor Laboratory Press, 2017.